Abstract
Object
The aim of our study was to enable automatic volumetry of the entire kidneys as well as their internal structures (cortex, medulla, and pelvis) from native magnetic resonance imaging (MRI) data sets.
Materials and methods
Segmentation of the entire kidneys and differentiation of their internal structures were performed in 12 healthy volunteers based on non-contrast-enhanced T1- and T2-weighted MR images. Two data sets (each acquired in one breath-hold) were co-registered using a rigid registration algorithm compensating for possible breathing-related displacements. An automatic algorithm based on thresholding and shape detection segmented the kidneys into their compartments and was compared to a manual labeling procedure.
Results
The resulting kidney volumes of the automated segmentation correlated well with those created manually (R 2 = 0.96). Average volume errors were determined to be 4.97 ± 4.08 % (entire kidney parenchyma), 7.03 ± 5.56 % (cortex), 12.33 ± 7.35 % (medulla), and 17.57 ± 14.47 % (pelvis). The variation of the kidney volume resulting from the automatic algorithm was found to be 4.76 % based on the measuring of one volunteer with three independent examinations.
Conclusion
The results demonstrate the feasibility of an accurate and repeatable automatic segmentation of the kidneys and their internal structures from non-contrast-enhanced magnetic resonance images.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Volumetry of the kidney is an important task in the early detection and monitoring of renal diseases like glomerular nephritis or hydronephrosis [1]. Furthermore, the segmentation of the internal renal structures as the cortex, medulla, and pelvis can be useful for functional and morphological assessments. It is known that the renal cortex volume tends to decrease over time in chronically injured kidneys. Therefore, a precise measurement of the cortical volume could be a helpful biomarker in follow-up examinations of chronic kidney diseases or transplanted kidneys [2].
A simple estimation for kidney volume can be obtained from renal length measurements using ultrasound imaging. However, this method has shown a tendency to underestimate the renal volume [3, 4]. Magnetic resonance imaging (MRI) provides spatially highly-resolved anatomical images, and therefore represents a more precise imaging modality for volumetric measurements. In recent years, dynamic contrast-enhanced measurements [5–7], e.g., using gadolinium-diethylene-triamine penta-acetic acid, have been used for the quantification of renal volume. The use of contrast agent leads to a considerable contrast between the renal cortex and medulla within the first minute after injection. In vivo measurements in animals [8] and humans [9, 10] have demonstrated a good differentiation between the entire kidneys and surrounding tissue. Contrast media helps to improve the contrast behavior between the renal compartments, but an optimal segmentation is based on at least two images recorded in different perfusion phases [6]. A well-known drawback of using contrast media—especially in patients with kidney insufficiency—is its tendency to cause nephrotoxic effects or even nephrogenic systemic fibrosis. For this reason, the approach used here works without the application of contrast media.
As mentioned above, MRI data sets acquired without contrast agent [10–12] usually result in a limited contrast between the kidneys and the surrounding tissue, as well as the inner structures. Thus, the potential and accuracy of non-contrast-enhanced MRI measurements in the context of an automatic renal segmentation remains to be proven. A precise determination of the entire volume of internal organs is executed rarely in clinical practice, because the manual segmentation over a large number of slices is a very time consuming procedure. Therefore, automatic segmentation algorithms are preferable for this task. Different methods like thresholding [8], clustering [13, 14], region growing [15], and contour detection [16, 17] or their combinations have been applied for segmentation.
For the separation of the internal renal structures (cortex, medulla, and pelvis), MR techniques providing a sufficient contrast between these compartments are required. Furthermore, for the identification of the entire kidneys, MR images must offer a high contrast between the kidneys and surrounding tissue (liver, spleen, gastrointestinal tract). Clinical abdominal imaging typically uses two-dimensional (2D) T1- and T2-weighted (w) sequences. The purpose of this study was to determine the accuracy of in vivo measurements of healthy human kidney volumes using non-contrast-enhanced T1- and T2-w MR images. First, we evaluated the feasibility of in vivo determination of the entire kidney volume in healthy humans by MR measurements without contrast agent. This step included an adaptation of the MR imaging parameters in the context of an automatic segmentation procedure. Second, an automatic algorithm was implemented for renal segmentation with a combined differentiation of the inner renal structures as the cortex, medulla, and pelvis based on non-contrast-enhanced MR images. Afterwards, the results of the automatic segmentation algorithm were compared to a manual segmentation (reference standard). Finally, the repeatability of the renal volume and sub-volume measurements was evaluated.
Materials and methods
Hardware and sequences
In vivo measurements of healthy volunteers were performed on a 1.5T whole-body MR scanner (Magnetom Sonata, Siemens Healthcare, Erlangen, Germany). The body coil was used for homogeneous RF transmission and 12-element anterior–posterior phased-array coils were applied for signal detection. This methodical study was approved by the local ethic committee and informed consent was obtained from all subjects.
Two clinical MR protocols were adapted in order to obtain a sufficient differentiation of the entire kidneys from the surrounding organs as well as to differentiate the internal renal structures. For the segmentation of the entire kidneys, a T2-w half-Fourier single-shot turbo spin echo (HASTE) sequence was used. For this sequence, the echo time (TE) was varied to achieve maximum kidney–liver and kidney–spleen contrast. For segmentation of the internal renal structures, a T1-w spoiled gradient echo (GRE) sequence was used, and the contrast between the renal cortex and the medulla was maximized by varying the flip angle (FA). The adaptation of these two parameters was performed during initial measurements of three healthy volunteers.
The HASTE sequence was applied with the following parameters: TR = 1,800 ms, bandwidth (BW) = 781 Hz/Pixel, FA = 150°, voxel size = 1.71 × 1.37 × 5.00 mm3, field-of-view = 350 × 350 mm2, parallel acquisition factor of 2. For accurate slice excitation, both kidneys were covered in a single breath-hold. To minimize the required number of slices, coronal slice orientation was chosen. Thirteen slightly tilted coronal slices (parallel to the orientation of the kidney axis) with a thickness of 5 mm were acquired without gaps in an interleaved manner, leading to a scan time of 23 s. The echo time TE was varied from 61 to 122 ms in order to determine the optimal contrast for the kidney segmentation.
The slice orientation and the voxel size in measurements with the GRE sequence were identical to those in the HASTE sequence. Other parameters of the GRE sequence were set to: TR/TE = 132/2.44 ms, BW = 260 Hz/Pixel, leading to a scan time of 19 s. In order to optimize the contrast between the cortex and the medulla, the excitation flip angle was varied between 30° and 90°. Fat saturation was applied to both T1-w and T2-w measurements to improve the overall image contrast and to avoid chemical shift artifacts.
With the adapted sequence parameters, measurements in 12 healthy volunteers (mean age 33 ± 11 years and body mass index of 25 ± 4.3 kg/m2) were performed to acquire the data for the kidney segmentation.
Image registration
Since the two data sets were acquired in different breath-holds, possible displacements of the kidneys between T1-w and T2-w data sets had to be compensated for. Thus, the T1-w images were co-registered slice-wise to the T2-w images using the registration program ELASTIX [18]. The following registration parameters were optimized in order to obtain high registration accuracy for the given images: transformation model (rigid, non-rigid), number of downsampling steps (1, 2, 3), polynomial order of the interpolation (1, 2, 3), and the number of iterations (200–1,000). As a merit function for the optimization, in two data sets the kidney areas were manually labeled in three medial T1-w slices and their T2-w counterparts. After each registration run, the intersection area of the kidneys in the registered T1- and T2-w images was calculated (separately for the left and right kidney) and expressed as a percentage of deviation from the kidney areas in the T2-w images. Furthermore, non-rigid distortions inside the kidneys (e.g., the medulla) were evaluated visually.
Segmentation algorithm
The image segmentation was performed in two steps using Matlab (Version R2011b, The MathWorks, Natick, USA). First, the entire kidneys were segmented from the surrounding tissues by thresholding the T2-w images and with a subsequent refinement step using prior knowledge about the kidney shape and location. In a second step, the T1-w and T2-w images were used to distinguish between the renal structures (cortex, medulla, and pelvis). All thresholds, specified in this algorithm, were determined and tested manually in several subjects for the same measurement setups and sequence parameters. The obtained optimal values were held constant for the automatic segmentation algorithm. The complete segmentation process is presented in Fig. 1 and explained in more detail in the following.
Entire kidney segmentation: A preliminary kidney mask was obtained for each slice of T2-w data separately by applying an empirically determined threshold at 32 % of the maximum pixel’s intensity in the slice (100 % corresponds to pure fluid). It was assumed that all pixels above this threshold belong to the kidney. Since the spleen, the vertebrae, and some parts of the gastrointestinal tract show similar signal intensity values as the kidney, this initial mask had to be further refined. All image segmentation steps were performed for the right and left kidney separately.
Because of the very small distance between the superior pole of left kidney and spleen, the threshold-based algorithm may generate a common area and splits the splenorenal recess into two parts. To remove this artificially generated connection between the kidney and spleen, the convex hull of the combined area was calculated. Then, two splenorenal regions could be identified by subtracting the connected area from the convex hull. The separation of the kidney from the spleen was performed along the shortest line connecting the splenorenal regions. Finally, the obtained binary mask was refined using active contours [19].
Another step isolated the gastrointestinal tract from the kidney, because it was also artificially detected as a kidney in the initial binary mask. The algorithm started at the central slice where the kidney has its maximum extension and is not impaired by partial volume effects. Afterwards, the algorithm propagated outwards in both directions. Since the gastrointestinal tract is located at the lower end of the kidneys, only the lower halves of the binary kidney masks were considered, thus maintaining the already well-segmented areas. Based on the currently processed mask (denoted as Mi), several test points along the contour were compared to corresponding points of the previous mask (denoted as Mi−1). If these test points are outside the segmented area of the previous contour, the form of the kidney outline of this mask was transferred to the current mask by taking the intersection of both masks (Mi ← Mi ∩ Mi−1).
Renal structure segmentation: Within the newly generated entire kidney mask, the renal cortex, medulla, and pelvis were subsequently separated. This separation algorithm utilizes assumptions of the renal anatomical structure, e.g., that the renal cortex surrounds parts of the medulla.
T1-w images were used to segment the renal cortex for the left and right kidney separately. Due to intensity inhomogeneities, a single threshold for the entire kidney was found to be inadequate. Instead, the classification of the cortex was realized by analyzing the signal distribution in each row of the kidney. All pixels with a value above a local threshold were assigned to the cortex. This threshold was calculated as the row’s mean intensity value inside the kidney mask minus the standard deviation. In addition, the first and last slices containing kidney tissue pixels (different for the right and left kidney) were labeled exclusively as cortex.
The algorithm for the segmentation of the pelvis used both T1- and T2-w images. The final pelvis mask was obtained by fusion of both segmented areas. In T1-w images the pelvis appears as a hypointense region of the kidney. Images with T2-w contrast are helpful in differentiation between the pelvis and ureter. Therefore, the pelvis segmentation was executed in two steps: fluid (urinary) areas showed lower signal intensity than renal parenchyma tissue in T1-w images. In the first step, the slices including parts of the renal pelvis were determined in T1-w images with a fixed threshold. This threshold was based on the darkest areas of the image. However, this leads to occasional misclassifications of fat pixels along the outer parenchyma in cases where the kidney mask occurs slightly larger than the kidney shape in T1-w images. Those undesired pixels were identified through their position (lateral) and deleted from the pelvis mask. In the second step, the segmentation of the pelvis was performed in T2-w images, where the pelvis appears as the brightest area of the entire kidney region. For this purpose, the pelvis was separated using a region-based threshold (pixel values higher than 80 % of the mean signal intensity of the entire kidney were assigned to the pelvis). Afterwards, the union pelvis mask was subtracted from the cortex mask to remove the erroneously identified pixels.
Finally, the renal medulla was obtained by subtracting the cortex and pelvis from the entire kidney mask. The final volumes of the entire kidneys, renal cortex, medulla, and pelvis were then calculated by voxel summation (voxel volume is 9.38 mm3).
Evaluation
A custom user interface based on Matlab was implemented to create reference standard masks for the entire kidneys (based on T2-w MR images), renal pelvis (based on T1- and T2-w images), and medulla (based on T1-w images). The reference standard masks for each volunteer were carefully drawn manually in each slice. The reference mask for the cortex was obtained by subtracting the pelvis and medulla masks from the entire kidney mask.
For the evaluation of the accuracy of the optimized algorithm, the volume error (ve) and overlap error (oe) of the automatic and manual segmentation were calculated using the following formulas:
Here, V M and V A represent the sets of voxels belonging to the manually and automatically segmented volumes, respectively, and the |V| operator is used to determine the cardinality of the sets. To compare the results with existing work, the volume overlap (vo, also known as Jaccard index) and Dice’s coefficient (dice) [20] were calculated according to
A repeatability study was performed in order to evaluate the variations between measurements of the same subject with the same sequence protocol and scanner. One volunteer was subsequently scanned three times after repositioning on the table. Then, for each data set the kidney volume was obtained by the proposed automatic segmentation algorithm, and the coefficient of variation (standard deviation divided by the mean) of the three volumes was calculated.
Results
MRI sequences
In order to be able to differentiate the entire kidneys from the surrounding organs (liver, spleen, and gastrointestinal tract) and their internal structures, two MR sequences were adapted. The contrast-to-noise ratios (CNR) between the cortex and medulla in the T1-w GRE images measured at different flip angles are shown in Table 1. Considering these CNR values as well as the contrast between the kidneys and surrounding tissues, an FA of 70° was found to be an acceptable trade-off.
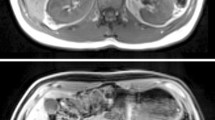
Table 2 shows CNR values between different tissues in T2-w HASTE images measured at varied echo times. The maximum CNR between the kidney and liver was at TE = 82 ms, while the maximum CNR between the kidney and spleen occurred at TE = 102 ms. Therefore, a TE of 95 ms was selected for the HASTE sequence. Corresponding T1- and T2-w images of one volunteer obtained with these adapted sequence parameters are shown in Fig. 2.
Image registration
To find the optimal registration algorithm, eleven different parameter sets were tested. For each parameterization, the percentage deviation of the manually labeled kidney areas between the T2-w images and the post-registered T1-w images is exhibited in Fig. 3. In all three slices, high deviations resulted when we employed the non-rigid deformation model (parameter sets 1–8). In some of these cases, the kidney’s inner structures showed even worse alignment than without registration, while the outer kidney boundary was well registered. This behavior was most likely caused by the lack of contrast between the cortex and medulla in the T2-w images. In contrast, using the rigid deformation model resulted in clearly better alignment both at the kidney surface and the boundaries inside the kidneys. The best alignment (mean deviation of 4.26 and 2.87 % for the left and right kidney, respectively) was obtained using a rigid deformation model with two resolution levels, cubic BSpline interpolation, normalized mutual information metric, and 200 iterations (parameter set 11). The computation time of this registration procedure was about 70 s per image, resulting in a processing time of approximately 15 min for the entire data set.
Percentage deviation in manually labeled kidney areas between T1- and T2-weighted images using different registration parameters. Rigid registrations (registration number 9–11) resulted in a higher internal agreement than non-rigid ones (1–8). Registration 11 showed the best results for all three slices
Segmentation algorithm
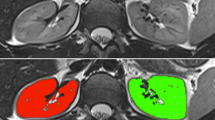
Figure 4a shows representative results for the kidney mask creation on the basis of the T2-w images for selected slices. Figure 4b demonstrates the corresponding successful segmentation of the cortex, medulla and pelvis. All determined coefficients are summarized in Table 3. The resulting ve of 4.97 ± 4.08 % for the entire kidney and 7.03 ± 5.56 % for the cortex in all 12 subjects shows a high agreement between manual and automatic segmentation.
The results of a slice-wise comparison of the manual and automatic segmentation are shown in Fig. 5. A high correlation of the segmented areas was observed with the determination coefficients (R 2) of 0.96, 0.88, 0.92, and 0.87 for the entire kidney, cortex, medulla, and pelvis, respectively. Furthermore, the lines obtained by linear regression (solid lines in Fig. 5) are in good accordance with the lines of identity (dashed lines), indicating correct tissue area quantification. Only the pelvis area quantification shows minor systematic underestimation.
In the repeatability study, the kidney volume between all three trials deviated by 4.76 %, calculated from the within-subject coefficient of variation. The internal structures showed deviations of 6.35 % (cortex), 1.74 % (medulla), and 16.33 % (pelvis).
The automatically (manually) obtained mean total volume of both kidneys over all volunteers was 404.8 ± 70.9 cm3 (412.1 ± 71.7 cm3). The mean volumes of the cortex, medulla and pelvis were determined to 252.1 ± 42.1 cm3 (263.9 ± 47.7 cm3), 108.9 ± 16.1 cm3 (104.0 ± 16.8 cm3), and 44.0 ± 24.7 cm3 (45.8 ± 18.6 cm3) respectively. The ratio between the cortex and the entire kidney volume was, on average, 64.0 ± 3.0 % (62.4 ± 2.5 %), while the ratio between medulla and the entire kidney volume was 25.4 ± 3.0 % (27.1 ± 3.1 %).
The computation time of the segmentation algorithm including mask generation, region determination, and export of the results was about 50 s for the entire data set.
Discussion
The presented technique provides a reliable automatic volumetric segmentation of the entire kidneys as well as the renal cortex, medulla, and pelvis in healthy volunteers based on non-contrast-enhanced MR images. A good agreement between automatic and manual segmentation of the entire kidneys was obtained.
In recent years, a number of studies of renal volumetric segmentation have been reported [1, 2, 13]. However, a straightforward comparison of their results is not always possible because some authors do not report the volume error values. The automatically calculated total kidney volume of 405 ± 71 cm3 for both kidneys in healthy subjects agrees with the range of values reported in literature, e.g., Cheong et al. [21] have shown a single kidney volume of 202 ± 36 ml for men and 154 ± 33 ml for women.
Tang et al. [13] proposed an automatic renal segmentation algorithm using contrast-enhanced MR images. They also compared their results to manual segmentation and calculated the overlap error. However, calculations have been performed with an equation that slightly differs from our definition of the volume overlap parameter. With an overlap of 66 % for the cortex and 76 % for the medulla, we are in the same range. For the renal pelvis, Tang et al. have reported a reasonable agreement of automatic to manual segmentation of nearly 90 %.
Gloger et al. [22] recently presented a fully automated kidney segmentation algorithm of 3D non-contrast-enhanced MR images acquired with a T1-w VIBE (volume-interpolated breath-hold examination) sequence. They have reported a volume error of 7.5 % for the right and 10.7 % for the left parenchyma, which is slightly worse compared to our results (ve ~5 %). Furthermore, the results of our method (O) and of Gloger et al. (G) for the overlap error (O: 20 %, G: 25 %), the volume overlap (O: 82 %, G: 78 %), as well as the dice coefficient (O: 90 %, G: 86 %) for the entire kidneys showed a high agreement to the manual segmentation. In contrast to Gloger et al., our algorithm segments also the medulla and pelvis. To the best of our knowledge, none of the approaches in the literature segmented the renal pelvis from native MR images. Additionally, our method paid attention to the combination of adapted MR images with an ordinary post-processing algorithm.
Slice orientation proved to be of crucial influence on kidney segmentation in MRI. Axial slice orientation provides only minor partial volume effects in the kidneys [23], but a higher number of slices have to be acquired covering the entire kidneys at the given slice thickness. This leads to an acquisition time potentially exceeding the breath-hold capacity. Using coronal or sagittal slice orientation, breathing movements showed the lowest through-plane component [24]. Since sagittal slice orientation requires a high number of slices to cover both kidneys and additionally showed worse partial volume effects, coronal slice orientation was considered the best choice regarding both measurement time and motion-related artifacts.
Our comparison of different approaches to image registration revealed that a non-rigid registration achieves a better adaptation of the outer structures of the kidney. Unfortunately, the medulla and pelvis regions showed strong deformations, which are not acceptable for the accurate volumetry of internal renal structures. In contrast, the rigid registration seems to be clearly beneficial for outer as well as internal structures. The still existing mismatch of the outline registration leads to a higher overlap error of the cortex, medulla, and pelvis in our study.
Study limitations
Limited quality of segmentation evolves from the border slices. Partial volume effects [23, 25] due to finite slice thickness and overlapping with other organs lead to erroneous calculation of the volume. Since scan time is limited due to the breath-hold phase, a relatively coarse voxel size of 1.71 × 1.37 × 5 mm3 was used in our study. A higher image resolution will improve the volumetric results and diminish the effects of the imprecise volumetric calculation of the first and last slices for both manual and automatic segmentation.
A potential problem that has to be accounted for is the separation of the spleen from the kidney. This problem increases in thin volunteers because the fat deposit between the spleen and kidney is almost completely lacking in those individuals. Parts of the kidney might be artificially removed by the algorithm, but this part was added (at least for the most part) automatically afterward by the snake.
Another limitation is the correct determination of the renal pelvis. The pelvis size depends on the fluid status of each volunteer and possibly on the actual flow conditions. However, those effects were not considered in this study. The overlap errors regarding the medulla and particularly the renal pelvis are more pronounced than for the renal cortex. The reason for this is an increase of the volume error due to geometrical features (e.g., large surface) in those compartments. It is well-known that a correct determination of the border of the pelvis in native MR images is critical. So the volume error of more than 17 % is in a reasonable range, but still stands to be improved. Some studies reported in the literature using dynamic contrast-enhanced approaches [13] have shown somewhat better results, but it should be kept in mind that our approach works without the administration of contrast medium.
Proposed sequence types and parameters can also be applied for studies on other MR scanners operating at 1.5T (especially if the used receiver coils have similar sensitivity and spatial characteristics). Suitable threshold values should be adapted if the measurement setup is changed, since the measured signal intensity depends on the hardware and software situation of the used MR unit. The optimization of the acquisition parameter was performed at a field strength of 1.5T. A change in field strength usually results in modified tissue contrast. Relaxation times of tissue do not behave linearly with field strength, and therefore sequence parameters should be optimized again, if measurements for renal segmentation are planned at different field strengths (e.g., 3 Tesla).
The presented segmentation algorithm has been proven on healthy volunteers. Lee et al. [26] recently reported that in some renal diseases the corticomedullary contrast is modified or even decreased. Similar to our approach, T1-w images were used for the differentiation of the cortex and medulla. It cannot be precisely stated yet how reliable the proposed algorithm will work in patients even under conditions with a weakened contrast between the cortex and medulla. In our work, we optimized the parameters of T1-w sequences regarding the relevant contrast between the medulla and cortex. With an inherent contrast-to-noise ratio of approximately 13 between the medulla and cortex obtained in healthy subjects, there remains some tolerance even in cases with slightly changed tissue properties. The proposed segmentation procedure is expected to work quite well in cases with normal signal characteristics of the renal compartments, but changes in respective volumes. It is clear that renal diseases that lead to strong changes in signal yield in T1- and T2-w MRI (e.g., tumors, cysts) will not allow the correct quantification of compartment volumes using the given approach.
Conclusion
In conclusion, the combination of adapted MR images, image registration, and automatic segmentation provides reliable and repeatable volumetric results of the entire kidney, renal cortex, medulla, and pelvis without applying contrast media. With a total image post-processing time of approximately 16 min, including registration and segmentation, the presented method is much faster than manual segmentation.
Implementing the proposed framework in clinical routine offers a noninvasive approach for the assessment and monitoring of morphological changes by calculating the ratio between the cortex and the entire kidney volume. This is especially interesting since cortical volume is known to decrease over time in some patients with affected kidneys [27, 28].
References
Jones RA, Easley K, Little SB, Scherz H, Kirsch AJ, Grattan-Smith JD (2005) Dynamic contrast-enhanced MR urography in the evaluation of pediatric hydronephrosis: Part I, functional assessment. AJR Am J Roentgenol 185(6):1598–1607
Karstoft K, Lodrup AB, Dissing TH, Sorensen TS, Nyengaard JR, Pederson M (2007) Different strategies for MRI measurements of renal cortical volume. J Magn Reson Imaging 26(6):1564–1571
Bakker J, Olree M, Kaatee R, de Lange EE, Moons KG, Beutler JJ, Beek FJ (1999) Renal volume measurements: accuracy and repeatability of US compared with that of MR imaging. Radiology 211(3):623–628
Bakker J, Olree M, Kaatee R, de Lange EE, Beek FJ (1998) In vitro measurement of kidney size: comparison of ultrasonography and MRI. Ultrasound Med Biol 24(5):683–688
Li S, Zoellner F, Merrem AD, Peng Y, Roervik J, Lundervold A, Schad LR (2012) Wavelet-based segmentation of renal compartments in DCE-MRI of human kidney: initial results in patients and healthy volunteers. Comput Med Imaging 36(2):108–118
Chevaillier B, Mandry D, Ponvianne Y, Collette Jl, Claudon M, Pietquin O (2008) Functional semi-automated segmentation of renal DCE-MRI sequences using a growing neural gas algorithm. In: 16th European Signal Processing Conference (EUSIPCO 2008), Lausanne
Grenier N, Basseau F, Ries M, Tyndal B, Jones R, Moonen C (2003) Functional MRI of the kidney. Abdom Imaging 28(2):164–175
Coulam CH, Bouley DM, Sommer FG (2002) Measurement of renal volumes with contrast-enhanced MRI. J Magn Reson Imaging 15(2):174–179
De Bazelaire CM, Duhamel GD, Rofsky NM, Alsop DC (2004) MR imaging relaxation times of abdominal and pelvic tissues measured in vivo at 3.0 T: preliminary results. Radiology 230(3):652–659
Di Leo G, Di Terlizzi F, Flor N, Morganti A, Sardanelli F (2011) Measurement of renal volume using respiratory-gated MRI in subjects without known kidney disease: intraobserver, interobserver, and interstudy reproducibility. Eur J Radiol 80(3):e212–e216
Kanki A, Ito K, Tamada T, Noda Y, Yamamoto A, Tanimoto D, Sato T, Higaki A (2013) Corticomedullary differentiation of the kidney: evaluation with noncontrast-enhanced steady-state free precession (SSFP) MRI with time-spatial labeling inversion pulse (time-SLIP). J Magn Reson Imaging 37(5):1178–1181
Cohen BA, Barash I, Kim DC, Sanger MD, Babb JS, Chandarana H (2012) Intraobserver and interobserver variability of renal volume measurements in polycystic kidney disease using a semiautomated MR segmentation algorithm. AJR Am J Roentgenol 199(2):387–393
Tang Y, Jackson HA, De Filippo RE, Nelson MD, Moats RA (2010) Automatic renal segmentation applied in pediatric MR urography. Int J Intell Inf Process 1(1):12–19
Borgelt C, Timm H, Kruse R (2000) Using fuzzy clustering to improve naive Bayes classifiers and probabilistic networks. In: Fuzzy Systems, The Ninth IEEE International Conference on, San Antonio, 1:53-58
Sun Y, Moura J, Yang D, Ye Q, Ho C (2002) Kidney segmentation in MRI sequences using temporal dynamics. IEEE Int Symp Biomed Imag 98–101
Vivier PH, Dolores M, Gardin I, Zhang P, Petitjean C, Dacher JN (2008) In vitro assessment of a 3D segmentation algorithm based on the belief functions theory in calculation renal volumes by MRI. AJR Am J Roentgenol 191(3):W127–W134
He L, Peng Z, Everding B, Wang X, Han CY, Weiss KL, Wee WG (2008) A comparative study of deformable contour methods on medical image segmentation. Image Vis Computing 26:141–163
Klein S, Staring M, Murphy K, Viergever MA, Pluim JP (2010) Elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 29(1):196–205
Kass M, Witkin A, Terzopoulos D (1988) Snakes: active contour models. Int J Comput Vis 1:321–331
Zöllner FG, Svarstad E, Munthe-Kaas AZ, Schad LR, Lundervold A, Rorvik J (2012) Assessment of kidney volumes from MRI: acquisition and segmentation techniques. Am J Roentgenol 199(5):1060–1069
Cheong B, Muthupillai R, Rubin MF, Flamm SD (2007) Normal values for renal length and volume as measured by magnetic resonance imaging. Clin J Am Soc Nephrol 2(1):38–45
Gloger O, Tönies KD, Liebscher V, Kugelmann B, Laqua R, Völzke H (2012) Prior shape level set segmentation on multistep generated probability maps of MR datasets for fully automated kidney parenchyma volumetry. IEEE Trans Med Imaging 31(2):312–325
González Ballester MA, Zissermann AP, Brady M (2002) Estimation of the partial volume effect in MRI. Med Image Anal 6(4):389–405
Pham D, KronT, Foroudi F, Schneider M, Siva S (2013) A review of kidney motion under free, deep and forced-shallow breathing conditions: implications for stereotactic ablative body radiotherapy treatment. Technol Cancer Res Treat
Gadeberg P, Gundersen HJ, Taagehoj F (1999) How accurate are measurements on MRI? A study on multiple sclerosis using reliable 3D stereological methods. J Magn Reson Imaging 10(1):72–79
Lee VS, Kaur M, Bokacheva L, Chen Q, Rusinek H, Thakur R, Moses D, Nazzaro C, Kramer EL (2007) What causes diminished corticomedullary differentiation in renal insufficiency? J Magn Reson Imaging 25(4):790–795
Mounier-Vehier C, Lions C, Devos P, Jaboureck O, Willoteaux S, Carre A, Beregi JP (2002) Cortical thickness: an early morphological marker of atherosclerotic renal disease. Kidney Int 61(2):591–598
Roger SD, Beale AM, Cattell WR, Webb JA (1994) What is the value of measuring renal parenchymal thickness before renal biopsy? Clin Radiol 49(1):45–49
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Will, S., Martirosian, P., Würslin, C. et al. Automated segmentation and volumetric analysis of renal cortex, medulla, and pelvis based on non-contrast-enhanced T1- and T2-weighted MR images. Magn Reson Mater Phy 27, 445–454 (2014). https://doi.org/10.1007/s10334-014-0429-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10334-014-0429-4