Abstract
Early diagnosis of Alzheimer’s disease (AD) is critical in preventing from irreversible damages to brain cognitive functions. Most computer-aided approaches consist of extraction of image features to describe the pathological changes and construction of a classifier for dementia identification. Deep learning technique provides a unified framework for simultaneous representation learning and feature classification, and thus avoids the troublesome hand-crafted feature extraction and feature engineering. In this paper, we propose an ensemble of AlexNets (EnAlexNets) algorithm for early diagnosis of AD using positron emission tomography (PET). We first use the automated anatomical labeling (AAL) cortical parcellation map to detect 62 brain anatomical volumes, then extract image patches in each kind of volumes to fine-tune a pre-trained AlexNet, and finally use the ensemble of those well-performed AlexNets as the classifier. We have evaluated this algorithm against seven existing algorithms on an ADNI dataset. Our results indicate that the proposed EnAlexNets algorithm outperforms those seven algorithms in differentiating AD cases from normal controls.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Alzheimer’s disease (AD) is a chronic and progressive decline in cognitive function due to the damage to brain cells. The number of patients suffering from AD worldwide is more than 30 million, and this number is expected to triple by 2050 due to the increasing life expectancy [1]. The onset of AD is typically insidious-cognitive decline generally begins years before reaching the threshold of clinical significance and functional impairment [2]. The prodromal stage of AD called mild cognitive impairment (MCI) is not bound to fall in the deterioration with treatments performed in time. The conversion rate from MCI to AD is estimated to be between 10 and 25% per year [3]. Although there are no current disease modifying agents to halt the progression of AD, there are a number of clinical trials underway in patients with pre-symptomatic disease. Thus, as effective therapies become available, the early identification of patients with MCI will be of tremendous benefit to patients and their families.
The pathology of AD includes cortical and subcortical atrophy together with the deposition of \( \beta \)-amyloid. Molecular medical imaging, such as the positron emission tomography (PET), single-photon emission computed tomography (SPECT), and magnetic resonance imaging (MRI), offers the ability to visualize hypometaboism or atrophy introduced by AD, and hence has led to a revolution in early diagnosis of AD [4]. In particular, functional PET and SPECT can detect subtle changes in cerebral metabolism prior to anatomical changes are evident or a symptomatological diagnosis of probable AD can be made with MR imaging [5]. Comparing to SPECT, PET is able to provide higher resolution information than SPECT in evaluating patients with suspected AD. 18-Fluorodeoxyglucose (FDG) is a widely used tracer in PET imaging for studying glucose metabolism. The comparative study made by Silverman [6] demonstrated that FDG-PET is superior to perfusion SPECT in identifying early changes associated with AD and other neurodegenerative dementias.
However, early diagnosis of AD remains a challenging task, since the pathological changes can be subtle in the early course of the disease and there can be some overlap with other neurodegenerative disorders [7]. Most computer-aided approaches consist of extraction of image features to describe the pathological changes and construction of a statistical model of the disease from a set of training examples using supervised methods [8]. The features that have been considered include global features, which are computed by applying the entire brain volume to a linear transform, and local features, such as the statistics, histograms, and gradients calculated from volumes of interest (VOIs). Most pattern classification techniques, including the k-mean clustering [9], artificial neural network (ANN) [10] and support vector machine (SVM) [5], have been successfully applied to this task.
Recently, the deep learning technique has shown proven ability to analysis medical images. It provides a unified framework for simultaneous learning image representation and feature classification, and thus avoids the troublesome hand-crafted feature extraction and feature engineering. Suk et al. [11] used the deep model learned image representation to differentiate AD subjects from MCI ones based on the assumption that those deep features inherent the latent high-level information. They also fused multiple sparse regression networks as the target-level representation [12]. Ortiz et al. [13] applied the deep belief network to the early diagnosis of the Alzheimer’s disease using both MRI and PET scans. Valliani et al. [14] employed the deep residual network (ResNet) that has been pre-trained on the ImageNet natural image dataset to identify dementia types.
In this paper, we propose an ensemble of AlexNets (EnAlexNets) model to diagnosis between AD and NC and differentiate between stages of MCI, mild MCI (mMCI) vs. severe MCI (sMCI). The uniqueness of our approach include: (1) using the automated anatomical labeling (AAL) cortical parcellation map to obtain 62 brain anatomical volumes; (2) extracting image patches from each cortical volume to train a candidate AlexNet, the champion model for the ILSVRC2012 image classification task; (3) selecting effective AlexNet models and ensemble them to generate more accurate diagnosis. The proposed EnAlexNets model was evaluated against seven methods on the ADNI dataset and achieved the state-of-the-art performance.
2 Dataset
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD.
We selected 962 FDG-PET studies from the ADNI database, including 241 AD cases, 306 mild MCI (mMCI) cases, 127 severe MCI (sMCI) cases and 288 normal controls (NCs). 185 MBq of 18F-Fludeoxyglucose (FDG) was injected to the subjects and PET scanning was commenced approximately 30 min after tracer injection that produced 3D scan consists of six 5 min frames. The scans used for this study are baseline or screening in the ADNI database, and had been through a pre-processing pipeline that includes co-registration, averaging, voxel normalization, and isotropic Gaussian smoothing [15]. This pre-processing work was done by the ADNI participants and it makes any subsequent analysis simpler as the data from different PET scanners are then uniform. The demographic information of the selected dataset is shown in Table 1.
3 Method
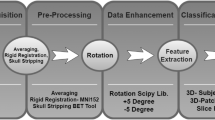
The proposed algorithm consists of four major steps: anatomical volume detection, data augmentation, patch classification, and dementia differentiation. A diagram that summarizes this algorithm was shown in Fig. 1.
3.1 Anatomical Volume Detection
The AAL cortical parcellation map [16], which provides 62 brain anatomical volumes including 54 symmetrical and 8 asymmetrical volumes, is well aligned with the template brain PET image supplied with the statistical parametric mapping (SPM, Version 2012) package [17].
To map the anatomical labels from the atlas onto each study and maintain the original brain image information, we spatially normalized the SPM brain PET template to FDG-PET images, which are self-aligned, using the spatial normalization procedure supplied with the SPM package. Thus each anatomical volume in the AAL atlas can be transferred to our FDG-PET images. For each volume, we located its geometry center on FDG-PET images and put a \( 67 \times 67 \) window on that center for image patch extraction.
3.2 Data Augmentation
Since number of extracted image patches is limited, we employed three data augmentation techniques to enlarge our image patch dataset. First, we shifted the PET image horizontally and vertically with a step of 5 voxels before patch extraction. Second, we move the patch extraction window to its adjacent transverse slices forwardly and backwardly. Third, rotate the PET image clockwise and counter-clockwise with 5 and 10°. With these operations, each image patch has nine augmented copies.
3.3 Patch Classification
Considering the limited image patches we have, too complex deep neural networks may suffer from over-fitting to our training data. Hence, we adopted the AlexNet, the champion model for the ILSVRC2012 image classification task, as our classifier, which consists of only 5 convolutional layers and 3 fully-connected layers (see Fig. 2).
It has been widely acknowledged that the image representation learned from large-scale datasets can be efficiently transferred to generic visual recognition tasks, which have limited training data [18, 19]. Therefore, the AlexNet used for this study was previously trained on the ImageNet training dataset, which is a 1000-category large-scale natural image database. To adapt this model to our problem, we replaced its last three layers with a fully connected layer, a softmax layer and a classification output layer, and adjust the learning rates such that those new layers can be trained quickly and other layers are trained slowly.
Since the AlexNet takes an input image of size \( 227 \times 227 \times 3 \), we duplicated each image patch three times and resize each copy to \( 227 \times 227 \) by the bilinear interpolation algorithm.
3.4 Dementia Differentiation
Since we obtained 62 cortical volumes on each PET scan, we can use the image patches extracted in each volume to fine-tune a pre-trained AlexNet. As we know, not all cortical volumes play equally critical roles in dementia diagnosis. Therefore, the diagnosis capacity of each fine-tuned AlexNet varies a lot. We chose 30% of the fine-tuned AlexNets, which performed best on the validation dataset, and adopted the majority voting scheme to combine their decisions for dementia differentiation. Such ensemble learning acts as the role of expert consultation aiming to get a more accurate diagnosis.
3.5 Evaluation
We adopted the 5-folder cross-validation scheme to evaluate the performance of the proposed algorithm. We randomly sampled the instances from each class to form five folds, aiming to ensure the distribution of data in each fold is as similar as that of the whole dataset. In each run, we used 80% of patches for training, 20% for validation and 20% for testing. The data augmentation procedure was not applied to test patches.
4 Results
Table 2 gives the accuracy of the proposed algorithm and other seven state-of-the-art algorithms when applying them to differentiate AD cases from NCs. It shows that our algorithm and the method reported in [13] achieved the highest classification accuracy. However, the other algorithm used both PET and MRI data. Meanwhile, it reveals that our algorithm achieved the highest specificity and the second best sensitivity among those eight algorithms.
Figure 3 depicts the accuracy of the proposed algorithm when applying it to the mMCI-sMCI classification. It shows that our algorithm achieved a classification accuracy of 85% in this problem.
5 Discussion
The proposed EnAlexNets algorithm is based on the pre-trained and fine-tuned AlexNets and ensemble of these models. Our experiments indicate that this algorithm is robust in securing more distinguish features and achieving better performance than that of using multi-modalities images. The EnAlexNets algorithm enables to effectively compensate for the less amount of information afforded by single image mode and reduce the complexity of diagnosis process requirements for multi-mode images. These investigations can be further improved by improving the CNN structure and enlarging the dataset. As for the time-complexity, the training time of multiple AlexNets summed up to 111,600 s same level as other state-of-art deep learning based methods. The diagnosis time of one patient case in our method was around 10 s. With better computation resources, this time consumed can be further diminished.
6 Conclusion
In this study, we proposed a multi-AlexNets based method for computer-aided diagnosis of AD and MCI. This method not only utilized the combination of different brain regions but also incorporated AlexNet with its strong feature representation ability. Better performance was observed on two classification tasks. Apart from the traditional diagnosis between AD and NC, we initially conducted the differentiation between different stages of MCI, mMCI vs. sMCI. Due to the latent progression from MCI to further stage, we intend to find the risk as early as possible, which is of great significance to both clinicians and patients. This study may be of great assistance to the computer-aided diagnosis of dementia and other biomedical fields.
References
Barnes, D.E., Yaffe, K.: The projected effect of risk factor reduction on Alzheimer’s disease prevalence. Lancet Neurol. 10(9), 819–828 (2011)
Ferreira, L.K., Rondina, J.M., Kubo, R., et al.: Support vector machine-based classification of neuroimages in Alzheimer s disease: direct comparison of FDG-PET, rCBF-SPECT and MRI data acquired from the same individuals. Revista Brasileira De Psiquiatria (2017)
Grand, J.H., Caspar, S., Macdonald, S.W.: Clinical features and multidisciplinary approaches to dementia care. Multidiscip. Healthc. 4, 125–147 (2011)
Gomez-Isla, T., Price, J.L., McKeel Jr., D., et al.: Profound loss of layer II entorhinal cortex neurons occurs in very mild Alzheimer’s disease. J. Neurosci. 16(14), 4491–4500 (1996)
Fung, G., Stoeckel, J.: SVM feature selection for classification of SPECT images of alzheimer’s disease using spatial information. Knowl. Inf. Syst. 11(2), 243–258 (2007)
Silverman, D.H.S.: Brain 18F-FDG PET in the diagnosis of neurodegenerative dementias: comparison with perfusion SPECT and with clinical evaluations lacking nuclear imaging. J. Nucl. Med. 45(4), 594–607 (2004)
Adeli, H., Ghosh-Dastidar, S., Dadmehr, N.: Alzheimer’s disease and models of computation: imaging, classification, and neural models. J. Alzheimers Dis. 7(3), 187–199 (2005)
Zheng, C., Xia, Y., Pan, Y., et al.: Automated identification of dementia using medical imaging: a survey from a pattern classification perspective. Brain Inform. 3(1), 17–27 (2016)
Pagani, M., Kovalev, V.A., Lundqvist, R., et al.: A new approach for improving diagnostic accuracy in Alzheimer’s disease and frontal lobe dementia utilising the intrinsic properties of the SPET dataset. Eur. J. Nucl. Med. Mol. Imaging 30(11), 1481–1488 (2003)
Nagao, M., Sugawara, Y., Ikeda, M., et al.: Heterogeneity of cerebral blood flow in frontotemporal lobar degeneration and Alzheimer’s disease. Eur. J. Nucl. Med. Mol. Imaging 31(2), 162–168 (2004)
Suk, H., Shen, D.: Deep learning-based feature representation for AD/MCI classification. In: Mori, K., Sakuma, I., Sato, Y., Barillot, C., Navab, N. (eds.) MICCAI 2013. LNCS, vol. 8150, pp. 583–590. Springer, Heidelberg (2013). https://doi.org/10.1007/978-3-642-40763-5_72
Suk, H., Shen, D.: Deep ensemble sparse regression network for Alzheimer’s disease diagnosis. In: Wang, L., Adeli, E., Wang, Q., Shi, Y., Suk, H. (eds.) MLMI 2016. LNCS, vol. 10019, pp. 113–121. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-47157-0_14
Ortiz, A., Munilla, J., Jorge, et al.: Ensembles of deep learning architectures for the early diagnosis of the Alzheimer’s disease. Int. J. Neural Syst. 26(7), 1650025 (2016)
Valliani, A., Soni, A.: Deep residual nets for improved Alzheimer’s diagnosis. In: Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics 2017, pp. 615–615 (2017)
ADNI/PET Pre-processing. http://adni.loni.usc.edu/methods/pet-analysis/pre-processing. Accessed 05 Feb 2016
Tzouriomazoyer, N., Landeau, B., Papathanassiou, D., et al.: Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI Single-Subject Brain. NeuroImage 15(1), 273–289 (2002)
Frackowiak, R.S.J.: Human brain function (2004)
Hu, J., Lu, J., Tan, Y.P., et al.: Deep transfer metric learning. IEEE Trans. Image Process. 25(12), 5576–5588 (2016)
Oquab, M., Bottou, L., Laptev, I., et al.: Learning and transferring mid-level image representations using convolutional neural networks. In: IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2014, pp. 1717–1724. IEEE (2014)
Karwath, A., Hubrich, M., Kramer, S.: Convolutional neural networks for the identification of regions of interest in PET scans: a study of representation learning for diagnosing Alzheimer’s disease. In: ten Teije, A., Popow, C., Holmes, J.H., Sacchi, L. (eds.) AIME 2017. LNCS (LNAI), vol. 10259, pp. 316–321. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-59758-4_36
Valliani, A., Soni, A.: Deep residual nets for improved Alzheimer’s diagnosis. In: ACM International Conference on Bioinformatics, Computational Biology and Health Informatics 2017, pp. 615–615 (2017)
Vu, T.D., Yang, H.J., Nguyen, V.Q., et al.: Multimodal learning using convolution neural network and Sparse Autoencoder. In: IEEE International Conference on Big Data and Smart Computing (BigComp) 2017, pp. 309–312. IEEE (2017)
Liu, S., Liu, S., Cai, W., et al.: Early diagnosis of Alzheimer’s disease with deep learning. In: 11th International Symposium on Biomedical Imaging (ISBI) 2014. IEEE (2014)
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China under Grants 61471297 and 61771397, in part by the China Postdoctoral Science Foundation under Grant 2017M623245, in part by the Fundamental Research Funds for the Central Universities under Grant 3102018zy031, and in part by the Australian Research Council (ARC) Grants.
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Zheng, C., Xia, Y., Chen, Y., Yin, X., Zhang, Y. (2018). Early Diagnosis of Alzheimer’s Disease by Ensemble Deep Learning Using FDG-PET. In: Peng, Y., Yu, K., Lu, J., Jiang, X. (eds) Intelligence Science and Big Data Engineering. IScIDE 2018. Lecture Notes in Computer Science(), vol 11266. Springer, Cham. https://doi.org/10.1007/978-3-030-02698-1_53
Download citation
DOI: https://doi.org/10.1007/978-3-030-02698-1_53
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-02697-4
Online ISBN: 978-3-030-02698-1
eBook Packages: Computer ScienceComputer Science (R0)