Abstract
Construct encoding self-complementary ‘hairpin’ RNA (hpRNA) has been shown as an effective trigger of post-translational gene silencing. Gene silencing can be effectively inhibited by virus-encoded silencing suppressors that have been acquired evolutionarily to surmount silencing and allow infection. Here, we show that AC2, the silencing suppressor protein of Mungbean Yellow Mosaic India Virus (MYMIV), can inhibit the hpRNA-induced gene silencing in planta. Phytoene desaturase (PDS)-silenced transgenic lines were generated using PDS-hairpin gene-silencing construct in Nicotiana. The photobleach phenotype was observed in different T1 and T2 PDS-silenced lines indicating that the hairpin gene-silencing construct can efficiently down-regulate or silence the endogenous gene activity, and that the silencing is stably inherited over generations. The analysis of abundance of the PDS transcripts showed that the degree of silencing ranged from 20 to 70 % in different T0 and T1 transgenic lines. Upon introduction of MYMIV-AC2 in the silenced PDS transgenic lines, by genetic hybridization, the PDS expression was restored in F1 hybrids. The F1 lines (♀PDS X ♂MYMIV-AC2) showed the normal green phenotype instead of the photobleached phenotype indicating that introgression of MYMIV-AC2 in PDS silenced T0 lines, suppresses the hairpin gene-silencing. Our findings suggest that hpRNA vector can efficiently and stably induce gene silencing in plants and that MYMIV-AC2 can inhibit this silencing. This is the first report where we put forward that the dual system of hpRNA and MYMIV-AC2 suppressor can serve as an efficient switch for turning off and turning on gene expression, with potential applications in gene function analysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gene silencing, often referred to as RNA silencing or RNA interference, is an evolutionarily conserved mechanism that protects the host genome against invasive nucleic acids, such as viruses, transposons, and transgenes (Hannon 2002). Both post-transcriptional gene silencing (PTGS) and transcriptional gene silencing (TGS) processes mediate effective defence mechanisms against invading genetic elements (Waterhouse et al. 2001). The PTGS mechanism detects double-stranded RNA (dsRNA) intermediates of the foreign nucleic acids and degrades them into 21- to 24-nucleotide small interfering RNAs (siRNAs). These small RNAs then act as sequence-based guides for specific protein complexes to target and destroy the homologous message-bearing nucleic acids. The dsRNA is thus an effective trigger of gene silencing which operates by sequence-specific RNA degradation (Waterhouse et al. 2001; Sharp 2001).
Introduction of hairpin RNA (hpRNA) can efficiently produce dsRNA in plants and animals. It has been demonstrated that gene constructs producing hpRNA are the most effective method for silencing gene expression in plants (Waterhouse et al. 1998; Smith et al. 2000). Many reports are now available for constructs specifically designed to deliver and express dsRNA in plants, in the form of hpRNA, eliciting a high degree and frequency of silencing of viral genes, transgenes, and endogenous genes (Waterhouse et al. 1998; Smith et al. 2000; Chuang and Meyerowitz 2000; Wang et al. 2000; Wesley et al. 2001). The phenotypic stability of the hpRNA-induced gene silencing over many generations has been considered as a crucial advantage in reliable application of gene silencing in plants and animals (Watson et al. 2005). Such hpRNA constructs have great potential as a tool for gene discovery and functional analysis (Wesley et al. 2001; Somerville 2000).
In plants, gene silencing also serves as an adaptive immune response that restricts accumulation or spread of invading viruses (Waterhouse et al. 2001). However, in what could be considered as an “evolutionary arms race”, the viruses encode suppressors of silencing to inhibit the gene silencing pathway, allowing infections in plants. The viral suppressor is a protein or an RNA element that blocks silencing of viral nucleic acid sequences guided by small RNAs, resulting in more severe pathogenic symptoms and diseases (Rahman et al. 2012). The discovery of the viral silencing suppressor itself played an important role in establishing RNA silencing as a natural antiviral response (Li and Ding 2006). Up to now, more than 50 viral silencing suppressors have been characterized from plant viruses and a few of them are also known from insect and animal viruses. Most of the suppressors act by distinct mechanisms and at discrete steps in the RNA silencing pathway and show no sequence relatedness. The AC2 protein of Mungbean yellow mosaic India virus (MYMIV), a member of the Geminiviridae family, genus Begomovirus, is one of the viral suppressors of RNA silencing (Rahman et al. 2012; Karjee et al. 2008; Singh et al. 2007; Bisaro 2006; Trinks et al. 2005).
AC2 is a multifunctional protein in Begomoviruses as it functions both as a transactivator of viral transcription and as a suppressor of RNA silencing (Trinks et al. 2005). AC2 protein is about ~15 kDa in size with three conserved domains, namely, a basic N-terminal nuclear localization domain, a core DNA binding domain with non-classical Zn finger motif (C37X1C39X7C47X6H54X4H59C60) in the middle, and a C terminus acidic transactivation domain (Trinks et al. 2005). In addition to its core role in viral transcription, AC2 is also known as a strong viral pathogenicity factor. Recently, we demonstrated that MYMIV-AC2 can overcome transgene-mediated gene silencing and enhance the transgene expression in planta by enhancing the expression of transgenically silenced GFP and TOPOII lines (Rahman et al. 2012).
Gene constructs encoding hpRNA are highly effective at inducing the silencing of both endogenous genes and transgenes that are homologous to the silencing effectors. These have been effectively employed in conferring virus resistance in plants (Smith et al. 2000; Chuang and Meyerowitz 2000; Wang et al. 2000; Wesley et al. 2001; Levin et al. 2000; Sijen et al. 2001; Wang and Waterhouse 2004). They have also been shown to induce RNAi in animals (Wang and Waterhouse 2002). In the present study, we investigated whether the suppressor activity of MYMIV-AC2 can overcome the hpRNA-induced gene silencing in planta. We used the phytoene desaturase (PDS) gene as a target of hpRNA because its silencing would result in photobleached leaves, a readily visible phenotype. To investigate whether AC2 can overcome this silencing, MYMIV-AC2 was stably introduced in the hpRNA-induced PDS-silenced tobacco lines by a genetic hybridization approach. It was observed that PDS expression was stably restored in F1 hybrid populations. The results thus reveal the potential of MYMIV-AC2 to suppress hpRNA-induced gene silencing in planta.
Materials and methods
Construction of PDS hairpin loop RNA plant binary vector
For the construction of pHANNIBAL-PDS hairpin loop vector, sense strand (5′–3′) of PDS was cloned into XhoI and EcoRI sites using forward 5′GCCCTCGAGGGCACTCAACTTTATAAACC3′ and reverse 5′GGAATTCCTTCAGTTTTCTGTCAAACC3′ primers. The PDS antisense (3′–5′) strand was cloned into BamHI and XbaI sites using forward 5′GCCTCTAGAGGCACTCAACTTTATAAACC3′ and reverse 5′AGGGATCCCTTCAGTTTTCT3′ primers of pHANNIBAL vector (CSIRO, Australia). PDS gene was amplified from the cDNA of Nicotiana tabacum cv. Xanthi by PCR using forward 5′CCGGATCCGGCACTCAACTTT3′ and reverse 5′AGGGATCCCTTCAGTTTTCT3′ primers using Platinum Taq Polymerase (Invitrogen). The whole cassette of PDS hairpin loop (PDS sense–intron–PDS antisense) was sandwiched between CaMV35S promoter and OCS terminator. To construct the pART27-PDS hairpin loop plant binary vector, a ~4.0-kb-long cassette (CaMV35S–PDS sense–intron–PDS antisense–OCS terminator) was recovered from PDS-pHANNIBAL plasmid by NotI enzyme to mobilize the cassette into the pART27 plant vector. This pART27-PDS-hairpin loop recombinant vector was subsequently used for Agrobacterium-mediated genetic transformation to silence PDS in tobacco plant.
Agrobacterium-mediated tobacco transformation to silence PDS
Agrobacterium-mediated genetic transformation of tobacco was done by using the leaf disc method. The pART27-PDS hairpin loop plant binary plasmid vector was transformed into Agrobacterium tumefaciens strain LBA4404. Agrobacterium cells containing the pART27-PDS hairpin RNAi construct were used for transformation of tobacco explants using the leaf disc transformation method (Horsch et al. 1985). The Agrobacterium culture was prepared in YEM broth at 28 °C and 200 rpm until the OD reached 0.6 at λmax 600 nm for plant transformation. Sterile leaf discs of about 3- to 4-week-old WT tobacco were infiltrated with the Agrobacterium containing the pART27-PDS hairpin RNA construct and were kept for co-cultivation for 2–3 weeks. Following co-cultivation, the explants were selected on regeneration MS medium containing kanamycin (150 mg/L) and cefotaxime (250 mg/L). Calli appeared from the co-cultivated leaf discs in selection and regeneration MS medium within 2 weeks of co-cultivation, while the control leaf discs (without co-cultivation) died down within this period. Shoots started to regenerate from green calli within 3–4 weeks. Shoots measuring approximately 2.0 cm were transferred to half-MS (Murashige and Skoog 1962) medium containing antibiotic for rooting. The rooted seedlings were hardened in vermiculite before being transferred to the soil in earthen pots.
Plant materials and growth condition
Nicotiana tabacum cv. Xanthi wild-type, MYMIV-AC2 transgenic T2 lines and PDS silenced transgenic lines plants were grown and maintained at 26 °C, 16 h photo-period, in a greenhouse. For germination of tobacco seeds, seeds were sterilized with 70 % ethanol for 1 min followed by thorough washes with water. Then, the seeds were kept in MS media containing appropriate antibiotics for 4 days in the dark, after which they were transferred to the light for further growth. Germinated seedlings were then transferred to vermiculite and then to soil pots for maintenance.
Self-fertilization and crossing experiment
Transgenic tobacco plants (AC2 lines and PDS silenced lines) were self-fertilized and only selected lines were advanced to the T2 generation. The selection of transgenic MYMIV-AC2 lines was based on phenotypic anomalies, where only the hypomorphic lines were advanced to T2 generation. Transgenically silenced PDS plants showing bleached phenotype were used in the subsequent crossing experiment. In the crossing experiment, transgenic PDS silenced T2 lines were used as recipient plants, whereas MYMIV-AC2 expressing T2 lines were used as donor plants. The F1 hybrid seeds were germinated in MS-medium containing kanamycin (250 mg/L), and subsequently the F1 population was grown and maintained in the greenhouse.
Genomic DNA isolation and genomic DNA PCR to screen transgenically silenced PDS lines
The genomic DNA of putative transgenically silenced PDS lines was isolated from the plant tissues using the standard CTAB-based genome-DNA extraction protocol (Sharma et al. 2002) and further used for molecular analysis. Transgenic analysis was carried out using promoter specific primer CaMV35S forward (5′AAGTTGACTGCCTGCAGGTC3′) and PDS sense strand reverse primer (5′AGGGATCCCTTCAGTTTTCT3′).
RNA isolation, Northern blot and RT-PCR analysis
Total RNA was isolated using Trizol reagent (Invitrogen) from young leaves following the manufacturer’s instructions. For northern blot analyses, 20 µg of total RNA was separated by electrophoresis on a 1 % formaldehyde agarose gel and blotted to a Hybond-N nylon membrane (Amersham Biosciences). The blots were hybridized with a [α-32P]dCTP-labeled PDS probe. For reverse transcriptase (RT), PCR 2 µg of DNase-treated (DNase I, amp grade; Invitrogen) total RNA was used to synthesize first strand cDNAs using SuperScriptTM III Reverse Transcriptase (Invitrogen). These cDNA templates were used for PCR amplification of PDS with forward 5′ CCGGATCCGGCACTCAACTTT3′ and reverse 5′AGGGATCCCTTCAGTTTTCT3′ primers. The PCR products were analyzed on a 1 % agarose gel. The relative intensities of the bands were estimated by densitometric intensity analysis using ImageJ software (http://rsbweb.nih.gov/ij/).
Results
Isolation and cloning of partial PDS gene
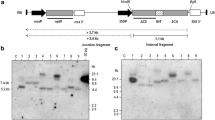
The partial mRNA of PDS was amplified from the cDNA of Nicotiana (cv. Xanthi) using specific primers (Fig. 1a) to construct the PDS-hpRNA vector as described in “Materials and methods”. It has been shown that a fragment as small as 98 bases is sufficient for effective gene silencing in hairpin vectors, such as the pHANNIBAL RNAi vector (Waterhouse and Helliwell 2003). The primers amplified a partial PDS cDNA of ~409 bp (Fig. 1b) which was then cloned into a TOPO-TA vector (Fig. 1 c, d) and the positive clones were sequenced. The sequence of the cloned partial-PDS fragment was aligned with the sequence of NCBI submitted partial-PDS gene (NCBI Accession: AJ616742.1; Tao and Zhou 2004), and the ClustalW alignment showed that the sequence obtained from the clone matched to the extent of ~99 % (Fig. 1e). This PDS clone was subsequently used to construct the PDS-hpRNA vector.
Isolation and cloning of partial mRNA of phytoene desaturase (PDS) from N. tabacum cv. Xanthi. a Ethidium bromide-stained agarose gel showing amplifications of a 409-bp fragment of PDS from tobacco cDNA. M indicates the 1-kb DNA ladder. b 409-bp PDS DNA eluted from gel was checked and used for cloning into the TOPO-TA vector. c Ethidium bromide-stained agarose gel showing colony PCR of PDS cloned into the TOPO-TA vector with PDS-specific primers. 1–14 positive clones, +ve cDNA used as template in PCR as positive control and –ve water used as template in PCR as negative control d EcoRI digestion of three PCR positive clones showing the 409 bp DNA fragment. D digested and UD undigested plasmid DNA. e ClustalW alignment of nucleotide sequences of AJ616742 (NCBI submitted partial mRNA of PDS) with cloned putative partial PDS gene
Construction of PDS hairpin RNA plant binary vector
To develop the PDS-hpRNA vector, the pHANNIBAL RNAi vector backbone was used (Fig. 2a). PDS sense strand (5′-3′) was cloned using the XhoI and EcoRI sites in the pHANNIBAL vector. The clones were checked by colony PCR using PDS-specific primers (data not shown) and two PCR positive clones were confirmed by XhoI and EcoRI restriction enzyme digestion (Fig. 2b) of their plasmid DNA. After cloning the PDS sense strand into the pHANNIBAL vector, the PDS sense-pHANNIBAL construct was further used for cloning of the PDS antisense (3′-5′) strand. The PDS antisense strand was cloned in BamHI and XbaI sites in the PDS sense-pHANNIBAL vector and the clones were confirmed by BamHI and XbaI restriction enzymes digestion (Fig. 2b) of isolated plasmid DNA. To confirm the final PDS-pHANNIBAL hairpin vector, the clones were digested with XhoI and XbaI restriction enzymes. As expected, a DNA fragment of ~1.6 kb, containing PDS-sense (409 bp)–intron (800 bp)–PDS-antisense (409 bp), was observed after digestion (Fig. 2b, lane d3).
Preparation of PDS hairpin RNAi binary construct to develop transgenically silenced PDS tobacco line. a Map of pHANNIBAL hairpin RNAi vector were used to prepare PDS hairpin RNAi construct. PDS sense strand has been cloned into XhoI and EcoRI sites, while PDS antisense has been cloned into BamHI and XbaI sites. b Ethidium bromide-stained agarose gel shows restriction enzymes digestion analysis of PDS-pHANNIBAL RNAi construct. Lane D1 shows 409-bp DNA fragment of sense strand after digestion with XhoI and EcoRI enzymes. D2 lane shows the DNA fragment of same length antisense strand digested with BamHI and XbaI enzymes, while lane D3 shows the DNA fragment of ~1.6-kb DNA (digestion with XhoI and XbaI enzymes) that contains sense (409 bp)–intron (800 bp)–antisense (409 bp) strands. UD and M represent the undigested plasmid and 1-kb DNA ladder, respectively. c Schematic representation of whole cassette of PDS hairpin loop (PDS sense–intron–PDS antisense) that is sandwiched between CaMV35 promoter and OCS terminator. This ~4.0-kb length of cassette (CaMV35S–sense–intron–antisense–OCS terminator) was recovered from the PDS-pHANNIBAL plasmid by NotI enzyme to mobilize the cassette into the pART27 vector (d), linearized by NotI enzyme. e Agarose gel showing colony PCR of PDS hairpin construct cloned into pART27 vector with PDS specific primers. 1–6 and 8–11 were positive clones, +ve (plasmid used as template in PCR) was positive control. f NotI restriction enzyme digestion of two PCR positive pART27-PDS hairpin loop clones shows ~4.0-kb DNA fragment of insert (CaMV35S–sense–intron–antisense–OCS terminator). D digested and UD undigested plasmids. This pART27–PDS-hairpin recombinant vector was subsequently used for Agrobacterium-mediated transformation to silence PDS in wild-type tobacco
To prepare the PDS hairpin RNA construct, we used pART27 vector (Fig. 2d). The PDS hairpin cassette (CaMV35S –PDS sense–Intron–PDS antisense–OCS terminator; Fig. 2c) was fished out from the PDS-pHANNIBAL construct by NotI restriction enzyme and cloned into the pART27 plant transformation vector, linearized by the NotI enzyme. A ~4.0-kb DNA fragment of PDS hairpin cassette (Fig. 2c) released by NotI digestion of the plasmid DNA confirmed the positive clones (Fig. 2f). The recombinant pART27-PDS hairpin plant construct was used for Agrobacterium-mediated transformation in tobacco to silence the PDS gene.
Development of PDS-silenced transgenic tobacco
PDS-silenced transgenic tobacco lines were developed by Agrobacterium-mediated genetic transformation in wild-type (WT) tobacco (cultivar Xanthi). Interestingly, we found some photo-bleached shoots regenerated from the calli of co-cultivated leaf discs within 3–4 weeks (Fig. 3a, b). About 50 full-grown bleached or partially bleached (Fig. 3b, c) shoots were obtained and transferred onto rooting medium. From these, 45 shoots gave rise to profuse roots in rooting medium. We observed a range of photo-bleaching phenotype, from high to medium, in the leaf area of regenerated plantlets (Fig. 3d–i). It was also observed that the putative transgenic lines were dwarf compared to WT control regenerated plants (Fig. 3g, i). The PDS hairpin construct, transformed into tobacco, possibly triggered the silencing of the endogenous PDS-mRNA so that the formation of PDS was disturbed in T0 lines. Therefore, it is logical to assume that the observed photo-bleaching and dwarf phenotypes in PDS-silenced T0 lines were due to the disturbance of carotenoid and gibberellic acid biosynthesis pathways.
Development of PDS-silenced transgenic tobacco after transformation with Agrobacterium containing PDS hairpin RNAi binary construct. a Bleached shoots regeneration from the calli of co-cultivated leaf discs within 3–4 weeks. b Regenerated bleached shoots in Petri plate. c Bleached shoots transferred to medium for rooting. d–f Three independent photo-bleached regenerated plantlets with roots. A range, from high to medium, of photo-bleached phenotype observed in the leaf area of regenerated plantlets. g, i Two putative PDS-silenced transgenic plantlets compared with WT (wild-type). Dwarf and photo-bleached phenotypes observed in transgenics compared to WT. j, k Six independent silenced or partially silenced transgenic PDS T1 lines showing photobleaching and less vigorous phenotypes
The dwarf and bleached regenerated plantlets with roots were subsequently transferred to vermiculite and then soil for further maintenance. Interesting, we observed that, after transferring the photo-bleached shoots in the vermiculite in the greenhouse, the bleaching phenotype in the PDS-silenced T0 transgenic lines persisted, though it was reduced as compared to that observed in the regeneration and rooting medium. Besides the observed photo-bleached phenotype, all the lines showed lean and feeble growth as compared to that of WT (Fig. 3j, k) in the greenhouse.
Screening and analysis of hairpin-induced silenced PDS lines
The putative PDS-silenced T0 transgenics were confirmed by molecular analysis. To confirm the presence of the PDS-hpRNA construct in the photo-bleached transgenic lines, PCR analysis was performed using CaMV35S forward and PDS reverse primers. gDNA from the transformed and WT lines was isolated and used as template in PCR reaction. Amplification of ~0.8-kb fragments from T0 tobacco lines 1, 2, 4, and 5 confirmed their transgenic nature, while no amplification was found from WT and line #3 (Fig. 4). The PCR-positive and photo-bleached T0 lines were subsequently advanced to T1 (Fig. 3j, k) level and used for further experimentation.
Screening of transgenically silenced PDS T0 lines. PCR of genomic DNA isolated from different T0 lines (lines 1–6) and from the control (WT) line using CaMV35S forward and PDS sense strand reverse primers; 0.8-kb amplification of PCR bands from lines 1, 2, 4, and 5 confirmed these transgenic tobacco T0 lines, while no amplification was observed in WT. +ve represents TOPO-PDS plasmid used as template in PCR and M indicates the 1-kb DNA ladder
To analyze the abundance of PDS transcripts in the different T0 transgenic lines, northern analysis was performed using total RNA isolated from different PDS-silenced T0 transgenic lines as well as WT tobacco. The analysis revealed that 30–70 % of PDS transcripts were silenced in different T0 transgenic lines compared to WT (Fig. 5a).
PDS transcripts abundance analysis in T0 and T1 transgenic tobacco lines. The relative abundance of PDS-RNA. The RNA level of the WT has been assigned as 100 % and all other values have been calculated relatively. All analysis of a and b were performed with ImageJ software. M indicates 1-kb DNA ladder. a Northern blot analysis showing the relative changes in abundance of PDS transcripts in different T0 transgenics (lines 1–14). Histogram data provide the relative densitometric intensity of the signal normalized against the intensity of the 28S rRNA from EtBr strained blots for the corresponding lane. b RT-PCR analysis showing the relative changes in abundance of PDS transcript in different T1 lines (lines 1–7). Histogram data provide the relative intensity of the signal as deduced by comparing the intensity to that of ACTIN for the corresponding lane
The T0 lines (lines 1–7, 10, 11, 1,2 and 14) which showed enhanced PDS silencing were self-fertilized and advanced to T1 level. The T1 lines which showed bleached or partial bleached phenotype were further analyzed to check for the reduced levels of PDS transcript through RT-PCR technique before using them in the crossing experiment. Our data suggest that silencing of PDS was stably transmitted to the T1 level and that the silencing ranged from 20 to 60 % in different T1 lines compared to WT (Fig. 5b). Among the T1 progenies of lines 1, 2, 4, 5, 6, and 7, the plants which exhibited better silencing were selected for subsequent crossing experiments.
Reversal of PDS silencing in F1 (♀PDS X ♂MYMIV-AC2) hybrids
To know whether the suppression activity of MYMIV-AC2 can induce the PDS expression from its silenced state, MYMIV-AC2 was introduced in the background of hpRNA-silenced PDS lines. Six different MYMIV-AC2-expressing transgenic T2 plants, that had been developed earlier in our laboratory (Rahman et al. 2012), were used as male parents (♂), while six different PDS hpRNA silenced transgenic T1 plants were used as the female parents (♀) to generate the cross-combinations. The self-hybridized PDS silenced lines (T1) and AC2 expressing lines were used as controls.
The F1 (♀PDS X ♂AC2) hybrids were analyzed to check for the induction of PDS expression in F1 hybrids. It was observed that all the F1 seedlings showed the normal green phenotype (Fig. 6c–e), while the selfed PDS-silenced line exhibited the photo-bleached phenotype (Fig. 6b). The reversal of PDS expression in the F1 hybrids was further confirmed by molecular analysis (Fig. 6f, upper panel: F1 lines: 3, 5, 9, 13, 20), where an appreciable amount of PDS-mRNA was detected. Next, we investigated whether the reversion of PDS in the F1 lines (Fig. 6c–e) was due to suppression activity of AC2, and the presence of the AC2 transcripts was also analzsed. The MYMIV-AC2 transcripts level corroborated well with the PDS expression profile. MYMIV-AC2 transcripts were present in all the PDS-expressing F1 lines (Fig. 6f, middle panel: F1 lines: 3, 5, 9, 13, 20) but were absent in the PDS-silenced T1 line (Fig. 6f, middle panel). This indicates that, after introgression of MYMIV-AC2 suppressor in the PDS-silenced background, the hpRNA-induced silencing of PDS is suppressed. The disappearance of the photo-bleached phenotype in F1 hybrids indicates that suppressor activity of MYMIV-AC2 is capable of blocking the RNAi effect that was induced by the PDS hpRNA vector.
Analysis of F1 (♀PDS X ♂MYMIV-AC2) hybrids. a Scheme of crossing. PDS-silenced T2 line was selected as the female (♀) parent and AC2 expressing T2 lines as the male (♂) parent to generate F1 hybrids. b hpRNA-silenced PDS lines showing the photo-bleached phenotype. c–e The F1 (♀PDS×♂AC2) hybrids showing the normal green phenotype due to reversal of PDS expression after introgression of MYMIV-AC2 suppressor in the PDS-silenced background. f RT-PCR analysis of PDS and AC2 transcripts abundance in different F1 hybrid lines (F1-3, F1-5, F1-9, F1-13, F1-20) and PDS-silenced T1-7 line. ACTIN was used as loading control (bottom). g The line chart shows correlation of transcripts abundance of PDS and AC2 in different F1 hybrids lines. The abundance of PDS and AC2 transcripts is presented after normalisation by comparing the ratio of densitometric intensity of the PDS and AC2 bands to that of ACTIN bands (as loading control). Densitomeric intensity was analyzed with ImageJ software. The trend lines of the PDS and AC2 transcripts obtained display the positive correlation between the transcript levels in different hybrids lines, as discussed in the text
A total of ~100 F1 seedlings obtained from 6 crosses were checked for PDS expression. We found ~29 % of F1 lines showed green, while ~45 % of F1 lines showed partial green or partial bleached (data not shown) phenotype indicating that a range of PDS reversion occurred in different F1 seedlings, while the mother PDS-silenced T1 lines appeared as the photo-bleached phenotype. Next, to study a correlation between the expression profiles of PDS and AC2 in F1 hybrids, we quantified the band intensity of PDS and AC2 transcripts shown in Fig. 6f and prepared a line chart (Fig. 6g). In general, we found that high AC2 expression led to high PDS expression (as evident in Fig. 6g, F1 lines 9 and 20), while low AC2 expression caused low PDS expression (as evident in Fig. 6g, F1 lines 3 and 13), indicating a correlation between reversion of PDS and MYMIV-AC2 expression in F1 hybrids. The lowest level of PDS expression was observed in the silenced T1 line where no AC2 expression was observed (Fig. 6f, g, lane T1-7). The trend lines of PDS and AC2 transcripts obtained display a positive correlation between the transcript levels in different hybrids lines (Fig. 6g). The green phenotype observed in the leaves of F1 hybrid lines indicated the reversal of PDS expression following the introduction of the MYMIV-AC2 suppressor in the PDS-silenced background. In the previous study, we found that introduction of MYMIV-AC2 in the silenced transgenic lines showed a high level expression of transgenes as well as a positive correlation between reversion of the silenced transgene and MYMIV-AC2 expression in F1 hybrid population and double transgenic lines (Rahman et al. 2012).
Discussion
RNA interference (RNAi) has been extensively used in various species to suppress the function of an endogenous gene and is becoming a common tool for the functional analysis of the genome (Hannon 2002). In plants, PTGS is triggered by small RNAs that are produced via a mechanism involving the production of dsRNA or self-complementary hpRNA (Waterhouse et al. 2001; Matzke et al. 2001; Vance and Vaucheret 2001). The constructs which would lead to the expression of such transcripts are thus an efficient way of inducing targeted gene silencing (Waterhouse et al. 1998). Transforming plants with hpRNA constructs typically generates a series of independent lines that have varying phenotypes and degrees of target mRNA reduction (Helliwell et al. 2002). In the present study, the endogenous PDS gene of tobacco was silenced by using the hpRNA construct. Stable transformation of this construct resulted in a range of photo-bleached PDS-silenced progenies in the T0 generation (Fig. 3). The molecular analysis revealed 30–70 % silencing of PDS transcripts in the T0 lines (Fig. 5a).
It has been reported earlier that the transformation of plants with hpRNA constructs leads to stable silencing that is inherited from generation to generation, thereby enabling the continued study of silencing phenomena (Waterhouse and Helliwell 2003). For example, the silencing of Arabidopsis fatty acid desaturase2 (FAD2) gene by a hpRNA has been maintained for five generations and has been found to be consistent in its effectiveness (Stoutjesdijk et al. 2002). In the present study, it was also found that the PDS silencing was efficiently propagated to the T1 generation and the silencing level remained at 30–70 % (Fig. 5b). Therefore, our results also support that the hairpin gene-silencing constructs can efficiently down-regulate or silence the endogene activity, and that such silencing is also stably inherited over generations.
The length of the target sequence used in the hpRNA construct strongly influences the effective silencing and sometimes has a role in stable and efficient gene silencing in plants and animals. Silencing appears to be most efficient when sequences of more than 300 base pairs are used in hpRNA constructs to target the transcripts (Wang and Waterhouse 2002). However, fragments as small as 98 bases have been shown to be sufficient for effective gene silencing in hairpin vectors, such as pHANNIBAL RNA vector (Waterhouse and Helliwell 2003). It has been suggested that the presence of an intron in the hpRNA vector may enhance its silencing efficiency (Wang and Waterhouse 2002), although the exact role of introns in gene silencing remains to be answered. Interestingly, the hpRNA transgene with a sequence stretch of only 19 base pairs identical to the target gene was also shown to be effective for gene silencing (Wang and Waterhouse 2002). In the present investigation, a 409-bp-long stretch of target PDS gene was used to construct the PDS-hpRNA vector. Transformation of this hpRNA vector in Nicotiana produced a stable and efficient silencing of endogenous PDS gene in T0 lines (Fig. 3, 5). Our study also demonstrated that the hairpin loop-induced silencing was stably inherited in the T1 generation.
Viruses encode proteins that have acquired the activity of suppressing small RNA-based gene silencing. This is normally used during infection to overcome the silencing-based first line of host plant defense. Different suppressors such as HC-Pro of Potyviruses, P19 of Tombusviruses, 2b of Cucumoviruses, AC2 of Begomovirus, etc. have been shown to suppress gene silencing, although the mechanisms of their action are still under investigation. This study is the first report on the involvement of AC2 in suppressing the hpRNA-induced gene silencing. On the introduction of the MYMIV-AC2 suppressor in the hpRNA-induced PDS-silenced background by crossing between PDS-silenced (♀) and MYMIV-AC2 (♂) lines we observed a reversal of the photo-bleached PDS phenotype in the F1 hybrid population (Fig. 6). The observed normal green phenotype in the F1 lines was comparable to that of WT indicating that the PDS silencing was blocked by the suppression activity of MYMIV-AC2.
Conclusion
Virus-encoded silencing suppressors have the potential to overcome post-transcriptional gene silencing. Each suppressor acts by distinct mechanisms and at discrete steps in the RNA silencing pathway. The findings obtained in this study suggest that MYMIV-AC2 can inhibit hpRNA-induced silencing. Our earlier studies have shown that MYMIV-AC2 can overcome transgene-mediated gene silencing to enhance its expression in planta (Rahman et al. 2012). Taken together, the results indicate that MYMIV-AC2 can be used efficiently to suppress both transgene-induced and hpRNA-induced siRNA-mediated gene silencing. This is the first report on the involvement of AC2 in suppressing hpRNA-induced gene silencing. We suggest that the dual system of hpRNA and MYMIV-AC2 suppressors can serve as an efficient switch for turning off and turning on gene expression, with potential applications in gene function analysis.
Abbreviations
- CaMV:
-
Cauliflower mosaic virus
- hpRNA:
-
Hairpin RNA
- MYMIV:
-
Mungbean Yellow Mosaic India Virus
- PTGS:
-
Post-translational gene silencing
- RNAi:
-
RNA interference
- TGS:
-
Transcriptional gene silencing
References
Bisaro DM (2006) Silencing suppression by geminivirus proteins. Virology 344(1):158–168
Chuang CF, Meyerowitz EM (2000) Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA 97:4985–4990
Hannon GJ (2002) RNA interference. Nature 418:244–251
Hartitz MD, Sunter G, Bisaro DM (1999) The geminivirus transactivator (TrAP) is a single-stranded DNA and zinc-binding phosphoprotein with an acidic activation domain. Virology 263:1–14
Helliwell CA, Wesley SV, Wielopolska AJ, Waterhouse PM (2002) High throughput vectors for efficient gene silencing in plants. Funct Plant Biol 29:1217–1225
Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231
Karjee S, Islam MN, Mukherjee SK (2008) Screening and identification of virus-encoded RNA silencing suppressors. Methods Mol Biol 442:187–203
Levin JZ, de Framond AJ, Tuttle A, Bauer MW, Heifetz PB (2000) Methods of double-stranded RNA-mediated gene inactivation in Arabidopsis and their use to define an essential gene in methionine biosynthesis. Plant Mol Biol 44:759–775
Li F, Ding S (2006) Virus counterdefense: diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol 60:503–531
Matzke MA, Matzke AJ, Pruss GJ, Vance VB (2001) RNA-based silencing strategies in plants. Curr Opin Genet Dev 2:221–227
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15(3):473–497
Rahman J, Karjee S, Mukherjee SK (2012) MYMIV-AC2, a geminiviral rnai suppressor protein, has potential to increase the transgene expression. Appl Biochem Biotechnol 167:758–775
Sharma AD, Gill PK, Singh P (2002) DNA isolation from dry and fresh samples of polysaccharide-rich plants. Plant Mol Biol Report 20:415a–415f
Sharp PA (2001) RNA interference-2001. Genes Dev 15:485–490
Sijen T, Vijn I, Rebocho A, van Blokland R, Roelofs D, Mol JNM, Kooter JM (2001) Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr Biol 11:436–440
Singh DK, Karjee S, Malik PS, Islam N, Mukherjee SK (2007) DNA replication and pathogenecity of MYMIV. In: Méndez-Vilas AB (ed) Communicating current research and educational topics and trends in applied microbiology, vol 1., FORMATEX microbiology series no 1FORMATEX, Spain, pp 155–162
Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM (2000) Total silencing by intron-spliced hairpin RNAs. Nature 407:319–320
Somerville C (2000) Genomics: plant biology 2010. Science 290:2077–2078
Stoutjesdijk PA, Singh SP, Liu Q, Hurlstone CJ, Waterhouse PA, Green AG (2002) hpRNA-mediated targeting of the Arabidopsis FAD2 gene gives highly efficient and stable silencing. Plant Physiol 129(4):1726–1731
Tao X, Zhou X (2004) A modified viral satellite DNA that suppresses gene expression in plants. Plant J 38(5):850–860
Trinks D, Rajeswaran R, Shivaprasad PV, Akbergenov R, Oakeley EJ, Veluthambi K, Hohn T, Pooggin MM (2005) Suppression of RNA silencing by geminivirus nuclear protein, AC2, correlates with transactivation of host gene. J Virol 79:2517–2527
Vance V, Vaucheret H (2001) RNA silencing in plants: defense and counter-defense. Science 292:2277–2280
Wang MB, Waterhouse PM (2002) Applications of gene silencing in plants. Curr Opin Plant Biol 5:146–150
Wang MB, Waterhouse PM (2004) High efficiency silencing of a β-glucuronidase gene in rice is correlated with repetitive transgene structure but is independent of DNA methylation. Plant Mol Biol 43:67–82
Wang MB, Abbott DC, Waterhouse PM (2000) A single copy of a virus derived transgene encoding hairpin RNA gives immunity to barley yellow dwarf virus. Mol Plant Pathol 1:347–356
Waterhouse PM, Helliwell CA (2003) Exploring plant genomes by RNA-induced gene silencing. Nat Rev Genet 4(1):29–38
Waterhouse PM, Graham MW, Wang MB (1998) Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA 95:13959–13964
Waterhouse PM, Wang MB, Lough T (2001) Gene silencing as an adaptive defence against viruses. Nature 411:834–842
Watson JM, Fusaro AF, Wang MB, Waterhouse PM (2005) RNA silencing platforms in plants. FEBS Lett 579:5982–5987
Wesley SV, Helliwell CA, Smith NA, Wang M, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, Robinson SP, Gleave AP, Green AG, Waterhouse PM (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27:581–590
Acknowledgments
This research was supported by grants from International Centre for Genetic Engineering and Biotechnology (ICGEB) and Department of Biotechnology (DBT), New Delhi, India. J.R. was pre-doctoral fellow of Plant Molecular Biology Group, ICGEB. The financial assistance of D.B.T. to N.S.M. and S.M. is highly acknowledged.
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rahman, J., Sanan-Mishra, N. & Mukherjee, S.K. MYMIV-AC2 protein suppresses hairpin-induced gene silencing in Nicotiana tabacum cv. Xanthi. Plant Biotechnol Rep 8, 337–347 (2014). https://doi.org/10.1007/s11816-014-0325-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-014-0325-4