Abstract
Plant small RNAs, namely si- and miRNAs, control a gamut of biological functions by regulating gene expressions. One of the major functions is to protect the host genome from molecular parasites, including the viruses. The virus-infected plants allow generating the siRNAs from all over the viral genomes that, in turn, control viral gene expressions post-transcriptionally leading to inhibition of viral growth and spread. In the case of DNA viruses, the siRNAs also exert transcriptional control of viral gene expression in an epigenetic manner by promoting methylation of the promoter of viral genes. Further, transcriptional gene silencing (TGS) mechanism has also been shown to be involved in symptom remission. DNA viruses also interfere with the methyl cycle to prevent the availability of methyl donor (S-adenosyl methionine) for methylating viral DNAs. However, in the battles between the host and viruses, the viruses have also evolved to encode few proteins from their genomes that counteract the RNAi-mediated host defense reactions. Such group of proteins is collectively known as RNAi suppressors which also participate in viral life cycle in manifold ways besides thwarting the host RNAi activities towards the viruses. In addition, these virus-encoded proteins also manipulate the components of TGS machinery such as histone and/or DNA methyl transferases, to combat the antiviral silencing mechanism. These are also called the pathogenicity factors as they principally govern the disease symptoms in the host. The mechanistic action of a few of the viral-encoded suppressors has been dealt in some detail within the text. These proteins deregulate the host miRNAs during the expression of disease. Several studies have now shown that transgenic expression of viral suppressors can alter the accumulation and/or functioning of miRNAs leading to developmental abnormalities. Molecules like HC-Pro, P19, etc. were shown to affect the processing and activity of miRNAs. Hence the antiviral strategies could be developed by silencing these viral suppressors. Our laboratories have developed tomato transgenics expressing miRNAs and tasiRNAs which can efficiently silence the RNAi suppressors of tomato leaf curl viruses and offer a high degree of tolerance towards the viruses. The future direction of research including the biotechnological usages of the viral suppressors has been discussed.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
RNA interference (RNAi) or RNA silencing is the natural process of switching off gene expression during fundamental processes like development, genome maintenance, and defense against foreign molecules like viruses. As a counter defense, viruses have also evolved to encode proteins to suppress RNA silencing mechanisms that are known as RNAi suppressors. With the rapid advancement in science, a lot of information has emerged regarding the mechanisms and machinery of RNA silencing and its suppression (Agrawal et al. 2003; Roth et al. 2004). These are being exploited as a new tool for developing antiviral products, which have large applications in field of medicine, agriculture, and basic biology. In this review, we have discussed the virus triggered RNAi response and the mechanisms evolved by viruses to suppress this pathway for their own advantage.
2 RNAi and the Suppressors
The RNAi science evolved with the serendipitous as well as the famous story of transgenic petunia flowers in 1990 (Napoli et al. 1990). Now the various forms and associated mechanisms of the effectors of gene silencing are well known and are still being represented in the literature. The three major forms of small RNAs, namely the small interfering RNAs (siRNAs), microRNAs (miRNAs), and picoRNAs (piRNAs), are well described in almost all eukaryotic creatures including many non-model organisms like parasites, fungi, etc. (Perrimon et al. 2010; Nicolas et al. 2013). Besides these three forms, many other forms of siRNAs like rasiRNA, tasiRNA, natsiRNAs, etc. are also reported in the literature. The biogenesis, functions, and cross-talks of these small RNAs requires the participation of many silencing factors, known collectively as the RNAi factors. The functions of these factors are well conserved across evolution; the characteristics motifs underlying the functions of many factors are well recognized. However, there are also many reported factors that exert their gene silencing effects in a system- and tissue-specific manner.
An evolutionarily conserved function of a subset of RNAi factors is to safeguard the host and its genome from invading molecular parasites like viruses and transposons. Following viral entry in the host, a pathogen-triggered immunity (PTI) will be invoked in the host. If the RNAi factors provide the PTI function, the pathogen effector-triggered sensitivity (ETS) will also come in play following the Z-model of PTI-ETI scheme of the host–pathogen arms race (Jones and Dang 2006). The viral-encoded suppressor of RNAi can straightforwardly fit the criterion of ETS. These are also known as RNA silencing suppressors (RSSs) or viral suppressors of RNA silencing (VSRs) and were initially brought into the limelight through a report by Voinnet et al. (1999). Till now more than 80 VSRs of plant, animal, and insect origins are documented; however, the mechanistic details of a few of VSRs are reported in atomistic details. These suppressors generally do not have common motifs but a subset of them has GW/WG repeats and RNA binding (RBS) motifs (Bivalkar-Mehla et al. 2011). It probably entails that the VSRs have multiple independent origins leading to high divergence in function and thus intercept at various steps of RNAi pathway.
3 Antiviral RNAi
3.1 Viral SiRNA Generation
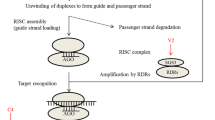
All viruses with either RNA or DNA genomes present genomic or sub-genomic forms of intracellular double-stranded (ds) RNA which are eventual sources of viral siRNAs (vi-siRNAs). The vi-siRNAs are commonly produced from three distinct processes in which the dsRNA precursors are formed and further subjected to Dicer or Dicer-like (DCL) mediated cleavage to switch on the RNA silencing mechanisms. The entire process starts with the utilization of available sources of dsRNA to yield the primary vi-siRNA, structure-associated vi-siRNA, and secondary vi-siRNA. Primary vi-siRNAs are the derivative of an intermediate of genome replication formed either due to the activity of virus-encoded RNA polymerases (encoded by RNA viruses) or through transcription of the viral genome in the case of DNA viruses. Apart from this, these structures are also produced by convergent transcription. Another class of siRNAs are structure-associated vi-siRNA, which are in fact the defectively base-paired viral transcripts forming an imperfect secondary structure. The third class includes the secondary vi-siRNA, which are produced from the ssRNA by the active participation of host RNA-dependent RNA polymerase (RDR) family (Ahlquist 2006). Biogenesis of vi-siRNA includes transcription, processing, modification and finally these vi-siRNAs load on to the RNA Induced Silencing complex (RISC) to silence the viral transcripts. Figure 1 displays the biogenesis and functions of the vi-siRNAs along with interference from the VSRs.
A schematic representation of vi-siRNA biogenesis and function. Double-stranded RNAs generated from RNA (viral mRNAs mainly) and DNA viruses (convergent, bidirectional, and replicative intermediate transcriptions) are cleaved with the specialized proteins DCLs and produce a variety of siRNA species (21, 22, and 24 nt). VSRs target these proteins to hinder siRNAs generation and subsequently impede both TGS and PTGS pathways. VSRs are shown in red stars. Black-dashed lines indicate the normal steps of silencing pathway, while red-dashed lines designate the process manipulated by the suppressors. CaMV P6, Cauliflower mosaic virus P6 protein; RYSV P6, Rice yellow stunt virus P6 protein; RYMV P1, Rice yellow mottle virus P1 protein; SPMMV P1, Sweet potato mild mottle ipomovirus P1 protein; ToRSV CP, Tomato ringspot virus coat protein
These viral dsRNAs are processed by endonucleolytic activity of DCL4 and DCL2 to produce 21 nt and 22 nt vi-siRNA, respectively. DCL2 acts as a substitute of DCL4 and its antiviral activity is initiated only in Arabidopsis plants lacking DCL4. Further, the duplex of vi-siRNA is stabilized by methylation at 2′ OH of 3′ terminal nucleotides by Hua Enhancer 1 (HEN1; Yu et al. 2005) to protect sRNA molecules against uridylation (Li et al. 2005) and against the exoribonuclease activity of small RNA degrading nucleases (SDN1-3) (Ramachandran and Chen 2008). Further, amplification and systematic RNA silencing occur through the activities of secondary siRNAs. RDR proteins facilitate production of elongated complementary viral RNAs (transitive RNA) which are subsequently subjected to DCL processing (Vance and Vaucheret 2001). Arabidopsis thaliana RDR6 contributes largely to the process of amplification. In case of DNA viruses, the 24 nt vi-siRNAs (product of DCL3) are also generated. These 24 nt vi-siRNAs initiate transcriptional gene silencing by inducing cytosine methylation of target DNA sequence (Lister et al. 2008). This phenomenon is discussed in detail in Sect. 3.2.
The formation, accumulation, and functional stages of vi-siRNAs are also subject to the inhibitory activities of VSRs as described in Sect. 4. The accumulation and activities of vi-siRNAs should, thus be viewed as the host defense response towards the viruses.
3.2 Transcriptional Control of Viral Genes
Transcriptional gene silencing (TGS) involves the production of siRNA homologous to the non-coding region of a target virus, which is also linked with the corresponding methylation of the virus genome, an event that controls viral gene expression. This process consists of three key steps: initiation, effector, and amplification/spreading of silencing. The 24 nt vi-siRNAs corresponding to viral non-coding regions are the key players in this process. In the RNA-directed DNA methylation (RdDM), vi-siRNAs are recruited to the RNA-Induced Transcriptional Silencing (RITS) complex leading to chromatin remodelling (Huang et al. 2007). Argonaute 4 (AGO4) protein promotes chromatin modification through cytosine as well as histone methylation. The downstream processing requires the contribution of various enzymes responsible for either de novo methylation (Domains Rearranged Methyltransferase 2, DRM2) or maintaining the methylation (Chromomethylase 3, CMT3; Methyltransferase 1, MET1; and Kryptonite, KYP2). Apart from this, RdDM entails the action of some chromatin remodelers such as Defective in RNA-Directed DNA Methylation1 (DRD1) and Decrease in DNA Methylation1 (DDM1), which are necessary to ensure viral DNA availability to RNA signals and the maintenance of symmetric methylation, respectively (Raja et al. 2010).
Production of vi-siRNAs and the consequent DNA methylation in various plant–virus interactions have been recently highlighted. These reports advocate that upon infection by DNA virus, the host activates TGS to suppress the transcription of the viral genome. For example, generation of a wide range of vi-siRNAs have been reported in Nicotiana–African cassava mosaic virus (ACMV) interaction as well as during Arabidopsis–Cabbage leaf curl virus (CaLCuV) interaction (Akbergenov et al. 2006; Vanderschuren et al. 2007). These results put forward that TGS is the key pathway, which is implicated in the plant defense against geminivirus. Later on, the association of TGS pathways with biological functions such as symptom remission phenotypes was revealed. For example, pepper–Pepper golden mosaic virus (PepGMV) interaction showed a recovery phenotype provided by the presence of vi-siRNAs (Carrillo-Tripp et al. 2007).
Moreover, it has been substantiated that the expression of virus genes is frequently targeted for the DNA methylation through the RdDM pathway (Yadav and Chattopadhyay 2011; Sahu et al. 2014). Methylation in the promoter region which is essential for the viral transcription can inhibit the accumulation of viral transcripts, thus reducing the infectivity in the infected plant. The 24 nt vi-siRNAs also cause methylation of the intergenic region of Mungbean yellow mosaic India virus (MYMIV; Yadav and Chattopadhyay 2011) as well as Tomato leaf curl New Delhi virus (ToLCNDV; Sahu et al. 2014). Scanning of the viral genome producing a higher level of siRNAs revealed that there was a strong correlation between the accumulation of small RNAs and genome methylation processes. Taken together, these studies suggest that the TGS and viral genome methylation act as a key regulatory process to minimize or limit the viral gene expression. The detailed study of the viral DNA methylation suggested that the expression of genes encoding enzymes linked with the cytosine methylation occurs in specific patterns (Yadav and Chattopadhyay 2011; Sahu et al. 2014). Upon ToLCNDV infection in a tolerant cultivar of tomato, higher expression of DRM1 had been observed leading to enhanced de novo methylation; moreover, higher expression in level of methylation maintenance genes CMT3 was also reported. Hence, we may infer that the change in the level of key methylation maintenance enzymes might be linked with RdDM, which is plausibly involved in the progression of siRNA-directed silencing pathway in a tolerant response against geminiviruses.
3.3 Post-transcriptional Control of Viral Proteins
3.3.1 Post-transcriptional Gene Silencing
Post-transcriptional gene silencing (PTGS) is one of the most efficient defense strategies that plants have devised against viral pathogens (Baulcombe 1999; Waterhouse et al. 2001). It is regarded as a form of immune system that operates at the nucleic acid level and can act against any cytoplasmic RNA species homologous with the small RNA molecules (Voinnet 2001). This defense is not host-programmed but depends on the genome sequence of the invading DNA or RNA virus (Ruiz et al. 1998; Matthew 2004), hence it can remarkably silence the expression of potentially any virus. The RNA silencing signals can propagate to distant parts of the plant, thus conferring immunity to non-infected parts of the plant (Palauqui et al. 1997; Voinnet and Baulcombe 1997; Palauqui and Vaucheret 1998; Voinnet et al. 1998; Sonoda and Nishiguchi 2000).
RNA silencing involves diversity in its mode of action as well as its components. Besides the vi-siRNAs, the host miRNAs also exert post-transcriptional control of viral transcripts. The biogenesis and function of these forms of small RNAs requires a number of different proteins. However, the two main players in the pathways are DCL and AGO, which require attention due to their commonality to all the small RNA pathways.
DCL belongs to a family of RNase III-like endoribonucleases which act on dsRNAs and cleave them into smaller fragments in a sequence-independent manner (Bernstein et al. 2001). In general, it contains a helicase-C, DExD-helicase, PAZ, Duf283, RNaseIII, and dsRNA-binding domain. All the DCLs contain two RNaseIII domains, which act simultaneously to cleave the dsRNA (Finnegan et al. 2003; Margis et al. 2006). The number of DCLs varies in organisms from single in humans and mice (Zhang et al. 2004) to four in A. thaliana (Finnegan et al. 2003; Liu et al. 2005). Mutation analysis of the four A. thaliana DCL (AtDCL) genes showed that the species and the corresponding functions of a small RNA depend on the type of DCL enzyme involved in its biogenesis. For example, AtDCL1 has been shown to generate miRNAs, while AtDCL2 is implicated in the production of siRNAs linked with virus defense and also the production of siRNAs from natural cis-acting antisense transcripts. On the other hand, AtDCL3 takes part in the siRNA generation that guides chromatin modification. AtDCL4 is essentially required to produce tasiRNAs which regulates variations associated with vegetative phase (Hutvágner et al. 2001; Llave et al. 2002). In rice, 8 DCL coding genes have been identified so far; however, their distinct roles and their effect in rice development are still unclear (Kapoor et al. 2008). Mutation analysis of DCL1 in rice showed great reduction in number of miRNAs as well as developmental arrest at seedling stage (Liu et al. 2005); however, in the same mutations, the production of siRNAs was not affected.
AGO is an evolutionarily conserved protein and the main slicer element of the RISC in plants and animals (Wu et al. 2009b). Its number significantly differs in each organism and 10 different AGO proteins are known in Arabidopsis (Vaucheret 2008), whereas 19 AGO proteins have been identified in rice. AGO1 is the major effector protein of miRNA-induced silencing (Mallory and Vaucheret 2010; Wu et al. 2009a; Wang et al. 2009). When a correct pairing of 2–8 nt between the miRNA and an RNA strand is detected, the catalytic machinery of RISC-AGO complex proceeds to the silencing of the target by either cleavage of the target or translational repression. The vi-siRNAs silence the viral transcripts in a similar post-transcriptional process using the siRNA-RISC pathways. The relative weights of TGS and PTGS are virus—as well as host tissue-specific. DNA viruses are more prone to TGS while the RNA viruses are subject more to PTGS processes.
3.3.2 Host MiRNA Control of Viral Genes
Animal miRNAs are well known to control viral genes but plant miRNAs doing the same job are not reported yet. However, bioinformatic predictions about plant miRNAs have shown that they have a role in plant–virus interactions by targeting the genomes of plant infecting viruses (Naqvi et al. 2010) and they are also thought to regulate the tissue tropism of virus in the host to some extent (Ghosh et al. 2009). In order to highlight the probable geminivirus targets for miRNAs encoded by the six plant genomes, we have carried out bioinformatics analysis in detail and the same is presented in tabular (Table 1) as well as figure form (Fig. 2).
(a) Number of potyvirus targets predicted for miRNAs encoded in the six plant genomes. The inset box shows one of the best representative miRNA:target alignment, between osa-miR5535 and Calla lily latent virus polyprotein gene (L594_gp1). (b) Number of geminivirus targets predicted for miRNAs encoded in the six plant genomes. The inset box shows one of the best representative miRNA:target alignments, osa-miR396f-5p:Sida golden
A total of 2527 miRNA sequences were downloaded from the miRBASE release 21 from six plants (Arabidopsis thaliana, Glycine max, Oryza sativa, Sorghum bicolor, Vitis vinifera, and Zea mays) (Griffiths-Jones et al. 2008). Complete genome sequences for two major families of plant infecting viruses, namely, geminiviruses and potyviruses, were obtained from Genbank (Benson et al. 2005) and these sequences were used to look for the targets of the above- mentioned miRNAs using a certain set of rules. A modified version of miRanda (ver. September 2008) was essentially used for target predictions (Enright et al. 2003). The miRanda scoring matrix allows G=U “wobble” pairs, important for the detection of RNA:RNA hybrid duplexes. The folding algorithm was based on the Vienna 1.3 RNA secondary structure programming protocols (Hofacker et al. 1994). Although miRanda was originally developed to look for animal miRNA targets, it can be modified and used to search for targets in other systems like viruses and plants (Hsu et al. 2007; Maziere and Enright 2007). The other criteria to consider a sequence as a putative miRNA target were: four or fewer mismatches overall, only one or none mismatches in the 5′ region of the miRNA (positions 1–12), no more than two consecutive mismatches in positions 13–21, and no mismatches in positions 10 and 11. Additionally, the miRNA:target pair should have low free-energy of bonding (maximum −20 kcal/mol) and parameter “strict” was also used to ensure no mismatches in seed region (Lin et al. 2009; Zhang et al. 2006; Schwab et al. 2005). The resultant hits in the viral targets have been summarized in the Table 1.
The number of targets found in each of the viral genomes has been displayed in Fig. 2a (for potyviruses) and Fig. 2b (for geminiviruses).
The viral targets include mostly the viral proteins and occasionally the intergenic regions. The targets are predicted to undergo slicing as well as translational repression.
4 Viral Counterstrategy
Viruses fight back the mechanism of host RNA silencing by encoding protein molecules known as RNA silencing suppressors (RSSs), and the RSSs encoded by viruses are also known as VSRs. The VSRs are known to interfere at different stages of RNA silencing pathways, thus helping in efficient infection and replication of virus in the host cell and spreading the infection systemically (Voinnet et al. 1999; Shi et al. 2002). These VSR molecules are generally usual viral proteins such as coat protein (CP), movement protein (MP), or proteases that carry the suppressor activity in the form of their secondary function (Hartitz et al. 1999). As a result, there is extensive assortment in the VSRs documented from the distinct viruses.
4.1 Earlier Experiments to Confirm RNA Silencing Suppression
The indications on the existence of the VSRs came from the early observations that certain specific proteins expressed by viruses played a significant role in their virulence. Subsequently it was highlighted that coinfection with a combination of viruses resulted in enhanced symptom severity rather than the single virus infection. One of the classical examples is Potato virus X (PVX) which, by itself, causes mild symptoms but multiplies vigorously during coinfection with the Potato virus Y (PVY) and Tobacco etch virus (TEV) (Pruss et al. 1997). This phenomenon was referred as synergism (Darnirdagh and Ross 1967) and it is now implicit that the enhanced synergism is mainly due to the weakening of host defense by VSR targeting the silencing pathway at multiple points (Pruss et al. 1997; Mlotshwa et al. 2005).
In the year 1998, the initial report on identification of virus-mediated RSS came exclusively on a potyvirus-encoded helper component proteinase (Hc-Pro). This protein was identified as a major component involved in the enhancement of replication of unrelated viruses. In one such report, it was shown that P1/Hc-Pro suppressed the PTGS of uidA gene coding for β-glucuronidase (GUS) reporter on a highly expressed locus (Kasschau and Carrington 1998). In a similar but independent study, Nicotiana tabacum post-transcriptionally silenced for uidA was crossed with four independent transgenic plants expressing TEV P1/HC-Pro. It was identified that silencing efficiency was boosted in the progenies (Anandalakshmi et al. 1998).
Another study by Brigneti and coworkers (1998) revealed that PTGS of a green fluorescent protein (GFP) transgene was repressed in Nicotiana benthamiana infected with Cucumber mosaic virus (CMV); however, this suppression was not evident with PVX infection. In the same experiment, they expressed HC-Pro of PVY and 2b protein of CMV-encoded proteins in a PVX vector and demonstrated that they act as VSRs. Their study also anticipated that HC-Pro acts by hindering the maintenance of PTGS process in the tissues where silencing had previously been established. On the other hand, the 2b protein had prevented the commencement of gene silencing at the growing parts of the plants (Brigneti et al. 1998). So the identification and understanding of VSRs provided evidence for reversal of silencing of RNA as a natural antiviral defense response (Voinnet 2001). Besides, the VSRs can suppress silencing in both animal and plant cells, regardless of their host preference due to the conserved nature of the silencing phenomenon.
Apart from this, there are few reports which confirm that single virus may code for multiple VSR proteins. For example, Citrus tristeza virus (CTV) was reported to code for three different proteins p20, p23, and the coat protein (CP) exhibiting RSS activity (Lu et al. 2004). These preliminary findings provided a novel insight to find more VSRs as it seemed to be a universal strategy used by viruses against one of the most potent induced immune system of plants. Since then, a large number of viral proteins have been discovered which show RSS activity of dissimilar potency depending upon the host.
4.2 Assays to Detect RNA Silencing Suppressors
Following the discovery of HC-Pro as a VSR, many other virus proteins exhibited the capability of inhibiting the host antiviral mechanism. This establishment was greatly accelerated due to the availability of several simple and efficient functional assays to detect the RSS activity. Identification and functional characterization of RSS in turn facilitated the understanding of the intricacies of the RNA silencing pathway. It also provided insight into the evolutionary arms race between the host and the pathogens during pathogenicity.
During the formative period of VSR concepts, the major bottleneck in the identification of RSS was probably the unavailability of large array of screening systems. In plants, however, a number of strategies have been exploited to analyze the RSS activities of a candidate viral protein. These are mainly based on monitoring the role of the viral protein in suppressing the RNA-mediated silencing of a reporter gene. The reporter gene may be silenced constitutively (Elmayan and Vaucheret 1996) or locally by infiltrating through the Ti plasmid via infection by Agrobacterium (Voinnet and Baulcombe 1997; Voinnet et al. 1998). In the subsequent sections, we have listed and briefly explained the commonly used assays.
4.2.1 Agrobacterium-Mediated Transient Assay
In this method, transgenic tobacco plants stably silenced for a reporter gene like GFP or GUS are used. The candidate RSS is locally introduced into the transgenic silenced plant through infiltration of an Agrobacterium strain carrying the putative VSR gene. This method is called agro-infiltration. If the ectopically expressed protein has capabilities of suppressing the RNA silencing, then localized reversal of silencing will lead to expression of the reporter gene in the infiltrated zone. This is one of the most widely used assays for RSS analysis, due to its simple protocol and rapid generation of result (Karjee et al. 2008). A modification of this method involves coinfiltrating the reporter gene into wild-type tobacco plants along with the VSR using two Agrobacterium strains and monitoring the reporter gene expression (Llave et al. 2000; Voinnet et al. 2000; Johansen and Carrington 2001). The infiltration of the reporter gene will eventually initiate RNA silencing and the reporter will be silenced after three to five days. In the presence of candidate RSS protein, there will be suppression of silencing and the reporter gene expression is retained to a high level or may even increase after 6 days. By means of different reporter constructs, for instance genes organized as inverted repeats, it is possible to evaluate at which step of RNA silencing the suppressor protein acts (Takeda et al. 2002).
4.2.2 Reversal of Transgene Induced Silencing
In this method, plants expressing a reporter gene are systemically silenced through introducing the Agrobacterium expressing the reporter gene or a fragment of it. The reporter gene expression is monitored after the infection of single or multiple viral constructs. Re-establishment of reporter gene expression designates that the tested virus construct contains a RSS activity. Nevertheless, PVX encoding a RSS, which is incapable to restore the reporter gene expression, has been utilized as a vector to evaluate the RSS capability of other viral proteins (Brigneti et al. 1998).
4.2.3 Crossing Assay
This assay exploits a cross between a silenced transgenic plant and a second transgenic plant expressing a candidate viral protein (Anandalakshmi et al. 1998; Kasschau and Carrington 1998). A substantial disadvantage of this method is that higher VSR activity develops various abnormal phenotypic defects in the plants (Anandalakshmi et al. 1998). Though, the assay had been successfully utilized in various studies (Kasschau et al. 2003; Chapman et al. 2004; Dunoyer et al. 2004). A better alternate to mitigate this risk would be the ectopic expression of target gene through a heterologous viral vector system inoculated onto the silenced transgenic plants.
4.2.4 Grafting Assay
The principle of this assay is based on the fact that silencing molecules or signals display systemic movement from a silenced rootstock to a non-silenced scion in a grafted plant. The silencing signal spreads via RNA-mediated processes and has been extensively studied and reviewed (Chitwood and Timmermans 2010; Kalantidis et al. 2008). Grafting experiment itself has been one of the most reliable strategies to study suppression of silencing in plants. For rootstock, a line silenced for a reporter transgene is selected and the candidate RSS is introduced in it with the help of genetic crossing experiments. Further, a scion expressing the same reporter transgene is grafted on this rootstock. If the candidate is not a RSS, the silencing signal will systemically spread from rootstock to scion and the reporter will be silenced in the scion. However, if the candidate has RSS activity, the reporter will be expressed in the scion. This assay is time consuming and needs raising transgenics as well as breeding experiments. The grafting itself requires lot of practice and expertise. However, the reliability of the assay compensates for its time. This assay has been quite helpful in the identification of suppressors with specific activity on local and systemic silencing.
4.2.5 Specific Biochemical Assays
There are specific biochemical assays in the RNA silencing pathway, namely the Dicing and RISC assays (Carbonell et al. 2012; Wang et al. 2015). These are stage-specific RNA silencing assays with specific final readouts. Dicing assays convert the dsRNA substrates in siRNAs and RISC assays produce sliced RNA transcripts from the input mRNA molecules. Exogenous additions of proteins in such assays impede the formation of the final readable products. In this way, proteins with RSS activities at the defined steps could be identified.
5 Functional Mechanism of Viral Suppressors of RNAi
RNAi-based immunity in plant against viruses entails a cascade of well-established molecular processes that enhances siRNA/miRNA production and promotes cleavage of targeted transcripts. As counter-defensive mechanisms, viruses may interfere and inhibit each steps of RNAi pathway, affecting the normal sRNA biogenesis. The RNAi suppressors can suppress the pathway of RNAi at different steps as mentioned below.
5.1 Interaction Between DsRNA-VSRs
Many VSRs have attributes of dsRNA-binding proteins. This possibly imitates the fact that every RNAi-mediated antiviral reaction consistently commence with DCL-mediated processing of virus-derived dsRNAs. Hence, targeting dsRNA which acts as a DCL substrate for protection would serve as common strategy for many VSRs. The VSRs are also known to bind siRNAs and consequently inhibit downstream activities of siRNAs.
5.2 Viral Suppressors Target RNAi Effectors
It has been revealed that VSRs may target key RNAi components such as targets AGO1 and DCLs for degradation. Since AGO1 is recognized as a prime component for miRNA function in plants, suppression of these RNAi effectors by VSRs leads to the inhibition of miRNA function. A few VSRs target DCL4 and suppress dicing. VSRs also target RDR proteins of the host to lessen biogenesis of dsRNA and amplification of siRNAs.
5.3 Suppression of Systemic RNAi by VSRs
Additionally, as a counter-defense mechanism, few VSRs are proficient in precise targeting of systemic silencing signal. For example, PVX-encoded P25 and CTV-encoded coat protein are well distinguished for their suppression action on systemic silencing.
5.4 Epigenetic Modifications
As mentioned earlier, cytosine methylation in DNA and histone methylation are the common epigenetic marks that could be brought in by small RNAs, mostly si- and miRNAs. These marks keep DNA unavailable for transcription by various mechanisms. The genomes of the viral DNAs are also known to be subject of this transcriptional control leading to TGS, an account of which is nicely dealt in a recent review (Pooggin 2013). However, a substantial portion of TGS can be reversed by VSRs. The AC2/C2 homologs of begomovirus and Curtovirus genera and the C1 protein of beta-satellite of some begomoviruses can cause reversal of TGS by various mechanisms. The AC2 protein of Mungbean yellow mosaic virus (MYMV), CaLCuV, Tomato golden mosaic virus (TGMV), etc. inactivate Adenosine Kinase (ADK), reducing production of SAM, the methyl donor, and thus cause release of TGS (Trinks et al. 2005; Buchmann et al. 2009). The C1 protein of beta-satellite of Tomato yellow leaf curl china virus (TYLCCNV) inactivates S-Adenosyl Homocysteine Hydrolase (SAHH), an enzyme required for synthesis of SAM, and thus reduces the level of cytosine methylation of viral DNA (Yang et al. 2011). The C2 protein of Beet severe curly top virus (BSCTV) causes reduction of vi-siRNA and decreases methylation of defense response genes so that defense proteins of salicylic acid pathway, the GST superfamily, etc. could be turned on (Yang et al. 2013). In a separate report, it has been shown that the same protein increases the life span of SAMDC1 and thus suppresses DNA methylation-mediated gene silencing in Arabidopsis (Zhang et al. 2011).
Besides DNA methylation, histone methylation is also targeted by the VSR, namely the AC2 protein (Sun et al. 2015). The AC2 protein of Indian cassava mosaic virus (ICMV) upregulates RAV2, which acts as a transcriptional repressor, inhibiting transcription of KYP, a histone methyl transferase. In this way, AC2 dampens TGS and allows viral survival in the infected host. Not only the DNA viruses, but the RNA viruses are also known to relieve TGS. The 2b protein, an RNAi suppressor of severe Shan-Dong (SD) isolate of Cucumber mosaic virus, suppresses RdDM by binding and sequestering siRNAs in a process involving AGO proteins in the nucleolus (Duan et al. 2012).
6 Few Representative VSRs
Even though many VSRs have been described, extensive research has been focused on a few following selected proteins.
6.1 HC-Pro of Potyviruses
The foremost-described VSR is the potyviral HC-Pro protein (Anandalakshmi et al. 1998). This protein is mainly found to affect the processes associated with vector transmission, polyprotein processing, replication of viral genome, and the systemic movement of the virus (Kasschau et al. 1997). It was also well characterized as a comprehensive pathogenicity enhancer assisting in the enhancement in the viral RNA accumulation and development of severe symptoms of virus infection during many distinct virus infections (Pruss et al. 1997), thus representing a direct and robust influence on the maintenance of RNA silencing.
Systemic infection by PVX carrying HC-Pro was capable of reversing the expression of GUS in the reporter gene silenced transgenic plants. It was demonstrated that cross between the GUS silenced lines and HC-Pro expressing plant may possibly reinstate GUS expression. This restoration was due to the action of HC-Pro which contributed to prevent the degradation of the gus mRNA (Anandalakshmi et al. 1998; Brigneti et al. 1998; Hamilton et al. 2002). This suggested that HC-Pro may perhaps impede an RNase III-like enzyme involved in the generation of siRNAs from dsRNA or an active component of the RISC. Interestingly, it was revealed later that HC-Pro did not interrupt the silencing signal cascade within a plant, albeit all siRNAs were eliminated (Mallory et al. 2001). Moreover, HC-Pro was displayed to capably avert the plant from retorting the silencing signal in a grafting experiment (Hamilton et al. 2002). Furthermore, there are few contradictory reports, which suggested the possible involvement of HC-Pro in the DNA methylation at the silenced transgene locus of genome (Llave et al. 2000; Mallory et al. 2001).
A step forward discovery on the mechanism of silencing suppression was the identification of interacting partner of P1/HC-Pro of TEV (Anandalakshmi et al. 2000). In this study, transgenic plants overexpressing rgs-CaM (regulator of gene silencing-calmodulin-like protein) showed phenotypic variations, which were found to be similar to HC-pro transgenic plants. Apart from these characteristics of HC-Pro, it has also been implicated in stimulating the miRNA-mediated gene regulation, thus supporting the previous observation of developmental defects detected in the transgenic plants (Mallory et al. 2002; Kasschau et al. 2003). Molecular structure of this protein revealed that the domain of HC-Pro possesses RNA-binding properties which is essential and prerequisite for silencing suppression (Kasschau and Carrington 2001). Further studies showed that it has the highly conserved FRNK box, which apparently provides a site of interaction with siRNA and miRNA duplex. This directly influences the miRNA abundance and associated regulatory functions, leading to the symptom development (Shiboleth et al. 2007). Overall, these studies suggested that HC-Pro potentially suppresses the RNA silencing downstream of dsRNA and miRNA generation. Conversely, it also alters the upstream process of the siRNA accumulation and probably impedes the systemic spread of silencing signal.
6.2 Cucumoviruses 2b (CMV-2b)
Previous studies have suggested that CMV-2b regulates systemic viral movement, and deficiency of this protein may reduce the pathogenicity of the virus (Ding et al. 1995a, b). Apart from this, CMV-2b protein was found to manipulate viral cell to cell movement in plants (Soards et al. 2002). First report of functional characterization of CMV-2b as a RSS revealed that the inhibition of 2b protein translation of the mild Q strain (Q-D2b) caused attenuation in Nicotiana glutinosa, along with the deficiency of systemic infection in cucumber plant (Ding et al. 1994). Several studies aimed to understand the function of the CMV-2b protein in virulence have been carried out, in last decade. In this context, Diaz-Pendon et al. (2007) identified that the Q-D2b mutant was competent of causing disease in Arabidopsis DCL2-4 mutants. These genes are the key components of RNA silencing component, hence provided a strong correlation between RNA silencing and CMV-2b function. Moreover, mutation in the D2b of severe Fny strain resulted in the restoration of virulence in the rdr1/6, ago1, and ago2 mutants of Arabidopsis (Wang et al. 2011). Interestingly, it was suggested that expression of CMV-2b protein from a mild strain may harmonize the infectivity in the developing tissues in response to the synergistic effect of Tobacco mosaic virus (Siddiqui et al. 2011).
CMV-2b was shown to avert the initiation of RNA silencing in newly emerging tissue but it cannot reverse established RNA silencing (Beclin et al. 1998; Brigneti et al. 1998). This result advocated that 2b might be potentially required for preventing the cell to cell spread of the silencing signal, from the locally infected parts to the rest of the plant to promote further virus spread (Goto et al. 2007). The CMV-2b exhibits dual cellular localization in the cytoplasm as well as nuclear foci (Lucy et al. 2000). Additionally, it was also revealed that CMV-2b possesses a monopartite nuclear localization signal (Lucy et al. 2000), hence may interfere with the restoration of transgene methylation, indicating functioning of 2b in the nucleus (Guo and Ding 2002).
CMV-2b has also been shown to affect the PTGS pathway by directly binding to siRNAs or long dsRNA (Guo and Ding 2002; Mitter et al. 2003), an activity, which differed from strain to strain of CMV. Exhaustive study done by Goto and coworkers (2007) revealed that 2b of severe strain (CM95R) of CMV binds in vitro to both chemically synthesized siRNAs and dsRNAs. Alternatively, 2b suppressor of an attenuated strain of CMV (CM95), which differs in single amino acid from the 2b CM95R, could barely bind to siRNAs. It signifies that the reduction in substantial RSS activity of the CM95 due to the single amino acid change may be responsible for the loss of siRNAs binding property of 2b.
It was also demonstrated that CMV-2b protein could inhibit the function of the siRNAs by directly interacting with AGO1. This interaction was studied in vitro and in vivo, and was found to be predominantly on one surface of the PAZ encompassing unit and part of the PIWI-box (Zhang et al. 2006; Ruiz-Ferrer and Voinnet 2007). This suggested that 2b specifically inhibited AGO1 cleavage activity in RISC reconstitution assays, thereby interfering with miRNA pathway and causing development abnormalities moderately phenocopying AGO1 mutant alleles.
Furthermore, 2b was revealed to be unable to inhibit the initiation of signal-independent RNA silencing of transgene and virus, by obstructing the RDR1-dependent viral siRNAs generation process (Diaz-Pendon et al. 2007). This stipulates that different mechanisms possibly will be involved in overcoming the antiviral defense by the infecting virus.
6.3 Tombusviruses P19
One of the robust VSRs is P19 of the tombusvirus, such as Cymbidium ringspot virus. It has the characteristic of recognizing the 2 nt extension at the 3′ end of 21 nt RNA duplexes for siRNA binding and thus inhibiting them from spreading systemically through the plant. It may also impede the activity of siRNA-primed RDR complex, which is assumed to modulate the establishment of the systemic signal (Voinnet 2001). Few reports also suggest that it has the capacity to interact and efficiently bind to variety of siRNA molecules, such as ss-siRNAs, long dsRNAs, and blunted 21 nt dsRNAs (Silhavy et al. 2002). Biochemical characterization of P19 in Drosophila cell extracts revealed that it might hinder the siRNAs loading into RISC effectors complexes (Lakatos et al. 2006).
Later on, the elucidation of the crystal structure of P19 binding a 21 nt siRNA duplex confirmed the physical interaction in between P19 and siRNAs. It helped the biologist to advance their understanding about how dimers of this protein are proficient in distinguishing RNA duplexes of 21 nt and also overhanging 3′ nt, which is a hallmark of the siRNAs (Vargason et al. 2003). Moreover, this siRNA binding characteristic of P19 was conserved among all the organisms containing silencing machinery, which also provided a base to develop P19 as widespread and potent tool to study RNA silencing process, Recently, inhibition of 3′ modification of small RNAs in Carnation Italian ringspot virus infected plants was studied and it was found that P19 binds to both 3′ modified and non-modified small RNAs in vivo. In general, 3′ modifications of viral siRNAs take place in cytoplasm, whereas in the case of miRNAs, this modification occurs in the nucleus. Hence, the P19 facilitated inhibition of the 3′ si/miRNAs alteration would entail spatial and sequential expression of both P19 and small RNAs. Finally, their data revealed that Hen1-like methyltransferase might account for the small RNA modification of their 3′-terminal nucleotide in N. benthamiana (Lozsa et al. 2008). Similar to HC-Pro, P19 has also been shown to interfere with the processing and activity of miRNAs by modulating the HEN-1-mediated methylation of miRNA.
Remarkably, the P19 protein of Tomato bushy stunt virus interacts with ALY proteins. These proteins have been shown to be associated with the export of RNAs from the nucleus and transcriptional co-activation in animal cells. P19 helps in the re-localization of a subset of these proteins from the nucleus to the cytoplasm. Co-expression of ALY proteins and P19 in N. benthamiana revealed that the subset of ALY proteins, which were not translocated from the nucleus significantly, altered the RNA silencing suppression ability of P19 (Canto et al. 2006).
6.4 Geminivirus AC2
Geminiviruses are characterized by small geminate particles (18–20 nm) containing either one or two single-stranded circular DNA molecules of around 2.7 kb (Stanley and Gay 1983). Based on genome organization, host range, and vector specificity, the members of the family Geminiviridae are classified into seven genera: Begomovirus, Mastrevirus, Curtovirus, Eragrovirus, Becurtovirus, Turncurtovirus, and Topocuvirus (Adams et al. 2013). The majority of begomoviruses have two components, referred to as DNA-A and DNA-B, both of which are essential for infectivity. Monopartite begomovirus such as isolates of Tomato yellow leaf curl Sardinia virus (Kheyr-Pour et al. 1991) has a single genomic component equivalent to DNA-A.
The protein encoded by the complementary strand of DNA-A component, named AC2, is one of the major pathogenicity factors. It is multifunctional protein encoded by all members of the genus Begomovirus. The protein has transactivation potential and is required for the expression of late viral genes AV1 and BV1 in at least some geminiviruses, thus also known as Transcriptional Activator Protein (TrAP) (Sunter and Bisaro 1991, 1992; Jeffrey et al. 1996). It binds to ssDNA in a non-specific way and only weakly to dsDNA, suggesting that it is not a canonical transcriptional factor, but probably interacts with host plant cellular proteins to trigger transcriptional activation (Hartitz et al. 1999).
In general, the AC2 protein has a modular structure consisting of three conserved domains: a basic domain with a nuclear localization signal at the N-terminus, a central DNA-binding Zn-finger motif, and C-terminal acidic activator domain (Hartitz et al. 1999). The AC2 or the C2 protein (a positional homolog of AC2 in TYLCCNV) encoded by monopartite and bipartite begomoviruses have been shown to possess strong RSS activity and are capable of suppressing TGS and or PTGS (Voinnet et al. 1999; van Wezel et al. 2002; Dong et al. 2003; Vanitharani et al. 2004; Trinks et al. 2005; Wang et al. 2005). It has been postulated that since AC2 protein of begomoviruses fails to bind any form of RNA, it thus needs to target host RNAi factors. AC2 protein was found to be directly interacting with RNA silencing pathway components like RDR6 and AGO1, which indicates its dual action site on the pathway to make the suppression more strong and effective (Kumar et al. 2015). Moreover, AC2 of CbLCV promotes the decapping activity of DCP2, which in turn accelerates mRNA turnover rate and also inhibits the siRNA accumulation (Ye et al. 2016).
AC2/C2 of TGMV (a begomovirus) and Beet curly top virus—BCTV (a curtovirus) have been shown to suppress PTGS by interacting and inactivating the SNF1 and adenosine kinases enzymes which appear to be involved in defense response (Hao et al. 2003; Wang et al. 2003). The adenosine kinase is known to be essential for the production of s-adenosyl methionine (SAM), an important cofactor for methyl transferases (Saze et al. 2003) and inhibition of its activity negatively affects methyl cycle (Wang et al. 2003, 2005).
In addition, C2/AC2 of the members of both begomovirus and curtovirus has been shown to be a suppressor of TGS (Buchmann et al. 2009; Zhang et al. 2011; Yang et al. 2012). Buchmann et al. (2009) first showed that Geminivirus C2 and AC2 proteins can be a TGS suppressor and demonstrated that they reduce the overall cytosine methylation. BSCTV acts as a TGS suppressor by interacting with SAM decarboxylase 1 (SAMDC1) and attenuating the degradation of SAMDC1, a key player in the methyl cycle (Zhang et al. 2011). Later, BSCTV C2 also has been shown to affect the generation of virus-derived siRNAs, a precursor for the initiation of RdDM, and thereby reducing the viral DNA methylation (Yang et al. 2012). More recently, AC2 of ICMV has been reported to inhibit kryptonite (KYP, a H3K9 methyl transferase) via the activation of transcription repressor RAV2 (RELATED TO ABI3 and VP1) (Sun et al. 2015). However, AC2 of TGMV and CbLCV has been shown to interact with the catalytic domain of KYP and further inhibits its methyl transferase activity in vitro (Castillo-Gonzalez et al. 2015). Furthermore, using TrAP protein lacking its transcription activation domain, a recent report revealed that this TrAP could reverse TGS in the reproductive plants, independent of ADK inactivation or transcription activation (Jackel et al. 2015).
6.5 Polerovirus P0
P0 adopts proteasome-mediated degradation of AGO1. Molecular analysis of Polerovirus P0 protein structure suggests that it encompasses F-box motif, which is essential to form the SCF-like complex, and also a prerequisite for P0’s RSS activity. Further studies of P0 suggested that it does not essentially affect the biogenesis of primary siRNAs; however, it may target the PAZ motif and adjacent upstream sequence of AGO1 to destabilize it and subsequently lead to proteasome-mediated degradation (Baumberger et al. 2007; Bortolamiol et al. 2007).
7 Disease or Pathogenicity: Host MicroRNA Dysregulation and Affected Functions
It has been shown that the cellular miRNAs are capable of regulating viral replication. The viruses at the same time may alter the expression of cellular miRNAs through the VSR molecules. The VSR-mediated changes in the profile of host miRNA abundance and activities are well known in literature. The VSRs might treat the chemically similar duplex-miRNAs and siRNAs in a more or less similar manner, even though the former groups of molecules are processed from hairpin loop RNA precursors transcribed from endogenous genes (Ambros et al. 2003). The processing and function of miRNA pathway involve common components including DCL1 and AGO1 (Bartel 2004). The VSR-mediated deviation of the normal miRNA profile of the host following the virus infection could be a major source of viral pathogenicity.
In plants, miRNAs target a wide range of mRNAs encoding transcription factors required for development (Park et al. 2002; Rhoades et al. 2002; Palatnik et al. 2003). These include factors required for meristem identity and maintenance, patterning, cell division, hormone signalling, and developmental timing. In addition, plant miRNAs also target mRNAs encoding miRNA metabolic factors and factors of unknown function (Rhoades et al. 2002; Xie et al. 2003). Loss of miRNA biogenesis or activity in Arabidopsis results in pleiotropic defects during embryonic, vegetative, and reproductive development (Park et al. 2002; Schauer et al. 2002; Kasschau et al. 2003).
It is proposed that most of the developmental defects triggered by virus infection are due to interference with pathways that depend on negative regulation by miRNAs. A study with TuMV in Arabidopsis demonstrated that P1/HC-Pro is the virus-encoded factor that mediates this interference. The suppression of miRNA-directed function and RNA silencing by P1/HC-Pro is likely due to interference with a common reaction, probably involving assembly or activity of RISC-like complexes. The consequence of virus infection is ectopic expression of some mRNAs that are normally negatively regulated by miRNA-guided cleavage. Infected plants, therefore, display a range of developmental abnormalities because the aberrantly expressed target mRNAs encode proteins belonging to families that control meristem identity (NAC domain and SBP-like proteins), organ identity and separation (AP2 domain and NAC domain proteins), radial patterning (SCL-like proteins), and hormone signalling (ARF proteins). Interference with leaf and flower formation and developmental timing; ectopic induction of cell division in non-meristematic tissues; and disruption of hormone production, signalling, and response are some of the well-characterized effects of different viruses in certain susceptible host plants (Hull 2001). Given that many of the miRNA target genes are expressed or repressed in specific cell-types in meristematic and organ primordium zones, we further propose that viruses triggering the most severe developmental defects are those that (1) invade meristematic and dividing cells and (2) encode potent RNA silencing suppressors. Indeed, although many viruses are known to be excluded from meristematic zones, in situ analysis revealed that meristems and organ primordia are effectively invaded by TuMV in Arabidopsis.
8 VSR-Targeted Antiviral Strategy
The VSRs are the pathogenicity factors and hence are very good targets for antiviral strategy. Many RNA viruses failed to cause disease in plants expressing siRNAs targeted to silence the VSRs of the infecting viruses. Similar strategy also works in mammalian systems. The non-human primates have been found protected against the deadly Ebola viruses when the animals are systemically injected with the siRNAs meant to silence the Ebola-VSR (Thi et al. 2016). The artificial miRNAs have also been used to silence the VSRs of RNA and DNA viruses of plants, and the transgenic plants expressing the miRNAs have been found tolerant/resistant against the viruses. The literature is replete with the information on siRNAs silencing the VSRs but the corresponding reports of artificial miRNAs (amiRNAs) are few. In the following, we give an account of the amiRNAs and tasiRNAs providing the antiviral strategy.
8.1 Artificial MiRNA Strategy
The amiRNA technology is being utilized to target the invading viral gene transcripts. In this regard, the VSR transcripts have been widely subjected to degradation (Tiwari et al. 2014). It was reported that miR156 and miR393 may inhibit the invasion of foreign genetic elements like plant viruses (Xing and Zhang 2010; Zhang et al. 2011). The ath-miR-159 based amiRNAs were designed to target viral sequences encoding P69, aVSR of Turnip yellow mosaic virus (TYMV) and HC-Pro of Turnip mosaic virus (TuMV). Transgenic Arabidopsis lines expressing amiR-P69 and amiR-HCPro were specifically resistant to TYMV and TuMV (Niu et al. 2006). The amiRNA sequences targeting the VSR, 2b of CMV, can efficiently confer effective resistance to CMV infection (Qu et al. 2007). Later amiRNA technology was used to confer virus resistance in transgenic tobacco and tomato (Ai et al. 2011; Zhang et al. 2011). The amiRNA targeting overlapping regions of geminiviruses genes AC1, AC2, and AC4 were used to generate transgenic tomato plants, that could resist infection by begomovirus, ToLCNDV (Yadava et al. 2010; Tien et al. 2013). There are also reports in literature on using the amiRNAs for generating resistance against Watermelon silver mottle virus in tobacco (Kung et al. 2012).
8.2 Artificial TasiRNA Strategy
Besides amiRNA, artificial tasiRNA technology has also been used to generate virus tolerant plants. A binary vector has been designed incorporating control elements such as the 5′ and 3′ binding sites of miR390 and keeping the VSR sequences sandwiched between the control elements. This vector when introduced in plants produces artificial tasiRNAs from the VSR sequences. These tasiRNAs slice the VSRs of the infecting ToLCVs. Thus the transgenics producing the artificial tasiRNAs was tolerant against the invading ToLCVs (Singh et al. 2015). Such strategy could in principle be adopted to develop plants tolerant for all viruses whose VSR sequences are known.
9 Future Perspectives
RNAi has been used extensively as a tool to study gene functions. The efficiencies of these processes are presumed to be subjects of several degrees and layers of modifications. The VSRs or RSSs can contribute largely to the modification processes. The VSRs, when overexpressed, can influence the outcomes of RNAi in several systems. In this connection it is important to reveal the identities of RSSs in all of the RNAi-competent organisms. A few of these are reported in host plants like tobacco and tomato but these class of RSSs from plant sources or other organisms have remained elusive so far. Hence appropriate assays need to be devised to trap RSSs from several nonviral pathogens and their hosts. Recently a class of proteins, namely RNase III-like proteins (RTLs), have been described from plants that act as general RNAi suppressors, which are induced in response to virus infection but are functionally repressed by plant VSRs (Shamandi et al. 2015). On the other hand, the β-C1 suppressor of TYLCCN virus collaborates with tomato rgs-CaM RNAi suppressor for efficient viral growth (Li et al. 2014). Thus along with the identification of RSSs from nonviral sources, the cross-talks between the RSSs are also very important to reveal the overall biology of RNAi.
VSRs could be used for multiple purposes, namely, reversal of siRNA-mediated disease, overcoming transgene silencing, enhancing expression of viral vectors and vaccine production, etc. Tobacco plants infected with TMV bearing the pathogenic satellite RNA show darkening effects in the leaves due to loss of chlorophyll biosynthesis encoding protein CHL1 which gets silenced by the siRNAs produced from the satellite RNA. This silencing effect is strongly inhibited by the P1/HC-PRO VSR of the potyvirus (Ricaño-Rodríguez et al. 2016). Many VSRs have been used to overcome transgene- or siRNA-mediated silencing (Rahman et al. 2012, 2014). However, there is an inherent difficulty in reversing such kind of RNAi as the presence of VSRs also interfere in the biogenesis and function of the plants hosting the VSRs, making the host plant developmentally retarded. Hence either the VSRs need to be modified or these should be chosen carefully such that the selected VSRs do not interfere in the miRNA pathways. A mutant form of HC-PRO has been used by Mallory et al. to enhance transgene expression in tobacco showing no developmental anomaly (Mallory et al. 2002). Similarly the VSR proteins of Beet yellow closterovirus like p64, p21, etc. might have minimal impact on the miRNA pathways (Til’kunova et al. 2004). A few VSRs when expressed in the heterologous systems remove the restrictions of RNAi but do not cause perturbations in the miR pathways. The VSR B2 of insect Flock House Virus (FHV) suppresses RNAi in C. elegans and also facilitates natural infection of Orsay virus in C. elegans but is inactive against miRNA-mediated silencing (Guo and Lu 2013). Thus FHV-B2 does not harm the C. elegans hosts. VSRs have been extensively used in improving replication and transcription of viral vectors used for gene therapy and vaccine production. Recently, P19 VSR from Tomato bushy stunt virus was stably expressed in human embryonic kidney cells (B6 cells) and the replication of Adenovirus shot up 100-fold in these cells. Adenoviruses are widely used viral vectors and along with p19 the oncolysis potential of the vector is increased five- to six fold in the tumor cells, raising the hope of translating these results in preclinical and clinical trials (Rauschhuber et al. 2012). Hence the selective usages of VSRs are very beneficial to remove the undesirable restrictions of RNAi.
The intertwined and multiple-layered arms race between host and pathogen must be interpretable in terms of “molecular arms race.” Both the host and viral components along with their cross-talks have been adequately described in literature (Ding and Voinnet 2007; Csorba et al. 2015). Viral genes evolve faster than host genes as the viruses want to combat host with novel winning designs, and in response, the antiviral silencing factors also evolve faster than other host genes to gain upper hand of the battle. Amidst all these, the VSRs evolve faster than any other known genes (Murray et al. 2013). Such changes impact strongly both on viruses and hosts. The diversity of VSRs’ structure and functions are partly accounted by such changes. Besides silencing RNAi, the VSRs also participate in other important aspects of viral life cycle (Csorba et al. 2015). So it would be important to assess how much of the viral life processes as well as their pathogenicity has changed over the evolutionary time scale. It would also be worthwhile to watch what new functions, besides RNAi silencing, like interfering with host hormone signalling, relocalizations of interacting host factors in subcellular structures etc., are being gained by the VSRs. The vi-siRNAs and VSRs interact directly as well as indirectly with many of the host factors that are involved in antiviral silencing pathways including rgs-CaM and RAV2 (Moissiard and Voinnet 2006; Nakahara et al. 2012; Endres et al. 2010). When VSRs undergo evolution, interacting host factors might also change, thus causing hosts to evolve. It would be very interesting to study the profile of changes in host evolutionary pattern in response to the evolution of VSRs. Besides RNAi factors, hosts also offer resistance to viruses by other antiviral pathways like R-gene-mediated hypersensitive response, hormone (SA/JA) mediated SAR pathways, etc. Another interesting area of research would be to follow how the evolving VSRs intersect these pathways.
References
Adams MJ, King AMQ, Carstens EB (2013) Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses. Arch Virol 158:2023–2030
Agrawal N, Dasarathi PVN, Mohmed A et al (2003) RNA interference biology, mechanism and applications. Microbiol Mol Biol Rev 67:657–685
Ahlquist P (2006) Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat Rev Micro Biol 4:371–382
Ai Y, Wang J, Johnson RE et al (2011) A novel ubiquitin binding mode in the S. cerevisiae translesion synthesis DNA polymerase? Mol BioSyst 7:1874–1882
Akbergenov R, Si-Ammour A, Blevins T et al (2006) Molecular characterization of geminivirus-derived small RNAs in different plant species. Nucleic Acids Res 34:462–471
Ambros V, Bartel B, Bartel DP (2003) A uniform system for microRNA annotation. RNA 9:277–279
Anandalakshmi R, Pruss GJ, Ge X et al (1998) A viral suppressor of gene silencing in plants. Proc Natl Acad Sci USA 95:13079–13084
Anandalakshmi R, Marathe R, Ge X et al (2000) A calmodulin-related protein that suppresses post-transcriptional gene silencing in plants. Science 290:142–144
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism and function. Cell 116:281–297
Baulcombe DC (1999) Fast forward genetics based on virus induced gene silencing. Curr Opin Plant Biol 2:109–113
Baumberger N, Tsai CH, Lie M et al (2007) The polerovirus silencing suppressor P0 targets Argonaute proteins for degradation. Curr Biol 17:1609–1614
Beclin C, Berthome R, Palauqui J et al (1998) Infection of tobacco or Arabidopsis plants by CMV counteracts systemic post-transcriptional silencing of nonviral (trans)genes. Virology 252:313–317
Benson DA, Karsch-Mizrachi I, Lipman DJ et al (2005) GenBank. Nucleic Acids Res 33(S1):D34–D38
Bernstein E, Caudy AA, Hammond SM et al (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363–366
Bivalkar-Mehla S, Vakharia J, Mehla R et al (2011) Viral RNA silencing suppressors (RSS): novel strategy of viruses to ablate the host RNA interference (RNAi) defense system. Virus Res 155:1–9
Bortolamiol D, Pazhouhandeh M, Marrocco K et al (2007) The poleovirus F Box protein P0 targets Argonaute1 to suppress RNA silencing. Curr Biol 17:1615–1621
Brigneti G, Voinnet O, Li WX et al (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J 17:6739–6746
Buchmann RC, Asad S, Wolf JN et al (2009) Geminivirus AL2 and L2 proteins suppress transcriptional gene silencing and cause genome-wide reductions in cytosine methylation. J Virol 83:5005–5013
Canto T, Uhrig JF, Swanson M et al (2006) Translocation of tomato bushy stunt virus P19 protein into the nucleus by ALY proteins compromises its silencing suppressor activity. J Virol 80:9064–9072
Carbonell A, Fahlgren N, Garcia-Ruiz H et al (2012) Functional analysis of three Arabidopsis ARGONAUTES using slicer-defective mutants. Plant Cell 24:3613–3629
Carrillo-Tripp J, Lozoya-Gloria E, Rivera-Bustamante RF (2007) Symptom remission and specific resistance of pepper plants after infection by pepper golden mosaic virus. Phytopathology 97:51–59
Castillo-González C, Liu X, Huang C et al (2015) Geminivirus-encoded TrAP suppressor inhibits the histone methyltransferase SUVH4/KYP to counter host defense. eLife 4:e06671
Chapman EJ, Prokhnevsky AI, Gopinath K et al (2004) Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes Dev 18:1179–1186
Chitwood DH, Timmermans MC (2010) Small RNAs are on the move. Nature 467:415–419
Csorba T, Kontra L, Burgyán J (2015) Viral silencing suppressors: tools forged to fine-tune host-pathogen coexistence. Virology 479–480:85–103
Darnirdagh IS, Ross AF (1967) A marked synergistic interaction of potato viruses X and Y in inoculated leaves of tobacco. Virology 31:296–307
Diaz-Pendon JA, Li F, Li W-X et al (2007) Suppression of antiviral silencing by Cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell 19:2053–2063
Ding SW, Voinnet O (2007) Antiviral immunity directed by small RNAs. Cell 130:413–426
Ding SW, Anderson BJ, Haase HR et al (1994) New overlapping gene encoded by the Cucumber mosaic virus genome. Virology 198:593–601
Ding S-W, Li W-X, Symons RH (1995a) A novel naturally occurring hybrid gene encoded by a plant RNA virus facilitates long distance virus movement. EMBO J 14:5762–5772
Ding SW, Rathjen JP, Li WX et al (1995b) Efficient infection from cDNA clones of cucumber mosaic cucumovirus RNAs in a new plasmid vector. J Gen Virol 76:459–464
Dong X, van Wezel R, Stanley J et al (2003) Functional characterization of the nuclear localization signal for a suppressor of posttranscriptional gene silencing. J Virol 77:7026–7033
Duan CG, Fang YY, Zhou BJ et al (2012) Suppression of Arabidopsis ARGONAUTE1-mediated slicing, transgene-induced RNA silencing, and DNA methylation by distinct domains of the cucumber mosaic virus 2b protein. Plant Cell 24:259–274
Dunoyer P, Lecellier CH, Parizotto EA et al (2004) Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16:1235–1250
Elmayan T, Vaucheret H (1996) Expression of single copies of a strongly expressed 35S transgene can be silenced post-transcriptionally. Plant J 9:787–797
Endres MW, Gregory BD, Gao Z et al (2010) Two plant viral suppressors of silencing require the ethylene-inducible host transcription factor RAV2 to block RNA silencing. PLoS Pathog 6:e1000729
Enright AJ, John B, Gaul U et al (2003) MicroRNA targets in Drosophila. Genome Biol 5:1
Finnegan EJ, Margis R, Waterhouse PM (2003) Posttranscriptional gene silencing is not compromised in the arabidopsis CARPEL FACTORY (DICER-LIKE1) mutant, a homolog of dicer-1 from drosophila. Curr Biol 13:236–240
Ghosh Z, Mallick B, Chakrabarti J (2009) Cellular versus viral microRNAs in host-virus interaction. Nucleic Acids Res 37:1035–1048
Goto K, Kobori T, Kosaka Y et al (2007) Characterization of silencing suppressor 2b of cucumber mosaic virus based on examination of its small RNA-binding abilities. Plant Cell Physiol 48:1050–1060
Griffiths-Jones S, Saini HK, van Dongen S et al (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36(S1):D154–D158
Guo HS, Ding SW (2002) A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J 21:398–407
Guo X, Lu R (2013) Characterization of virus-encoded RNAi suppressors in Caenorhabditis elegans. J Virol 87:5414–5423
Hamilton A, Voinnet O, Chappell L et al (2002) Two classes of short interfering RNA in RNA silencing. EMBO J 21:4671–4679
Hao L, Wang H, Sunter G et al (2003) Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell 15:1034–1048
Hartitz MD, Sunter G, Bisaro DM (1999) The tomato golden mosaic virus transactivator (TrAP) is a single-stranded DNA and zinc-binding phosphoprotein with an acidic activation domain. Virology 263:1–14
Hofacker IL, Fontana W, Stadler PF et al (1994) Fast folding and comparison of RNA secondary structures. Monatsh Chem 125:167–188
Hsu PWC, Lin LZ, Hsu SD et al (2007) ViTa: prediction of host microRNAs targets on viruses. Nucleic Acids Res 35(S1):D381–D385
Huang Y, Greene E, Murray Stewart T et al (2007) Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proc Natl Acad Sci USA 104:8023–8028
Hull R (2001) Matthew’s plant virology, 4th edn. Academic, San Diego
Hutvágner G, McLachlan J, Pasquinelli AE et al (2001) A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293:834–838
Jackel JN, Buchmann RC, Singhal U et al (2015) Analysis of geminivirus AL2 and L2 proteins reveals a novel AL2 silencing suppressor activity. J Virol 89:3176–3187
Jeffrey JL, Pooma W, Petty ITD (1996) Genetic requirements for local and systemic movement of tomato golden mosaic virus in infected plants. Virology 223:208–218
Johansen LK, Carrington JC (2001) Silencing on the spot induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol 126:930–938
Jones JDG, Dang JL (2006) The plant immune system. Nature 444:323–329
Kalantidis K, Schumacher HT, Alexiadis T et al (2008) RNA silencing movement in plants. Biol Cell 100:13–26
Kapoor M, Arora R, Lama T et al (2008) Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genomics 9:451
Karjee S, Islam MN, Mukherjee SK (2008) Screening and identification of virus encoded silencing suppressors. Methods Mol Biol 442:187–203
Kasschau KD, Carrington JC (1998) A counter defensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461–470
Kasschau KD, Carrington JC (2001) Long-distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology 285:71–81
Kasschau KD, Cronin S, Carrington JC (1997) Genome amplification and long-distance movement functions associated with the central domain of Tobacco etch potyvirus helper component-proteinase. Virology 228:251–262
Kasschau KD, Xie Z, Allen E et al (2003) P1/HC-Pro a viral suppressor of RNA silencing interferes with Arabidopsis development and miRNA function. Dev Cell 4:205–217
Kheyr-Pour A, Bendahmane M, Matzeit V et al (1991) Tomato yellow leaf curl virus from Sardinia is a whitefly-transmitted monopartite geminivirus. Nucleic Acids Res 19:6763–6769
Kumar V, Mishra SK, Rahman J et al (2015) Mungbean yellow mosaic Indian virus encoded AC2 protein suppresses RNA silencing by inhibiting Arabidopsis RDR6 and AGO1 activities. Virology 486:158–172
Kung Y-J, Lin S-S, Huang Y-L et al (2012) Multiple artificial microRNAs targeting conserved motifs of the replicase gene confer robust transgenic resistance to negative-sense single-stranded RNA plant virus. Mol Plant Pathol 13:303–317
Lakatos L, Csorba T, Pantaleo V et al (2006) Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J 25:2768–2780
Li J, Yang Z, Yu B et al (2005) Methylation protects miRNAs and siRNAs from a 3′ end uridylation activity in Arabidopsis. Curr Biol 15:1501–1507
Li R, Weldegergis BT, Li J et al (2014) Virulence factors of geminivirus interact with MYC2 to subvert plant resistance and promote vector performance. Plant Cell 26:4991–5008
Lin SS, Wu HW, Elena SF et al (2009) Molecular evolution of a viral non-coding sequence under the selective pressure of amiRNA-mediated silencing. PLoS Pathog 5:e1000312
Lister R, O’Malley RC, Tonti-Filippini J et al (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133:523–536
Liu B, Li P, Li X et al (2005) Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol 139:296–305
Llave C, Kasschau KD, Carrington JC (2000) Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc Natl Acad Sci USA 97:13401–13406
Llave C, Kasschau KD, Rector MA et al (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell 14:1605–1619
Lozsa R, Csorba T, Lakatos L et al (2008) Inhibition of 3′ modification of small RNAs in virus-infected plants requires spatial and temporal co-expression of small RNAs and viral silencing-suppressor proteins. Nucleic Acids Res 36:4099–4107
Lu R, Folimonov A, Shintaku M et al (2004) Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc Natl Acad Sci USA 101:15742–15747
Lucy AP, Guo HS, Li WX et al (2000) Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J 19:1672–1680
Mallory A, Vaucheret H (2010) Form, functionand regulation of ARGONAUTE proteins. Plant Cell 22:3879–3889
Mallory AC, Ely L, Smith TH et al (2001) HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13:571–583
Mallory AC, Reinhart BJ, Bartel D et al (2002) A viral suppressor of RNA silencing differentially regulates the accumulation of short interfering RNAs and micro-RNAs in tobacco. Proc Natl Acad Sci USA 99:15228–15233
Margis R, Fusaro AF, Smith NA et al (2006) The evolution and diversification of Dicers in plants. FEBS Lett 580:2442–2450
Matthew L (2004) RNAi for plant functional genomics. Comp Funct Genomics 5:240–244
Maziere P, Enright AJ (2007) Prediction of microRNA targets. Drug Discov Today 12:452–458
Mitter N, Sulistyowati E, Dietzgen RG (2003) Cucumber mosaic virus infection transiently breaks dsRNA-induced transgenic immunity to Potato virus Y in tobacco. Mol Plant-Microbe Interact 16:936–944
Mlotshwa S, Schauer SE, Smith TH et al (2005) Ectopic DICER-LIKE1 expression in P1/HC-Pro Arabidopsis rescues phenotypic anomalies but not defects in microRNA and silencing pathways. Plant Cell 17:2873–2288
Moissiard G, Voinnet O (2006) RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc Natl Acad Sci USA 103:19593–19598
Murray GGR, Kosakovsky Pond SL, Obbard DJ (2013) Suppressors of RNAi from plant viruses are subject to episodic positive selection. Proc R Soc B Biol Sci 280:20130965
Nakahara KS, Masuta C, Yamada S et al (2012) Tobacco calmodulin-like protein provides secondary defense by binding to and directing degradation of virus RNA silencing suppressors. Proc Natl Acad Sci USA 109:10113–10118
Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2:279–289
Naqvi AR, Haq QM, Mukherjee SK (2010) MicroRNA profiling of tomato leaf curl new delhi virus (ToLCNDV) infected tomato leaves indicates that deregulation of mir159/319 and mir172 might be linked with leaf curl disease. Virol J 7:1
Nicolas FE, Torres-Martínez S, Ruiz-Vazquez RM (2013) Loss and retention of RNA interference in fungi and parasites. PLoS Pathog 9:e1003089
Niu QW, Lin SS, Reyes JL et al (2006) Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol 24:1420–1428
Palatnik JF, Allen E, Wu X et al (2003) Control of leaf morphogenesis by microRNAs. Nature 425:257–263
Palauqui J-C, Vaucheret H (1998) Transgenes are dispensable for the RNA degradation step of cosuppression. Proc Natl Acad Sci USA 95:9675–9680
Palauqui JC, Elmayan T, Pollien JM et al (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J 16:4738–4745
Park W, Li J, Song R et al (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12:1484–1149
Perrimon N, Ni JQ, Perkins L (2010) In vivo RNAi: today and tomorrow. Cold Spring Harb Perspect Biol 2:a003640
Pooggin MM (2013) How can plant DNA viruses evade siRNA-directed DNA methylation and silencing? Int J Mol Sci 14:15233–15259
Pruss G, Ge X, Shi XM et al (1997) Plant viral synergism: the potyviral genome encodes a broad-range pathogenicity enhancer that transactivates replication of heterologous viruses. Plant Cell 9:859–868
Qu J, Ye J, Fang RX (2007) Artificial microRNA-mediated virus resistance in plants. J Virol 81:6690–6699
Rahman J, Karjee S, Mukherjee SK (2012) MYMIV-AC2, a geminiviral RNAi suppressor protein, has potential to increase the transgene expression. Appl Biochem Biotechnol 167:758–775
Rahman J, Sanan-Mishra N, Mukherjee SK (2014) MYMIV-AC2 protein suppresses hairpin-induced gene silencing in Nicotiana tabacum cv. Xanthi. Plant Biotechnol Rep 8:337–347
Raja P, Wolf JN, Bisaro DM (2010) RNA silencing directed against geminiviruses: post-transcriptional and epigenetic components. Biochim Biophys Acta 1799:337–351
Ramachandran V, Chen X (2008) Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321:1490–1492
Rauschhuber C, Noske N, Ehrhardt A (2012) New insights into stability of recombinant adenovirus vector genomes in mammalian cells. Eur J Cell Biol 91:2–9
Rhoades MW, Reinhart BJ, Lim LP et al (2002) Prediction of plant microRNA targets. Cell 110:513–520
Ricaño-Rodríguez J, Adame-García J, Portilla-Vázquez S et al (2016) The extraordinary nature of RNA interference in understanding gene downregulation mechanism in plants. In: Abdurakhmonov IY (ed) Plant genomics. InTech
Roth BM, Pruss GJ, Vance VB (2004) Plant viral suppressors of RNA silencing. Virus Res 102:97–108
Ruiz MT, Voinnet O, Baulcombe DC (1998) Initiation and maintenance of virus-induced gene silencing. Plant Cell 10:937–946
Ruiz-Ferrer V, Voinnet O (2007) Viral suppression of RNA silencing: 2b wins the Golden Fleece by defeating Argonaute. Bioessays 29:319–323
Sahu PP, Sharma N, Puranik S, Prasad M (2014) Post-transcriptional and epigenetic arms of RNA silencing: a defense machinery of naturally tolerant tomato plant against tomato leaf curl New Delhi virus. Plant Mol Biol Rep 32:1015–1029
Saze H, Mittelsten Scheid O, Paszkowski J (2003) Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet 34:65–69
Schauer SE, Jacobsen SE, Meinke DW et al (2002) DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci 7:487–491
Schwab R, Palatnik JF, Riester M et al (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8:517–527
Shamandi N, Zytnicki M, Charbonnel C et al (2015) Plants encode a general siRNA suppressor that is induced and suppressed by viruses. PLoS Biol 13:e1002326
Shi BJ, Palukaitis P, Symons RH (2002) Differential virulence by strains of Cucumber mosaic virus is mediated by the 2b gene. Mol Plant-Microbe Interact 15:947–955
Shiboleth YM, Haronsky E, Leibman D et al (2007) The conserved FRNK box in HC-Pro a plant viral suppressor of gene silencing is required for small RNA binding and mediates symptom development. J Virol 81:13135–13148
Siddiqui SA, Valkonen JP, Rajamäki M et al (2011) The 2b silencing suppressor of a mild strain of Cucumber mosaic virus alone is sufficient for synergistic interaction with Tobacco mosaic virus and induction of severe leaf malformation in 2b-transgenic tobacco plants. Mol Plant-Microbe Interact 24:685–693
Silhavy D, Molnar A, Lucioli A et al (2002) A viral protein suppresses RNA silencing and binds silencing-generated 21- to 25-nucleotide double-stranded RNAs. EMBO J 21:3070–3080
Singh RK, Rai N, Kumar P et al (2015) Inheritance study in tomato (Solanum lycopersicum) for Tomato leaf curl virus (ToLCV) resistance. Indian J Agri Sci 85:896–901
Soards AJ, Murphy AM, Palukaitis P et al (2002) Virulence and differential local and systemic spread of cucumber mosaic virus in tobacco are affected by the CMV 2b protein. Mol Plant-Microbe Interact 15:647–653
Sonoda S, Nishiguchi M (2000) Graft transmission of post-transcriptional gene silencing: target specificity for RNA degradation is transmissible between silenced and non-silenced plants, but not between silenced plants. Plant J 21:1–8
Stanley J, Gay MR (1983) Nucleotide sequence of cassava latent virus DNA. Nature 301:260–262
Sun YW, Tee CS, Ma YH et al (2015) Attenuation of histone methyltransferase KRYPTONITE-mediated transcriptional gene silencing by Geminivirus. Sci Rep 5:16476
Sunter G, Bisaro DM (1991) Transactivation in a geminivirus: AL2 gene product is needed for coat protein expression. Virology 180:416–419
Sunter G, Bisaro DM (1992) Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 4:1321–1331
Takeda A, Sugiyama K, Nagano H et al (2002) Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus. FEBS Lett 532:75–79
Thi EP, Lee ACH, Geisbert JB et al (2016) Rescue of non-human primatesfrom advanced Sudan ebolavirus infection with lipid encapsulated siRNA. Nat Microbiol 1:16142
Tien VV, Choudhury NR, Mukherjee SK (2013) Transgenic tomato plants expressing artificial microRNAs for silencing the pre-coat and coat proteins of a begomovirus, Tomato leaf curl New Delhi virus, show tolerance to virus infection. Virus Res 172:35–45
Til’kunova T, Zinovkin RA, Agranovskiĭ AA (2004) RNA-binding properties of proteins of the beet yellows closterovirus. Mol Biol (Mosk) 38:553–558
Tiwari M, Sharma D, Trivedi PK (2014) Artificial microRNA mediated gene silencing in plants: progress and perspectives. Plant Mol Biol 86:1
Trinks D, Rajeswaran R, Shivaprasad PV et al (2005) Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J Virol 79:2517–2527
Vance V, Vaucheret H (2001) RNA silencing in plants – defense and counter defense. Science 292:2277–2280
Vanderschuren H, Akbergenov R, Pooggin MM et al (2007) Transgenic cassava resistance to African cassava mosaic virus is enhanced by viral DNA-A bidirectional promoter-derived siRNAs. Plant Mol Biol 64:549–557
Vanitharani R, Chellappan P, Pita JS et al (2004) Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J Virol 78:9487–9498
Vargason JM, Szittya G, Burgyan J et al (2003) Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115:799–811
Vaucheret H (2008) Plant argonautes. Trends Plant Sci 13:350–358
Voinnet O (2001) RNA silencing as a plant immune system against viruses. Trends Genet 17:449–459
Voinnet O, Baulcombe DC (1997) Systemic signalling in gene silencing. Nature 389:553
Voinnet O, Vain P, Angell S et al (1998) Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoter less DNA. Cell 95:177–187
Voinnet O, Pinto YM, Baulcombe DC (1999) Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci USA 96:14147–14152
Voinnet O, Lederer C, Baulcombe DC (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:157–167
Wang H, Hao L, Shung CY et al (2003) Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. Plant Cell 15:3020–3032
Wang H, Buckley KJ, Yang X et al (2005) Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J Virol 79:7410–7418
Wang B, Li S, Qi HH et al (2009) Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat Struct Mol Biol 16:1259–1266
Wang X-B, Jovel J, Udomporn P et al (2011) The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell 23:1625–1638
Wang T, Castillo-González C, You L et al (2015) In vitro reconstitution assay of miRNA biogenesis by Arabidopsis DCL1. Bio-protocol 5:e1454
Waterhouse PM, Wang MB, Lough T (2001) Gene silencing as an adaptive defence against viruses. Nature 411:834–842
van Wezel R, Liu H, Tien P et al (2002) Mutation of three cysteine residues in tomato yellow leaf curl virus-China C2 protein causes dysfunction in pathogenesis and posttranscriptional gene silencing-suppression. Mol Plant-Microb Interact 15:203–208
Wu G, Park MY, Conway SR et al (2009a) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138:750–759
Wu L, Zhang Q, Zhou H et al (2009b) Rice microRNA effector complexes and targets. Plant Cell 21:3421–3435
Xie Z, Kasschau KD, Carrington JC (2003) Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol 13:784–789
Xing Y, Zhang Q (2010) Genetic and molecular bases of rice yield. Annu Rev Plant Biol 61:421–442
Yadav RK, Chattopadhyay D (2011) Enhanced viral intergenic region-specific short interfering RNA accumulation and DNA methylation correlates with resistance against a geminivirus. Mol Plant-Microbe Interact 24:1189–1197
Yadava P, Suyal G, Mukherjee SK (2010) Begomovirus DNA replication and pathogenicity. Curr Sci 98:360–368
Yang X, Xie Y, Raja P et al (2011) Suppression of methylation-mediated transcriptional gene silencing by βC1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog 7:e1002329
Yang L, Wu G, Poethig RS (2012) Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proc Natl Acad Sci USA 109:315–320
Yang LP, Fang YY, An CP et al (2013) C2-mediated decrease in DNA methylation, accumulation of siRNAsand increase in expression for genes involved in defense pathways in plants infected with beet severe curly top virus. Plant J 73:910–917
Ye R, Chen Z, Lian B et al (2016) A Dicer-independent route for biogenesis of siRNAs that direct DNA methylation in Arabidopsis. Mol Cell 61:222–235
Yu B, Yang Z, Li J et al (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307:932–935
Zhang H, Kolb FA, Jaskiewicz L et al (2004) Single processing center models for human Dicer and bacterial RNase III. Cell 118:57–68
Zhang X, Yuan Y-R, Pei Y et al (2006) Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev 20:3255–3268
Zhang Z, Chen H, Huang X et al (2011) BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 2(3):273–288
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Sanan-Mishra, N., Chakraborty, S., Gupta, D., Mukherjee, S.K. (2017). RNAi Suppressors: Biology and Mechanisms. In: Rajewsky, N., Jurga, S., Barciszewski, J. (eds) Plant Epigenetics. RNA Technologies. Springer, Cham. https://doi.org/10.1007/978-3-319-55520-1_11
Download citation
DOI: https://doi.org/10.1007/978-3-319-55520-1_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-55519-5
Online ISBN: 978-3-319-55520-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)