Abstract
Machine learning is widely used to analyze biological sequence data. Non-sequential models such as SVMs or feed-forward neural networks are often used although they have no natural way of handling sequences of varying length. Recurrent neural networks such as the long short term memory (LSTM) model on the other hand are designed to handle sequences. In this study we demonstrate that LSTM networks predict the subcellular location of proteins given only the protein sequence with high accuracy (0.902) outperforming current state of the art algorithms. We further improve the performance by introducing convolutional filters and experiment with an attention mechanism which lets the LSTM focus on specific parts of the protein. Lastly we introduce new visualizations of both the convolutional filters and the attention mechanisms and show how they can be used to extract biologically relevant knowledge from the LSTM networks.
S.K. Sønderby and C.K. Sønderby—These authors contributed equally.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Deep neural networks have gained popularity for a wide range of classification tasks in image recognition and speech tagging [9, 20] and recently also within biology for prediction of exon skipping events [30]. Furthermore a surge of interest in recurrent neural networks (RNN) has followed the recent impressive results shown on challenging sequential problems like machine translation and speech recognition [2, 14, 27]. Within biology, sequence analysis is a very common task used for prediction of features in protein or nucleic acid sequences. Current methods generally rely on neural networks and support vector machines (SVM), which have no natural way of handling sequences of varying length. Furthermore these systems rely on highly hand-engineered input features requiring a high degree of domain knowledge when designing the algorithms [11, 24]. RNNs are a type of neural networks that naturally handles sequential data. In an RNN the input to the network’s hidden layers is both the input features at the current timestep and the hidden activation from the previous time step. Hence an RNN corresponds to placing neural networks with shared identical weights at each timestep and letting information flow across the sequence by connecting the networks with (recurrent) weights between the hidden layers. In bioinformatics RNNs have previously been used for contact map prediction [10], and protein secondary structure predition [3, 21]. However standard RNNs have been shown to be difficult to train with backpropagation through time due to both vanishing and exploding gradients [5]. To mitigate this problem, Hochreiter et al. [17] introduced the long short term memory (LSTM) that uses a gated memory cell instead of the standard sigmoid or hyperbolic tangent hidden units used in standard RNNs. In the LSTM the value of each memory cell is controlled with input, modulation, forget and output gates, which allow the LSTM network to store analog values for many time steps by controlling access to the memory cell. In practice this architecture have proven easier to train than standard RNN.
In this paper LSTMs are used to analyze biological sequences and predict to which subcellular compartment a protein belongs. This prediction task, known as protein sorting or subcellular localization, has attracted large interest in the bioinformatics field [11]. We show that an LSTM network, using only the protein sequence information, has significantly better performance than current state of the art SVMs and furthermore have nearly as good performance as large hand engineered systems relying on extensive metadata such as GO terms and evolutionary phylogeny, see Fig. 1 [6, 7, 18]. These results show that LSTM networks are efficient algorithms that can be trained even on relatively small datasets of around 6000 protein sequences. Secondly we investigate how an LSTM network recognizes the sequence. In image recognition, convolutional neural networks (CNN) have shown state of the art performance in several different tasks [8, 20]. Here the lower layers of a CNN can often be interpreted as feature detectors recognizing simple geometric entities, see Fig. 2. We develop a simple visualization technique for convolutional filters trained on either DNA or amino acid sequences and show that in the biological setting filters can be interpreted as motif detectors, as visualized in Fig. 2. Thirdly, inspired by the work of Bahdanau et al. [2], we augment the LSTM network with an attention mechanism that learns to assign importance to specific parts of the protein sequence. Using the attention mechanism we can visualize where the LSTM assigns importance, and we show that the network focuses on regions that are biologically plausible. Lastly we show that the LSTM network learns a fixed length representation of amino acids sequences that, when visualized, separates the sequences into clusters with biological meaning. The contributions of this paper are:
-
1.
We show that LSTM networks combined with convolutions are efficient for predicting subcellular localization of proteins from sequence.
-
2.
We show that convolutional filters can be used for amino acid sequence analysis and introduce a visualization technique.
-
3.
We investigate an attention mechanism that lets us visualize where the LSTM network focuses.
-
4.
We show that the LSTM network effectively extracts a fixed length representation of variable length proteins.
Schematic showing how MultiLoc combines predictions from several sources to make predictions whereas the LSTM networks only rely on the sequence [18].
Left: first layer convolutional filters learned in [20], note that many filters are edge detectors or color detectors. Right: example of learned filter on amino acid sequence data, note that this filter is sensitive to positively charged amino acids (Color figure online).
2 Materials and Methods
This section introduces the LSTM cell and then explains how a regular LSTM (R-LSTM) can produce a single output. We then introduce the LSTM with attention mechanism (A-LSTM), and describe how the attention mechanism is implemented.
2.1 LSTM NETWORKS
The memory cell of the LSTM networks is implemented as described in [15] except for peepholes, because recent papers have shown good performance without [27, 32, 33]. Figure 3 shows the LSTM cell. Equations (1-10) state the forward recursions for a single LSTM layer.
where all quantities are given as row-vectors and activation and dropout functions are applied element-wise. Note that for the first hidden layer \(h_t^{1}\) the input \(x_t\) are the amino acid features. In the memory cell \(i_t\), \(f_t\) and \(o_t\) can are gating functions controlling input, storage and output of the value \(c_t\) stored in each cell. Due to the logistic squashing function used for each gate, the value are always in the interval (0,1) and information can flow through the gate if the value is close to one. If dropout is used it is only applied to non-recurrent connections in the LSTM cell [31]. In a multilayer LSTM \(h_t\) is passed upwards to the next layer.
LSTM memory cell. i: input gate, f: forget gate, o: output gate, g: input modulation gate, c: memory cell. The Blue arrow heads refers to \(c_{t-1}\). The notation corresponds to Eqs. 1 to 10 such that \(W_{xo}\) denotes wights for x to output gate and \(W_{hf}\) denotes weights for \(h_{t-1}\) to forget gates etc. Adapted from [33].
A: Unidirectional LSTM predicting a single target. B: Unrolled single layer BLSTM. The forwards LSTM (red arrows) starts at time 1 and the backwards LSTM (blue arrows) starts at time T, then they go forwards and backwards respectively. The errors from the forward and backward nets are combined and a prediction is made for each sequence position. Adapted from [13]. C: Unidirectional LSTM for predicting a single target. Squares are LSTM layers (Color figure online).
2.2 Regular LSTM Networks for Predicting Single Targets
When used for predicting a single target for each input sequence, one approach is to output the predicted target from the LSTM network at the last sequence position as shown in Fig. 5A. A problem with this approach is that the gradient has to flow from the last position to all previous positions and that the LSTM network has to store information about all previously seen data in the last hidden state. Furthermore a regular bidirectional LSTM (BLSTM, 5B)[26] is not useful in this setting because the backward LSTM will only have seen a single position, \(x_T\), when the prediction has to be made. We instead combine two unidirectional LSTMs, as shown in Fig. 5C, where the backward LSTM has the input reversed. The output from the two LSTMs are combined before predictions.
2.3 Attention Mechanism LSTM Netowork
Bahdanau et al. [2] have introduced an attention mechanism for combining hidden state information from a encoder-decoder RNN approach to machine translation. The novelty in their approach is that they use an alignment function that for each output word finds important input words, thus aligning and translating at the same time. We modify this alignment procedure such that only a single target is produced for each sequence. The developed attention mechanism can be seen as assigning importance to each position in the sequence with respect to the prediction task. We use a BLSTM to produce a hidden state at each position and then use an attention function to assign importance to each hidden state as illustrated in Fig. 4. The weighted sum of hidden states is used as a single representation of the entire sequence. This modification allows the BLSTM model to naturally handle tasks involving prediction of a single target per sequence. Conceptually this corresponds to adding weighted skip connections (green arrow heads Fig. 4) between any \(h_t\) and the output network, with the weight on each skip connection being determined by the attention function. Each hidden state \(h_t\), \(t=1,\ldots ,T\) is used as input to a Feed Forward Neural Network (FFN) attention function:
where \(W_a\) is an attention hidden weight matrix and \(v_a\) is an attention output vector. From the attention function we form softmax weights:
that are used to produce a context vector c as a convex combination of T hidden states:
The context vector is then used is as input to the classification FFN f(c). We define f as a single layer FFN with softmax outputs.
2.4 Subcellular Localization Data
The model was trained and evaluated on the dataset used to train the MultiLoc algorithm published by Höglund et al. [18]Footnote 1. The dataset contains 5959 proteins annotated to one of 11 different subcellular locations. To reduce computational time the protein sequences were truncated to a maximum length of 1000. We truncated by removing from the middle of the protein as both the N- and C-terminal regions are known to contain sorting signals [11]. Each amino acid was encoded using 1-of-K encoding, the BLOSUM80 [16] and HSDM [25] substitution matrices and sequence profiles, yielding 80 features per amino acid. Sequence profiles where created with PROFILpro [22]Footnote 2 using 3 blastpgp [1]Footnote 3 iterations on UNIREF50.
2.5 Visualizations
Convolutional filters for images can be visualized by plotting the convolutional weights as pixel intensities as shown in Fig. 2. However a similar approach does not make sense for amino acid inputs due to the 1-of-K vector encoding. Instead we view the 1D convolutions as a position specific scoring matrix (PSSM). The convolutional weights can be reshaped into a matrix of \(l_{filter}\)-by-\(l_{enc}\), where the amino acid encoding length is is 20. Because the filters show relative importance we rescale all filters such that the height of the highest column is 1. Each filter can then be visualized as a PSSM logo, where the height of each column can be interpreted as position importance and the height of each letter is amino acid importance. We use Seq2Logo with the PSSM-logo setting to create the convolution filter logos [28].
We visualize the importance the A-LSTM network assigns to each position in the input by plotting \(\alpha \) from Eq. 12. Lastly we extract and plot the hidden representation from the LSTM networks. For the A-LSTM network we use c from Eq. 13 and for the R-LSTM we use the last hidden state, \(h_t\). Both c and \(h_t\) can be seen as fixed length representation of the amino acid sequences. We plot the representation using t-SNE [29].
2.6 Experimental Setup
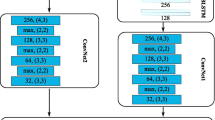
All models were implemented in Theano [4] using a modified version of the Lasagne libraryFootnote 4 and trained with gradient descent. The learning rate was controlled with ADAM (\(\alpha = 0.0002\), \(\beta _1 = 0.1\), \(\beta _2 = 0.001\), \(\epsilon = 10^{-8}\) and \(\lambda = 10^{-8}\)) [19]. Initial weights were sampled uniformly from the interval \([-0.05, 0.05]\). The network architecture is a 1D convolutional layer followed by an LSTM layer, a fully connected layer and a final softmax layer. All layers use 50 % dropout. The 1D convolutional layer uses convolutions of sizes 1, 3, 5, 9, 15 and 21 with 10 filters of each size. Dense and convolutional layers use ReLU activation [23] and the LSTM layer uses hyperbolic tangent. For the A-LSTM model the size of the first dimension of \(W_a\) was 400. We used 4/5 of the data for training and the last 1/5 of the data for testing. The hyperparameters were optimized using 5-fold cross validation on the training data. The cross validation experiments showed that the model converged after 100 epochs. Using the established hyperparameters the models were retrained on the complete training data and the test performance were reported after epoch 100.
3 Results
Table 1 shows accuracy for the R-LSTM and A-LSTM models and several other models trained on the same dataset. Comparing the performance of the R-LSTM, A-LSTM and MultiLoc models, utilizing only the sequence information, the R-LSTM model (0.879 Acc.) performs better than the A-LSTM model (0.854 Acc.) whereas the MultiLoc model (0.767 Acc.) performs significantly worse. Furthermore the 10-ensemble R-LSTM model further increases the performance to 0.902 Acc. Comparing this performance with the other models, combining the sequence predictions from the MultiLoc model with large amounts of metadata for the final predictions, only the Sherloc2 model (0.930 Acc.) performs better than the R-LSTM ensemble. Figure 6 shows a plot of the attention matrix from the A-LSTM model. Figure 8 shows examples of the learned convolutional filters. Figure 7 shows the hidden state of the R-LSTM and the A-LSTM model.
Examples of learned filters. Filter A captures proline or trypthopan stretches, (B) and (C) are sensitive to positively and negatively charged regions, respectively. Note that for C, negative amino acids seems to suppress the output. Lastly we show a long filter which captures larger sequence motifs in the proteins.
4 Discussion and Conclusion
In this paper we have introduced LSTM networks with convolutions for prediction of subcellular localization. Table 1 shows that the LSTM networks perform much better than other methods that only rely on information from the sequence (LSTM ensemble 0.902 vs. MultiLoc 0.767). This difference is all the more remarkable given the simplicity of our method, only utilizing the sequences and their localization labels, while MultiLoc incorporates specific domain knowledge such as known motifs and signal anchors. One explanation for the performance difference is that the LSTM networks are able to look at both global and local sequence features whereas the SVM based models do not model global dependencies. The LSTM networks have nearly as good performance as methods that use information obtained from other sources than the sequence (LSTM ensemble 0.902 vs. SherLoc2 0.930). Incorporating these informations into the LSTM models could further improve the performance of these models. However, it is our opinion that using sequence alone yields the biologically most relevant prediction, while the incorporation of, e.g., GO terms limits the usability of the prediction requiring similar proteins to be already annotated to some degree. Furthermore, as we show below, a sequence-based method potentially allows for a de novo identification of sequence features essential for biological function.
Fig. 6 shows where in the sequence the A-LSTM network assigns importance. Sequences from the compartments ER, extracellular, lysosomal, and vacuolar all belong to the secretory pathway and contain N-terminal signal peptides, which are clearly seen as bars close to the left edge of the plot. Some of the ER proteins additionally have bars close to the right edge of the plot, presumably representing KDEL-type retention signals. Golgi proteins are special in this context, since they are type II transmembrane proteins with signal anchors, slightly further from the N-terminus than signal peptides [18]. Chloroplast and mitochondrial proteins also have N-terminal sorting signals, and it is apparent from the plot that chloroplast transit peptides are longer than mitochondrial transit peptides, which in turn are longer than signal peptides [11]. For the plasma membrane category we see that some proteins have signal peptides, while the model generally focuses on signals, presumably transmembrane helices, scattered across the rest of the sequence with some overabundance close to the C-terminus. Some of the attention focused near the C-terminus could also represent signals for glycosylphosphatidylinositol (GPI) anchors [11]. Cytoplasmic and nuclear proteins do not have N-terminal sorting signals, and we see that the attention is scattered over a broader region of the sequences. However, especially for the cytoplasmic proteins there is some attention focused close to the N-terminus, presumably in order to check for the absence of signal peptides. Finally, peroxisomal proteins are known to have either N-terminal or C-terminal sorting signals (PTS1 and PTS2) [11], but these do not seem to have been picked up by the attention mechanism.
In Fig. 8 we investigate what the convolutional filters in the model focus on. Notably the short filters focus on amino acids with specific characteristics, such as positively or negatively charged, whereas the longer filters seem to focus on distributions of amino acids across longer sequences. The arginine-rich motif in Fig. 7C could represent part of a nuclear localization signal (NLS), while the longer motif in Fig. 7D could represent the transition from transmembrane helix (hydrophobic) to cytoplasmic loop (in accordance with the “positive-inside” rule). We believe that the learned filters can be used to discover new sequence motifs for a large range of protein and genomic features.
In Fig. 7 we investigate whether the LSTM models are able to extract fixed length representations of variable length proteins. Using t-SNE we plot the LSTMs hidden representation of the sequences. It is apparent that proteins from the same compartment generally group together, while the cytoplasmic and nuclear categories tend to overlap. The corresponds with the fact that these two categories are relatively often confused, see Table 2. The categories form clusters which make biological sense; all the proteins with signal peptides (ER, extracellular, lysosomal, and vacuolar) lie close to each other in t-SNE space in all three plots, while the proteins with other N-terminal sorting signals (chloroplasts and mitochondria) are close in the R-LSTM plots (but not in the A-LSTM plot). Note that the lysosomal and vacuolar categories are very close to each other in the plots, this corresponds with the fact that these two compartments are considered homologous [18].
In summary we have introduced LSTM networks with convolutions for subcellular localization. By visualizing the learned filters we have shown that these can be interpreted as motif detectors, and lastly we have shown that the LSTM network can represent protein sequences as a fixed length vector in a representation that is biologically interpretable.
References
Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.J.: Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic acids Res. 25(17), 3389–3402 (1997)
Bahdanau, D., Cho, K., Bengio, Y.: Neural Machine Translation by Jointly Learning to Align and Translate. arXiv preprint arXiv:1409.0473 (Sep 2014)
Baldi, P., Brunak, S., Frasconi, P.: Exploiting the past and the future in protein secondary structure prediction. Bioinformatics 15(11), 937–946 (1999)
Bastien, F., Lamblin, P., Pascanu, R., Bergstra, J., Goodfellow, I., Bergeron, A., Bouchard, N., Warde-Farley, D., Bengio, Y.: Theano: new features and speed improvements, November 2012. arXiv preprint arXiv:1211.5590
Bengio, Y., Simard, P., Frasconi, P.: Learning long-term dependencies with gradient descent is difficult. IEEE Trans. Neural Netw. 5(2), 157–166 (1994)
Blum, T., Briesemeister, S., Kohlbacher, O.: MultiLoc2: integrating phylogeny and Gene Ontology terms improves subcellular protein localization prediction. BMC bioinform. 10, 274 (2009)
Briesemeister, S., Blum, T., Brady, S., Lam, Y., Kohlbacher, O., Shatkay, H.: SherLoc2: a high-accuracy hybrid method for predicting subcellular localization of proteins. J. Proteome Res. 8(11), 5363–5366 (2009)
Cunn, Y.L., Boser, B., Denker, J., Henderson, D., Howard, R., Hubbard, W., Jackel, L.: Handwritten digit recognition with a back-propagation network. In: Lippmann, R., Moody, J., Touretzky, D. (eds.) Advances in neural information processing systems. pp. 396–404 (1990)
Dahl, G., Yu, D., Deng, L., Acero, A.: Context-dependent pre-trained deep neural networks for large-vocabulary speech recognition. IEEE Trans. Audio Speech Lang. Process. 20(1), 30–42 (2012)
Di Lena, P., Nagata, K., Baldi, P.: Deep architectures for protein contact map prediction. Bioinformatics 28(19), 2449–2457 (2012)
Emanuelsson, O., Brunak, S., von Heijne, G., Nielsen, H.: Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2(4), 953–971 (2007)
Goldberg, T., Hamp, T., Rost, B.: LocTree2 predicts localization for all domains of life. Bioinformatics 28(18), i458–i465 (2012)
Graves, A.: Supervised sequence labelling with recurrent neural networks. Springer, Heidelberg (2012)
Graves, A., Jaitly, N.: Towards end-to-end speech recognition with recurrent neural networks. In: Proceedings of the 31st International Conference on Machine Learning (ICML-14), pp. 1764–1772 (2014)
Graves, A.: Generating sequences with recurrent neural networks, (2013). arXiv preprint arXiv:1308.0850
Henikoff, S., Henikoff, J.G.: Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89, 10915–10919 (1992)
Hochreiter, S., Schmidhuber, J., Elvezia, C.: Long short-term memory. Neural Comput. 9(8), 1735–1780 (1997)
Höglund, A., Dönnes, P., Blum, T., Adolph, H.W., Kohlbacher, O.: MultiLoc: prediction of protein subcellular localization using N-terminal targeting sequences, sequence motifs and amino acid composition. Bioinformatics 22(10), 1158–1165 (2006)
Kingma, D., Ba, J.: Adam: a method for stochastic optimization, December 2014. arXiv preprint arXiv:1412.6980
Krizhevsky, A., Sutskever, I., Hinton, G.: Imagenet classification with deep convolutional neural networks. In: Pereira, F., Burges, C.J.C., Bottou, L., Weinberger, K. (eds.) Advances in neural information processing systems, pp. 1097–1105 (2012)
Magnan, C., Baldi, P.: SSpro/ACCpro 5: almost perfect prediction of protein secondary structure and relative solvent accessibility using profiles, machine learning, and structural similarity. Bioinformatics 30(18), 1–6 (2014)
Magrane, M. et al.: UniProt Consortium: Uniprot knowledgebase: a hub of integrated protein data. Database 2011, bar009 (2011)
Nair, V., Hinton, G.E.: Rectified linear units improve restricted boltzmann machines. In: Proceedings of the 27th International Conference on Machine Learning (ICML-10), pp. 807–814 (2010)
Petersen, T., Brunak, S., von Heijne, G., Nielsen, H.: SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8(10), 785–786 (2011)
Prlić, A., Domingues, F.S., Sippl, M.J.: Structure-derived substitution matrices for alignment of distantly related sequences. Protein Eng. 13, 545–550 (2000)
Schuster, M., Paliwal, K.: Bidirectional recurrent neural networks. Signal Process. 45(11), 2673–2681 (1997)
Sutskever, I., Vinyals, O., Le, Q.: Sequence to sequence learning with neural networks. In: Advances in Neural Information Processing Systems, pp. 3104–3112 (2014)
Thomsen, M.C.F., Nielsen, M.: Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Res. 40, W281–W287 (2012)
Van Der Maaten, L.J.P., Hinton, G.E.: Visualizing high-dimensional data using t-sne. J. Mach. Learn. Res. 9, 2579–2605 (2008)
Xiong, H.Y., Alipanahi, B., Lee, L.J., Bretschneider, H., Merico, D., Yuen, R.K.C., Hua, Y., Gueroussov, S., Najafabadi, H.S., Hughes, T.R., Morris, Q., Barash, Y., Krainer, A.R., Jojic, N., Scherer, S.W., Blencowe, B.J., Frey, B.J.: The human splicing code reveals new insights into the genetic determinants of disease. Science 347, 1254806 (2014)
Zaremba, W., Sutskever, I., Vinyals, O.: Recurrent neural network regularization (2014). arXiv preprint arXiv:1409.2329
Zaremba, W., Kurach, K., Fergus, R.: Learning to Discover Efficient Mathematical Identities. In: Ghahramani, Z., Welling, M., Cortes, C., Lawrence, N., Weinberger, K. (eds.) Advances in Neural Information Processing Systems, pp. 1278–1286, June 2014
Zaremba, W., Sutskever, I.: Learning to Execute, October 2014. arXiv preprint arXiv:1410.4615
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Sønderby, S.K., Sønderby, C.K., Nielsen, H., Winther, O. (2015). Convolutional LSTM Networks for Subcellular Localization of Proteins. In: Dediu, AH., Hernández-Quiroz, F., Martín-Vide, C., Rosenblueth, D. (eds) Algorithms for Computational Biology. AlCoB 2015. Lecture Notes in Computer Science(), vol 9199. Springer, Cham. https://doi.org/10.1007/978-3-319-21233-3_6
Download citation
DOI: https://doi.org/10.1007/978-3-319-21233-3_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-21232-6
Online ISBN: 978-3-319-21233-3
eBook Packages: Computer ScienceComputer Science (R0)