Abstract
Small RNA molecules play a crucial regulatory role in maintaining genome stability as well as developmental regulations through a set of complex and partially overlapping pathways in a wide range of eukaryotic organisms. Active in both cytoplasm and nucleus, RNA interference regulates eukaryotic gene expression through transcriptional repression by epigenetic modification and interaction with transcription machinery. Small interfering RNAs (siRNAs/miRNAs) of 21–24 nucleotides constitute the innate defence arm against a variety of pathogens, especially viruses. Plant viruses with either DNA or RNA genomes are subjected to small RNA-directed RNA degradation. Additionally, DNA viruses are subjected to another line of defence through ‘RNA-directed DNA methylations’ (RdDM). On the other hand, viral-encoded proteins, called silencing suppressors (VSRs), are known to counter the defence machinery, and therefore the virus can evade the host surveillance system. Some plant viruses additionally adopt certain strategies like acquiring silencing resistant structures (some RNA virus) to evade the RNA silencing machinery and thereby shaping the viral as well as the host genome. Recently, it has been reported that particular viral proteins and viral siRNAs contribute directly to pathogenicity by interacting with certain host proteins or RNAs. Transcriptional regulation of host gene by small RNA of viral origin plays important role in pathogenesis and symptom development. Small regulatory RNAs of cellular rather than pathogen origin have also been found to play a broad role in improving the basal defence in the case of plant–virus interaction. This chapter provides key insights into the complex intricate machinery of diverse RNA silencing mechanisms, describes various evolutionary diverse strategies of viral silencing suppressors at various steps, offers a broader view of host recovery following virus infection and finally suggests the possible applications of RNA silencing to generate virus resistant plants.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
RNA silencing is an evolutionary conserved gene regulatory mechanism active in a majority of eukaryotic system (e.g. plants, animals, yeast and insects). The versatile mechanism involves inhibition both at translational level (through the degradation of the target mRNA and/or by inhibiting its translation in a sequence-specific manner, i.e. posttranscriptional gene silencing, PTGS) and during transcription of transposons and repetitive DNA elements (transcriptional gene silencing, TGS). The concept of RNA silencing was born in early 1990 when transgenic plants expressing an extra copy of chalcone synthase (CHS) unexpectedly resulted in the suppression of both transgene and endogenous CHS mRNA giving birth to a phenomenon called co-suppression in scientific landscape (Napoli et al. 1990; van der Krol et al. 1990). Subsequently, in 1996, in the fungus Neurospora crassa, a similar phenomenon was noticed (Cogoni et al. 1996). The direct study of RNA interference was performed in C. elegans, where delivery of exogenous dsRNA resulted in sequence-specific degradation of cognate cellular mRNAs (Fire et al. 1998). Similar effects were observed in the majority of other eukaryotes including mouse, Drosophila and human (Elbashir et al. 2001; Billy et al. 2001). From the inception of the concept, the intricacy and importance of this process channelized persistent efforts in detailed exploration of the mechanism.
Three classes of small RNA regulate gene expression in cytoplasm. These are microRNAs (miRNAs), small RNAs which have imperfect complementarity with target and are generated from long RNA with hairpin structure causing translational repression of target mRNA; small interfering RNAs (siRNAs), with perfect complementarity to targets and cause transcript degradation; and PIWI-interacting RNAs (piRNAs) targeting transcripts in animal germ lines. Plants produce miRNA and siRNA but no piRNA. The term RNAi is conventionally used for siRNA-mediated silencing but convergence of different small RNA pathway prompted us to use RNAi as an umbrella term in this chapter to describe small RNA-dependent silencing. In plants, the RNA silencing machinery includes at least three complex yet partially overlapping pathways: (1) siRNA-mediated cytoplasmic RNA silencing (PTGS), (2) silencing mediated by miRNAs and (3) DNA methylation-dependent silencing at the transcriptional level (Baulcombe 2004).
Viruses, one of the most important causative agents of infectious diseases in both plants and animals, encode few multitasking proteins to support their life cycle. For successful replication of the genome, viruses use self-encoded replicase along with various replication factors (Mori et al. 1992) or reverse transcriptase (Laco and Beachy 1994) in case specific manner. Interestingly, viruses are also known to efficiently use host-encoded RNA-dependent RNA polymerase (Dalmay et al. 2000; Mourrain et al. 2000). Most plant viruses have a narrow host range and capable of manipulating developmental pathways of the hosts leading to striking and elaborate array of symptoms formation (Hull 2002). Plant viral symptoms were also observed and commercially exploited long before the concept of virus came into existence. For example, the flame-like streaks of tulip flower caused by infection with tulip-breaking virus (Dekker et al. 1993) achieved high prices. The autumnal yellow appearance of eupatorium plants caused by Eupatorium yellow vein virus and its cognate betasatellite was praised by a Chinese princess (Saunders et al. 2003).
Recent studies have demonstrated ‘RNA silencing’ as a major contributor of plant defence response against viruses (Wang and Metzlaff 2005; Pallas and Garcia 2011; Waterhouse et al. 2001). Plant viruses induce PTGS in a homology-dependent manner (Baulcombe 2004; Meister and Tuschl 2004; Eamens et al. 2008). During the initial stage of RNA silencing, long double-stranded RNAs (dsRNAs) (produced by the transcripts of inverted repeat sequence as in transposons or by the transcription from convergent promoters or by host RDRs (either in primer-dependent or primer-independent mechanism) (Dalmay et al. 2000; Mourrain et al. 2000; Sijen et al. 2001)) are cleaved into small interfering RNAs (siRNAs) of 21–24 nucleotides (Bernstein et al. 2001; Hamilton et al. 2002) which play diverse and redundant functions (Xie et al. 2004; Gasciolli et al. 2005; Blevins et al. 2006). Duplex siRNAs then undergo unwinding in ATP-dependent manner (Nykanen et al. 2001) and one of the two strands called guide strand gets incorporated into complex machinery of proteins called RNA-induced silencing complex (RISC) to carry out sequence-specific degradation of complementary target mRNA (Khvorova et al. 2003). A similar strategy has been adopted by both plants and animals to generate endogenous microRNAs (miRNAs). siRNA-directed transcriptional and heterochromatic gene silencing (Lippman and Martienssen 2004) constitute the third branch where the siRNAs (24–26 nt) of slightly larger size can be generated from transcripts of inverted repeats or tandem-repeated sequence and by ectopically expressed RNAs corresponding to the promoter region (Jones et al. 1999; Mette et al. 2000) and function in association with AGO4 and RDR2 (Hamilton et al. 2002; Qi et al. 2005; Xie et al. 2005; Zilberman et al. 2003). Methylation of cytosine residue in DNA (RNA-dependent DNA methylation, RdDM) or at the 9th position of histone H3 (H3K9) also results in suppression of gene expression (Blander and Guarente 2004). This RdDM pathway has been reported to maintain the genome integrity at both centromeric and telomeric repeat regions and suppress the transcription of transposons and other invasive DNAs (Matzke et al. 2009; Haag and Pikaard 2011). On the other hand, viruses in turn develop several strategies to interfere with host defence machinery to establish successful pathogenesis. One of the most fascinating strategies is the evolution of viral suppressors of RNA silencing which interfere with various steps of RNA silencing pathway of host (Anandalakshmi et al. 1998; Brigneti et al. 1998; Kasschau and Carrington 1998; Llave et al. 2000). Additionally, viruses also adopt different strategies to bypass RNA silencing machinery. Bromoviruses protect their RNA genome from host ribonucleases by accumulating inside the membrane vesicle (Schwartz et al. 2002). Members of the family Avsunviroidae undergo chloroplastic replication and thereby protect the viroid genome from RNA silencing (Tabler and Tsagris 2004). Again, the extensive intramolecular fold structures of viroids make them inaccessible to RISC complex (Wang et al. 2004). Defective interfering RNAs, which are devoid of target sequences also help to escape RNA silencing as observed in case of tombusviruses (Dalmay et al. 1995). Transfected siRNAs specific for either influenza A or HIV virus failed to target viral genome because of the occurrence of quasi species by spontaneous mutations in the target region (Ge et al. 2003; Boden et al. 2003; Das et al. 2004). Viruses (e.g. respiratory syncytial virus) also interact with certain cytoplasmic proteins (Biltko and Barik 2001) for encapsidation, high rate of replication and spread that may finally aid the viruses to escape RNA silencing machinery.
Components of RNA Silencing Machinery
RNA silencing is an ancient defence mechanism. While coming down the ladder of evolution, the process has incorporated several unique and functionally diverse proteins as important contributors in the complexity and specificity of the pathway. Proteins like Dicer-like enzymes (DCLs), ARGONAUTEs (AGOs), HYPONASTIC LEAVES (HYL1) and other dsRNA-binding proteins, HUA ENHANCER1 (HEN1) and RNA-DEPENDENT RNA POLYMERASEs are providing specificity to the RNA silencing machinery of plant.
Dicer-Like Proteins (DCLs): In comparison to other eukaryotes (mammals, worms, flies and fungi) A. thaliana encodes different DCLs. DCLs/DICERs are multidomain RNAse III-like ribonucleases which include evolutionary conserved N-terminal RNA helicase domain, central Piwi/Argonaute/Zwille (PAZ) domain and C-terminal dual catalytic and dsRNA-binding domains (Bernstein et al. 2001; Schauer et al. 2002). DCL1, the first RNAse III-like ribonucleases discovered, processes endogenous dsRNAs to miRNAs (21–22 nt) which in turn control diverse set of mRNAs of various transcription factors (Park et al. 2002; Xie et al. 2005). DCL2 produces less abundant 22-nt siRNAs population, and DCL3 produces 24-nt hc-siRNAs to carry out RdRM and to modify cis- and trans-elements of the gene, DNA repeats and transposons loci (Xie et al. 2004; Bouche et al. 2006). DCL4 is involved in the biogenesis of endogenous tasiRNAs (Reinhart et al. 2002; Gasciolli et al. 2005). Recently, DICER-LIKE 4 (DCL4) has been shown to terminate transcription of Arabidopsis endogenous FCA gene (a nuclear RNA-binding protein which controls the flowering time in Arabidopsis) by promoting cleavage of the aberrant RNA produced from the locus (Liu et al. 2012). In rice, DCL4 and DCL3 homolog DCL3b are likely to be involved in the generation of phased siRNAs of 21 and 24 nucleotides, respectively (Song et al. 2012). It has been proposed that formation of differently sized siRNAs is probably mediated by a PAZ domain of the DCLs, which configures the single-cleavage centre with respect to long N-terminal RNase III domain (Schauer et al. 2002).
Argonautes (AGOs): The uniqueness of RISC complex is provided by the AGO proteins. They can bind to both siRNA and miRNA. Ten different AGO proteins are encoded by Arabidopsis genome (Baulcombe 2004) of which AGO1, AGO2, AGO5 and AGO7 reportedly contribute to the antiviral defence in plant (Wang et al. 2011). Typically RISC-containing miRNAs and 21-nt siRNAs (produced by DCL1 and DCL4, respectively) associate with either AGO1, AGO2, AGO7 or AGO10 to cause posttranscriptional gene silencing (PTGS) of target mRNA by translational repression (Brodersen et al. 2008) or slicing (Baumberger and Baulcombe 2005). In contrast, 24-nt siRNAs, produced by DCL3, associate with AGO4, AGO6 or AGO9 and initiate transcriptional gene silencing (TGS) (Brosnan and Voinnet 2011).
AGO contains four functionally distinct domains to interact extensively with small RNA molecules. PAZ domain recognizes the 3′ end (Lingel et al. 2003, 2004; Ma et al. 2004); PIWI domain adopts an RNase-H fold and confers targeted endonucleolytic activity to certain AGOs to cleave the target mRNA in the region complementary to the guide RNA (Song et al. 2003; Yuan et al. 2005; Kawamura et al. 2008); MID domains of AGOs interact with the 5′ end of small RNAs and can direct the sorting of different classes of small RNAs into the appropriate AGO family members (Frank et al. 2012, 2010; Ma et al. 2005; Parker et al. 2005). Recently, the N-terminal domain of AGO has been proposed to be involved in unwinding of duplex siRNAs/miRNAs (Kwak and Tomari 2012). These AGOs vary in terms of catalytic triad present either in them or in residues that are involved in 5′ phosphate binding (Liu et al. 2004; Rivas et al. 2005). AGOs have been found to coimmunoprecipitate with viral small RNAs, but AGO1, largely considered as the principal slicer, has been found to bind to miRNAs and certain class of siRNAs of endogenous origin but not with the viral siRNAs (Hunter et al. 2003; Fagard et al. 2000; Baumberger et al. 2007), while AGO4 is required for RdDM mediated by 24-nt siRNAs (Zilberman et al. 2004). AGO1 and AGO5 preferentially bind to small RNAs containing 5′ terminal U or C residues, respectively whereas AGO2 and AGO4 have a strong strand bias for small RNAs with 5′ terminal A. 5′ terminal nucleotide of small RNAs determines strand selection into AGO complexes. Nevertheless, this 5′ end-dependent incorporation is not exclusive (Mi et al. 2008; Montgomery et al. 2008; Takeda et al. 2008; Havecker et al. 2010). DDH residues of AGO1 have been shown to possess cleavage activity (Elbashir et al. 2001; Mallory et al. 2004).
RNA-Dependent RNA Polymerases (RDRs): Host-encoded RNA-dependent RNA polymerase (RDR) uses viral primary siRNA molecules as primers to convert (aberrant) RNA target sequences into new long dsRNAs which in turn are processed into secondary siRNAs. These RDR-dependent amplified pools of viral siRNAs are originated from the entire target RNA sequence causing transitive silencing Sijen et al. (2001)). In Arabidopsis 6 RDRs have been reported. Three of them, i.e. RDR1, RDR2 and RDR6, belong to RDRα group containing a catalytic DLDGD motif. Arabidopsis RDR1, RDR2 and RDR6, and orthologs of these genes, are involved in the amplification, and plants from which these genes have been knocked out are highly susceptible to various plant viruses. RDR3, RDR4 and RDR5 contain DFDGD motif and are characterized as members of RDRγ group (Wassenegger and Krczal 2006). RDR2 and RDR6 are found to be the most important members that contribute significantly in endogenous small RNA pathway by converting the ssRNA templates to dsRNA in a primer-independent manner (Curaba and Chen 2008). The role of RDR 3, 4 and 5 are not well explored yet. They are found as tandemly repeated clusters on chromosome II. RDR1 has been found to be induced in response to either viral infection or salicylic acid (Yu et al. 2003). Interplay of different RDRs is important in regulating antiviral response of host. RDR1 from N. tabacum suppresses RDR6-mediated antiviral silencing and enhances viral infection in N. benthamiana where it is reported to be truncated due to insertion of inframe mutation (Ying et al. 2010). RDR2 also antagonizes the production of RDR6-dependent siRNAs in sense PTGS (Jauvion et al. 2012).
Nonfamily dsRNA-Binding Proteins: HYPONASTIC LEAVES1 (HYL1) and dsRNA-binding proteins (DRB 2–4) bind to DCLs and assist cleavage of double stranded RNAs. HYL1 interacts with and DCL1 and colocalizes in the nuclear bodies along with C2H2 Zn finger protein, Serrate and this complex is required for miRNA processing (Han et al. 2004; Vazquez et al. 2004; Fang and Spector 2007; Song et al. 2007). DRB4 interacts specifically with DCL4 (Hiraguri et al. 2005). R2D2 in Drosophila (Liu et al. 2003) and RDE4 in C. elegans (Tabara et al. 2002) are two important DRBs helping Dicers to deliver duplex small RNAs to downstream effector complexes.
HUA ENHANCER 1 (HEN1): HEN1, a small methylase, is unique to plants which methylates the 2′ OH of the terminal nucleotide at 3′ end of the small RNAs (Yang et al. 2006; Yu et al. 2005). The small RNA duplexes with 3′ 2-nt overhangs are preferred substrate for HEN1 and get methylated immediately after DCL-mediated cleavage to provide stability and protection to the small RNAs against oligouridylation (Yang et al. 2006).
Origin of Viral siRNAs
Genome of plant viruses interestingly can serve as both the target and trigger of RNA silencing. Earlier it was speculated by the scientific fraternity that double-stranded RNA intermediate generated during replication of positive-strand RNA virus could trigger the production of vsiRNA (Ahlquist 2002) which, if true, would generate equal amount of siRNA from both positive and negative RNA strand. But Molnar and associates found that the genomic strand of the viruses gave rise to greater amount of vsiRNA. Consequently, it was proposed that highly structured, single-stranded viral RNAs could be processed into vsiRNAs to trigger RNA silencing (Molnar et al. 2005). Moreover, certain regions on the viral genome were identified as ‘hot’ which had greater potential of producing vsiRNA over ‘non-hot’ regions. It was further suggested that single-stranded viral RNA with stable secondary structure is more likely the probable source of vsiRNA than dsRNA replication intermediates (Szittya et al. 2010; Donaire et al. 2009). In the case of plant DNA viruses which replicate through dsDNA intermediate also produce vsiRNAs from foldback structures of RNA transcription units (Moissiard and Vionnet 2006; Vanitharani et al. 2005).
Role of vsiRNAs in Attenuating Expression of Host Transcript
Detailed studies with vsiRNA have indicated that vsiRNAs can posttranscriptionally regulate the host transcripts expression. Bioinformatics study with Potato spindle tuber viroid (PSTVd-RG1) revealed presence of stretches of 19–20-nt sequences from various plant species that share sequence identity with the viroid. Interestingly, most of these sequences corresponded to virulence-regulating region of the pathogen. Analysing the plant sequence divulged presence of number of transcription factors and chromo-domain helicase DNA-binding protein that shared sequence homology with the viral sequences (Wang et al. 2004). This result suggested putative role of vsiRNA in regulating expression of host regulatory genes. Study with Cauliflower mosaic virus revealed that the CaMV infection greatly reduced the expression of one mRNA from Arabidopsis sharing 18–25nt microhomology with 35S RNA leader sequence (Moissiard and Vionnet 2006). The functionality and efficiency of vsiRNA in regulating host genes depends on many cellular factors including activity of the silencing suppressors of viral origin and abundance of vsiRNAs. Recently it has been reported that siRNAs derived from viral satellite RNA can directly regulate the expression of a host gene and hence attenuate the disease symptoms. A 24-nt region of CMV-Y satellite RNA (Y-Sat), called the ‘yellow domain’, was shown to be responsible for yellow symptoms induced in Y-Sat-infected tobacco plants (Kuwata et al. 1991). Smith et al. (2011) observed that a 22-nt complementary region of this yellow domain was present in the sequence of subunit I of magnesium chelatase, an enzyme involved in chlorophyll biosynthesis. Extensive study revealed that Y-Sat-induced symptoms are elicited by the vsiRNAs-mediated silencing of CHLI (2011). This result was further confirmed by Shimura et al. (2011), who showed that N. benthamiana plants expressing inverted repeat sequence of Y-Sat also develop yellow symptom mimicking the virus-infected phenotype. Downregulation of CHLI expression in both transgenic and Y-Sat-infected plants further proved the role of Y-Sat-derived vsiRNA in affecting host expression. Finally, it was demonstrated that Y-Sat-derived vsiRNAs could specifically target the 22-nt sequence in CHLI mRNA and therefore downregulate CHLI mRNA, thus inducing the yellowing symptom by impairing the chlorophyll biosynthesis pathway.
Cell-to-Cell and Long-Distance Movement of Virus-Derived siRNAs in Plants
In plants, RNAi acts non-cell autonomously and spreads in transacting manner. Grafting experiments with transgene-induced rootstocks with non-silenced target shoots or scion showed that a sequence-specific silencing signal is transmitted from rootstocks into shoots (Palauqui et al. 1997). Mobile RNA silencing was found to have two distinct arms in plants: cell-to-cell (through plasmodesmata) (Himber et al. 2003; Dunoyer et al. 2010) and long-distance movement through phloem (Palauqui et al. 1997; Voinnet and Baulcombe 1997; Yoo et al. 2004; Kalantidis et al. 2008). The siRNAs (21 to 24 nt) generated from the processing of the long dsRNAs act as mobile silencing signal for both the cases. Few of the components of this signal transduction pathway including number of small RNAs, proteins and protein channels have been identified.
Local Movement of Silencing Signal From Cell to Cell: Local movement of silencing signal occurs through specialized intercellular channels called plasmodesmata (Lucas and Lee 2004; Oparka 2004; Kim and Zambryski 2005; Maule 2008). In the absence of signal amplification triggered by cellular RDRs, the spread of silencing signal is limited to 10–15 cells beyond the site of signal initiation. Plasmodesmata can allow the transfer of up to 27 kDa protein (Kobayashi and Zambryski 2007). However, plasmodesmata, upon binding to various virus-encoded proteins, can undergo significant change in their size exclusion limit (Imlau et al. 1999) and thereby allowing larger molecules like viral ribonucleoproteins and transcription factors (e.g. KNOTTED 1) to pass through it (Lucas et al. 1995; Carrington et al. 1996). Spread of ‘local’ or ‘limited’ cell-to-cell silencing depends on the 21-nt siRNAs generated by DCL4 in AGO1-dependent cleavage of target endogenous genes (Himber et al. 2003; Parizotto et al. 2004). When SULPHUR gene fragment was expressed using phloem companion cell-specific promoter, mutation in RDR6 failed to interfere with the yellowing of the companion and its adjacent 10–15 cells (Himber et al. 2003) indicating little role of RDR6 in the local silencing process. Mutation of DCL4 leads to loss of non-cell-autonomous silencing indicating that 21-nt but not 24-nt siRNAs are sufficient for non-cell-autonomous RNAi (Hamilton et al. 2002; Dunoyer et al. 2005, 2010).
Extensive Long-Distance Movement of Silencing Signals: Spread of silencing signal beyond 10–15 cells is termed as extensive silencing. The larger siRNAs (24–26 nt) rather than 21 nt are essential for spread of long-distance silencing signals (Himber et al. 2003) which is dependent on signal amplification by RDR6, SGS3 and a putative RNA helicase (SDE3) (Mourrain et al. 2000; Vaistij et al. 2002; Himber et al. 2003) either in primer-dependent (5′–3′ transitivity) or primer-independent way (3′–5′ transitivity). This amplification is predominantly carried out by the secondary siRNAs generated from cleaved dsRNAs that function as repetitive wave of local cell-to-cell signalling of 10–15 cells at a time (Himber et al. 2003).Two important proteins NRPD1a (a component of RNA Pol IV) (Herr et al. 2005; Kanno et al. 2005; Onodera et al. 2005; Pontier et al. 2005) and RDR2 (an RNA-dependent RNA polymerase 2) (Xie et al. 2004; Herr et al. 2005; Pikaard 2006) function as essential components of non-cell-autonomous RNA silencing (Dunoyer et al. 2007; Smith et al. 2007). Phloem acts as a specific highway for transport of long-distance systemic silencing signals through specialized sieve tube cells from source to sink (Palauqui et al. 1997). Additionally, long-distance spread of silencing signal requires high amount of target transcripts (Garcia-Perez et al. 2004; Schwach et al. 2005). Spreading of miR166 expression in phloem tissues during leaf development indicates its involvement in long-distance signal movement to act at distance (Juarez et al. 2004). Abundance of PHO2 and miR399 in the phloem with regard to inorganic phosphate (Pi) alteration and coexpression suggests their involvement in systemic silencing (Lin et al. 2008). miR172 was found to be present in sRNA libraries prepared from phloem exudates and likely to play important role in long-distance signalling (Zeevaart 2008). miR319 gets transported from leaves to roots where it targets a subset of the TCP family of transcription factors that regulates LOX2 expression (Yoo et al. 2004; Schommer et al. 2008; Buhtz et al. 2010). Phloem small-RNA-binding protein 1 (PSRP1) was subsequently shown to bind and facilitate movement of single-stranded sRNA molecules between cells (Ham et al. 2009). CmPP16 protein from Cucurbita maxima was shown to possess properties similar to those of viral movement proteins (Aoki et al. 2005).
Antiviral Defence Pathways in Plants: RNA Silencing
In 1992, Lindbo and Dougherty observed that transgenic plant expressing non-translatable coat protein of tobacco etch virus (TEV) was resistant to cognate virus. Taking clue from this observation, it was rather sagaciously proposed that the resistant phenotype was the consequence of a mechanism active in cytoplasm which target and destroy the mRNA in sequence-specific manner (Lindbo et al. 1993). The hypothesis was the first step in building the concept of RNA silencing as antiviral defence. Later on it was also observed that an infectious viral cDNA clone engineered to carry a part of a host gene, when mobilized inside the plant, caused silencing of both the specific host gene and the viral sequence. Antiviral RNA silencing in plant has turned out to be an integrated network of at least 3 different mechanisms, namely, cytoplasmic RNA silencing, endogenous mRNA silencing by microRNAs and DNA methylation-dependent silencing at transcriptional level (Baulcombe 2004). These mechanisms not only provide antiviral resistance but also are crucial for cellular functions such as regulation of gene expression, maintenance of genome integrity and stress response. The basic processes of RNA silencing have been well documented in several reviews (Meister and Tuschl 2004; Eamens et al. 2008; Ruiz-Ferrer and Vionnet 2009; Ding 2010; Llave 2010).
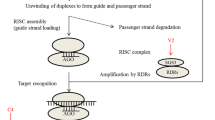
Cytoplasmic RNA silencing (Fig. 1a) starts through a process known as virus-induced gene silencing (VIGS). dsRNAs or hpRNAs are targeted by DCLs to produce small RNAs of varying length (21–24 nt). The resulting small RNAs are unwound with the help of an ATP-dependent RNA helicase and subsequently incorporated into RISC-containing AGO1 to perform degradation of viral mRNA and translational repression or methylation of the homologous target genes. Transcriptional gene silencing (Fig. 1c) initiates in the nucleus following infection with viruses or subviral elements which are gradually subjected to inactivation through DNA methylation (TGS). RNA-directed DNA methylation (RdDM) plays a very important role in terms of silencing transposons as well as repetitive DNA elements to maintain genome integrity as well as stability (Matzke et al. 2009; Haag and Pikaard 2011). In RdDM, dsRNAs are synthesized by a DNA-dependent RNA polymerase called RNA polymerase IV (Pol IV) and RDR2 specific to plant and then processed by DCL3 to produce 24-nt siRNAs. These 24-nt siRNAs form an AGO4-containing RISC and interact with the nascent transcript prepared by RNA Pol V (another plant-specific RNA polymerase). This interaction facilitates recruitment of various methylation factors like DRM2, and ultimately de novo cytosine methylation of the target DNA takes place. Therefore, in general RdDM has been known to contribute to plant defence by transcriptional repression of genes from DNA viruses.
Anti-viral RNA silencing pathways in plants and their suppression by plant viral encoded suppressors. Three major RNA silencing pathways include (a) cytoplasmic RNA silencing, (b) endogenous mRNA silencing by mRNAs and (c) transcriptional gene silencing through RdDM. Plant VSRs are represented by various shapes, while various steps of inhibition is represented by block arrows
The miRNA pathway (Fig. 1b) starts when the miRNA genes are transcribed by RNA polymerase II, and the resulting transcripts contain complementary regions that form short imperfect hairpins. These imperfect hairpins are processed by DCL1 in the nucleus into 21-nt miRNAs with the aid of several other proteins like zinc-finger protein SERRATE and the dsRNA-binding proteins DRB1 and HYL1. The miRNAs play a decisive role in plant development by either repressing or optimizing the expression of various transcription factors associated with developmental processes. miRNAs in plants function through homology-dependent RNA degradation as well as through translational repression (Brodersen et al. 2008) unlike animal miRNAs which usually bind to 3′ UTR. In cytoplasmic siRNA-dependent RNA silencing pathway, the exogenous or endogenous long dsRNAs or short hpRNAs are degraded by either DCL4 or DCL2 to generate 21- and 22-nt siRNAs, respectively. These siRNAs then recruited onto AGO1-containing RISC and RISC-containing guide siRNA cleave target mRNAs. RDR6, one among the six reported RDRs of Arabidopsis, then synthesizes long double-stranded RNA using ssRNAs as template to give rise to trans-acting siRNAs of 21 nt which also have been found to play a role in various stress responses as well as plant developmental processes. Another type of siRNAs called natural antisense siRNAs have been reported in many eukaryotes including plants which are produced from cis-natural antisense transcripts (cis-NATs) in response to various biotic and abiotic stresses (Zhang et al. 2012).
Counter-Defence Response of Virus: Plant Viral Suppressors (VSRS)
Plant viral synergism is defined by a situation, where a plant infected with two or more unrelated viruses shows symptoms much severe than that caused by either of the virus alone. Majority of the synergistic interaction between two viruses usually involves potyvirus as one of the co-infecting virus. With the discovery of importance of helper component proteinase (Hc-Pro) of potyvirus in synergism, the concept of viral suppressor working as a counter-defence tool was born. Hc-Pro was designated as viral suppressor of gene silencing (Vance et al. 1995). Viral suppressors (VSRs) have emerged as one of the most potent tricks available with the viruses to invade hosts’ defence for establishing successful pathogenesis. VSRs are found in almost all the plant viruses though few reports of suppressors from insects and mammalian viruses are also available at present. Plant VSRs have been found to evolve as diverse group of proteins and share very less sequence homology across the genera. VSRs are presumably evolved to counter the host silencing machinery-mediated defence response and therefore to suppress the host surveillance system. Different suppressors are reported to inhibit different steps of RNA silencing machinery by interacting with different effector components by any of the following ways: (i) through interaction with dsRNA to inhibit their processing by Dicers, (ii) binding to siRNAs and sequestering them to make them unavailable for the RISC, (iii) interacting either directly or indirectly with AGO to degrade or inactivate them and thus preventing functional RISC assembly and (iv) inhibition of systemic silencing by interacting with either host RDR or DCL4 or DBR4. In addition, some other mechanisms have also been proposed. Some VSRs activate specific group of miRNAs and thereby inhibit some of the important effector molecule of RNA silencing machinery. In other cases, VSRs outcompetes HEN1 for binding to siRNA duplex having 2-nt overhang at 3′ end. There are VSRs, specially found in DNA viruses, which have been shown to inactivate TGS either by transactivating a set of host genes, which in turn act as suppressor or inactivate some of the host methyl transferases. Role of the individual group of suppressors and their mode of action is given in Table 1.
In addition to their primary role in suppression of antiviral RNA silencing, VSRs can act as potent mediator of plant viral diseases by affecting the intrinsic function of essential host factors through direct or indirect interactions. For example, P2 protein, encoded in Rice dwarf virus (RDV), interacts directly with rice ent-kaurene oxidases (Zhu et al. 2005) and interferes with gibberellin biosynthesis. Reduced hormone accumulation results in stunting of the infected rice plants. CMV-2b protein has been shown to interact with Arabidopsis catalase (CAT3) and interfere with its scavenging activity (Inaba et al. 2011). At least 10 host proteins were reported to contribute to pathogenicity of tombusviruses (Ishibashi et al. 2010).
The Plant Fights Back: Phenomenon of Host Recovery
The idea of ‘host recovery’ phenomenon came as a converging mechanism of both natural resistance and host PTGS where the plants infected by virus showed initial symptom on inoculated leaves, but the newly emerging leaves were completely asymptomatic. The systemic and newly emerged leaves provided complete sequence-specific resistance against the virus. The incident of virus-induced symptom recovery was observed for the first time in 1928 when the initial leaves of tobacco plants infected with tobacco ring spot virus showed necrosis and disease symptom. The upper systemic leaves were asymptomatic and consequently showed resistance to secondary infection by the homologous virus (Wingard 1928). Further study suggests that methylation-dependent gene silencing is also associated with host recovery. Such type of recovery has been well documented in geminivirus-infecting host plants. Wild-type Arabidopsis and N. benthamiana plants inoculated with Beet curly top virus (BCTV) L2- mutant showed recovery (L2 interferes with methylation by interacting with host methyl transferase) owing to the recovery of host from the mutant virus infection (Hormuzdi and Bisaro 1995; Wang et al. 2003). Geminivirus-induced symptom recovery has also been reported in watermelon, cassava and pepper following infection with cucurbit leaf crumple virus, African cassava mosaic Cameroon virus [ACMV-CM] and pepper golden mosaic virus, respectively (Hagen et al. 2008; Chellappan et al. 2004; Rodriguez-Negrete et al. 2009). Transient expression of dsRNA corresponding to viral IR showed enhanced symptom recovery in Zucchini plants (Hagen et al. 2008). Recovery of plants from virus infection was linked with RNA silencing machinery, and particularly in geminiviruses recovery was correlated with reduced viral titre followed by increased viral siRNA accumulation (Chellappan et al. 2004). Natural recovery of host can also be observed following infection with nepovirus (Ratcliff et al. 1997) and caulimovirus (Covey et al. 1997). In contrary to previous studies as observed in DNA viruses, it was reported that the recovery of N. benthamiana carrying functional RDR1 orthologue of Medicago truncatula was associated with RNA silencing but not with reduced viral titre from a necrotic response induced by a nepovirus, Tomato ring spot virus (ToRSV). The disappearance of symptoms was not accompanied by reduction of viral mRNA (Jovel et al. 2007). Mutation in AV2 also leads to recovery (Basu et al. unpublished data) because of its inability to bind to SGS3, and therefore, allowing the amplification of silencing signal in presence of RDR6–SGS3 interaction and the secondary siRNA so produced degrade the viral mRNA and ultimately led to recovery in the newly emerging systemic leaves.
The Role of Plant miRNA in Plant–Virus Interaction
Recently it has been reported that plant miRNAs are responsive to developmental cues and environmental stresses. Tomato plants after infection with Cucumovirus and Tobamovirus showed significant differential expression in 85 % of its total miRNA pool (Fang and Spector 2007). All the differentially expressed miRNA were classified into 25 families. Among all these families, miR159 and miR171 contained most number of miRNAs. Most of these miRNAs were targeted to control expression of transcription factors, plant flower and leaf and height development and reproductive growth. High-throughput sequencing revealed a set of conserved miRNAs. Earlier it was also shown that infection with Tobamoviridae, Potyviridae, and Potexviridae families caused altered accumulation of certain miRNA in Nicotiana tabucum in which miRNAs 156, 164, 165 and 167 accumulated to higher levels compared to noninfected tissues (Bazzini et al. 2007). Silencing suppressors of various plant viruses have been reported to change target mRNA level through directly altering the accumulation of endogenous miRNA levels inducing changes also in target mRNA accumulations (Kasschau et al. 2003; Dunoyer et al. 2004; Zhang et al. 2006). Other workers have established the correlation between enhanced expression of miR168 and AGO1 mRNA in virus-infected plants (Zhang et al. 2006; Csorba et al. 2007; Havelda et al. 2008). These reports also make room to develop a novel strategy where manipulating the host miRNA level holds promise to combat with the viral stress.
Application of RNA Silencing Towards Plant Virus Resistance
Plant pathogens especially viruses are responsible for severe loss in crop production every year throughout the world. Earlier these pathogens were controlled using conventional measures including crop rotation, use of insecticides and breeding with resistant varieties. During 1986, Beachy and his associates demonstrated for the first time the use of pathogen-derived sequence (using TMV coat protein) to engineer resistance in the host (Powell et al. 1986). Since then various strategies based on either protein-mediated or RNA-mediated resistance have been developed. The actual mechanism of protein-mediated resistance is still not clear, and several pathways may be involved based on the type of gene used for engineering resistance. On the other hand, the mechanism of RNA silencing is well understood. During the last two decades, substantial effort has been channelized based on siRNA-mediated RNA silencing to engineer resistance in plants. These approaches differed in varied precursor sequence like pathogen-derived sequences in sense or antisense orientation, shRNA constructs, intron hairpin constructs and artificial miRNA sequences that were used to generate siRNA in plants. The use of intron hairpin RNAi constructs has been shown to be highly effective and caused nearly 100 % silencing of the target gene as compared to sense, antisense or hpRNAi constructs (Smith et al. 2000). It is also possible to target multiple viruses using single-RNAi constructs containing sequences from multiple viruses to generate broad-spectrum resistance (Jan et al. 2000; Bucher et al. 2006). One important hindrance to employ RNAi for engineering resistance is the selection of target and the minimum length of the target sequence for effective silencing. Hutvagner et al. (2000) showed that siRNAs generated by silencing of GUS gene mainly correspond to two-third region of 3′end of mRNA. Now a number of computational algorithms are freely available online for the rational design of siRNA and selection of target sequence to generate effective silencing of the target gene using several parameters.
Increased knowledge of microRNA (miRNA) biogenesis machinery and their role in regulation of transcript expression has helped to develop synthetic or artificial miRNAs (amiRNAs) to direct efficient silencing of any target transcript. amiRNA-mediated approach is one of the recently developed strategies with wide range of applications especially for conferring viral resistance in crop plants. Several studies have established potential of amiRNAs to target and degrade mRNAs of both viral and plant origin and thereby specifically degrading the target mRNA (Schwab et al. 2006, Niu et al. 2006, Qu et al. 2007, Zhang et al. 2012). Recently, amiRNAs targeting different ORFs of Tomato leaf curl virus AC1 along with AC2/AC4 (Yadava and Mukherjee 2010), the middle region of the AV1 (coat protein) transcript (amiR-AV1-3) and the overlapping region of the AV1 and AV2 (pre-coat protein) transcripts (amiR-AV1-1) (Vu et al. 2013) were designed and expressed in transgenic tomato plants to confer resistance and tolerance to ToLCV, respectively. Seemingly, amiRNA approach has several advantages over conventional siRNA-mediated strategy. In the hairpin RNAi approach, multiple siRNAs are formed from single precursor, and off-target genes are often silenced, while in amiRNA approach only one mature miRNA is produced targeting the specific gene. In amiRNAs mismatches can be introduced to avoid signal amplification and transitivity. siRNA-based gene silencing has been shown to be temperature dependent, while miRNA biogenesis has been shown to occur in various conditions and under extreme temperatures and therefore has wider scope of applications.
Conclusion
RNA silencing is an evolutionary conserved mechanism, which operates in several eukaryotic organisms across kingdoms and involves highly sequence-specific degradation of complementary RNA and transcriptional gene silencing. sRNAs of 21–24 nts in length are the key players of RNA silencing. The major components (players) of RNA silencing machinery in plants include AGO1, RDRs, DCLs, HEN1 and HYL1. These components are required for processing of dsRNA into siRNA and maintenance of RNA silencing. The mobile silencing signal moves from initiating cell to neighbouring cells through plasmodesmata and to long distance through phloem. Viruses are one of the most devastating pathogens of plants causing substantial crop loss every year. Viruses are both inducers and targets of RNA silencing. dsRNAs generated during replication of RNA viruses or transcription of overlapping sequences in DNA viruses induce RNA silencing which leads to sequence-specific degradation of target RNA into 21–24-nt siRNAs. Viruses in turn evolved suppressors of RNA silencing as powerful weapon to counter the host defence machinery. The suppressors encoded by different plant viruses act at different steps of RNA silencing thus inhibiting RNA silencing pathways in plants. Occasionally, infected plants show recovery from virus infection leading to remission of symptoms. Recovered plants remain immune to subsequent infection by a homologous virus through RNA silencing mechanism. Based upon the knowledge of RNA silencing mechanism, it is possible to engineer virus resistance in plants based on RNA silencing using viral-derived sequences as target.
References
Ahlquist P (2002) RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 296:1270–1273
Anandalakshmi R, Pruss GJ, Ge X, Marathe R, Mallory AC, Smith TH, Vance VB (1998) A viral suppressor of gene silencing in plants. Proc Natl Acad Sci USA 95:13079–13084
Aoki K, Suzui N, Fujimaki S, Dohmae N, Yonekura-Sakakibara K, Fujiwara T, Hayashi H, Yamaya T, Sakakibara H (2005) Destination-selective long-distance movement of phloem proteins. Plant Cell 17:1801–1814
Baulcombe D (2004) RNA silencing in plants. Nature 431:356–363
Baumberger N, Baulcombe DC (2005) Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA 102:11928–11933
Baumberger N, Tsai CH, Lie M, Havecker E, Baulcombe DC (2007) The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr Biol 17:1609–1614
Bazzini AA, Hopp HE, Beachy RN, Asurmendi S (2007) Infection and coaccumulation of tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development. Proc Natl Acad Sci USA 104:12157–12162
Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363–366
Billy E, Brondani V, Zhang H, Muller U, Filipowicz W (2001) Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci USA 98:14428–14433
Biltko V, Barik S (2001) Phenotypic silencing of cytoplasmic genes using sequence specific double stranded interfering RNA and its application in the reverse genetics of wild type negative strand RNA viruses. Biomed Center Microbiol 1:34–55
Bisaro DM (2006) Silencing suppression by geminivirus proteins. Virology 344:158–168
Blander G, Guarente L (2004) The Sir2 family of protein deacetylases. Ann Rev Biochem 73:417–435
Blevins T, Rajeswaran R, Shivaprasad PV, Beknazariants D, Si-Ammour A, Park HS, Vazquez F, Robertson D, Meins F, Hohn T, Pooggin MM (2006) Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res 34:6233–6246
Boden D, Pusch O, Lee F, Tucker L, Shank PR, Ramratnam B (2003) Promoter choice affects the potency of HIV-1 specific RNA interference. Nucleic Acids Res 31:5033–5038
Bortolamiol D, Pazhouhandeh M, Marrocco K, Genschik P, Ziegler-Graff V (2007) The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr Biol 17:1615–1621
Bouche N, Lauressergues D, Gasciolli V, Vaucheret H (2006) An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. The EMBO J 25:3347–3356
Brigneti G, Voinnet O, Li WX, Ji LH, Ding SW, Baulcombe DC (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J 17:6739–6746
Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320:1185–1190
Brosnan CA, Vionnet O (2011) Cell to cell and long-distance siRNA movement in plants: mechanisms and biological implications. Curr Opin Plant Biol 14:580–587
Bucher E, Sijen T, De Haan P, Goldbach R, Prins M (2003) Negative-strand tospoviruses and tenuiviruses carry a gene for a suppressor of gene silencing at analogous genomic positions. J Virol 77:1329–1336
Bucher E, Lohuis D, van Poppel PMJA, Geerts-Dimitriadou C, Goldbach R, Prins M (2006) Multiple virus resistance at a high frequency using a single transgene construct. J Gen Virol 87:3697–3701
Buhtz A, Pieritz J, Springer F, Kehr J (2010) Phloem small RNAs, nutrient stress responses, and systemic mobility. BMC Plant Biol 10:64–76
Cao XS, Zhou P, Zhang XM, Zhu SF, Zhong XH, Xiao Q, Ding B, Li Y (2005) Identification of an RNA silencing suppressor from a plant double-stranded RNA virus. J Virol 79:13018–13027
Carrington JC, Kasschau KD, Mahajan SK, Schaad MC (1996) Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8:1669–1681
Chellappan P, Vanitharani R, Pita J, Fauquet CM (2004) Short interfering RNA accumulation correlates with host recovery in DNA virus-infected hosts, and gene silencing targets specific viral sequences. J Virolol 78:7465–7477
Chellappan P, Vanitharani R, Fauquet CM (2005) MicroRNA-binding viral protein interferes with Arabidopsis development. Proc Natl Acad Sci USA 102:10381–10386
Chen J, Li WX, Xie DX, Peng JR, Ding SW (2004) Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of MicroRNA in host gene expression. Plant Cell 16:1302–1313
Chiba M, Reed JC, Prokhnevsky AI, Chapman EJ, Mawassi M, Koonin EV, Carrington JC, Dolja VV (2006) Diverse suppressors of RNA silencing enhance agroinfection by a viral replicon. Virology 346:7–14
Cogoni C, Irelan JT, Schumacher M, Schmidhauser T, Selker EU, Macino G (1996) Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J 15:3153–3163
Covey SN, AlKaff NS, Langara A, Turner DS (1997) Plants combat infection by gene silencing. Nature 385:781–782
Csorba T, Bovi A, Dalmay T, Burgyan J (2007) The p122 subunit of tobacco mosaic virus replicase is a potent silencing suppressor and compromises both small interfering RNA- and MicroRNA-mediated pathways. J Virol 81:11768–11780
Cuellar WJ, Kreuze JF, Rajamaki ML, Cruzado KR, Untiveros M, Valkonen JPT (2009) Elimination of antiviral defense by viral RNase III. Proc Natl Acad Sci USA 106:10354–10358
Curaba JL, Chen XM (2008) Biochemical activities of Arabidopsis RNA-dependent RNA polymerase 6. J Biol Chem 283:3059–3066
Dalmay T, Szittya G, Burgyan J (1995) Generation of defective interfering RNA dimers of cymbidium ringspot tombusvirus. Virology 207:510–517
Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101:543–553
Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R, Berkhout B (2004) Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol 78:2601–2605
Dekker EL, Derks AF, Asjes CJ, Lemmers ME, Bol JF, Langeveld SA (1993) Characterization of potyviruses from tulip and lily which cause flower-breaking. J Gen Virol 74(Pt 5):881–887
Ding SW (2010) RNA-based antiviral immunity. Nat Rev Immunol 10:632–644
Donaire L, Wang Y, Gonzalez-Ibeas D, Mayer KF, Aranda MA, Llave C (2009) Deep-sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology 392:203–214
Dunoyer P, Ritzenthaler C, Hemmer O, Michler P, Fritsch C (2002) Intracellular localization of the Peanut clump virus replication complex in tobacco BY-2 protoplasts containing green fluorescent protein-labelled endoplasmic reticulum or golgi apparatus. J Virol 76:865–874
Dunoyer P, Lecellier CH, Parizotto EA, Himber C, Voinnet O (2004) Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant CelI 16:1235–1250
Dunoyer P, Himber C, Voinnet O (2005) DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet 37:1356–1360
Dunoyer P, Himber C, Ruiz-Ferrer V, Alioua A, Voinnet O (2007) Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat Genet 39:848–856
Dunoyer P, Brosnan CA, Schott G, Wang Y, Jay F, Alioua A, Himber C, Voinnet O (2010) An endogenous systemic RNAi pathway in plants. EMBO J 29:1699–1712
Eamens A, Wang MB, Smith NA, Waterhouse PM (2008) RNA silencing in plants: yesterday, today, and tomorrow. Plant Physiol 147:456–468
Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T (2001) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J 20:6877–6888
Fagard M, Boutet S, Morel JB, Bellini C, Vaucheret H (2000) AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc Natl Acad Sci USA 97:11650–11654
Fang Y, Spector DL (2007) Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol 17:818–823
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811
Frank F, Sonenberg N, Nagar B (2010) Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465:818–822
Frank F, Hauver J, Sonenberg N, Nagar B (2012) Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs. EMBO J 31:3588–3595
Garcia-Perez RD, Houdt HV, Depicker A (2004) Spreading of post-transcriptional gene silencing along the target gene promotes systemic silencing. Plant J 38:594–602
Gasciolli V, Mallory AC, Bartel DP, Vaucheret H (2005) Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol 15:1494–1500
Ge Q, McManus MT, Nguyen T, Shen CH, Sharp PA, Eisen HN, Chen J (2003) RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc Natl Acad Sci USA 100:2718–2723
Glick E, Zrachya A, Levy Y, Mett A, Gidoni D, Belausov E, Citovsky V, Gafni Y (2008) Interaction with host SGS3 is required for suppression of RNA silencing by tomato yellow leaf curl virus V2 protein. Proc Natl Acad Sci USA 105:157–161
Goswami S, Sahana N, Pandey V, Doblas P, Jain RK, Palukaitis P, Canto T, Praveen S (2012) Interference in plant defense and development by non-structural protein NSs of Groundnut bud necrosis virus. Virus Res 163(1):368–373
Haag JR, Pikaard CS (2011) Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol 12:483–492
Hagen C, Rojas MR, Kon T, Gilbertson RL (2008) Recovery from Cucurbit leaf crumple virus (Family Geminiviridae, Genus Begomovirus) infection is an adaptive antiviral response associated with changes in viral small RNAs. Virology 98:1029–1037
Ham BK, Brandom JL, Xoconostle-Cazares B, Ringgold V, Lough TJ, Lucas WJ (2009) A polypyrimidine tract binding protein, pumpkin RBP50, forms the basis of a phloem-mobile ribonucleoprotein complex. Plant Cell 21:197–215
Hamilton A, Voinnet O, Chappell L, Baulcombe DC (2002) Two classes of short interfering RNA in RNA silencing. EMBO J 21:4671–4679
Han MH, Goud S, Song L, Fedoroff N (2004) The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci USA 101:1093–1098
Hao L, Wang H, Sunter G, Bisaro DM (2003) Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell 15:1034–1048
Harries PA, Palanichelvam K, Yu W, Schoelz JE, Nelson RS (2009) The Cauliflower mosaic virus protein p6 forms motile inclusions that traffic along actin microfilaments and stabilize microtubules. Plant Physiol 149:1005–1016
Hass G, Azevedo J, Moissiard G, Geldreich A, Himber C, Bureau M, Fukuhara T, Keller M, Voinnet O (2008) Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4. EMBO J 27:2102–2112
Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC (2010) The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22:321–334
Havelda Z, Varallyay E, Valoczi A, Burgyan J (2008) Plant virus infection-induced persistent host gene downregulation in systemically infected leaves. Plant J 55:278–288
Herr AJ, Jensen MB, Dalmay T, Baulcombe DC (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308:118–120
Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O (2003) Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J 22:4523–4533
Hiraguri A, Itoh R, Kondo N, Nomura Y, Aizawa D, Murai Y, Koiwa H, Seki M, Shinozaki K, Fukuhara T (2005) Specific interactions between Dicer-like proteins and HYL1/DRB family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol Biol 57:173–188
Hormuzdi SG, Bisaro DM (1995) Genetic analysis of beet curly top virus: examination of the roles of L2 and L3 genes in viral pathogenesis. Virology 206:1044–1054
Hull R (2002) Mathews plant virology, 4th edn. Academic, San Diego, pp 47–74
Hunter C, Sun H, Poethig RS (2003) The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr Biol 13:1734–1739
Hutvagner G, Mlynarova L, Nap JP (2000) Detailed characterization of the posttranscriptional gene-silencing-related small RNA in a GUS gene-silenced tobacco. RNA 6:1445–1454
Imlau A, Truernit E, Sauer N (1999) Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11:309–322
Inaba J, Kim BM, Shimura H, Masuta C (2011) Virus-induced necrosis is a consequence of direct protein-protein interaction between a viral RNA-silencing suppressor and a host catalase. Plant Physiol 156:2026–2036
Ishibashi K, Nishikiori M, Ishikawa M (2010) Interactions between tobamovirus replication proteins and cellular factors: their impacts on virus multiplication. Mol Plant Microbe Interact 23:1413–1419
Jan FJ, Pang SZ, Tricoli DM, Gonsalves D (2000) Evidence that resistance in squash mosaic comovirus coat protein-transgenic plants is affected by plant developmental stage and enhanced by combination of transgenes from different lines. J Gen Virol 81:2299–2306
Jauvion V, Rivard M, Bouteiller N, Elmayan T, Vaucheret H (2012) RDR2 partially antagonizes the production of RDR6-dependent siRNA in sense transgene-mediated PTGS. PLoS One 7:e29785
Jia D, Guo N, Chen H, Akita F, Xie L, Omura T, Wei T (2012) Assembly of the viroplasm by viral non-structural protein Pns10 is essential for persistent infection of rice ragged stunt virus in its insect vector. J Gen Virol 93(Pt 10):2299–2309
Jones L, Hamilton AJ, Voinnet O, Thomas CL, Maule AJ, Baulcombe DC (1999) RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 11:2291–2301
Jovel J, Walker M, Sanfacon H (2007) Recovery of Nicotiana benthamiana plants from a necrotic response induced by a nepovirus is associated with RNA silencing but not with reduced virus titer. J Virol 81:12285–12297
Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MCP (2004) microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428:84–88
Kalantidis K, Schumacher HT, Alexiadis T, Helm JM (2008) RNA silencing movement in plants. Biol Cell 100:13–26
Kalinina NO, Rakitina DV, Solovyev AG, Schiemann J, Morozov SY (2002) RNA helicase activity of the plant virus movement proteins encoded by the first gene of the triple gene block. Virology 296:321–329
Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ (2005) Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet 37:761–765
Kasschau KD, Carrington JC (1998) A counterdefensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95:461–470
Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC (2003) P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev Cell 4:205–217
Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H (2008) Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453:793–795
Khvorova A, Reynolds A, Jayasena SD (2003) Functional siRNAs and miRNAs exhibit strand bias. Cell 115:209–216
Kim I, Zambryski PC (2005) Cell-to-cell communication via plasmodesmata during Arabidopsis embryogenesis. Curr Opin Plant Biol 8:593–599
Kobayashi K, Zambryski P (2007) RNA silencing and its cell-to-cell spread during Arabidopsis embryogenesis. Plant J 50:597–604
Kubota K, Tsuda S, Tamai A, Meshi T (2003) Tomato mosaic virus replication protein suppresses virus-targeted posttranscriptional gene silencing. J Virol 77:11016–11026
Kuwata S, Masuta C, Takanami Y (1991) Reciprocal phenotype alterations between two satellite RNAs of cucumber mosaic virus. J Gen Virol 72(Pt 10):2385–2389
Kwak PB, Tomari Y (2012) The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat Struct Mol Biol 19:145–151
Laco GS, Beachy RN (1994) Rice tungro bacilliform virus encodes reverse transcriptase, DNA polymerase, and ribonuclease H activities. Proc Natl Acad Sci USA 91:2654–2658
Li H, Li WX, Ding SW (2002) Induction and suppression of RNA silencing by an animal virus. Science 296:1319–1321
Lin SI, Chiang SF, Lin WY, Chen JW, Tseng CY, Wu PC, Chiou TJ (2008) Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol 147:732–746
Lindbo JA, Silvarosales L, Proebsting WM, Dougherty WG (1993) Induction of a highly specific antiviral state in transgenic plants – implications for regulation of gene-expression and virus-resistance. Plant Cell 5:1749–1759
Lingel A, Simon B, Izaurralde E, Sattler M (2003) Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426:465–469
Lingel A, Simon B, Izaurralde E, Sattler M (2004) Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat Struct Mol Biol 11:576–577
Lippman Z, Martienssen R (2004) The role of RNA interference in heterochromatic silencing. Nature 431:364–370
Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X (2003) R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301:1921–1925
Liu JD, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ (2004) Argonaute2 is the catalytic engine of mammalian RNAi. Science 305:1437–1441
Liu FQ, Bakht S, Dean C (2012) Cotranscriptional role for Arabidopsis DICER-LIKE 4 in transcription termination. Science 335:1621–1623
Llave C (2010) Virus-derived small interfering RNAs at the core of plant-virus interactions. Trends Plant Sci 15:701–707
Llave C, Kasschau KD, Carrington JC (2000) Virus-encoded suppressor of posttranscriptional gene silencing targets a maintenance step in the silencing pathway. Proc Natl Acad Sci USA 97:13401–13406
Love AJ, Geri C, Laird J, Carr C, Yun BW, Loake GJ, Tada Y, Sadanandom A, Milner JJ (2012) Cauliflower mosaic virus protein P6 inhibits signaling responses to salicylic acid and regulates innate immunity. PLoS One 7(10):e47535
Lozsa R, Csorba T, Lakatos L, Burgyan J (2008) Inhibition of 3′ modification of small RNAs in virus-infected plants require spatial and temporal co-expression of small RNAs and viral silencing-suppressor proteins. Nucleic Acids Res 36:4099–4107
Lu R, Folimonov A, Shintaku M, Li WX, Falk BW, Dawson WO, Ding SW (2004) Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc Natl Acad Sci USA 101:15742–15747
Lucas WJ, Lee JY (2004) Plant cell biology – Plasmodesmata as a supracellular control network in plants. Nat Rev Mol Cell Biol 5:712–726
Lucas WJ, Bouche-Pillon S, Jackson DP, Nguyen L, Baker L, Ding B, Hake S (1995) Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270:1980–1983
Ma JB, Ye KQ, Patel DJ (2004) Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429:318–322
Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ (2005) Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434:666–670
Mallory AC, Reinhart BJ, Jones-Rhoades MW, Tang G, Zamore PD, Barton MK, Bartel DP (2004) MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 59 region. EMBO J 23:3356–3364
Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ (2009) RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol 21:367–376
Maule AJ (2008) Plasmodesmata: structure, function and biogenesis. Curr Opin Cell Biol 11:680–686
Mayers CN, Palukaitis P, Carr JP (2000) Subcellular distribution analysis of the cucumber mosaic virus 2b protein. J Gen Virol 81:219–226
Meister G, Tuschl T (2004) Mechanisms of gene silencing by double-stranded RNA. Nature 431:343–349
Merai Z, Kerenyi Z, Molnar A, Barta E, Valoczi A, Bisztray G, Havelda Z, Burgyan J, Silhavy D (2005) Aureusvirus P14 is an efficient RNA silencing suppressor that binds double-stranded RNAs without size specificity. J Virol 79:7217–7226
Mette MF, Aufsatz W, van der Winden J, Matzke MA, Matzke AJM (2000) Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J 19:5194–5201
Mi SJ, Cai T, Hu YG, Chen Y, Hodges E, Ni FR, Wu L, Li S, Zhou H, Long CZ et al (2008) Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133:116–127
Moissiard G, Voinnet O (2006) RNA silencing of host transcripts by cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc Natl Acad Sci USA 103:19593–19598
Molnar A, Csorba T, Lakatos L, Varallyay E, Lacomme C, Burgyan J (2005) Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J Virol 79:7812–7818
Montgomery TA, Yoo SJ, Fahlgren N, Gilbert SD, Howell MD, Sullivan CM, Alexander A, Nguyen G, Allen E, Ahn JH, Carrington JC (2008) AGO1-miR173 complex initiates phased siRNA formation in plants. Proc Natl Acad Sci USA 105:20055–20062
Mori M, Mise K, Okuno T, Furusawa I (1992) Expression of brome mosaic virus-encoded replicase genes in transgenic tobacco plants. J Gen Virol 73(Pt 1):169–172
Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, Remoue K, Sanial M, Vo TA, Vaucheret H (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101:533–542
Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2:279–289
Niu QW, Lin SS, Reyes JL, Chen KC, Wu HW, Yeh SD, Chua NH (2006) Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat Biotechnol 24:1420–1428
Nykanen A, Haley B, Zamore PD (2001) ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107:309–321
Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, Pikaard CS (2005) Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120:613–622
Oparka KJ (2004) Getting the message across: how do plant cells exchange macromolecular complexes? Trends Plant Sci 9:33–41
Palauqui JC, Elmayan T, Pollien JM, Vaucheret H (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J 16:4738–4745
Pallas V, Garcia JA (2011) How do plant viruses induce disease? Interactions and interference with host components. J Gen Virol 92:2691–2705
Parizotto EA, Dunoyer P, Rahm N, Himber C, Voinnet O (2004) In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev 18:2237–2242
Park W, Li JJ, Song RT, Messing J, Chen XM (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12:1484–1495
Parker JS, Roe SM, Barford D (2005) Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature 434:663–666
Pazhouhandeh M, Dieterle M, Marrocco K, Lechner E, Berry B, Brault V, Hemmer O, Kretsch T, Richards KE, Genschik P, Ziegler-Graff V (2006) F-box-like domain in the polerovirus protein P0 is required for silencing suppressor function. Proc Natl Acad Sci USA 103:1994–1999
Pikaard CS (2006) Cell biology of the Arabidopsis nuclear siRNA pathway for RNA-directed chromatin modification. Cold Spring Harbour Symp Quant Biol 71:473–480
Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T (2005) Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev 19:2030–2040
Powell AP, Nelson RS, De B, Hoffmann N, Rogers SG, Fraley RT, Beachy RN (1986) Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Science 232:738–743
Qi Y, Denli AM, Hannon GJ (2005) Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell 19:421–428
Qu F, Ren T, Morris TJ (2003) The coat protein of turnip crinkle virus suppresses posttranscriptional gene silencing at an early initiation step. J Virol 77:511–522
Qu J, Ye J, Fang R (2007) Artificial microRNA-mediated virus resistance in plants. J Virol 81:6690–6699
Ratcliff F, Harrison BD, Baulcombe DC (1997) A similarity between viral defense and gene silencing in plants. Science 276:1558–1560
Reed JC, Kasschau KD, Prokhnevsky AI, Gopinath K, Pogue GP, Carrington JC, Dolja VV (2003) Suppressor of RNA silencing encoded by beet yellows virus. Virology 306:203–209
Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16:2313–2313
Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu JD, Hannon GJ, Joshua-Tor L (2005) Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struc Mol Biol 12:340–349
Rodriguez-Negrete EA, Carrillo-Tripp J, Rivera-Bustamante RF (2009) RNA silencing against geminivirus: complementary action of posttranscriptional gene silencing and transcriptional gene silencing in host recovery. J Virol 83:1332–1340
Ruiz-Ferrer V, Voinnet O (2009) Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol 60:485–510
Saunders K, Bedford ID, Yahara T, Stanley J (2003) Aetiology: the earliest recorded plant virus disease. Nature 422:831
Schauer SE, Jacobsen SE, Meinke DW, Ray A (2002) DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci 7:487–491
Schommer C, Palatnik JF, Aggarwal P, Chetelat A, Cubas P, Farmer EE, Nath U, Weigel D (2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6:1991–2001
Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18(5):1121–1133
Schwach F, Vaistij FE, Jones L, Baulcombe DC (2005) An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol 138:1842–1852
Schwartz M, Chen J, Janda M, Sullivan M, den Boon J, Ahlquist P (2002) A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol Cell 9:505–514
Shimura H, Pantaleo V, Ishihara T, Myojo N, Inaba J, Sueda K, Burgyan J, Masuta C (2011) A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog 7:e1002021
Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RHA, Fire A (2001) On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107:465–476
Silhavy D, Molnar A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyan J (2002) A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J 21:3070–3080
Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM (2000) Gene expression – total silencing by intron-spliced hairpin RNAs. Nature 407:319–320
Smith LM, Pontes O, Searle I, Yelina N, Yousafzai FK, Herr AJ, Pikaard CS, Baulcombe DC (2007) An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell 19:1507–1521
Smith NA, Eamens AL, Wang MB (2011) Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog 7:e1002022
Song JJ, Liu JD, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L (2003) The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol 10:1026–1032
Song L, Han MH, Lesicka J, Fedoroff N (2007) Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci USA 104:5437–5442
Song XW, Li PC, Zhai JX, Zhou M, Ma LJ, Liu B, Jeong DH, Nakano M, Cao SY, Liu CY, Chu CC, Wang XJ, Green PJ, Meyers BC, Cao XF (2012) Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J 69:462–474
Szittya G, Moxon S, Pantaleo V, Toth G, Rusholme Pilcher RL, Moulton V, Burgyan J, Dalmay T (2010) Structural and functional analysis of viral siRNAs. PLoS Pathog 6:e1000838
Tabara H, Yigit E, Siomi H, Mello CC (2002) The dsRNA binding protein RDE-4 interacts with RDE-1, DCR- 1, and a DexH-box helicase to direct RNAi in C. elegans. Cell 109:861–871
Tabler M, Tsagris M (2004) Viroids: petite RNA pathogens with distinguished talents. Trends Plant Sci 9:339–348
Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y (2008) The mechanism selecting the guide strand from small RNA duplexes is different among Argonaute proteins. Plant Cell Physiol 49:493–500
Thomas CL, Leh V, Lederer C, Maule AJ (2003) Turnip crinkle virus coat protein mediates suppression of RNA silencing in Nicotiana benthamiana. Virology 306:33–41
Vaistij FE, Jones L, Baulcombe DC (2002) Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14:857–867
van der Krol AR, Mur LA, Beld M, Mol JN, Stuitje AR (1990) Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2:291–299
Vance VB, Berger PH, Carrington JC, Hunt AG, Shi XM (1995) 5′ proximal potyviral sequences mediate potato virus X/potyviral synergistic disease in transgenic tobacco. Virology 206:583–590
Vanitharani R, Chellappan P, Fauquet CM (2005) Geminiviruses and RNA silencing. Trends Plant Sci 10:144–151
Vazquez F, Gasciolli V, Crete P, Vaucheret H (2004) The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 14:346–351
Vogler H, Akbergenov R, Shivaprasad PV, Dang V, Fasler M, Kwon MO, Zhanybekova S, Hohn T, Heinlein M (2007) Modification of small RNAs associated with suppression of RNA silencing by tobamovirus replicase protein. J Virol 81:10379–10388
Voinnet O, Baulcombe DC (1997) Systemic signalling in gene silencing. Nature 389:553
Vionnet O, Lederer C, Baulcombe DC (2000) A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:157–167
Vu TV, Choudhury NR, Mukherjee SK (2013) Transgenic tomato plants expressing artificial microRNAs for silencing the pre-coat and coat proteins of a begomovirus, tomato leaf curl New Delhi virus, show tolerance to virus infection. Virus Res 172(1–2):35–45
Wang MB, Metzlaff M (2005) RNA silencing and antiviral defense in plants. Curr Opin Plant Biol 8:216–222
Wang H, Hao LH, Shung CY, Sunter G, Bisaro DM (2003) Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. Plant Cell 15:3020–3032
Wang MB, Bian XY, Wu LM, Liu LX, Smith NA, Isenegger D, Wu RM, Masuta C, Vance VB, Watson JM, Rezaian A, Dennis ES, Waterhouse PM (2004) On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc Natl Acad Sci USA 101:3275–3280
Wang XB, Jovel J, Udomporn P, Wang Y, Wu Q, Li WX, Gasciolli V, Vaucheret H, Ding SW (2011) The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell 23:1625–1638
Wassenegger M, Krczal G (2006) Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci 11:142–151
Waterhouse PM, Wang MB, Lough T (2001) Gene silencing as an adaptive defence against viruses. Nature 411:834–842
Wei T, Kikuchi A, Moriyasu Y, Suzuki N, Shimizu T, Hagiwara K, Chen H, Takahashi M, Ichiki-Uehara T, Omura T (2006) The spread of Rice dwarf virus among cells of its insect vector exploits virus-induced tubular structures. J Virol 80:8593–8602
Wingard SA (1928) Hosts and symptoms of ring spot, a virus disease of plants. J Agric Res 37:127–153
Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2:E104
Xie ZX, Allen E, Wilken A, Carrington JC (2005) DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA 102:12984–12989
Yadava P, Mukherjee SK (2010) Engineering geminivirus resistance in tomatoes using artificial microRNAs. Keystone symposium on RNA silencing mechanisms in plants, Santa Fe, 21–26 Feb 2010
Yang Z, Ebright YW, Yu B, Chen X (2006) HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 29 OH of the 39 terminal nucleotide. Nucleic Acids Res 34:667–675
Yang XL, Xie Y, Raja P, Li SZ, Wolf JN, Shen QT, Bisaro DM, Zhou XP (2011a) Suppression of methylation-mediated transcriptional gene silencing by beta C1-SAHH protein interaction during geminivirus-betasatellite infection. PLoS Pathog 7(10):e1002329
Yang X, Tan SH, The YJ, Yuan YA (2011b) Structural implications into dsRNA binding and RNA silencing suppression by NS3 protein of Rice Hoja Blanca Tenuivirus. RNA 17(5):903–911
Ye K, Patel DJ (2005) RNA silencing suppressor p21 of Beet yellows virus forms an RNA binding octameric ring structure. Structure 13:1375–1384
Yelina NE, Savenkov EI, Solovyev AG, Morozov SY, Valkonen JP (2002) Long-distance movement, virulence, and RNA silencing suppression controlled by a single protein in hordei- and potyviruses: complementary functions between virus families. J Virol 76:12981–12991
Ying XB, Dong L, Zhu H, Duan CG, Du QS, Lv DQ, Fang YY, Garcia JA, Fang RX, Guo HS (2010) RNA-dependent RNA polymerase 1 from Nicotiana tabacum suppresses RNA silencing and enhances viral infection in Nicotiana benthamiana. Plant Cell 22:1358–1372
Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee YM, Lough TJ, Lucas WJ (2004) A systemic small RNA signaling system in plants. Plant Cell 16:1979–2000
Yu DQ, Fan BF, MacFarlane SA, Chen ZX (2003) Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol Plant Microbe Interact 16:206–216
Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307:932–935
Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ (2005) Crystal structure of A-aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell 19:405–419
Zeevaart JA (2008) Leaf-produced floral signals. Curr Opin Plant Biol 11:541–547
Zhang X, Yuan YR, Pei Y, Lin SS, Tuschl T, Patel DJ, Chua NH (2006) Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes De 20:3255–3268
Zhang X, Li H, Zhang J, Zhang C, Gong P, Ziaf K, Xiao F, Ye Z (2011a) Expression of artificial microRNAs in tomato confers efficient and stable virus resistance in a cell-autonomous manner. Transgenic Res 20(3):569–581
Zhang Z, Chen H, Huang X, Xia R, Zhao Q, Lai J, Teng K, Li Y, Liang L, Du Q, Zhou X, Guo H, Xie Q (2011b) BSCTV C2 attenuates the degradation of SAMDC1 to suppress DNA methylation-mediated gene silencing in Arabidopsis. Plant Cell 23:273–288
Zhang X, Xia J, Lii YE, Barrera-Figueroa BE, Zhou X, Gao S, Lu L, Niu D, Chen Z, Leung C, Wong T, Zhang H, Guo J, Li Y, Liu R, Liang W, Zhu JK, Zhang W, Jin H (2012) Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome Biol 13:R20
Zhu S, Gao F, Cao X, Chen M, Ye G, Wei C, Li Y (2005) The rice dwarf virus P2 protein interacts with entkaurene oxidases in vivo, leading to reduced biosynthesis of gibberellins and rice dwarf symptoms. Plant Physiol 139:1935–1945
Zilberman D, Cao X, Jacobsen SE (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299:716–719
Zilberman D, Cao XF, Johansen LK, Xie ZX, Carrington JC, Jacobsen SE (2004) Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr Biol 14:1214–1220
Acknowledgement
We acknowledge the support for the Department of Biotechnology (DBT), Govt. of India, for financial assistance.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer India
About this chapter
Cite this chapter
Basu, S., Sharma, V.K., Bhattacharyya, D., Chakraborty, S. (2014). An Overview of Antiviral RNA Silencing in Plant: Biogenesis, Host–Virus Interaction and Potential Applications. In: Gaur, R., Sharma, P. (eds) Approaches to Plant Stress and their Management. Springer, New Delhi. https://doi.org/10.1007/978-81-322-1620-9_18
Download citation
DOI: https://doi.org/10.1007/978-81-322-1620-9_18
Published:
Publisher Name: Springer, New Delhi
Print ISBN: 978-81-322-1619-3
Online ISBN: 978-81-322-1620-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)