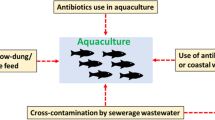
Abstract
It is well known that antibiotics are playing leading roles in several areas such as human and animal health and farm production. Unfortunately, the use of antimicrobial compounds as a panacea is coming to an end. More and more antibiotic-resistant bacterial strains are listed in all spheres of activity where they were used. Aquaculture, which is a key industry in providing animal protein needs to an exponentially growing human population, is undergoing the attack of pathogens that are increasingly difficult to control due to antibiotic resistance. In this chapter, we will explore, from an aquaculture perspective, the discovery and use of antibiotics, explain the major mechanisms of antibiotic resistance and the adverse consequences of using broad-spectrum antibiotics, and finally discuss briefly on alternatives to traditional antimicrobial agents. We will also give a concrete example with the bacterium Aeromonas salmonicida subsp. salmonicida, which is a worrisome pathogen for the aquaculture industry and for which crucial discoveries have been recently made thanks to the advances in sequencing technologies.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 A Brief History of Antibiotics
1.1 Discovery of Antibiotics
The term “antibiotic” was introduced in the scientific literature by Selman A. Waksman to describe “[…] a chemical substance, produced by micro-organisms, which has the capacity to inhibit the growth of and even to destroy bacteria and other micro-organisms” (Waksman 1947). Although the use of antibiotics may predate actual knowledge of their existence several hundred years ago (Aminov 2010), it is generally accepted that the beginning of the antibiotic era, as we know it, corresponds to the discovery of penicillin (produced by the ascomycete Penicillium) by Sir Alexander Fleming in 1929 (Fleming 1929). The Second World War was a major conflict that motivated scientific research and technological advances, including the antibiotic research field (Quinn 2013). At the end of the Second World War, penicillin was made available for public use on a large scale and sparked a moment of great excitement where several new antibiotic compounds have been discovered and various synthetic processes developed (Nicolaou and Rigol 2018). Undoubtedly, antibiotics have played a leading role in modern medicine, and it is widely accepted that their use has saved countless lives and increased life expectancy.
1.2 First Steps in the Use of Antibiotics in Aquaculture
Aquaculture is an increasing worldwide industry that accounted for 44.1% of the total fish production in 2014 (FAO 2016). Fish and seafood farmers therefore resort on intensive farming conditions to sustain this increasing demand for fish protein and polyunsaturated fatty acids. By its very nature, intensive fish farming exposes organisms to various stressors such as high stock density and poor water quality, thus creating a favorable environment for infection by pathogens (Sundberg et al. 2016). Obviously, fighting against fish diseases is crucial to secure production.
Shortly after their introduction for human medicine, new antibiotic compounds were made available for aquaculture (Austin and Austin 2016). One of the first well-documented usages of antibiotics in aquaculture was to treat sick brook trout (Salvelinus fontinalis) from furunculosis, a disease caused by Aeromonas salmonicida (previously named Bacterium salmonicida), using sulfonamides (Gutsell 1946). A major breakthrough was the discovery that combinations of sulfonamides and diaminopyrimidines (sometimes designated potentiated sulfonamides) can have a synergic effect since they both inhibit different steps of the folic acid pathway (Campbell 1999). This combination of antibiotics was found to be very effective against major fish pathogens, including Aeromonas caviae, A. salmonicida, Vibrio anguillarum, and Yersinia ruckeri (McCarthy et al. 1974; Horsberg et al. 1997).
2 The Phenomenon of Antibiotic Resistance
Despite the undeniable fact that antibiotics have helped shape modern medicine, for both humans and animals, the fight against pathogenic bacteria is far from behind us (Holmes et al. 2016). In this sense, bacteria can use various mechanisms of protection against antibiotic molecules.
2.1 Why Does a Bacterium Become Resistant?
Bacteria living in communities in the same ecological niche compete for resources. It is known that certain bacteria will themselves produce antimicrobial compounds to increase their competitiveness (Hibbing et al. 2010). Consequently, it is expected that bacterial strains resistant to antibiotics are naturally present in the environment (Martinez 2009). However, a balance will exist between resistant and sensitive strains. The overuse of antibiotics in the human (Goossens et al. 2005), veterinary (Wayne et al. 2011), and food (livestock, fish, and crop farming) contexts has created a major problem by disrupting this equilibrium (Cabello 2006; Martin et al. 2015). Although some countries have banned the use of antibiotics as growth promoters, including the European Union in 2006 (Martin et al. 2015), it is estimated that 80% of the antibiotics in the United States are used for agriculture and aquaculture, often to stimulate livestock growth or administrated as prophylactic treatments (Hollis and Ahmed 2013). Although it is difficult to clearly define the sector that has contributed the most to the amplification of the antibiotic resistance phenomenon (Chang et al. 2015), this overuse has—and continues to—generate selective pressure for bacterial cells resistant to these molecules, hence making them become dominant (Fig. 1).
2.2 How Does a Bacterium Become Resistant?
Bacteria develop resistance to antibiotics through two main strategies (Munita and Arias 2016). The first one is by mutation of key genes (usually those whose products are targeted by the antibiotic). A well-known example of resistance produced by mutation of a gene is against rifampicin. This antibiotic binds to the β-subunit of the RNA polymerase (encoded by the rpoB gene), thus blocking transcription of bacterial DNA (Campbell et al. 2001). Some non-synonymous mutations in the rpoB gene can decrease the affinity of rifampicin to its target, resulting in resistance (Floss and Yu 2005).
The second strategy is by acquisition of resistance genes through horizontal transfers. Compared to gene mutations (first strategy) that occur in one generation and are transmitted vertically to offspring, horizontal transfers involve the acquisition of exogenous DNA. Bacteria exchange genetic material through three main mechanisms (Fig. 2): transformation (incorporation of environmental DNA), transduction (transfer by a phage), and conjugation (contact between cells) (Holmes et al. 2016).
Although some cases of acquisition of antibiotic resistance genes by transformation or transduction have been documented, conjugation is the mechanism that contributes most to the spread of these genes (von Wintersdorff et al. 2016). In general, conjugation uses plasmids, defined as self-replicating extrachromosomal genetic elements (Actis et al. 1999), as vectors to promote the flow of genes. In fact, the biological functions of plasmids are extremely diverse and can help improve the fitness of cells by inducing, among others, antimicrobial resistance, increasing metabolic capacity, and providing virulence factors (Srivastava 2013). Finally, it is also possible to acquire antibiotic resistance genes with integrons (Fig. 2), which are site-specific recombination systems capable of recruiting genes (especially antibiotic resistance) (Deng et al. 2015).
Genes causing antibiotic resistance are divided into two groups: those that alter or destroy the antibiotic molecule and those that either decrease the influx or increase the expulsion of the compound (mainly by efflux pumps) out of the cell (Munita and Arias 2016). The two mechanisms of resistance (modification of the antibiotic or alteration of its flow) are not mutually exclusive. This is the case, for example, of chloramphenicol, for which resistance can be provided by acetylation of the molecule by a chloramphenicol acetyltransferase making it ineffective or by a decrease of its intracellular concentration by efflux pumps (Schwarz et al. 2004).
2.3 The Role of Aquaculture in Antibiotic Resistance
Aquaculture plays a central role in sustaining the demand for protein by the increasing human population (Diana 2009). To maintain the pace and remain economically profitable, fish farms need to implement intensive production conditions. This type of aquaculture, however, creates a conducive environment for the spread of disease and also promotes the emergence of pathogenic bacteria (Pulkkinen et al. 2010; Sundberg et al. 2016). As a consequence, antimicrobial agents are often used in a prophylactic manner in intensive fish farming (Cabello et al. 2013). In major producing countries like Chile, the amount of antibiotics authorized and administrated in a veterinary context can surpass the amount used in human medicine (Cabello et al. 2013).
Although this way of using antibiotics may seem attractive in the short term, the effects can be disastrous in the long term. As stated earlier, the use of an antibiotic promotes selection for resistant strains impossible to treat with the same compound. It is estimated that 80% of the administrated drugs (usually either as additives in food or by balneation) persist in the environment as active compounds (Cabello et al. 2016). A good example is oxytetracycline, which was found to persist in sediment after administration, thus causing a significant increase in bacteria resistant to this antibiotic (Samuelsen et al. 1992). A similar correlation has been made between the use of florfenicol in the Province of Quebec (Canada) and the observed amount of resistant A. salmonicida subsp. salmonicida strains to this antibiotic (Morin 2010).
Aquatic environments are ideal reservoirs of antibiotic resistance for two reasons: (1) by its high capacity of infiltration, unsanitary water containing antimicrobial compounds or other pollutants can easily contaminate clean water, and (2) water is a favorable environment for horizontal gene transfers between bacteria (Lupo et al. 2012). To this respect, using the zebrafish model, Fu and collaborators demonstrated that aquatic animal guts significantly contribute to the spread of antibiotic resistance genes in water environments (Fu et al. 2017).
The exchange of genes involved in antibiotic resistance is even more worrying in the sense that resistant bacteria that are usually not pathogenic to humans can transfer their resistance genes to some human pathogens. For example, fish pathogen A. salmonicida subsp. salmonicida and human pathogen Salmonella enterica might have exchanged plasmid-bearing resistance genes directly or indirectly through intermediate bacteria (McIntosh et al. 2008; Vincent et al. 2014; Trudel et al. 2016).
In order to understand the phenomenon of antibiotic resistance, it is important to consider bacterial communities in a given environment as a network, a coherent and dynamic system, and not as isolated and static individuals. Bacteria with a central position (hubs) are of paramount importance since they can play a relay role between several bacteria that cannot directly exchange genetic material (Fig. 3).
3 Adverse Effects of Antibiotherapy on Fish Microbiota
3.1 Roles of the Microbiota in Fish Health
Like all pluricellular organisms, fish live in close association with resident microbial communities (hereby called microbiota) composed of hundreds of microbial species. It is known that the rainbow trout microbiota is composed of 52 core bacterial lineages (Wong et al. 2013) with significant variation with respect to body site. For example, up to 199 genera can be found on rainbow trout skin (Lowrey 2014). The number of unique functional genes harbored by the microbiota surpasses the number of host genes by a 100-fold order of magnitude (Tsai and Coyle 2009). This large set of genes provided by the microbiota can complement (or even provide) metabolic pathways for nutrient metabolism (Enjalbert et al. 2017), host immunity (Belkaid and Hand 2014), and even cognitive and behavioral modulation (Carabotti et al. 2015). Those beneficial contributions of the microbiota may be disrupted following antibiotic treatment through collateral targeting of key symbionts.
3.2 Collateral Targeting of the Microbiota by Antimicrobial Compounds
When an infection occurs, it can be laborious and time-consuming to identify the strain that causes the disease. Consequently, antimicrobial agents having a broad spectrum (i.e., targeting a wide range of bacterial species) are usually prioritized. Although the bacterial strain that caused the infection may be correctly targeted, a wide range of other bacteria (including commensal or mutualistic symbionts) may also be affected in a collateral manner (Fig. 4). Therefore, the intensive use of antibiotics does not only promote resistance to these compounds; it also leads to deleterious side effects. A study conducted on zebrafish revealed that long-term use of legal aquaculture concentrations of oxytetracycline and sulfamethoxazole caused adverse effects on fish gut health (Zhou et al. 2018a, b) such as a decrease in goblet cell number and antioxidative enzymes and loss of intestinal microbiota diversity. Systemic effects such as decreased resistance to infection and higher oxygen consumption rate were also observed. In mosquitofish (Gambusia affinis), 1-week exposure to rifampicin caused a drop in viable counts from the skin microbiota (0.02% resistant) but led to >70% resistance following less than 2 days recovery after antibiotic treatment (Carlson et al. 2017). This increase was attributed to rifampicin-mediated selection for bacteria of the Comamonadaceae family.
Collateral targeting of bacterial symbionts by a broad-spectrum antibiotic treatment. In panel 1: Host-microbiota interactions allow mitigation of pathogens either by direct antagonism by the microbiota or by modulating the host’s immune response. In panel 2: Application of a broad-spectrum antibiotic treatment targets pathogens as well as nonpathogenic symbionts from the microbiota, thereby leading to dysbiosis, i.e., a disruption of normal host-microbiota interactions. A few antimicrobial resistant (AMR) bacteria (pictured here with blue cell walls) survive, including a pathogen that acquired resistance genes from another symbiont prior to treatment (see panel 1). In panel 3: AMR bacteria thrive and occupy the gut niche previously occupied by symbionts killed by the antimicrobial treatment. As a result, the host does not adequately respond to infection
3.3 Over-elicitation of Inflammatory Responses
Symbionts from the microbiota produce natural antigens that continuously induce mucosal immune tolerance to innocuous antigens such as food proteins and molecular components of commensal bacteria (Pabst and Mowat 2012). The use of broad-spectrum antimicrobials may therefore lead to a depletion of symbionts, which help prevent active immune responses directed against the rest of the microbiota. In humans, the lack of proper immune tolerance is a contributing factor to several inflammatory diseases such as ulcerative colitis and allergy (Chistiakov et al. 2015).
3.4 Permanent Alteration of the Microbiota
The microbiota, although depending on the surrounding environment of the host, is strongly influenced by developmental and reproductive factors. In human infants, gut microbiota colonization is mainly controlled by the maternal skin microbiota, the mode of delivery, and the initial diet (i.e., colostrum and maternal milk) (Mackie et al. 1999; Fernández et al. 2013). An analogous situation was found in discus fish (Symphysodon discus), whose progeny feeds exclusively on maternal skin mucus in early stages of life (Sylvain and Derome 2017). In the case of human neonates, deprivation from vertical transmission routes may cause irreversible alteration of microbiota composition (Neu and Rushing 2011). In the case of discus fish, maternal mucus feeding was essential for the offspring to obtain a normal adult-like microbiota (Sylvain and Derome 2017).
Certain symbionts are only acquired during a definite part of the host’s life cycle. The use of broad-spectrum antimicrobials may therefore lead to permanent changes in the host microbiota. This is exemplified by the study of Carlson and collaborators, demonstrating in Gambusia affinis lasting effects on mucosal microbiomes following antibiotic exposure, including the persistence of drug-resistant organisms and the inability of those microbiomes to return to a pre-treatment state (Carlson et al. 2017).
Severe consequences may arise from such a disrupted microbiota throughout development, ranging from metabolic deficiencies to increased susceptibility toward opportunistic infections (Langdon et al. 2016). In Atlantic salmon (Salmo salar), farmed juveniles (parrs) have a substantially lower survival rate when introduced in nature than their wild counterparts (Saloniemi et al. 2004). Interestingly, wild and farmed parrs also encountered a tremendous mismatch regarding gut microbiota composition (Lavoie et al. 2018). Furthermore, 6 months after being released into the river, stocked parrs still had a hatchery imprinting of their microbiota (Lavoie et al. 2018). Those striking differences suggest that microbiota could be another factor that could impact survival, due to its close relationship with host physiology.
3.5 Increased Carrying Capacity for Resistant and/or Pathogenic Bacteria
The intensive use of antibiotics can increase the carrying capacity of the host for pathogenic bacteria via three mechanisms. The first one, as explained above (see Sects. 3.1 and 3.2), results from the simultaneous enrichment of resistant bacteria and depletion of sensitive ones (which leaves more “room” to be occupied by resistant bacteria). The second mechanism results from the ability of bacteria to easily exchange genetic material between cells (i.e., horizontal gene transfer). Bacteria that are not pathogenic but resistant can transfer their resistance genes to sensitive but pathogenic bacteria, thereby increasing the carrying capacity of the host for pathogenic bacteria. The third mechanism is the depletion of bacteria that enhance colonization resistance. It is a broad mechanism including (1) production of mucins and defensins which prevent adherence of pathogens to mucosal tissues (Chairatana and Nolan 2017), (2) production of bactericidal and/or bacteriostatic compounds (Buffie and Pamer 2013), and (3) competition with pathogenic bacteria for the acquisition of nutrients and cofactors (Hibbing et al. 2010).
In summary, the use of antimicrobials may mitigate an outbreak in the short term but may favor the emergence of antibiotic-resistant pathogens in the long term. Furthermore, interactions between host and microbiota may be disrupted, thereby exacerbating the nefarious effects of those resistant pathogens on fish health.
4 Aquatic Pathogens Resistant to Antibiotics: The Case of Aeromonas salmonicida
Although it would be impossible to make a complete and exhaustive list of the aquatic pathogens for which antibiotic-resistant strains exist, there are some key problematic bacteria for the aquaculture industry. For example, several strains from the genera Aeromonas, Yersinia, Photobacterium, Edwardsiella, and Vibrio were listed as resistant to antibiotics, mainly through acquisition of resistance genes mediated by mobile elements (Miller and Harbottle 2018). One of the first cases of plasmid-mediated antibiotic resistance in a fish pathogen was reported in 1971 from an A. salmonicida strain isolated in 1959 in the United States (Aoki et al. 1971). The strain was described as resistant to sulfathiazole and tetracycline. The same study reported that these resistance phenotypes could be transferred to a strain of Escherichia coli. They found 15 years later that a conjugative plasmid, pAr-32, was responsible for the observed resistance (Aoki et al. 1986). It has been inferred that pAr-32 is identical to plasmid pRA3, which is the reference for IncU plasmids in addition to being found in the human and fish pathogen Aeromonas hydrophila (Bradley et al. 1982). In 1983, a striking correlation between the use of antimicrobial compounds and the observed resistance in strains recovered from outbreaks was reported (Aoki et al. 1983). They also found that strains of A. salmonicida isolated from cultured fish were more prone to have plasmid-bearing resistance genes than those from wild fish, which were shown to be mostly sensitive to the tested antibiotics.
Since then, several plasmids were found in many strains of A. salmonicida (Table 1) but also in other species of the same genus (Piotrowska and Popowska 2015). It is well known that advances in DNA sequencing technologies allowed us to discover and to classify genetic elements, such as plasmids, at an unprecedented pace and at relatively low cost (Vincent et al. 2017; Orlek et al. 2017). In recent years, a myriad of plasmids has been discovered and characterized in A. salmonicida through sequencing, many of which cause antibiotic resistance. A major discovery was the ability of A. salmonicida to exchange plasmids and other mobile DNA with pathogenic bacteria, such as Aeromonas bestiarum (plasmid pAB5S9b) and S. enterica (plasmid pSN254b and a class 1 integron) (Vincent et al. 2014; Trudel et al. 2016). This is even more worrying in a veterinary context given that both pAB5S9b and pSN254b cause resistance to all antibiotics approved by the Canadian Ministry of Health’s Veterinary Drugs Directorate to treat infected fish (oxytetracycline, florfenicol [a chloramphenicol analog], sulfadimethoxine/ormetoprim, and sulfadiazine/trimethoprim).
Although several plasmids found in A. salmonicida confer resistance to antimicrobial agents or even provide virulence factors, there are also plasmids without any known biological function. They are consequently considered as cryptic. Commonly, A. salmonicida subsp. salmonicida has three small cryptic plasmids ranging from 5.2 to 5.6 kbp and bearing either a ColE1- or a ColE2-type replicon: pAsa1 (ColE2), pAsa2 (ColE1), and pAsa3 (ColE2). These small plasmids only bear genes involved in their replication, maintenance, and mobilization. Although their presence in A. salmonicida subsp. salmonicida isolates is known since 1983 (Toranzo et al. 1983), it was in 1989 that they were named (Belland and Trust 1989), and their DNA was completely sequenced 14 years later (Boyd et al. 2003). Until recently, there was no evidence or even a clue on why these plasmids are so highly conserved throughout the isolated A. salmonicida strains around the world.
The plasmid pAsal1 shares high homology with pAsa3 while bearing two additional elements: the aopP gene, encoding a virulence factor related to the type three secretion system, and an insertion sequence (Fehr et al. 2006; Attéré et al. 2015). This plasmid is found in the majority of A. salmonicida subsp. salmonicida isolates (Attéré et al. 2015). For a long period of time, this plasmid was the sole example of a non-cryptic plasmid putatively derived from a cryptic plasmid. The situation became clearly different recently. In 2016, a plasmid named pAsa7 and bearing a gene causing resistance to chloramphenicol was published (Vincent et al. 2016). One of the salient features of this plasmid was not only the high resistance to chloramphenicol that it provides but that pAsa7 is highly similar to pAsa2. This observation reinforced the hypothesis that small cryptic plasmids, pAsa2 in the case of pAsa7, could be free high-copy receptacles for A. salmonicida to acquire new genes, such as for antibiotic resistance. The work of Attéré et al. in 2017 described two new plasmids, pAsaXI and pAsaXII, putatively derived from the cryptic plasmids pAsa3 and pAsa2, respectively (Attéré et al. 2017). The fact that these plasmids harbor genes involved in virulence and resistance to formaldehyde, often used as a disinfectant in aquaculture (Leal et al. 2016), combined to the existence of pAsal1 and pAsa7 let Attéré et al. state that small cryptic plasmids could be moldable vectors allowing the strains to quickly face off harsh conditions by acquiring genes involved in new functions. Additional plasmids derived from cryptic plasmids will likely be discovered in the future.
Regulation of antibiotics usages differs between countries and generates distinct pressure on the bacterial strains. This was exemplified by a recent study reporting the geographic distribution of antibiotic resistance genes and plasmids in A. salmonicida subsp. salmonicida strains from eastern Canada (Trudel et al. 2016). This study reported that several strains had plasmids, such as pSN254b and pAB5S9b, encoding genes involved in resistance to florfenicol and tetracycline, two antibiotics widely used in Canadian aquaculture. A similar observation was made by a study that investigated the epidemiology aspects of furunculosis in Denmark (Bartkova et al. 2017). They found that the resistant strains of A. salmonicida subsp. salmonicida isolated in Denmark were resistant to trimethoprim and sulfonamide, two of the few antibiotics approved to treat fish infection in this country.
5 Antibiotic Alternatives and the One Health Perspective
It is now clear that in addition to better diagnostic tools, we need new treatments to fight antibiotic-resistant bacteria (Allen et al. 2014; Reardon 2015; Czaplewski et al. 2016). A group of 24 academic and industry scientists identified ten alternatives to antibiotics with enough clinical data and independent studies to believe in their approval by 2025 (Czaplewski et al. 2016). These alternatives include, for example, the use of probiotics, phages (or products derived therefrom), and antimicrobial peptides. However, this same team indicates that a budget of one and a half billion sterling pounds is needed to test and develop these ten alternatives to antibiotics. In addition, clinical trials focus primarily on human pathogens including Clostridium difficile, Pseudomonas aeruginosa, and Staphylococcus aureus bacteria. Therefore, it is very difficult to predict the success of these alternatives against other pathogenic and antibiotic-resistant bacteria especially in a veterinary context.
In addition to antibiotic alternatives, experts agree that antibiotic resistance must be considered in the concept of the One Health initiative (Fig. 5). This concept states that humans, animals, and the environment must be considered as a whole (Queenan et al. 2016). A failure in one of these three components will inevitably affect the other two. In addition, there are some key environments that are closely related to animals and humans, as those that are aquatic (Gormaz et al. 2014). Indeed, from a “One Health” point of view, mitigating fish diseases solely through antibiotherapy without considering the rearing environment would be insufficient. Many fish pathogens such as A. salmonicida, Flavobacterium spp., and Vibrio spp. are opportunistic, i.e., their virulence emerges when their host’s homeostasis is challenged by other abiotic and/or biotic factors (Derome et al. 2016). Therefore, one cannot truly cure an opportunistic disease without first addressing its root cause (namely, the inadequate sanitary and environmental conditions that triggered the outbreak).
6 Conclusion
Antibiotics were, and still continue to be, a crucial weapon against pathogenic bacteria. However, in addition to usually generate dysbiosis in the natural microbial flora, their introduction as a prime choice to treat bacterial infections caused the selection of strains resistant to these compounds. This is worrying in the context that several strains are now multidrug-resistant. In aquaculture, this is even more problematic since, at least for some countries, only a few antibiotics are approved as therapeutic agents.
It is increasingly important to find alternatives to antibiotics. Several options were explored in aquaculture, such as the use of bacteriophages (viruses specifically infecting bacteria), probiotics, and essential oils (Boutin et al. 2012; Romero 2012; Martínez Cruz et al. 2012; Sutili et al. 2017; Seghouani et al. 2017; Gon Choudhury et al. 2017). Fortunately, alternatives to antibiotics are also being explored for human pathogens, and it is reasonable to believe that the aquaculture industry will benefit from the discoveries that will be made in this area (Czaplewski et al. 2016) and vice versa.
In all cases, although antibiotics are undoubtedly crucial to cure infections, they should be used reasonably and with caution. Stanislas F. Snieszko, a pioneer in fish parasites and disease (Mitchell 2001), recommended to never rely exclusively on antibiotics, especially in the long term, to fight infections. What is essential in the context of aquaculture is to reduce the potential sources of contamination, to increase the standards of hygiene, and to have good hatchery practices (Snieszko and Bullock 1957). The recent discoveries on the importance of host-microbiota interactions on host health strongly suggest that a better hygiene does not mean eradicating microbes, but rather improving microbial homeostasis in hatcheries.
References
Actis LA, Tolmasky ME, Crosa JH (1999) Bacterial plasmids: replication of extrachromosomal genetic elements encoding resistance to antimicrobial compounds. Front Biosci 4:D43–D62
Adams C, Austin B, Meaden P, McIntosh D (1998) Molecular characterization of plasmid-mediated oxytetracycline resistance in Aeromonas salmonicida. Appl Environ Microbiol 64:4194–4201
Allen HK, Trachsel J, Looft T, Casey TA (2014) Finding alternatives to antibiotics. Ann N Y Acad Sci 1323:91–100. https://doi.org/10.1111/nyas.12468
Aminov RI (2010) A brief history of the antibiotic era: lessons learned and challenges for the future. Front Microbiol 1:134. https://doi.org/10.3389/fmicb.2010.00134
Aoki T, Egusa S, Kimura T, Watanabe T (1971) Detection of R factors in naturally occurring Aeromonas salmonicida strains. Appl Microbiol 22:716–717
Aoki T, Kitao T, Iemura N, Mitoma Y, Nomura T (1983) The susceptibility of Aeromonas salmonicida strains isolated in cultured and wild salmonids to various chemotherapeutics. Bull Jpn Soc Sci Fish 49:17–22. https://doi.org/10.2331/suisan.49.17
Aoki T, Mitoma Y, Crosa JH (1986) The characterization of a conjugative R-plasmid isolated from Aeromonas salmonicida. Plasmid 16:213–218. https://doi.org/10.1016/0147-619X(86)90059-4
Attéré SA, Vincent AT, Trudel MV, Chanut R, Charette SJ (2015) Diversity and homogeneity among small plasmids of Aeromonas salmonicida subsp. salmonicida linked with geographical origin. Front Microbiol 6:1274. https://doi.org/10.3389/fmicb.2015.01274
Attéré SA, Vincent AT, Paccaud M, Frenette M, Charette SJ (2017) The role for the small cryptic plasmids as moldable vectors for genetic innovation in Aeromonas salmonicida subsp. salmonicida. Front Genet 8:211. https://doi.org/10.3389/fgene.2017.00211
Austin B, Austin DA (2016) Control. In: Austin B, Austin DA (eds) Bacterial fish pathogens: disease of farmed and wild fish. Springer, Cham, pp 693–709. https://doi.org/10.1007/978-3-319-32674-0_14
Bartkova S, Leekitcharoenphon P, Aarestrup FM, Dalsgaard I (2017) Epidemiology of Danish Aeromonas salmonicida subsp. salmonicida in fish farms using whole genome sequencing. Front Microbiol 8:2411. https://doi.org/10.3389/fmicb.2017.02411
Belkaid Y, Hand TW (2014) Role of the microbiota in immunity and inflammation. Cell 157:121–141. https://doi.org/10.1016/j.cell.2014.03.011
Belland RJ, Trust TJ (1989) Aeromonas salmonicida plasmids: plasmid-directed synthesis of proteins in vitro and in Escherichia coli minicells. Microbiology 135:513–524. https://doi.org/10.1099/00221287-135-3-513
Boutin S, Bernatchez L, Audet C, Derome N (2012) Antagonistic effect of indigenous skin bacteria of brook charr (Salvelinus fontinalis) against Flavobacterium columnare and F. psychrophilum. Vet Biol 155:355–361
Boyd J et al (2003) Three small, cryptic plasmids from Aeromonas salmonicida subsp. salmonicida A449. Plasmid 50:131–144. https://doi.org/10.1016/S0147-619X(03)00058-1
Bradley DE, Aoki T, Kitao T, Arai T, Tschäpe H (1982) Specification of characteristics for the classification of plasmids in incompatibility group U. Plasmid 8:89–93. https://doi.org/10.1016/0147-619X(82)90045-2
Buffie CG, Pamer EG (2013) Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol 13:790–801. https://doi.org/10.1038/nri3535
Cabello FC (2006) Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ Microbiol 8:1137–1144. https://doi.org/10.1111/j.1462-2920.2006.01054.x
Cabello FC et al (2013) Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Environ Microbiol 15:1917–1942. https://doi.org/10.1111/1462-2920.12134
Cabello FC, Godfrey HP, Buschmann AH, Dölz HJ (2016) Aquaculture as yet another environmental gateway to the development and globalisation of antimicrobial resistance. Lancet Infect Dis 16:e127–e133. https://doi.org/10.1016/S1473-3099(16)00100-6
Campbell KL (1999) Sulphonamides: updates on use in veterinary medicine. Vet Dermatol 10:205–215. https://doi.org/10.1046/j.1365-3164.1999.00181.x
Campbell EA et al (2001) Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell 104:901–912. https://doi.org/10.1016/S0092-8674(01)00286-0
Carabotti M, Scirocco A, Maselli MA, Severi C (2015) The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann Gastroenterol 28:203–209. https://doi.org/10.1038/ajgsup.2012.3
Carlson JM, Leonard AB, Hyde ER, Petrosino JF, Primm TP (2017) Microbiome disruption and recovery in the fish Gambusia affinis following exposure to broad-spectrum antibiotic. Infect Drug Resist 10:143–154. https://doi.org/10.2147/IDR.S129055
Chairatana P, Nolan EM (2017) Defensins, lectins, mucins, and secretory immunoglobulin A: microbe-binding biomolecules that contribute to mucosal immunity in the human gut. Crit Rev Biochem Mol Biol 52:45–56. https://doi.org/10.1080/10409238.2016.1243654
Chang Q, Wang W, Regev-Yochay G, Lipsitch M, Hanage WP (2015) Antibiotics in agriculture and the risk to human health: how worried should we be? Evol Appl 8:240–247. https://doi.org/10.1111/eva.12185
Chistiakov DA, Bobryshev YV, Kozarov E, Sobenin IA, Orekhov AN (2015) Intestinal mucosal tolerance and impact of gut microbiota to mucosal tolerance. Front Microbiol 5:781. https://doi.org/10.3389/fmicb.2014.00781
Czaplewski L et al (2016) Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect Dis 16:239–251. https://doi.org/10.1016/S1473-3099(15)00466-1
Deng Y et al (2015) Resistance integrons: class 1, 2 and 3 integrons. Ann Clin Microbiol Antimicrob 14:45. https://doi.org/10.1186/s12941-015-0100-6
Derome N, Gauthier J, Boutin S, Llewellyn M (2016) Bacterial opportunistic pathogens of fish. In: Hurst CJ (ed) The rasputin effect: when commensals and symbionts become parasitic. Springer, Cham, pp 81–108. https://doi.org/10.1007/978-3-319-28170-4_4
Diana JS (2009) Aquaculture production and biodiversity conservation. Bioscience 59:27–38. https://doi.org/10.1525/bio.2009.59.1.7
Enjalbert F, Combes S, Zened A, Meynadier A (2017) Rumen microbiota and dietary fat: a mutual shaping. J Appl Microbiol 123:782–797. https://doi.org/10.1111/jam.13501
FAO (2016) The state of world fisheries and aquaculture 2016. FAO, Rome
Fehr D et al (2006) AopP, a type III effector protein of Aeromonas salmonicida, inhibits the NF-kappaB signalling pathway. Microbiology 152:2809–2818. https://doi.org/10.1099/mic.0.28889-0
Fernández L, Langa S, Martín V, Maldonado A (2013) The human milk microbiota: origin and potential roles in health and disease. Phiarmacol Res 69:1–10. https://doi.org/10.1016/j.phrs.2012.09.001
Fleming A (1929) On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br J Exp Pathol 10:226–236. https://doi.org/10.1038/146837a0
Floss HG, Yu T-W (2005) Rifamycin—Mode of action, resistance, and biosynthesis. Chem Rev 105:621–632. https://doi.org/10.1021/cr030112j
Fu J et al (2017) Aquatic animals promote antibiotic resistance gene dissemination in water via conjugation: role of different regions within the zebra fish intestinal tract, and impact on fish intestinal microbiota. Mol Ecol 26:5318–5333. https://doi.org/10.1111/mec.14255
Gon Choudhury T, Tharabenahalli Nagaraju V, Gita S, Paria A, Parhi J (2017) Advances in bacteriophage research for bacterial disease control in aquaculture. Rev Fish Sci Aquac 25:113–125. https://doi.org/10.1080/23308249.2016.1241977
Goossens H, Ferech M, Vander Stichele R, Elseviers M (2005) Outpatient antibiotic use in Europe and association with resistance: a cross-national database study. Lancet 365:579–587. https://doi.org/10.1016/S0140-6736(05)17907-0
Gormaz JG, Fry JP, Erazo M, Love DC (2014) Public health perspectives on aquaculture. Curr Environ Health Rep 1:227–238. https://doi.org/10.1007/s40572-014-0018-8
Gutsell JS (1946) Sulfa drugs and the treatment of furunculosis in trout. Science 104:85–86
Hibbing ME, Fuqua C, Parsek MR, Peterson SB (2010) Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8:15–25. https://doi.org/10.1038/nrmicro2259
Hollis A, Ahmed Z (2013) Preserving antibiotics, rationally. N Engl J Med 369:2474–2476. https://doi.org/10.1056/NEJMp1311479
Holmes AH et al (2016) Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387:176–187. https://doi.org/10.1016/S0140-6736(15)00473-0
Horsberg TE, Martinsen B, Sandersen K, Zernichow L (1997) Potentiated sulfonamides: in vitro inhibitory effect and pharmacokinetic properties in Atlantic salmon in seawater. J Aquat Anim Health 9:203–210
L’Abée-Lund TM, Sørum H (2000) Functional Tn5393-like transposon in the R plasmid pRAS2 from the fish pathogen Aeromonas salmonicida subspecies salmonicida isolated in Norway. Appl Environ Microbiol 66:5533–5535. https://doi.org/10.1128/AEM.66.12.5533-5535.2000
L’Abée-Lund TM, Sørum H (2001) Class 1 integrons mediate antibiotic resistance in the fish pathogen Aeromonas salmonicida worldwide. Microb Drug Resist 7:263–272. https://doi.org/10.1089/10766290152652819
L’Abée-Lund TM, Sørum H (2002) A global non-conjugative Tet C plasmid, pRAS3, from Aeromonas salmonicida. Plasmid 47:172–181. https://doi.org/10.1016/S0147-619X(02)00001-X
Langdon A, Crook N, Dantas G (2016) The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med 8. https://doi.org/10.1186/s13073-016-0294-z
Lavoie C, Courcelle M, Redivo B, Derome N (2018) Structural and compositional mismatch between captive and wild Atlantic salmon (Salmo salar) parrs gut microbiota highlights the relevance of integrating molecular ecology for management and conservation methods. Evol Appl 11:1671–1685. https://doi.org/10.1111/eva.12658
Leal JF, Neves MGPMS, Santos EBH, Esteves VI. (2016) Use of formalin in intensive aquaculture: properties, application and effects on fish and water quality. Rev Aquac 0:1–15. doi: https://doi.org/10.1111/raq.12160
Lowrey LT (2014) The microbiome of rainbow trout (Oncorhynchus mykiss). University of New Mexico, Albuquerque
Lupo A, Coyne S, Berendonk TU (2012) Origin and evolution of antibiotic resistance: the common mechanisms of emergence and spread in water bodies. Front Microbiol 3:18. https://doi.org/10.3389/fmicb.2012.00018
Mackie RI, Sghir A, Gaskins HR (1999) Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr 69:1035S–1045S
Martin MJ, Thottathil SE, Newman TB (2015) Antibiotics overuse in animal agriculture: a call to action for health care providers. Am J Public Health 105:2409–2410. https://doi.org/10.2105/AJPH.2015.302870
Martinez JL (2009) Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ Pollut 157:2893–2902. https://doi.org/10.1016/j.envpol.2009.05.051
Martínez Cruz P, Ibáñez AL, Monroy Hermosillo OA, Ramírez Saad HC (2012) Use of probiotics in aquaculture. ISRN Microbiol 2012:1–13. https://doi.org/10.5402/2012/916845
McCarthy DH, Stevenson JP, Salsbury AW (1974) Combined in-vitro activity of trimethoprim and sulphonamides on fish-pathogenic bacteria. Aquaculture 3:87–91. https://doi.org/10.1016/0044-8486(74)90100-8
McIntosh D et al (2008) Transferable, multiple antibiotic and mercury resistance in Atlantic Canadian isolates of Aeromonas salmonicida subsp. salmonicida is associated with carriage of an IncA/C plasmid similar to the Salmonella enterica plasmid pSN254. J Antimicrob Chemother 61:1221–1228. https://doi.org/10.1093/jac/dkn123
Miller R, Harbottle H (2018) Antimicrobial drug resistance in fish pathogens. Microbiol Spectr 6:1–20. https://doi.org/10.1128/microbiolspec.ARBA-0017-2017
Mitchell AJ (2001) Finfish health in the United States (1609–1969): historical perspective, pioneering researchers and fish health workers, and annotated bibliography. Aquaculture 196:347–438. https://doi.org/10.1016/S0044-8486(01)00547-6
Morin R (2010) L’utilisation des antibiotiques pour combattre la furonculose chez l’omble de fontaine génère de l’antibiorésistance chez Aeromonas salmonicida. L’aquicole 15:1–6
Munita JM, Arias CA (2016) Mechanisms of antibiotic resistance. Microbiol Spectr 4:1–24. https://doi.org/10.1128/microbiolspec.VMBF-0016-2015
Neu J, Rushing J (2011) Cesarean versus vaginal delivery: long-term infant outcomes and the hygiene hypothesis. Clin Perinatol 38:321–331. https://doi.org/10.1016/j.clp.2011.03.008
Nicolaou KC, Rigol S (2018) A brief history of antibiotics and select advances in their synthesis. J Antibiot (Tokyo) 71:153–184. https://doi.org/10.1038/ja.2017.62
Orlek A et al (2017) Plasmid classification in an era of whole-genome sequencing: application in studies of antibiotic resistance epidemiology. Front Microbiol 8:182. https://doi.org/10.3389/fmicb.2017.00182
Pabst O, Mowat AM (2012) Oral tolerance to food protein. Mucosal Immunol 5:232–239. https://doi.org/10.1038/mi.2012.4
Piotrowska M, Popowska M (2015) Insight into the mobilome of Aeromonas strains. Front Microbiol 6:494. https://doi.org/10.3389/fmicb.2015.00494
Pulkkinen K et al (2010) Intensive fish farming and the evolution of pathogen virulence: the case of columnaris disease in Finland. Proc R Soc B Biol Sci 277:593–600. https://doi.org/10.1098/rspb.2009.1659
Queenan K, Häsler B, Rushton J (2016) A One Health approach to antimicrobial resistance surveillance: is there a business case for it? Int J Antimicrob Agents 48:422–427. https://doi.org/10.1016/j.ijantimicag.2016.06.014
Quinn R (2013) Rethinking antibiotic research and development: World War II and the penicillin collaborative. Am J Public Health 103:426–434. https://doi.org/10.2105/AJPH.2012.300693
Reardon S (2015) Antibiotic alternatives rev up bacterial arms race. Nature 521:402–403. https://doi.org/10.1038/521402a
Reith ME et al (2008) The genome of Aeromonas salmonicida subsp. salmonicida A449: insights into the evolution of a fish pathogen. BMC Genomics 9:427. https://doi.org/10.1186/1471-2164-9-427
Romero J (2012) Antibiotics in aquaculture – use, abuse and alternatives. In: Navarrete P, Carvalho E (eds) Health and environment in aquaculture. InTech, Rijeka, pp 159–198. https://doi.org/10.5772/28157
Saloniemi I, Jokikokko E, Kallio-Nyberg I, Jutila E, Pasanen P (2004) Survival of reared and wild Atlantic salmon smolts: size matters more in bad years. ICES J Mar Sci 61:782–787. https://doi.org/10.1016/j.icesjms.2004.03.032
Samuelsen OB, Torsvik V, Ervik A (1992) Long-range changes in oxytetracycline concentration and bacterial resistance towards oxytetracycline in a fish farm sediment after medication. Sci Total Environ 114:25–36. https://doi.org/10.1016/0048-9697(92)90411-K
Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A (2004) Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev 28:519–542. https://doi.org/10.1016/j.femsre.2004.04.001
Seghouani H, Garcia-Rangel CE, Füller J, Gauthier J, Derome N (2017) Walleye autochthonous bacteria as promising probiotic candidates against Flavobacterium columnare. Front Microbiol 18:1349
Snieszko SF, Bullock GL (1957) Treatment of sulfonamide-resistant furunculosis in trout and determination of drug sensitivity. Fish Bull 57:555–564
Sørum H, L’Abée-Lund TM, Solberg A, Wold A (2003) Integron-containing IncU R plasmids pRAS1 and pAr-32 from the fish pathogen Aeromonas salmonicida. Antimicrob Agents Chemother 47:1285–1290. https://doi.org/10.1128/AAC.47.4.1285-1290.2003
Srivastava S (2013) Plasmids: their biology and functions. In: Genetics of bacteria. Springer, New Delhi, pp 125–151. https://doi.org/10.1007/978-81-322-1090-0_6
Sundberg L-R et al (2016) Intensive aquaculture selects for increased virulence and interference competition in bacteria. Proc Biol Sci 283:20153069. https://doi.org/10.1098/rspb.2015.3069
Sutili FJ, Gatlin DM, Heinzmann BM, Baldisserotto B (2017) Plant essential oils as fish diet additives: benefits on fish health and stability in feed. Rev Aquac 10:716–726. https://doi.org/10.1111/raq.12197
Sylvain FÉ, Derome N (2017) Vertically and horizontally transmitted microbial symbionts shape the gut microbiota ontogenesis of a skin-mucus feeding discus fish progeny. Sci Rep 7:5263. https://doi.org/10.1038/s41598-017-05662-w
Tanaka KH et al (2016) The mosaic architecture of Aeromonas salmonicida subsp. salmonicida pAsa4 plasmid and its consequences on antibiotic resistance. PeerJ 4:e2595. https://doi.org/10.7717/peerj.2595
Toranzo AE, Barja JL, Colwell RR, Hetrick FM (1983) Characterization of plasmids in bacterial fish pathogens. Infect Immun 39:184–192
Trudel MV et al (2016) Diversity of antibiotic-resistance genes in Canadian isolates of Aeromonas salmonicida subsp. salmonicida: dominance of pSN254b and discovery of pAsa8. Sci Rep 6:35617. https://doi.org/10.1038/srep35617
Tsai F, Coyle WJ (2009) The microbiome and obesity: is obesity linked to our gut flora? Curr Gastroenterol Rep 11:307–313. https://doi.org/10.1007/s11894-009-0045-z
Vincent AT et al (2014) Detection of variants of the pRAS3, pAB5S9, and pSN254 plasmids in Aeromonas salmonicida subsp. salmonicida: multidrug-resistance, interspecies exchanges, and plasmid reshaping. Antimicrob Agents Chemother 58:7367–7374. https://doi.org/10.1128/AAC.03730-14
Vincent AT et al (2016) Antibiotic resistance due to an unusual ColE1-type replicon plasmid in Aeromonas salmonicida. Microbiology 162:942–953. https://doi.org/10.1099/mic.0.000286
Vincent AT, Derome N, Boyle B, Culley AI, Charette SJ (2017) Next-generation sequencing (NGS) in the microbiological world: how to make the most of your money. J Microbiol Methods 138:60–71. https://doi.org/10.1016/j.mimet.2016.02.016
von Wintersdorff CJH et al (2016) Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Front Microbiol 7:173. https://www.frontiersin.org/article/10.3389/fmicb.2016.00173
Waksman SA (1947) What is an antibiotic or an antibiotic substance? Mycologia 39:565–569. https://doi.org/10.2307/3755196
Wayne A, Mccarthy R, Lindenmayer J (2011) Therapeutic antibiotic use patterns in dogs: observations from a veterinary teaching hospital. J Small Anim Pract 52:310–318. https://doi.org/10.1111/j.1748-5827.2011.01072.x
Wong S et al (2013) Aquacultured rainbow trout (Oncorhynchus mykiss) possess a large core intestinal microbiota that is resistant to variation in diet and rearing density. Appl Environ Microbiol 79:4974–4984. https://doi.org/10.1128/AEM.00924-13
Zhou L, Limbu SM, Shen M et al (2018a) Environmental concentrations of antibiotics impair zebrafish gut health. Environ Pollut 235:245–254. https://doi.org/10.1016/j.envpol.2017.12.073
Zhou L, Limbu SM, Qiao F, Du Z-Y, Zhang M (2018b) Influence of long-term feeding antibiotics on the gut health of zebrafish. Zebrafish 15:340–348. https://doi.org/10.1089/zeb.2017.1526
Acknowledgments
The authors are grateful to Sammy Nyongesa (INRS-Institut Armand-Frappier, Laval, Canada) for his critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Vincent, A.T., Gauthier, J., Derome, N., Charette, S.J. (2019). The Rise and Fall of Antibiotics in Aquaculture. In: Derome, N. (eds) Microbial Communities in Aquaculture Ecosystems. Springer, Cham. https://doi.org/10.1007/978-3-030-16190-3_1
Download citation
DOI: https://doi.org/10.1007/978-3-030-16190-3_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-16189-7
Online ISBN: 978-3-030-16190-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)