Abstract
The neurobiology of Alzheimer’s disease (AD) has been extensively studied by applying network analysis techniques to activation patterns in fMRI images. However, the structure of the directed networks representing the activation patterns, and their differences in healthy and Alzheimer’s people remain poorly understood. In this paper, we aim to identify the differences in fMRI activation network structure for patients with AD, late mild cognitive impairment (LMCI) and early mild cognitive impairment (EMCI). We use a directed graph theoretical approach combined with entropic measurements to distinguish subjects falling into these three categories and the normal healthy control (HC) group. We explore three methods. The first is based on applying linear discriminant analysis to vectors representing the in and out degree statistics of different anatomical regions. The second uses an entropic measure of node assortativity to gauge the asymmetries in the node with in and out degree. The final approach selects the most salient anatomical brain regions and uses the degree statistics of the connecting directed edges.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Functional magnetic resonance imaging (fMRI) provides a sophisticated means of studying the neuropathophysiology associated with Alzheimer’s disease (AD) [1]. Specifically, the blood oxygen level-dependent (BOLD) signal in fMRI indicates the activation potential of different brain regions, and neuronal activity between the various brain regions can be determined by measuring the correlation between activation signals. The resulting network representation of region activity has proved useful in understanding the functional working of the brain [2]. Functional neuroimaging has also proved useful in understanding Alzheimer’s disease (AD) via the analysis of intrinsic brain connectivity [3]. Abnormal brain function in AD is characterized by progressive impairment of episodic memory and other cognitive domains, resulting in dementia and, ultimately, death [5]. Although there is converging evidence about the identity of the affected regions in fMRI, it is not clear how this abnormality affects the functional organization of the whole brain.
Tools from complex network analysis provide a convenient approach for understanding the functional association of different regions in the brain [3]. The approach is to characterize the topological structures present in the brain and to quantify the functional interaction between brain regions, using the mathematical study of networks and graph theory. Graph theory offers an attractive route since it provides effective tools for characterizing network structures together with their intrinsic complexity. This approach has led to the design of several practical methods for characterizing the global and local structure of undirected graphs [4]. Features based on the global and local measures of connectivity are widely used in functional brain analysis [6]. By comparing the structural and functional network topologies between different populations of subjects, graph theory provides meaningful and easily computable measurements to reveal connectivity abnormalities in both neurological and psychiatric disorders [5].
Unfortunately, there is relatively little literature aimed at studying structural network features using directed graphs. The reason for that is the vast majority of techniques suggested by graph theory pertain to undirected rather than directed graphs. However, directed graphs are a more natural representation for brain structure, since they allow the temporal causality of activation signals for different anatomical structures in the brain. Moreover, Granger causality provides a powerful tool that can be used to investigate the direction of information flow between different brain regions [6]. When combined with machine learning algorithms, classification exhibited from directed graphs provides an effective way of detecting functional regions associated with Alzheimer’s disease [6]. By explicitly defining anatomical and functional connections in a directed manner between brain regions, fMRI data may be analyzed in a more detailed way and used to identify the different stages of neurodegenerative diseases [5, 6].
This paper is motivated by the need to fill this important gap in the literature, and to establish effective methods for measuring the structural properties of directed graphs representing inter-regional casual networks extracted from fMRI brain data. In particular, in order to characterize the functional organization of the brain, our approach uses as its starting point the von Neumann entropy for directed graphs. In a recent paper, Ye et al. [4] have derived an approximation of the Neumann entropy of a directed graph that depends on the in and out degrees of nodes in a directed graph. Thus it provides a natural way of capturing the flow of information across a directed network, based on the asymmetry of edges entering and exiting its nodes. We aim to use the directed network entropy to develop graph analytical methods to measure the degree of functional connectivity in brain networks.
We demonstrate that the resulting techniques can be used to distinguish the fMRI data from healthy controls and AD objects. The AD subjects exhibit significantly lower regional connectivity and exhibit disrupted the global functional organization when compared to healthy controls. Moreover, we apply linear discriminant analysis to brain network data from two groups of subjects with early mild cognitive impairment (EMCI) and late mild cognitive impairment (LMCI). Our results indicate that the node in and out degree statistics together with their associated von Neumann entropy may be useful as a graph-based indicator to distinguish Alzheimer’s disease subjects from normal healthy control population.
2 Directed Graphs in fMRI Networks
2.1 Preliminaries
Let G(V, E) be a directed graph with node set V and directed edge set \(E \subseteq V \times V\). Each edge \(e=(u,v) \in E\), has a start vertex u and end-vertex v. The adjacency matrix A of the directed graph is defined as
For the node u the in-degree and out-degree of node are
and the total degree of node in the directed graph is \(d_u = d_u^{in} + d_u^{out}\). An edge is said to be unidirectional if \(A_{uv}=1\) and \(A_{vu}=0\), and bidirectional if \(A_{uv}=1\) and \(A_{vu}=1\).
2.2 Von Neumann Entropy for Directed Graphs
For an undirected graph the von Neumann entropy [7] computed from the normalised Laplacian spectrum has been shown to be effective for network characterization. In fact, Han et al. [8] have shown how to approximate the calculation of von Neumann entropy in terms of simple degree statistics. Their approximation allows the cubic complexity of computing the von Neumann entropy from the Laplacian spectrum, to be reduced to one of quadratic complexity using simple edge degree statistics, i.e.
This expression for the von Neumann entropy has been shown to be an effective tool for characterizing structural properties of networks. Moreover, it has extremal values for cycles and fully connected graphs. Ye et al. [4] have extended this result to directed graphs by distinguishing between the in-degree and out-degree of nodes, giving the following expression for the entropy
where the edge set E is partitioned into two disjoint subsets \(E_1\) and \(E_2\), which respectively contain the unidirectional and directional edges.
The two subsets \(E_1\) and \(E_2\) satisfy the conditions that \(E_1 = \{(u,v)|(u,v)\in E \cap (v,u) \notin E\}\), \(E_2 = \{(u,v)|(u,v)\in E \cap (v,u) \in E\}\). \(E_1 \cup E_2 = E\), \(E_1 \cap E_2 = \emptyset \). If most of the edges in the graph are unidirectional, i.e., \(|E_1|\gg |E_2|\), then the graph is said to be strongly directed. In this case we can ignore the entropy associated with the summation over \(E_2\), giving the approximate entropy for strongly directed graphs as
There are thus two factors determining the entropy. The first is the ratio of the in to out degree of the start node u of the directed edge, i.e. \(\rho _u= \frac{d_u^{in}}{d_u^{out}}\), while the second is the directed version of the edge entropy, i.e. \( \frac{1}{d_u^{out} d_v^{in}} \). The former weights the contributions of the entropy associated with the directed edges exiting node u. The contributions to the entropy are thus large if the ratio \(\rho _u\) is small, and directed edge connects nodes with large both out and in degree.
2.3 Entropic Edge Assortativity for Directed Graphs
For undirected graphs, the assortativity is the tendency of nodes to connect to those of similar degree. This concept can be extended to directed graphs if we measure the tendency of nodes to connect to those nodes of similar in and out degree. Foster et al. [11] define the directed assortativity as
where \(\alpha , \beta \in \{in, out\}\) is the incoming and outgoing direction for a directed edge. \(\bar{d_u^{\alpha }} = |E|^{-1} \sum _{(u,v)\in E} d_u^{\alpha }\) and \(\sigma ^\alpha = \sqrt{|E|^{-1} \sum _{(u,v)\in E} (d_u^{\alpha } - \bar{d_u^{\alpha }})^2}\). The similar definitions are for \(\bar{d_v^{\beta }}\) and \(\sigma ^\beta \).
Ye [10] adopts a different approach to defining degree assortativity for directed graphs based on von Neumann entropy decomposition. The method is based on the observation that edges associated with high degree nodes have large entropy and preferentially attach to clusters in a graph. The entropic assortativity measurement provides a novel way to analyze the graph structure. For instance, with the approximation for the von Neumann entropy for directed graph \(S_D\), the coefficient of directed edge assortativity is given by [10]
where \(S_{uv}^{u}\) associate the entropy of all the outgoing edges from vertex u, and \(S_{uv}^{v}\) are all the incoming edges of vertex v.
3 Experiments and Evaluations
In this section, we describe the application of the above methods to the analysis of interregional connectivity structure for fMRI activation networks for normal and Alzheimer subjects. We first examine the differences in degree distribution for the four groups of subjects. Then we apply the entropy-based analysis to distinguish Early Mild Cognitive Impairment (EMCI) and Late Mild Cognitive Impairment (LMCI).
3.1 fMRI Data Set
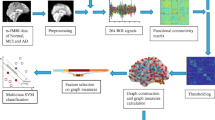
The fMRI data comes from the ADNI initiative [9]. Each image volume is acquired every two seconds with Blood-Oxygenation-Level-Dependent (BOLD) signals. The fMRI voxels here have been aggregated into larger regions of interest (ROIs). The different ROI’s correspond to different anatomical regions of the brain and are assigned anatomical labels to distinguish them. There are 96 anatomical regions in each fMRI image. The correlation between the average time series in different ROIs represents the degree of functional connectivity between regions which are driven by neural activities [12].
A directed graph with 96 nodes is constructed for each patient based on the magnitude of the correlation and the sign of the time-lag between the time-series for different anatomical regions. To model causal interaction among ROIs, the directed graph uses the time lagged cross-correlation coefficients for the average time series for pairs of ROIs. We detect directed edges by finding the time-lag that results in the maximum value of the cross-correlation coefficient. The direction of the edge depends on whether the time lag is positive or negative. We then apply a threshold to the maximum values to retain directed edges with the top 40% of correlation coefficients. This yields a binary directed adjacency matrix for each subject, where the diagonal elements are set to zero. Those ROIs which have missing time series data are discarded.
Subjects fall into four categories according to their degree of disease severity. The classes are full Alzheimer’s (AD), Late Mild Cognitive Impairment (LMCI), Early Mild Cognitive Impairment (EMCI) and Normal Healthy Controls (HC). The LMCI subjects are more severely affected and close to full Alzheimer’s, while the EMCI subjects are closer to the healthy control group (Normal). We have fMRI data for 30 AD subjects, 34 LMC subjects, 47 EMCI subjects, and 38 normal healthy control subjects.
3.2 Alzheimer’s Classification
We first investigate the in and out degree distribution of the data by showing a scatter plot in-degree versus out-degree for each directed edge in the data. In order to extract potential structural difference, the distribution of points in the scatter plot is analyzed using a general linear model. Figure 1 shows the scatter plots of in-degree versus out-degree, comparing the first AD vs. Normal and secondly EMCI vs. LMCI respectively. The obvious difference is that normal subjects exhibit a high degree of interregional connection compared to Alzheimer’s subjects. A similar effect is shown by Early and Late detection groups. Table 1 shows the coefficients of a linear model with 95% confidence bounds and root mean square error.
The results of fitting the linear model show that the in and out degree distributions for the nodes in the AD and LMCI groups of subjects have a greater slope than those of the Normal and Early groups. This implies that there is a greater imbalance in in-degree and out-degree in the Alzheimers and late detection groups. In other words, the nodes in the fMRI inter-regional connectivity graphs for these two groups tend to have larger in-degree than out-degree. Moreover, the small value of RMSE in these two groups reveals that for Alzheimer’s subjects the scatter about the regression lines is smallest. By contrast, for the normal and early control subjects the scatter is significantly higher. This underlines the imbalance in in-degree for the subjects belonging to the diseased groups.
The in-degree/out-degree distribution for edges in the directed graphs in Normal Healthy Control and Alzheimer’s groups (left), Early Mild Cognitive Impairment(EMCI) and Late Mild Cognitive Impairment (LMCI) (right). The blue stars represent the edges in normal patients’ graphs which occupy the high degree region with large variance. The red cycles show the AD patients’ graphs with narrow and low degree occupation. (Color figure online)
We can explore this asymmetry of in and out degree in more detail using Ye’s entropy assortativity measure. This gauges the extent to which nodes to connect to others with similar in-degree or out-degree [6]. To represent the structural difference regarding the entropy associated with degree of each node, we plot the histogram of edge entropy assortativity in Fig. 2. It shows the difference in entropy of the directed edges for subjects in AD vs. Normal, and EMCI vs. LMCI. By comparing the directed edges in the AD and normal groups, we conclude that the edges in the directed graphs for Alzheimer’s subjects tend to have a higher value of entropy, and this reveals the structure is weakly connected with a lower average in out to in degree ratio. A similar effect is shown in the EMCI and LMCI subject groups. For late Alzheimer’s subjects, the shift in entropy to the right represents the weak degree connection in the nodes. This clearly reveals the loss of interregional connection for directed edges in Alzheimer’s.
Finally, the in-degree and out-degree of nodes are used as the features to distinguish the different group of subjects. For each edge, we construct four dimensional feature vectors with two nodes and in and out degree measurements on each node. So the graph can be represented by these directed edges associated with four-dimensional feature vectors. We perform the linear discriminant analysis (LDA) on the Alzheimer’s (AD) and Normal healthy control groups as the training process to find the decision boundary. Then the LDA model is applied on the EMCI and LMCI groups to classify patients. We compare the results and the labels to get classification accuracy.
Table 2 shows the classification accuracy of linear discriminant analysis(LDA). The directed graphs for the AD and Normal subjects are used as the training data to find the decision boundary. The performance of the resulting LDA classier is high with an accuracy of 87.87% when computed using 10-fold cross-validation. We randomly divide the AD and Normal subjects into 10 disjoint subsets of equal size. Remove one subset, train the LDA model using the other nine subsets. This process is repeated by removing each of the ten subsets once at a time and then average the classification accuracy. In order to evaluate the performance of classification, we provide results for sensitivity and specificity for LDA classifier. The sensitivity indicates the percentage of Alzheimer’s people who are correctly identified. It reaches 88.59% which represents the high percentage of correctly classified. In addition, the specificity shows the true negative that is the healthy people correctly identified as healthy. It is 87.10% revealing most normal healthy people are correctly identified in the Normal group. Similarly to the LDA in AD and Normal classier, for the discrimination of subjects belonging to the EMCI and LMCI groups, we obtain a classification accuracy of 80.47%. Although this result is acceptable, the sensitivity is reduced to 75.85% indicating some percentage of patients are not correctly classified in LMCI groups.
3.3 Identifying Salient Nodes for Disease Classification
Identifying diseased regions in the brain is also important in the study in Alzheimer’s analysis. Several studies have shown that in anatomical structures the corresponding ROIs are important for understanding brain disorders [1, 3]. Here we compute the difference of out-degree and in-degree in our study and investigate the method for identification of the disease nodes in patients with Alzheimer’s.
Directed edge entropy difference between Alzheimer’s (AD) and Normal Healthy Controls (HC) groups (left). The ratio of out-degree and in-degree difference corresponding to each ROI in two groups of AD and Normal patients (right). The significant changes of degree ratio in each nodes associate to the similar pattern in edge entropy plot, which illustrates the disease area in the brain.
We first compute the histogram of degree imbalance, i.e. out-degree minus in-degree for each node. Figure 3 compares histograms obtained for AD and HC, and for EMCI and LMCI. The obvious feature is that the directed graphs for HC (normal) and EMCI (early development) groups give a much broader range of degree difference compared to that for the AD (fully developed disease) and LMCI (late development) groups. In other words for subjects with fully developed AD, there is a loss of connection between brain regions and gives rise to a narrowing of the distribution of degree difference.
We now plot the difference in directed edge entropy between corresponding regions (nodes) in the directed graphs for the AD and HC groups. We find a similar feature pattern of the degree difference in both plots as shown in Fig. 4. The entropic measurements associated with degree difference in the brain areas, such as the Temporal Gyrus, Parahippocampal Gyrus, Operculum Cortex and Lingual Gyrus, suggest that subjects with AD experience loss of interconnection in their brain network during the progression of the disease.
As listed in Table 3, the ten anatomical regions with the largest entropy differences for subjects with full AD are right Parahippocampal Gyrus, left Inferior Temporal Gyrus, left Paracingulate Gyrus, right Temporal Fusiform Cortex, right Heschl’s Gyrus, left Parietal Operculum Cortex, right Paracingulate Gyrus, left Temporal Fusiform Cortex, left Central Opercular Cortex and left Inferior Frontal Gyrus. This result is consistent with the previous study [5, 6], which suggested that the middle temporal gyrus is an important region in AD pathology [3]. The parahippocampal gyrus has consistently been reported as being an affected region in EMCI and AD [11]. The loss of connection between these brain regions results in significant functional impairment between healthy subjects and patients with AD.
We now repeat our LDA analysis using just the salient regions listed in Table 3, since it is the impairment of connections to these anatomical structures that appears to determine the onset of AD. We perform LDA on the 4 vectors representing the pairs of listed anatomical regions. The classification accuracy is shown in Table 4. In comparison to the previous results in Table 2, the accuracy increases by about 3% in AD/Normal groups and 6% in the EMCI/LMCL groups. All other performances are also improved with these selected degree features.
4 Conclusions
In conclusion, this paper is motivated by filling the gap in the literature of analyzing fMRI regional brain interaction networks using directed graphs. We commence from the recently developed simplified approximations to the von Neumann entropy of directed graphs, which are dependent on the graph size and the in and out degree statistics of vertices. In order to characterize the functional organization of the brain, assortativity of nodes in directed graphs provides insights into the neuropathology of Alzheimer’s disease. Entropic measurements associated with node degree identifies the edge connection features which offer high discrimination between subjects suffering from AD and normal subjects.
References
van den Heuvel, M., Pol, H.E.H.: Exploring the brain network: a review on resting-state fMRI functional connectivity. J. Eur. Neuropsychopharmacol. 20, 519–534 (2010)
Anwar, A.R., et al.: Complex network analysis of resting-state fMRI of the brain. In: 2016 IEEE 38th Annual International Conference of the Engineering in Medicine and Biology Society (EMBC). IEEE (2016)
Rubinov, M., Sporns, O.: Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52(3), 1059–1069 (2010)
Ye, C., Wilson, R.C., Comin, C.H., Costa, L.D.F., Hancock, E.R.: Approximate von Neumann entropy for directed graphs. Phys. Rev. E 89(5), 052804 (2014)
Rombouts, S.A., Barkhof, F., Goekoop, R., Stam, C.J., Scheltens, P.: Altered resting state networks in mild cognitive impairment and mild Alzheimer’s disease: an fMRI study. Hum. Brain Mapp. 26(4), 231–239 (2005)
Khazaee, A., Ebrahimzadeh, A., Babajani-Ferem, A.: Classification of patients with MCI and AD from healthy controls using directed graph measures of resting-state fMRI. Behav. Brain Res. 322, 339–350 (2016)
Passerini, F., Severini, S.: The von neumann entropy of networks. Int. J. Agent Technol. Syst. 1, 58–67 (2008)
Han, L., Escolano, F., Hancock, E.R., Wilson, R.C.: Graph characterizations from von neumann entropy. Pattern Recogn. Lett. 33, 1958–1967 (2012)
Alzheimer’s Disease Neuroimaging Initiative (ADNI). http://adni.loni.usc.edu/
Ye, C., Wilson, R.C., Hancock, E.R.: An entropic edge assortativity measure. In: Liu, C.-L., Luo, B., Kropatsch, W.G., Cheng, J. (eds.) GbRPR 2015. LNCS, vol. 9069, pp. 23–33. Springer, Cham (2015). doi:10.1007/978-3-319-18224-7_3
Foster, J.G., Foster, D.V., Grassberger, P., Paczuski, M.: Edge direction and the structure of networks. Proc. Natl. Acad. Sci. U.S.A. 107(24), 10815–10820 (2010)
Wang, J., Wilson, R.C., Hancock, E.R.: fMRI activation network analysis using bose-einstein entropy. In: Robles-Kelly, A., Loog, M., Biggio, B., Escolano, F., Wilson, R. (eds.) S+SSPR 2016. LNCS, vol. 10029, pp. 218–228. Springer, Cham (2016). doi:10.1007/978-3-319-49055-7_20
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Wang, J., Wilson, R.C., Hancock, E.R. (2017). Detecting Alzheimer’s Disease Using Directed Graphs. In: Foggia, P., Liu, CL., Vento, M. (eds) Graph-Based Representations in Pattern Recognition. GbRPR 2017. Lecture Notes in Computer Science(), vol 10310. Springer, Cham. https://doi.org/10.1007/978-3-319-58961-9_9
Download citation
DOI: https://doi.org/10.1007/978-3-319-58961-9_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-58960-2
Online ISBN: 978-3-319-58961-9
eBook Packages: Computer ScienceComputer Science (R0)