Abstract
Spermatogenesis represents a complex succession of cell division and differentiation events resulting in the continuous formation of spermatozoa. Such a complex program requires precise expression of enzymes and structural proteins which is effected not only by regulation of gene transcription and translation, but also by targeted protein degradation. In this chapter, we review current knowledge about the role of the ubiquitin–proteasome system in spermatogenesis, describing both proteolytic and non-proteolytic functions of ubiquitination. Ubiquitination plays essential roles in the establishment of both spermatogonial stem cells and differentiating spermatogonia from gonocytes. It also plays critical roles in several key processes during meiosis such as genetic recombination and sex chromosome silencing. Finally, in spermiogenesis, we summarize current knowledge of the role of the ubiquitin–proteasome system in nucleosome removal and establishment of key structures in the mature spermatid. Many mechanisms remain to be precisely defined, but present knowledge indicates that research in this area has significant potential to translate into benefits that will address problems in both human and animal reproduction.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Ubiquitin
- Proteasome
- Deubiquitinating enzymes
- Gonocytes
- Spermatogonia
- Histones
- Ubiquitinated histones
- Meiotic sex chromosome inactivation
- Recombination
- Nucleosome
- Acrosome
General Features and Functions of the Ubiquitin–Proteasome System (UPS)
Ubiquitination is the process by which ubiquitin, an 8.5 kDa peptide highly conserved throughout evolution, becomes covalently linked to its target proteins (rev. in [1]). This linkage usually occurs between the carboxy-terminal end of the ubiquitin peptide and the ε-amino group of the side chain of a lysine residue on the protein substrate. Rarely, the ubiquitin is attached to the α-amino group at the N-terminus of the protein [2, 3] or the thiol group of a cysteine residue [4]. Often a chain of ubiquitin moieties is attached to the substrate protein in which each distal moiety of the chain is linked to one of the seven lysine residues found in the more proximal ubiquitin moiety. The ubiquitin chains, particularly when the number of ubiquitin moieties is greater than four and linked via the lysine 48 residue, are recognized by the 26S proteasome complex, resulting in the degradation of the protein substrate [5]. The 26S proteasome consists of a 20S catalytic core particle bound at one or both ends by a 19S cap particle [6]. The 20S core has a cylindrical structure formed by the stacking of four rings, each composed of seven distinct subunits [7]. The two outer rings consist of subunits α1–7 and the two inner rings consist of subunits β1–7. These latter subunits contain the active sites of the protease. The 19S caps cover the ends of the 20S cylindrical core particle, thereby acting as gates to prevent uncontrolled proteolysis. The 19S particles contain subunits that recognize and bind ubiquitin chains or adaptor proteins which in turn bind ubiquitinated proteins, targeting them to the proteasome (rev. in [8]). The 19S particles also contain deubiquitinating enzymes that remove the ubiquitin from the targeted protein to recycle the ubiquitin for new conjugation (rev. in [9]). Subunits with ATPase activity are present and likely involved in unfolding the protein, opening of the gate of the 20S core particle, and subsequent translocation of the substrate into the channel for hydrolysis. The 20S core particle can also interact with three other regulators. PA28α/β is a heptameric complex induced by interferon, which can bind to the ends of the core particle and is involved in adaptive immunity [10]. PA28γ is a structurally related protein whose function remains unclear. PA200 is a monomeric protein (rev. in [11]) that can also bind to the 20S core and has been suggested to play roles in DNA repair, mitochondrial inheritance and, as will be reviewed here, in spermatogenesis.
The UPS has numerous substrates—perhaps as many as half of all cellular proteins [12]. Many of these proteins are regulatory proteins and include transcription factors such as Myc [13], cell cycle proteins such as p27 [14], cyclins [15], and signaling pathway molecules such as IκBα [16, 17]. By regulating the cellular levels of proteins, the UPS is able to modulate a variety of physiological processes in the cell (Fig. 9.1). In addition, ubiquitination can serve nonproteolytic functions, particularly when the target proteins are monoubiquitinated or ubiquitinated by chains formed by non-Lys 48 linkages (rev. in [18]). In these situations, the ubiquitination can alter the structure and function of the target protein or serve to recruit other proteins. For example, ubiquitination of plasma membrane proteins can target them for internalization and trafficking through the endocytic pathway to multivesicular bodies, for subsequent degradation in lysosomes (rev. in [19]). Ubiquitination can also result in the activation of signaling pathways, as seen downstream of the inflammatory cytokine receptors for TNFα or IL-1, in which the ubiquitination of RIP1 or TRAF6 respectively results in the activation of NFκB (rev. in [20]). As detailed later in this chapter, monoubiquitination of histones can result in recruitment of other proteins/enzymes that mediate other chromatin modifications.
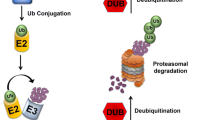
Outline of the ubiquitin–proteasome system. Ubiquitination is the process by which the peptide ubiquitin (Ub) is conjugated at its C-terminal end to the side chain of a lysine residue in the target protein. Proteins can be either monoubiquitinated or conjugated to polyubiquitin chains formed by the linkage of the more distal Ub moiety to a lysine residue in the more proximal Ub moiety. Ub contains seven lysine residues allowing different types of chains to be formed depending on which lysine is used to form the chain. Classically, chains formed using lysine 48 on Ub target the attached protein for recognition and degradation by the 26S proteasome. Proteins conjugated with chains formed using other lysines can mediate non-proteolytic functions of ubiquitination such as DNA repair or signal transduction. Monoubiquitination of proteins generally does not lead to degradation, but is involved in processes such as trafficking of plasma membrane proteins through the endocytic pathway or regulation of gene transcription (through monoubiquitination of histones). Recent studies indicate, though, that these distinctions in functions depending on type of ubiquitination are not absolute. Ubiquitination can also be reversed through the action of deubiquitinating enzymes
Conjugation of ubiquitin is mediated by a sophisticated three-step enzyme cascade (Fig. 9.2) [21, 22]. In the first step, the E1 ubiquitin activating enzyme activates ubiquitin in an ATP-dependent manner and charges it onto the cysteine residue of the active site of an E2 ubiquitin conjugating enzyme. The E2 then conjugates the ubiquitin to protein substrates in concert with a third protein, the E3 ubiquitin ligase that is essential for binding the substrate. There are two major classes of E3s, namely the RING (Really Interesting New Gene) and HECT (Homologous to E6-AP Carboxy-Terminus—named after E6AP, the first E3 described in this class) that can be recognized by specific conserved protein domains/motifs. In addition, there are U-box containing E3s presenting a distinct sequence motif, but whose tertiary structure is similar to a RING domain. There are two E1s in mammalian cells—Uba1, Uba6—with the former being the dominant E1 activity in most cell types. Approximately 30 genes encode E2s, each of which can interact with a subset of the approximately 800 E3s in the mammalian genome.
Mechanism of action of enzymes involved in conjugating ubiquitin (Ub) to proteins. (a) Ub is first activated by Ub activating enzyme (E1) in a reaction that involves ATP hydrolysis and results in the formation of a high energy thiolester linkage between Ub and the active site cysteine residue. The E1 then transfers the activated Ub onto the active site cysteine residue of one of a family of Ub conjugating enzymes (E2s or UBCs). The E2 then conjugates Ub to protein substrates in concert with a third protein, Ub protein ligase (E3) that plays the critical role of binding the substrate. (b) Ub activating enzyme (E1, 2 genes in mammals) supplies activated Ub to a family of Ub conjugating enzymes (E2s, ~30 genes in mammals). Each E2 can interact with a subset of Ub ligases (E3s, ~800 in mammals) to mediate conjugation of Ub to specific protein substrates
Protein ubiquitination may be reversed by deubiquitinating enzymes. These enzymes number approximately 100 and can be divided into five distinct classes, based on sequence conservation [23]. Four of these classes—USP (Ubiquitin Specific Proteases, largest class with ~60 genes), Otubains, UCH (Ubiquitin C-terminal Hydrolases), and MJD (Machado Joseph Disease) enzymes - are cysteine proteases. The fifth class contains the JAMM (JAB1/MPN/Mov34) domain containing proteins which are metalloproteinases. In addition to reversing the effects of ubiquitination, these enzymes also function to recycle ubiquitin after their protein targets have been committed to degradation by the proteasome or within the lysosome (Fig. 9.3). There are four genes that encode ubiquitin [24]. Two are polyubiquitin genes which encode linear polymers of multiple copies of ubiquitin and the other two encode a single copy of ubiquitin fused at its carboxy terminal end to a ribosomal subunit. Deubiquitinating enzymes are responsible for processing the products of these four genes into the monomeric proteins and this appears to occur co-translationally.
The large families of enzymes involved in ubiquitination allow for highly selective and precisely regulated ubiquitination of target proteins. The number of potential cellular functions regulated by ubiquitination is extensive. In this chapter, we will review the current knowledge about some of the functions of the UPS in spermatogenesis, highlighting particular examples that have been well defined, as well as identifying important gaps in our current knowledge.
UPS: Essential for Gonocyte and Spermatogonial Development
Spermatogenesis represents a complex succession of cell division and differentiation events resulting in the continuous formation of spermatozoa. This process relies on a reservoir of spermatogonial stem cells (SSCs) that can undergo self-renewal or differentiation [25, 26]. The SSCs arise from precursor cells known as gonocytes (also called pre- or pro-spermatogonia), 1 week after birth in rodents, and a few weeks after birth in humans [27]. Neonatal gonocyte development involves three main processes: proliferation, migration, and differentiation [27]. Proliferation is regulated by platelet-derived growth factor (PDGF) and 17β-estradiol [28, 29], while differentiation is induced by retinoic acid [30]. During these processes, a major reorganization of cellular structure takes place [27].
The role of the UPS in cell remodeling and protein turnover in gonocytes was unknown until recently. Isolated rat gonocytes can be induced, upon exposure to retinoic acid, to express the differentiation marker Stra8 (stimulated by retinoic acid gene 8) [30] which is expressed in differentiating spermatogonia and pre-meiotic germ cells [31–33]. This retinoic acid dependent induction can be blocked by the presence of the proteasome inhibitors lactacystin or bortezomib, indicating that proteasome activity is necessary for gonocyte differentiation [34].
Gene expression analysis using microarrays identified 91 UPS genes that are significantly expressed in post natal day (PND) 3 rat gonocytes. Of these, at least five genes have a higher expression level in PND3 gonocytes compared to PND8 spermatogonia, suggesting that down-regulation of these genes may be involved in this developmental transition. These genes include ubiquitin activating enzymes Uba1 and Uba6, and ubiquitin ligases Huwe1, Trim47, and Rnf149. Uba1 has previously been shown to be expressed in adult germ cells and is needed for both capacitation and fertilization [35]. Uba6 was suggested to play a role in the transition between mitosis to meiosis [36, 37]. Huwe1 is highly expressed in gonocytes, spermatogonia and spermatocytes [38, 39]. Interestingly, it translocates from the cytoplasm to the nucleus during the gonocyte to spermatogonia transition [34]. We have recently inactivated the gene specifically in germ cells and our preliminary results demonstrate a defect in SSC renewal and differentiation confirming an important role for Huwe1 in this early stage of spermatogenesis. No literature on the function of Trim47 is available, while a recent study showed that Rnf149 is the ubiquitin ligase for the serine/threonine kinase BRAF [40]. When these five genes were analyzed in gonocytes treated with or without retinoic acid, only Rnf149 was significantly decreased upon retinoic acid treatment. This indicates that Rnf149 is actively down-regulated during both in vitro and in vivo differentiation.
There are other genes highly expressed in gonocytes that may also be involved in differentiation based on their functions in other systems. Among these is the conjugating enzyme Ube2e3 which is essential for epithelial cell proliferation and is down-regulated during differentiation [41]. Other UPS genes abundant in gonocytes and spermatogonia include the ubiquitin ligases Mdm2, Huwe1 and Ubr5, reported to interact with p53 in other cell types [34]. These genes could potentially regulate cell cycle progression in proliferating gonocytes or spermatogonia.
The SCF βTrCP ubiquitin ligase is a well characterized ubiquitin ligase that plays important roles in regulating cell cycle and apoptosis. There are two isoforms of the βTrCP substrate recognition subunit, βTrCP1 and βTrCP2, both of which are expressed in spermatogonia. Gene inactivation of βTrCP1 has no effect on spermatogonial development but insufficiency of both isoforms results in disrupted organization of these cells due to loss of adherens junctions and E-cadherin. This effect appears to be due to stabilization of a single βTrCP substrate Snail1, a transcription factor that induces E-cadherin expression, as silencing Snail1 in the βTrCP deficient testis was able to reverse the defect in spermatogenesis [42].
The deubiquitinating enzyme Uchl1 (alt. name PGP9.5) is highly expressed in spermatogonia. It has been proposed that spermatogonia undergo both symmetrical and asymmetrical division to maintain the stem cell pool and differentiation of progeny [43]. Interestingly, Uchl1 protein segregates asymmetrically to the two daughter cells in asymmetric division. The cells that inherit a high level of Uchl1 express the undifferentiated spermatogonia marker Plzf, whereas those with a low level of Uchl1 express the differentiated spermatogonia marker c-Kit. This suggests that Uchl1 may be an intrinsic determinant for spermatogonial self-renewal or differentiation [44]. P97/VCP, a chaperonin that can bind ubiquitinated proteins and target them to the UPS, may through this function act as a regulator of signaling pathways such as the BMP pathway in gonocytes and spermatogonia in neonatal testes [45, 46]. Further analysis of these and other relevant genes using loss of function approaches in the testis is required to confirm the roles of these proteins. Specific functions of the UPS in this early mitotic stage of spermatogenesis will likely be found in view of the known importance of the system in proliferation of other cell types.
UPS: An Important Regulator of Meiosis
Since the UPS has a critical role in mitotic division (rev. in [47, 48]), it was anticipated that it would also be involved in the meiotic division leading to the development of haploid round spermatids during spermatogenesis. Indeed, over the past decade, considerable work has been done to elucidate the role of the UPS in meiosis, which is marked by highly regulated events such as meiotic entry, genetic recombination of homologous chromosomes, meiotic sex chromosome inactivation (MSCI), and meiotic exit. Defects in these important events often lead to developmental arrest in germ cells and consequent infertility. In this section, we will describe a few general observations regarding the importance of ubiquitin and the proteasome, and then highlight examples that demonstrate the highly efficient and selective ability of the UPS to tightly regulate some of these processes, thereby promoting adequate production of normal spermatozoa.
The importance of the UPS during meiosis is illustrated by defects observed upon depletion of ubiquitin or impairment of the proteasome. For example, disruption of the polyubiquitin gene Ubb in mice leads to infertility with a germ cell arrest at the prophase stage of meiosis I [49]. Ubb is one of the two polyubiquitin genes and is therefore important for de novo synthesis of ubiquitin. Inactivation of Ubb results in a significant decrease in free ubiquitin in the testis, which, associated with the meiotic defect, implies that insufficient levels of ubiquitin in the mutant testis disrupt the ubiquitination required during meiosis. The spermatocytes in the ubb null mice are marked by the absence of discrete foci of γH2AX, a histone modification normally associated with the transcriptionally silenced XY body in mid-pachytene spermatocytes. This absence of γH2AX foci could be due to either a delay in the progression of the pachynema in mutant spermatocytes or to a delay in the formation of the XY body. Despite decreased ubiquitin levels in mutant testes, histone H2A was still ubiquitinated (uH2A) and the uH2A remained associated with the XY body, but the levels of uH2A were about 50 % lower than those found in normal testes [49, 50]. The uH2A has been suggested to be essential for maintaining silencing of the unsynapsed chromosomes. Microarray analysis of the testicular transcriptome did indeed reveal many more upregulated genes than downregulated in the mutant testes. There was abnormal expression of many testis-specific/meiosis-specific genes which may explain in part the meiotic defect seen in these mice [50].
The requirement for ubiquitin in meiosis appears conserved in eukaryotic evolution. Mutants of the polyubiquitin gene Ubi4(+) in S. pombe abort meiosis at the first or second division and are characterized by short, condensed non-separated chromosomes, abnormal spindle and disintegrated spindle pole bodies. Ubi4(+) mRNA is normally strongly induced prior to meiosis, with a slight increase in protein levels. However, in these mutants, there is a significant decrease in ubiquitin levels when the cells enter meiosis, suggesting that this impaired availability of ubiquitin is the cause of the aborted meiosis [51].
With respect to the proteasome, silencing of two subunits of the catalytic core of the proteasome PBS-4 and PAS-5, in C. elegans, results in impaired entry into meiosis, suggesting that the UPS regulates the switch from proliferation to meiosis by degrading proteins necessary for proliferation [52]. In mice, inactivation of a proteasome activator, PA200, which is highly expressed in the testis, results in sub-fertility and significant histological defects in spermatocytes, including a high level of apoptosis. Given that both the proteasome and PA200 have previously been suggested to regulate repair of DNA double strand breaks (DSB) [53, 54], this study hinted at a role for the proteasome in the repair of DSBs during meiosis, possibly by regulating the metabolism of repair proteins or histones [55].
UPS and Meiotic Recombination
Meiotic recombination, one of the key steps in the prophase of the first meiotic division, ensures efficient exchange of genetic material supporting the production of germ cells with genetic variability, whilst maintaining genomic integrity. During this time, double strand breaks (DSBs), which are a prerequisite for homologous recombination, are introduced in a controlled manner. A role of the UPS in this process has been best elucidated in the yeast S. cerevisiae in which mutations of the ubiquitin conjugating enzyme Rad6 (Ubc2) result in defective sporulation and DNA repair [56–59]. Mutation of the Rad6 gene leads to meiotic prophase arrest, with only 10-20 % of the rad6 mutant cells entering meiosis I. These mutants are characterized by an overall decrease in the frequency of DSBs in the cell, which could be explained by a delay in their entry into the pre-meiotic S-phase. Rad6 interacts with the Bre1 ubiquitin ligase and together, they mediate ubiquitination of histone H2B. Disruption of Bre1 or substitution of the lysine residue of H2B that is ubiquitinated, also leads to a reduction in DSB formation during meiosis, indicating that these effects are due to Rad6/Bre1 mediated ubiquitination of H2B. The uH2B may be required for recruitment of the DSB-forming machinery to DSB hotspots [59].
Roles for these enzymes in meiotic recombination appear to be conserved in the mammalian testis. The ubiquitin conjugating enzymes Hr6a and Hr6b are mammalian orthologs of Rad6/Ubc2. Hr6b knockout mice showed increased apoptosis of primary spermatocytes in the first wave of spermatogenesis, with longer synaptonemal complexes in the pachytene spermatocyte nuclei, depletion of synaptonemal complex proteins from near telomeric regions and increased foci containing the mismatch DNA repair protein MLH1, indicating a high crossing-over frequency [60, 61]. The knockout mice, however, have no defect in overall ubiquitinated H2A levels, indicating the existence of other ubiquitin-conjugating enzymes that can perform this function.
The Hr6a/Hr6b ubiquitin conjugating enzymes interact with a number of E3s, which may be involved in their functions in meiotic recombination. In particular, they interact with the Ubr family of ligases which function in the N-end rule pathway (rev. in [62]). This pathway was initially described as a set of enzymes that are involved in recognizing ubiquitination substrates by their N-terminal amino acid. Loss of the Ubr2 ligase leads to infertility in males with degeneration of the postnatal testis beyond 3 weeks. This is due to an arrest at the leptotene/zygotene and pachytene stages of spermatocytes, which are conspicuous by their lack of intact synaptonemal complexes and eventually die through apoptosis [63]. This phenotype could be explained by a role of Ubr2 in the maintenance of genomic integrity and homologous recombination repair of double strand breaks, as was observed in mouse fibroblast cells derived from Ubr2−/− embryos [64]. The exact substrates of Ubr2 that mediate these effects are unknown. Ubr2 has been shown to interact with TEX19.1, a germ cell-specific protein. However, TEX19.1 does not appear to be a substrate of Ubr2 as it is unexpectedly destabilized in Ubr2 knockout mice and the loss of TEX19.1 phenocopies loss of Ubr2. Thus, the role of Ubr2 may be to stabilize TEX19.1 by preventing the ability of other ubiquitin ligases to interact and ubiquitinate this protein [65]. Clinically, single nucleotide polymorphisms in the Ubr2 gene have been associated with azoospermia in men who presented meiotic stage arrest in spermatogenesis, suggesting a molecular basis for the infertility in this type of non-obstructive azoospermia [66].
Another ubiquitin ligase that has an essential role during homologous recombination is Rnf4. Rnf4 ubiquitinates specifically the SUMO-modified proteins. Inactivation of Rnf4 results in the persistence of DNA damage signaling following an ionizing radiation insult. Mechanistically, Rnf4 ubiquitinates SUMO-modified MDC1 (Mediator of DNA damage checkpoint protein 1) and SUMO-modified BRCA1 (Breast cancer type I susceptibility protein, involved in DNA damage response), both events being required for the loading of RAD51, a component of the homologous recombination repair machinery [67]. Mice lacking Rnf4 have defective spermatogenesis characterized by increased apoptosis and depletion of spermatocytes [67]. Mouse mutants of a putative ubiquitin ligase Mei4, a mouse ortholog of Human Enhancer of Invasion 10 (Hei10), show defective assembly of chromosomes at the metaphase plate during meiosis I, leading to arrest and apoptosis [68]. Late meiotic prophase in these germ cells is marked by the absence of cyclin-dependent kinase 2 and mismatch repair proteins at the chromosome cores, correlating with immature chromosome separation due to lack of chiasmata formation.
UPS and Meiotic Sex Chromosome Inactivation
During the pachytene stage of prophase I, the X and Y chromosomes are partially synapsed at their pseudoautosomal regions, and undergo transcriptional silencing, a process called meiotic sex chromosome inactivation (MSCI), which is vital for the progression of meiosis and production of spermatozoa. The sex body containing the heterochromatic X and Y chromosomes is enriched in ubiquitinated H2A and levels normally peak in pachytene spermatocytes [69]. Ubiquitination of H2A has been linked to gene repression in both Drosophila and mammals [70, 71] and therefore may be essential for MSCI. Supporting this idea, mice deficient in the previously mentioned ubiquitin ligase Ubr2 are also characterized by a failure in H2A ubiquitination and impaired chromosome-wide silencing of genes linked to unsynapsed X and Y chromosomes. Ubr2 interacts with the ubiquitin conjugating enzyme Hr6b, which can conjugate ubiquitin to histone H2A in vitro, suggesting that these enzymes may be responsible for generating the uH2A in the pachytene spermatocytes. The lack of MSCI probably activates a pachytene checkpoint system and results in a consequent arrest at meiotic prophase I [72, 73]. MSCI is also marked by decreased H3K4 dimethylation which results in persistent gene silencing in round spermatids. Mediators of this process appear to be the Hr6b conjugating enzyme along with another of its interacting ubiquitin ligases, Rad18. Loss of function of either Hr6b or Rad18 leads to increased level of H3K4 dimethylation on the XY body and the whole nucleus at the diplotene stage, an indication of derepression of silenced genes [74]. Rad18 has previously been shown to be an orchestrator of homologous recombination repair after DNA damage by binding to the recombinase RAD51C [75]. Silencing Rad18 in mice leads to subfertility, and reduced body weight and testis size. The mice show increased asynapsis of the X and Y-chromosomes in the pachytene spermatocytes, as well as increased dimethylation of H3K4 and corresponding derepression of X-linked genes that are normally silenced [74].
The importance of uH2A for MSCI has been questioned by findings in mice deficient in the ubiquitin ligase Rnf8. This ubiquitin ligase has multiple roles including involvement in DSB repair. Rnf8 recognizes ATM–mediated phosphorylated MDC1 bound to γ-H2AX, which allows histone ubiquitination and the ubiquitin-dependent recruitment of 53BP1 and Brca1, helping in the repair of DSB [76–79]. Another ubiquitin ligase, Rnf168 may act in concert with Rnf8 to propagate the ubiquitination signal at the DSBs [80]. Mice lacking Rnf8 are sterile and present defective ubiquitination of the XY body in pachytene spermatocytes [81–83]. However, MSCI occurred normally indicating that uH2A on the XY body is not essential for this silencing [83]. More recently, it has been shown that Rnf8 dependent ubiquitination of H2A is instead essential for the establishment of H3K4 dimethylation which is required for escape gene activation in post meiotic stages of spermatogenesis [81, 84].
Other components of the UPS have been reported to associate with the sex chromosomes during this phase of meiosis, but their functions are still unclear. The ubiquitin ligase Ret finger protein, a transcriptional repressor that is able to interact with nuclear matrix associated proteins and double stranded DNA, plays a possible role in the positioning and attachment of the XY body to the nuclear lamina next to the nuclear membrane in pachytene spermatocytes [85]. This seclusion of the XY body from the rest of the chromosomes is critical to handle the asynchronous synapsis of the sex chromosomes post homologous recombination during meiosis (rev. in [86, 87]).
UPS and Meiotic Progression
There are a number of UPS genes that are required at various time-points of meiotic progression such as during the transition from mitosis to meiosis, the preparatory stage for homologous recombination, transitions from metaphase to anaphase during meiosis I, chromosome segregation, transitions during meiosis II and the exit from meiosis. The anaphase promoting complex or cyclosome (APC/C) is a multisubunit ubiquitin ligase that targets various proteins for degradation, notably securin, to facilitate chromosome segregation during mitosis (rev. in [88, 89]). The APC/C has also been demonstrated to play a role in meiosis and this function is conserved amongst several organisms. Null mutations in the APC5 subunit in yeast [90–92] and loss of function of APC/C subunits in C. elegans embryos cause an arrest at metaphase of meiosis I [93–95]. However, the role of the APC/C in male meiosis of mammals is yet to be explored. Moreover, in C. elegans, a temperature sensitive mutation of the ubiquitin ligase Cul2 results in inhibition of the first steps of meiotic prophase I progression because of the accumulation of HTP-3, a member of the HORMA (Hop1–Rev1–Mad2) domain-containing proteins, required for preparing chromosomes for meiosis. The protein is needed for loading of cohesion REC8 and components of the synaptonemal complex, mainly SYP-1 and HIM-3. In the absence of Cul2, there is premature binding of the synaptonemal complex members, probably leading to the activation of a DNA replication checkpoint [96].
Furthermore, there are a few mutation studies in higher organisms that indicate the importance of the UPS in other aspects of meiotic progression. Among these, gene targeting of the ubiquitin ligase Cullin4A in mice leads to increased accumulation of DNA licensing protein CDT1, phospho-p53 and MLH1 with disrupted meiosis II and increased cell death in pachytene–diplotene cells [97]. Inactivation of another murine RING ubiquitin ligase, Siah1a was found to cause abnormal meiotic division in spermatocytes, with an increased accumulation and then consequent degeneration of the metaphase and anaphase cells. There were no post-meiotic round spermatids and the cells that progressed beyond metaphase were binucleated/multinucleated, indicating defective chromosome segregation. The accumulation and death of the spermatocytes from the metaphase I to telophase I transition may be due to the accumulation of KID, an anaphase I inhibitor protein and a substrate for Siah1A [98]. Besides its role in spermatogonial development, the β-TrCP ligase is also involved in meiosis. The β-TrCP1 mRNA is upregulated in spermatocytes and indeed, mice lacking this isoform are subfertile with marked decreased production of spermatids and increased number of metaphase I spermatocytes consistent with a defect in meiosis. The mutant testis also showed increased levels of the cell cycle regulators and β-TrCP substrates, Emi1 and cyclin A, providing a possible molecular mechanism for this defect [99]. Rat100/EDD/HYD/UBR5, a UBC-4 dependent HECT ubiquitin ligase, is induced during postnatal development, peaking around PND 25 in rats. It is highly expressed in spermatocytes with low expression in the round and elongating spermatids [100]. It is a homolog of the Drosophila hyperplastic discs gene, point mutants of which cause male infertility due to a lack of progression of the germ cells past the primary spermatocyte stage [101].
Ubiquitin C-terminal hydrolases or UCHs are believed to be essential for maintaining ubiquitination activity by releasing ubiquitin from its substrates. Overexpressing Uchl1 in the testis of male mice results in sterility due to blockage of spermatogenesis at the pachytene stage of spermatocytes and increased apoptosis in these cells. PCNA (proliferating cell nuclear antigen) was strongly expressed in the primary spermatocytes with little expression in spermatogonia and Sertoli cells [102]. Knocking out Uchl1 led to an increased number of premeiotic germ cells between PND 7 and 14, concomitant with increased levels of apoptotic proteins TRP53, Bax, and caspase-3 [103]. A study looking at the profiles of genes exhibiting expression patterns similar to that of retinoic acid target gene Stra8, an initiator of meiosis known to peak at PND 10, identified ubiquitin-activating enzyme Uba6 [37]. Uba6 transcript levels are highest in the human and mouse testis compared to other tissues [36] with the protein expression pattern coinciding with its mRNA expression. Besides being expressed in neonatal gonocytes [34, 37], it is highly expressed in the cytoplasm of spermatogonia and preleptotene spermatocytes at PND 10, with localization shifting to the nuclei of preleptotene, leptotene and zygotene spermatocytes in PND 20 mice. These observations suggest a possible role of Uba6 in meiotic initiation [37].
It is evident from the aforementioned studies that the UPS is indispensible for the progression of major meiotic events during spermatogenesis. Collectively, several loss-of function studies clearly associated the absence of UPS genes with profound defects in homologous recombination, meiotic sex chromosome inactivation and metaphase I-anaphase I transitions. Ubiquitination of histones appears to be particularly important in mediating some of the key events in homologous recombination and regulation of gene expression from the sex chromosomes. As an important regulator of these events, the UPS ensures accurate exchange of genetic information between homologous chromosomes, chromosome assembly/segregation and expression/silencing of genes thus setting the foundation for the germ cells to undergo successful transition through the last stage of spermatogenesis.
UPS: A Modulator of Spermiogenesis
The haploid round spermatids derived at the end of meiosis II undergo extensive nuclear and morphological remodeling, leading to the formation of elongated, compact, transcriptionally quiescent and highly specialized spermatozoa [104]. This remodeling is mediated by a wide array of highly regulated molecular events. The UPS appears to play important roles during spermiogenesis by removing proteins that are no longer needed in the more differentiated spermatids, as well as in modulating the regulatory mechanisms that control these processes.
A role for the UPS in spermiogenesis was suggested early on by observations that levels of ubiquitinated proteins are markedly increased during the first wave of spermatogenesis in the rat at times when haploid spermatids first appear and are becoming the prominent germ cell in the testis. This is likely due to an activation of ubiquitin conjugating activity, which is dependent mostly on the Ubc4 family of ubiquitin conjugating enzymes whose levels are dramatically increased in spermatids [105]. Proteasome activity is also high in elongating spermatids as well as in mature spermatozoa [105, 106].
Studies in Drosophila support a role for the UPS during spermiogenesis. In D. melanogaster, 12 of the 33 subunits of the 26S proteasome are represented by multiple paralogous genes [107–109], and in each case, one of the paralogs is testis-specific [110]. Analyses of the testis specific α3T, α6T and Rpt3R subunits revealed that their expression is induced at different stages, but they are prominent in spermatids [110]. The functional significance of α6T and Rpt3R in spermiogenesis is evident from their inactivation, which results in a male sterile phenotype [108, 110]. Knocking out α6T results in defective actin cone movement during sperm individualization (terminal differentiation), and abnormal nuclear morphology and maturation, the first defect being either due to improper degradation of important components of the actin cone movement machinery or due to the defective regulation of the caspase activation which is essential for sperm individualization [108]. In addition, two testis-specific subunits of the 20S core proteasome, Pros28.IA and Pros28.1B are expressed during spermatid differentiation [107] suggestive of proteasomal functions that are unique to spermiogenesis.
These results demonstrate a general implication of ubiquitination and proteasome mediated degradation during spermiogenesis, and the conservation of this function across species. In the following sections, we will highlight examples that illustrate how specific components of the UPS play essential roles in spermiogenesis in a carefully regulated and selective manner.
UPS Genes and Nucleosome Remodeling
The round spermatid nuclei initially house a transcriptionally active genome consisting of DNA and histones. The nucleosome, the basic subunit of chromatin, is formed by a 147 bp segment of DNA wrapped around a histone octamer core consisting of two copies of histones H2A, H2B, H3 and H4 [111]. During spermiogenesis, these histones are replaced initially by transition proteins 1 and 2, and then finally by protamines 1 and 2 that give rise to a more compact and hence transcriptionally silent genome [82, 112, 113]. Although the exact reason behind this replacement is not clear yet, data suggests that this is necessary for DNA condensation and packaging into the sperm head nucleus and to protect the DNA cargo from damage. Indeed, aberrations in the process are associated with male infertility [113–117]. Ubiquitinated H2A as well as [69] polyubiquitinated forms of H3, TH3 (testis-specific H3) and H2B have been observed in the elongating spermatids, suggesting that this post-translational modification might be essential for loosening up the chromatin for accurate nucleosome removal, or may simply target these histones to the proteasome [118]. In Drosophila, histones are lost prior to individualization and this is preceded by H4 hyperacetylation and H2A monoubiquitination. The spermatid nuclei are also marked by high levels of ubiquitin, UbcD6 (drosophila homolog of yeast Rad6 and mammalian Hr6a and Hr6b), SUMO and DNA strand breaks [119].
Studies using mice deficient in the RING ubiquitin ligase Rnf8 [82] and the PA200 activator of the proteasome [120] have provided insights into the roles of uH2A and H4 acetylation in nucleosome removal. Both of these mutant mice show defective removal of nucleosomes in spermatids. As described in the previous section, Rnf8 mutant testes are deficient in sex body associated uH2A in meiosis. Rnf8 dependent ubiquitination of H2A was shown to recruit MOF, an acetyltransferase required for H4K16 acetylation [82]. The PA200 activator of the proteasome can recognize and bind this acetylated histone, leading to its degradation in the 20S core particle [120]. Together, these studies provide a model for nucleosome removal. However, it should be pointed out that other investigators did not find any changes in H4K16 acetylation levels or in histone–protamine exchange in Rnf8 inactivated mice [84]. Instead, they observed that Rnf8-dependent H2A ubiquitination was shown to be important for the establishment of active epigenetic modifications on the sex chromosomes during meiosis such as dimethylation of H3K4. This in turn, led to the establishment of other active epigenetic marks on the XY body in post-meiotic spermatids such as trimethylation of H3K4, histone lysine crotonylation, and incorporation of the histone variant H2AFZ. These modifications regulate escape gene activation and hence epigenetic programming in the sperm.
UPS and Acrosome Biogenesis
One of the critical structures that are formed during spermiogenesis is the acrosome, which is believed to arise from the fusion of Golgi-derived vesicles. Ubiquitinated proteins have been detected in every step of acrosome formation during rat spermiogenesis [121]. Using electron microscopy, ubiquitin signals were shown to exist in the matrix of transport vesicles between the acrosome and the Golgi apparatus, with strong expression in different regions of the developing acrosome after vesicle enlargement and fusion with the nucleus. Additionally, there are a number of UPS enzymes that have been functionally linked to sperm morphogenesis. TMF/ARA160, a Golgi associated protein that was shown to have ubiquitin ligase activity, is detected in the Golgi of spermatocytes and spermatids but disappears in the mature spermatozoa. Homozygous null male mice are sterile, showing absence of homing of Golgi-derived proacrosomal vesicles to the perinuclear surface in the spermatids. Other defects in these mice include improper removal of the cytoplasm, misshapen sperm head, coiled tail and lack of motility indicating a broader role of TMF/ARA160 in the regulation of sperm differentiation [122].
A number of other UPS components are localized in compartments suggestive of a role in acrosome biogenesis. Usp8 (mUBPy) is a deubiquitinating enzyme highly expressed in the mouse testis [123]. While the localization in round spermatids is diffuse and scattered within the perinuclear zone, a region that holds both the Golgi and the centrosome, the protein marks every step of the developing acrosomal vesicle in the differentiating spermatids, and then relocates to the cytoplasmic surface of the acrosome and to the centrosomal region in the mature sperm. This expression pattern is very similar to that of the molecular chaperone MSJ1 and subunits of the 20S proteasome [123], suggesting that Usp8 may have a functional association with MSJ1 and the proteasome, acting together to balance protein folding and degradation during the time of acrosome biogenesis. The role of Usp8 in acrosome biogenesis was further studied by protein–protein interaction assays and immunolabeling experiments where it was found to interact with components of the early endosomal machinery [124]. Antibodies to Usp8 or ESCRT-0 (endosomal-sorting complex required for transport-0) co-labeled vesicles contributing to acrosome formation. Usp8, through its MIT (microtubule interacting and trafficking/transport) domain, links the labeled vesicles with microtubules, which participate in acrosome formation by promoting the migration of proacrosomic vesicles [124, 125].
UPS and Sperm Tail Biogenesis
Sperm tail structuring is vital to the process of spermiogenesis as it ensures proper mitochondrial arrangement and microtubule assembly essential for generation of motile spermatozoa. Different UPS enzymes have been implicated in the structuring of the flagellum/tail of the developing spermatozoa. Male mice homozygous null for the ubiquitin ligase Herc4 sired litters that were 50 % smaller and associated with a 50 % reduction in sperm motility. Many of the mature spermatozoa had angulated tails with cytoplasmic droplets being retained at the region of angulation, indicating the essential role of Herc4 in removal of cytoplasmic droplets and efficient post-testicular sperm maturation [126]. It would be apt to mention here that the sperm cytoplasmic droplet contains several components of the UPS such as ubiquitin, ubiquitin conjugating enzyme E2, the ubiquitin C-terminal hydrolase Uchl1/PGP9.5 and various proteasome core subunits types α and β MECL-1/b2i [127] that may regulate the processing of the cytoplasmic droplet contents.
Usp14 is a proteasome associated ubiquitin specific protease whose expression is reduced in homozygous ataxia (axJ) mice leading to neurological defects and death within 2 months of age. These mice are sterile. To study non-neuronal functions of Usp14, the enzyme was expressed trangenically only in the brain of the mutant mice. Deficiency of Usp14 in the testis resulted in reduced testis size, decreased sperm production, morphologically abnormal spermatids and infertility. In general, Usp14 is diffusely distributed in the cytoplasm of the round and elongating spermatids and becomes associated with the post-acrosomal segment of the spermatid nucleus in steps 14–16. In step 16, Usp14 is restricted to the redundant nuclear envelopes (a region at the base of the elongating spermatid nucleus arising due to removal of nuclear pore complexes) and the cytoplasmic droplets in elongating spermatids and differentiated spermatozoa, which appear to be sites of ubiquitin dependent proteolysis. Usp14 deficiency may lead to low availability of free monoubiquitin necessary for the degradation of substrates or may lead to decreased deubiquitinating activity of the proteasome. The mutant phenotype was also characterized by the up-regulation of other deubiquitinating enzymes, namely Uchl1, Uchl3, Uchl5 and Usp5, possibly a cellular response to cope with decreased levels of free circulating monoubiquitin [128].
Other UPS enzymes that may be involved in sperm tail remodeling based on their localization include the MARCH10 ubiquitin ligases. These enzymes are expressed in elongating and elongated spermatids, and specifically localize to the cytoplasmic lobes, the principal piece and the annulus of the flagella. Overexpression of MARCH10a in COS7 cells showed that it is associated with microtubules, while in sperm MARCH10b was distributed in the cytoplasm [129]. In COS7 cells, MARCH 10a was found to have auto-ubiquitinating activity that was abolished upon microtubule disassembly, suggesting a possible similar function in sperm flagellum formation. MARCH7 is highly expressed in spermiogenic cells from round spermatids to elongated spermatids and spermatozoa. The localization of the protein on the neck, midpiece, cytoplasmic lobes and acroplaxome of the elongated spermatid suggests that this ligase participates in spermiogenesis by regulating the structural and functional integrity of these different parts of the developing spermatozoon [130]. ZNF645, a human RING finger protein, is localized to the post-acrosomal perinuclear theca and the entire tail of the mature sperm, suggesting a possible role of the UPS in development of these structures [131].
UPS and Overall Spermatid Maturation
Some components of the UPS appear important for multiple facets of spermatid development. Deficiency of the ubiquitin ligase Itch is associated with an increase in germ cell apoptotic index during the peri-pubertal stage at PND 28, as well as in adults at PND 56. The mutant mice are characterized by a developmental delay in spermatogenesis at PND28 and spermatid head defects and disorganization of the spermatids at PND56 [132]. Apart from the already mentioned defects in meiosis, the Hr6b/Ubc2 null mutants are also characterized by an increased flagellar diameter, abnormal periaxonemal structures and decreased motility [133]. The spermatids and spermatozoa also show head shape defects such as an increased space between the nucleus and acrosome cap, association of flat membranous saccule-like structures with the manchette, manchette associated with nucleus with an irregular outline, and nuclear evagination/invagination [134, 135]. Inactivation of the deubiquitinating enzyme CYLD in mice leads to sterility with a drastic testicular atrophy at PND 28. Round and elongating spermatids are scarce in these animals, with complete absence of spermatozoa in the epididymis. The mutant elongating spermatids have an incorrectly formed acrosome. Other defects include a lack of radial testicular organization and a significant increase in apoptosis. CYLD was shown to interact and negatively regulate ubiquitin-dependent NF-κB activator RIP1. Loss of CYLD leads to constitutive activation of NF-κB signaling with aberrant expression of anti-apoptotic genes [136].
Some of the structures that are established during spermiogenesis include the acrosome–acroplaxome complex and the head–tail coupling apparatus (HTCA) in the sperm head. The microtubular manchette formed on the caudal end of the acrosome–acroplaxome complex is a transitory structure suggested to be required for the transport of cargo between the nucleus and cytoplasm and towards the HTCA and tail. The RING finger ligase Rnf19A was shown to interact with Psmc3, a component of the 19S regulatory cap of the 26S proteasome; both co-localized on the Golgi-derived proacrosomal vesicles, and subsequently on the cytosolic side of the acrosome and acroplaxome and at the acroplaxome ring–manchette perinuclear ring region, thus suggesting a role for the UPS in acrosome biogenesis, sperm head shaping and sperm tail development [137]. The transmembrane ubiquitin ligase MARCH-XI, expressed predominantly in developing spermatids of rats, is localized on the trans-Golgi network (TGN) and multivesicular bodies (MVB). It forms complexes with the adaptor protein complex I and ubiquitinated/non ubiquitinated forms of fucose-containing glycoproteins, which are transported to the MVB from the TGN during acrosome biogenesis. This association suggests that the ubiquitin ligase has a ubiquitin-mediated role in the sorting of cargo in the TGN-MVB body transport pathway involved in spermiogenesis [138].
There are also studies hinting at the role of the UPS in centrosome reduction, a hallmark of spermatid elongation [139, 140]. After generating the sperm axoneme, the centrosome is either completely removed or reduced to a single inactive centriole in mammals [140]. Ubiquitin immune-reactivity was detected in the centrosomal part of the human and rhesus sperm tail, and proteasome subunits were also detected inside or near the sperm centriole [141, 142]. Deubiquitinating enzyme Usp8 also associates with gamma tubulin, a centrosomal protein marker suggesting a role in centrosome function [143].
These studies have shown that the UPS is a vital mediator of spermiogenesis, regulating events such as chromatin remodeling, acrosome biogenesis, sperm head shaping and structuring of the flagellum with consequences on fertility. While some functions are speculative based on localization and association studies, especially the ones looking at the role of UPS in acrosome biogenesis and flagellum formation, a significant number of functions are well established based on gene inactivation models.
Summary and Future Perspectives
From the studies cited in this review, it is clear that the UPS plays important roles at every step of spermatogenesis starting from gonocytes to differentiated spermatids, ready to be released into the epididymis. To date, there is a large volume of evidence for the regulation of various enzymes of the UPS at different stages of spermatogenesis. This is particularly true for regulation of levels of expression of the enzymes or their subcellular localization. Though less abundant, there are still plentiful examples of loss-of-function studies, especially gene inactivation models, that identify clear physiological functions of the UPS in spermatogenesis (Table 9.1). Often, these are critical functions as the inactivation of these genes results in abnormal progression or the developmental arrest of germ cells leading to deficits in infertility. Although the number of physiological functions of the UPS identified in the testis is increasing quickly, in most cases, there is still limited information on the molecular mechanisms by which the system exerts these effects. Specifically, the exact substrates of the enzymes involved and therefore the molecular pathways affected are often unknown. Such information is important to confirm that the genes implicated from loss of function studies play a direct role in the physiological process. In developmental processes that require multiple sequential steps, such as occurs in spermatogenesis, loss of function at an upstream step may impact indirectly a downstream step.
The extent of our knowledge on the roles of the UPS in spermatogenesis is also variable depending on the stage considered. The functions of this system in gonocytes and spermatogonial stem cell renewal and differentiation remain largely to be evaluated. A more detailed understanding of the roles of the UPS in regulating germ cell proliferation and differentiation will likely be useful in identifying new approaches to enhance the preservation of gonocytes and spermatogonial stem cells for translational purposes such as fertility preservation in prepubertal boys subjected to sterilizing chemotherapy, transgenesis and improved animal husbandry.
The most detailed understanding of the functions of the UPS in spermatogenesis is in meiosis. As detailed above, many loss of function studies have demonstrated impaired meiotic progression. Indeed, it is in early meiosis that spermatogenesis is arrested when the supply of free ubiquitin is limited [49, 50], suggesting that a critical increase in ubiquitinating activity occurs at this stage. There are examples in which molecular mechanisms have been delineated at a reasonably high resolution. This is particularly true for the roles of ubiquitination of histones in the repair of double strand breaks in DNA. Ubiquitinated H2A also appears important in meiotic sex chromosome inactivation [49, 50], but controversy as to whether it is essential remains to be resolved [81, 84].
A number of functions of the UPS in spermiogenesis have been proposed, but most remain to be confirmed through loss of function approaches. The relative paucity of such data may be related to technical limitations. In gene knockout studies, effects in spermiogenesis may be masked if the inactivated gene also plays critical roles earlier in spermatogenesis and causes arrest of development in spermatogonia or spermatocytes. Histones in spermatids are known to be ubiquitinated, but whether this ubiquitination plays an essential role in their replacement by protamines, in the epigenetic regulation of gene expression in these cells or in the resulting embryo remain intriguing and important questions at this time. Much additional work is required to establish firmly the role of ubiquitination in acrosome biogenesis and tail formation. In other cellular systems, ubiquitination has been shown to be important in mitochondrial remodeling and turnover [144]. Its potential role in the critical mitochondrial rearrangement in the mid-piece during spermiogenesis is largely unexplored.
Although emphasis is often placed on the proteolytic functions of ubiquitination, it is now well established that this post-translational modification serves many nonproteolytic functions. This is particularly true in the case of chromatin proteins such as histones. It is clear that the monoubiquitination of histone H2B or H2A recruits other proteins involved in DNA repair and studies cited in this review suggest that ubiquitination of histones plays important roles in regulating gene expression. Such modifications in the spermatozoa may have important epigenetic functions if they alter gene expression in the resulting embryo. Since spermatozoa with disordered nuclear condensation are often used in intracytoplasmic sperm injection in assisted reproduction, abnormalities in chromatin ubiquitination may be important in the developmental defects that have been observed at increased frequency in the offspring born from these procedures (rev in [145]).
As the number of loss of function studies with clear defects in spermatogenesis continues to increase (Table 9.1), it provides an opportunity to test whether abnormalities in these genes may be responsible for similar phenotypes in infertile men. This will enhance the possibilities for precise molecular diagnoses which are lacking in this clinical syndrome and may one day lead to specific therapies. Perhaps of more obvious translational potential is that this growing catalog of genes whose loss results in infertility are potential targets for the development of male contraceptives. Since they are mostly enzymes, they are in theory amenable to high throughput assays for screening of candidate drug inhibitors. Some are also germ cell specific and so inhibition may be less likely to result in toxicity in other cell types of the body.
The vast numbers of UPS genes that are expressed in the testis but have not been studied in this context indicate that we are still in the early phases of discovery in this area. Nonetheless, current insights confirm that the UPS plays important physiological roles in spermatogenesis and holds translational potential that will surely be revealed in the years to come.
References
Glickman MH, Ciechanover A. The ubiquitin–proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82(2):373–428.
Breitschopf K, Bengal E, Ziv T, Admon A, Ciechanover A. A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J. 1998;17(20):5964–73.
Ciechanover A, Ben-Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 2004;14(3):103–6.
Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309(5731):127–30.
Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–83.
Lander GC, Estrin E, Matyskiela ME, Bashore C, Nogales E, Martin A. Complete subunit architecture of the proteasome regulatory particle. Nature. 2012;482(7384):186–91.
Seemuller E, Lupas A, Stock D, Lowe J, Huber R, Baumeister W. Proteasome from thermoplasma acidophilum: a threonine protease. Science. 1995;268(5210):579–82.
Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513.
Lee MJ, Lee BH, Hanna J, King RW, Finley D. Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes. Mol Cell Proteomics. 2011;10(5):R110 003871.
Sijts EJ, Kloetzel PM. The role of the proteasome in the generation of MHC class I ligands and immune responses. Cell Mol Life Sci. 2011;68(9):1491–502.
Savulescu AF, Glickman MH. Proteasome activator 200: the heat is on. Mol Cell Proteomics. 2011;10(5):R110 006890.
Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78(5):761–71.
Salghetti SE, Kim SY, Tansey WP. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 1999;18(3):717–26.
Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat Cell Biol. 1999;1(4):193–9.
Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–8.
Alkalay I, Yaron A, Hatzubai A, Orian A, Ciechanover A, Ben-Neriah Y. Stimulation-dependent I kappa B alpha phosphorylation marks the NF-kappa B inhibitor for degradation via the ubiquitin–proteasome pathway. Proc Natl Acad Sci U S A. 1995;92(23):10599–603.
Chen Z, Hagler J, Palombella VJ, Melandri F, Scherer D, Ballard D, et al. Signal-induced site-specific phosphorylation targets I kappa B alpha to the ubiquitin–proteasome pathway. Genes Dev. 1995;9(13):1586–97.
Hochstrasser M. Ubiquitin signalling: what’s in a chain? Nat Cell Biol. 2004;6(7):571–2.
Clague MJ, Urbe S. Ubiquitin: same molecule, different degradation pathways. Cell. 2012;143(5):682–5.
Chen ZJ. Ubiquitination in signaling to and activation of IKK. Immunol Rev. 2012;246(1):95–106.
Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–33.
Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37(Pt 5):937–53.
Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–97.
Ozkaynak E, Finley D, Solomon MJ, Varshavsky A. The yeast ubiquitin genes: a family of natural gene fusions. EMBO J. 1987;6:1429–39.
de Rooij DG. Proliferation and differentiation of spermatogonial stem cells. Reproduction. 2001;121(3):347–54.
Russell LD, Ettlin R, Hikim APS, Clegg ED. Histological and histopathological evaluation of the testis. Clearwater: Cache River Press; 1990.
Culty M. Gonocytes, the forgotten cells of the germ cell lineage. Birth Defects Res C Embryo Today. 2009;87(1):1–26.
Li H, Papadopoulos V, Vidic B, Dym M, Culty M. Regulation of rat testis gonocyte proliferation by platelet-derived growth factor and estradiol: identification of signaling mechanisms involved. Endocrinology. 1997;138(3):1289–98.
Thuillier R, Mazer M, Manku G, Boisvert A, Wang Y, Culty M. Interdependence of platelet-derived growth factor and estrogen-signaling pathways in inducing neonatal rat testicular gonocytes proliferation. Biol Reprod. 2010;82(5):825–36.
Wang Y, Culty M. Identification and distribution of a novel platelet-derived growth factor receptor beta variant: effect of retinoic acid and involvement in cell differentiation. Endocrinology. 2007;148(5):2233–50.
Oulad-Abdelghani M, Bouillet P, Decimo D, Gansmuller A, Heyberger S, Dolle P, et al. Characterization of a premeiotic germ cell-specific cytoplasmic protein encoded by Stra8, a novel retinoic acid-responsive gene. J Cell Biol. 1996;135(2):469–77.
Zhou Q, Nie R, Li Y, Friel P, Mitchell D, Hess RA, et al. Expression of stimulated by retinoic acid gene 8 (Stra8) in spermatogenic cells induced by retinoic acid: an in vivo study in vitamin A-sufficient postnatal murine testes. Biol Reprod. 2008;79(1):35–42.
Zhou Q, Li Y, Nie R, Friel P, Mitchell D, Evanoff RM, et al. Expression of stimulated by retinoic acid gene 8 (Stra8) and maturation of murine gonocytes and spermatogonia induced by retinoic acid in vitro. Biol Reprod. 2008;78(3):537–45.
Manku G, Wing SS, Culty M. Expression of the ubiquitin proteasome system in neonatal rat gonocytes and spermatogonia: role in gonocyte differentiation. Biol Reprod. 2012;87(2):44, 1–18.
Yi YJ, Zimmerman SW, Manandhar G, Odhiambo JF, Kennedy C, Jonakova V, et al. Ubiquitin-activating enzyme (UBA1) is required for sperm capacitation, acrosomal exocytosis and sperm–egg coat penetration during porcine fertilization. Int J Androl. 2012;35(2):196–210.
Pelzer C, Kassner I, Matentzoglu K, Singh RK, Wollscheid HP, Scheffner M, et al. UBE1L2, a novel E1 enzyme specific for ubiquitin. J Biol Chem. 2007;282(32):23010–4.
Hogarth CA, Mitchell D, Evanoff R, Small C, Griswold M. Identification and expression of potential regulators of the mammalian mitotic-to-meiotic transition. Biol Reprod. 2011;84(1):34–42.
Liu Z, Oughtred R, Wing SS. Characterization of E3Histone, a novel testis ubiquitin protein ligase which ubiquitinates histones. Mol Cell Biol. 2005;25(7):2819–31.
Liu Z, Miao D, Xia Q, Hermo L, Wing SS. Regulated expression of the ubiquitin protein ligase, E3(Histone)/LASU1/Mule/ARF-BP1/HUWE1, during spermatogenesis. Dev Dyn. 2007;236(10):2889–98.
Hong SW, Jin DH, Shin JS, Moon JH, Na YS, Jung KA, et al. Ring finger protein 149 is an E3 ubiquitin ligase active on wild-type v-Raf murine sarcoma viral oncogene homolog B1 (BRAF). J Biol Chem. 2012;287(28):24017–25.
Plafker KS, Farjo KM, Wiechmann AF, Plafker SM. The human ubiquitin conjugating enzyme, UBE2E3, is required for proliferation of retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2008;49(12):5611–8.
Kanarek N, Horwitz E, Mayan I, Leshets M, Cojocaru G, Davis M, et al. Spermatogenesis rescue in a mouse deficient for the ubiquitin ligase SCF{beta}-TrCP by single substrate depletion. Genes Dev. 2010;24(5):470–7.
Oatley JM, Brinster RL. Regulation of spermatogonial stem cell self-renewal in mammals. Annu Rev Cell Dev Biol. 2008;24:263–86.
Luo J, Megee S, Dobrinski I. Asymmetric distribution of UCH-L1 in spermatogonia is associated with maintenance and differentiation of spermatogonial stem cells. J Cell Physiol. 2009;220(2):460–8.
Cayli S, Ocakli S, Erdemir F, Tas U, Aslan H, Yener T, et al. Developmental expression of p97/VCP (Valosin-containing protein) and Jab1/CSN5 in the rat testis and epididymis. Reprod Biol Endocrinol. 2011;9:117.
Cayli S, Erdemir F, Ocakli S, Ungor B, Kesici H, Yener T, et al. Interaction between Smad1 and p97/VCP in rat testis and epididymis during the postnatal development. Reprod Sci. 2012;19(2):190–201.
Vodermaier HC. Cell cycle: waiters serving the destruction machinery. Curr Biol. 2001;11(20):R834–7.
Harper JW. A phosphorylation-driven ubiquitination switch for cell-cycle control. Trends Cell Biol. 2002;12(3):104–7.
Ryu KY, Sinnar SA, Reinholdt LG, Vaccari S, Hall S, Garcia MA, et al. The mouse polyubiquitin gene Ubb is essential for meiotic progression. Mol Cell Biol. 2008;28(3):1136–46.
Sinnar SA, Small CL, Evanoff RM, Reinholdt LG, Griswold MD, Kopito RR, et al. Altered testicular gene expression patterns in mice lacking the polyubiquitin gene Ubb. Mol Reprod Dev. 2011;78(6):415–25.
Okazaki K, Okayama H, Niwa O. The polyubiquitin gene is essential for meiosis in fission yeast. Exp Cell Res. 2000;254(1):143–52.
Macdonald LD, Knox A, Hansen D. Proteasomal regulation of the proliferation vs. meiotic entry decision in the Caenorhabditis elegans germ line. Genetics. 2008;180(2):905–20.
Ustrell V, Hoffman L, Pratt G, Rechsteiner M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002;21(13):3516–25.
Krogan NJ, Lam MH, Fillingham J, Keogh MC, Gebbia M, Li J, et al. Proteasome involvement in the repair of DNA double-strand breaks. Mol Cell. 2004;16(6):1027–34.
Khor B, Bredemeyer AL, Huang CY, Turnbull IR, Evans R, Maggi Jr LB, et al. Proteasome activator PA200 is required for normal spermatogenesis. Mol Cell Biol. 2006;26(8):2999–3007.
Madura K, Prakash S, Prakash L. Expression of the Saccharomyces cerevisiae DNA repair gene RAD6 that encodes a ubiquitin conjugating enzyme, increases in response to DNA damage and in meiosis but remains constant during the mitotic cell cycle. Nucleic Acids Res. 1990;18(4):771–8.
Robzyk K. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287(5452):501–4.
Game JC, Chernikova SB. The role of RAD6 in recombinational repair, checkpoints and meiosis via histone modification. DNA Repair. 2009;8(4):470–82.
Yamashita K, Shinohara M, Shinohara A. Rad6–Bre1-mediated histone H2B ubiquitylation modulates the formation of double-strand breaks during meiosis. Proc Natl Acad Sci U S A. 2004;101(31):11380–5.
Baarends WM, Wassenaar E, Hoogerbrugge JW, van Cappellen G, Roest HP, Vreeburg J, et al. Loss of HR6B ubiquitin-conjugating activity results in damaged synaptonemal complex structure and increased crossing-over frequency during the male meiotic prophase. Mol Cell Biol. 2003;23(4):1151–62.
Liebe B, Petukhova G, Barchi M, Bellani M, Braselmann H, Nakano T, et al. Mutations that affect meiosis in male mice influence the dynamics of the mid-preleptotene and bouquet stages. Exp Cell Res. 2006;312(19):3768–81.
Tasaki T, Kwon YT. The mammalian N-end rule pathway: new insights into its components and physiological roles. Trends Biochem Sci. 2007;32(11):520–8.
Kwon YT, Xia Z, An JY, Tasaki T, Davydov IV, Seo JW, et al. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol Cell Biol. 2003;23(22):8255–71.
Ouyang Y, Kwon YT, An JY, Eller D, Tsai SC, Diaz-Perez S, et al. Loss of Ubr2, an E3 ubiquitin ligase, leads to chromosome fragility and impaired homologous recombinational repair. Mutat Res. 2006;596(1–2):64–75.
Yang F, Cheng Y, An JY, Kwon YT, Eckardt S, Leu NA, et al. The ubiquitin ligase Ubr2, a recognition E3 component of the N-end rule pathway, stabilizes Tex19.1 during spermatogenesis. PLoS One. 2010;5(11):e14017.
Miyamoto T, Tsujimura A, Miyagawa Y, Koh E, Namiki M, Horikawa M, et al. Single nucleotide polymorphism in the UBR2 gene may be a genetic risk factor for Japanese patients with azoospermia by meiotic arrest. J Assist Reprod Genet. 2011;28(8):743–6.
Vyas R, Kumar R, Clermont F, Helfricht A, Kalev P, Sotiropoulou P, et al. RNF4 is required for DNA double-strand break repair in vivo. Cell Death Differ. 2013;20(3):490–502.
Ward JO, Reinholdt LG, Motley WW, Niswander LM, Deacon DC, Griffin LB, et al. Mutation in mouse hei10, an e3 ubiquitin ligase, disrupts meiotic crossing over. PLoS Genet. 2007;3(8):e139.
Baarends WM, Hoogerbrugge JW, Roest HP, Ooms M, Vreeburg J, Hoeijmakers JH, et al. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev Biol. 1999;207(2):322–33.
Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in polycomb silencing. Nature. 2004;431(7010):873–8.
Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell. 2004;16(4):631–9.
An JY, Kim EA, Jiang Y, Zakrzewska A, Kim DE, Lee MJ, et al. UBR2 mediates transcriptional silencing during spermatogenesis via histone ubiquitination. Proc Natl Acad Sci U S A. 2010;107(5):1912–7.
An JY, Kim E, Zakrzewska A, Yoo YD, Jang JM, Han DH, et al. UBR2 of the N-end rule pathway is required for chromosome stability via histone ubiquitylation in spermatocytes and somatic cells. PLoS One. 2012;7(5):e37414.
Inagaki A, Sleddens-Linkels E, Wassenaar E, Ooms M, van Cappellen WA, Hoeijmakers JH, et al. Meiotic functions of RAD18. J Cell Sci. 2011;124(Pt 16):2837–50.
Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, et al. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol. 2009;11(5):592–603.
Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131(5):901–14.
Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318(5856):1637–40.
Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131(5):887–900.
Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci U S A. 2007;104(52):20759–63.
Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136(3):435–46.
Santos MA, Huen MS, Jankovic M, Chen HT, Lopez-Contreras AJ, Klein IA, et al. Class switching and meiotic defects in mice lacking the E3 ubiquitin ligase RNF8. J Exp Med. 2010;207(5):973–81.
Lu LY, Wu J, Ye L, Gavrilina GB, Saunders TL, Yu X. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev Cell. 2010;18(3):371–84.
Li L, Halaby MJ, Hakem A, Cardoso R, El Ghamrasni S, Harding S, et al. Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer. J Exp Med. 2010;207(5):983–97.
Sin HS, Barski A, Zhang F, Kartashov AV, Nussenzweig A, Chen J, et al. RNF8 regulates active epigenetic modifications and escape gene activation from inactive sex chromosomes in post-meiotic spermatids. Genes Dev. 2012;26(24):2737–48.
Gillot I, Matthews C, Puel D, Vidal F, Lopez P. Ret finger protein: an E3 ubiquitin ligase juxtaposed to the XY body in meiosis. Int J Cell Biol. 2009;2009:524858.
Hoyer-Fender S. Molecular aspects of XY body formation. Cytogenet Genome Res. 2003;103(3–4):245–55.
Handel MA. The XY, body: a specialized meiotic chromatin domain. Exp Cell Res. 2004;296(1):57–63.
Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7(9):644–56.
Thornton BR, Toczyski DP. Precise destruction: an emerging picture of the APC. Genes Dev. 2006;20(22):3069–78.
Yu H, Peters JM, King RW, Page AM, Hieter P, Kirschner MW. Identification of a Cullin homology region in a subunit of the anaphase-promoting complex. Science. 1998;279:1219–22.
Zachariae W, Shevchenko A, Andrews PD, Ciosk R, Galova M, Stark MJ, et al. Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to Cullins. Science. 1998;279(5354):1216–9.
Ors A, Grimaldi M, Kimata Y, Wilkinson CR, Jones N, Yamano H. The transcription factor Atf1 binds and activates the APC/C ubiquitin ligase in fission yeast. J Biol Chem. 2009;284(36):23989–94.
Stein KK, Nesmith JE, Ross BD, Golden A. Functional redundancy of paralogs of an anaphase promoting complex/cyclosome subunit in Caenorhabditis elegans meiosis. Genetics. 2010;186(4):1285–93.
Furuta T, Tuck S, Kirchner J, Koch B, Auty R, Kitagawa R, et al. EMB-30: an APC4 homologue required for metaphase-to-anaphase transitions during meiosis and mitosis in Caenorhabditis elegans. Mol Biol Cell. 2000;11(4):1401–19.
Golden A, Sadler PL, Wallenfang MR, Schumacher JM, Hamill DR, Bates G, et al. Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J Cell Biol. 2000;151(7):1469–82.
Burger J, Merlet J, Tavernier N, Richaudeau B, Arnold A, Ciosk R, et al. CRL2(LRR-1) E3-ligase regulates proliferation and progression through meiosis in the Caenorhabditis elegans germline. PLoS Genet. 2013;9(3):e1003375.
Yin Y, Lin C, Kim ST, Roig I, Chen H, Liu L, et al. The E3 ubiquitin ligase Cullin 4A regulates meiotic progression in mouse spermatogenesis. Dev Biol. 2011;356(1):51–62.
Dickins RA, Frew IJ, House CM, O’Bryan MK, Holloway AJ, Haviv I, et al. The ubiquitin ligase component Siah1a is required for completion of meiosis I in male mice. Mol Cell Biol. 2002;22(7):2294–303.
Guardavaccaro D, Kudo Y, Boulaire J, Barchi M, Busino L, Donzelli M, et al. Control of meiotic and mitotic progression by the F box protein beta-Trcp1 in vivo. Dev Cell. 2003;4(6):799–812.
Oughtred R, Bedard N, Adegoke OA, Morales CR, Trasler J, Rajapurohitam V, et al. Characterization of rat100, a 300-kilodalton ubiquitin-protein ligase induced in germ cells of the rat testis and similar to the Drosophila hyperplastic discs gene. Endocrinology. 2002;143(10):3740–7.
Mansfield E, Hersperger E, Biggs J, Shearn A. Genetic and molecular analysis of hyperplastic discs, a gene whose product is required for regulation of cell proliferation in Drosophila melanogaster imaginal discs and germ cells. Dev Biol. 1994;165(2):507–26.
Wang YL, Liu W, Sun YJ, Kwon J, Setsuie R, Osaka H, et al. Overexpression of ubiquitin carboxyl-terminal hydrolase L1 arrests spermatogenesis in transgenic mice. Mol Reprod Dev. 2006;73(1):40–9.
Kwon J, Mochida K, Wang YL, Sekiguchi S, Sankai T, Aoki S, et al. Ubiquitin C-terminal hydrolase L-1 is essential for the early apoptotic wave of germinal cells and for sperm quality control during spermatogenesis. Biol Reprod. 2005;73(1):29–35.
Tang EI, Mruk DD, Cheng CY. MARKs (MAP/microtubule affinity-regulating kinases), microtubule dynamics and spermatogenesis. J Endocrinol. 2013;217:R13–23.
Rajapurohitam V, Morales CR, El-Alfy M, Lefrancois S, Bedard N, Wing SS. Activation of a UBC4-dependent pathway of ubiquitin conjugation during postnatal development of the rat testis. Dev Biol. 1999;212(1):217–28.
Tipler CP, Hutchon SP, Hendil K, Tanaka K, Fishel S, Mayer RJ. Purification and characterization of 26S proteasomes from human and mouse spermatozoa. Mol Hum Reprod. 1997;3(12):1053–60.
Yuan X, Miller M, Belote JM. Duplicated proteasome subunit genes in Drosophila melanogaster encoding testes-specific isoforms. Genetics. 1996;144(1):147–57.
Zhong L, Belote JM. The testis-specific proteasome subunit Prosalpha6T of D. melanogaster is required for individualization and nuclear maturation during spermatogenesis. Development. 2007;134(19):3517–25.
Ma J, Katz E, Belote JM. Expression of proteasome subunit isoforms during spermatogenesis in Drosophila melanogaster. Insect Mol Biol. 2002;11(6):627–39.
Belote JM, Zhong L. Duplicated proteasome subunit genes in Drosophila and their roles in spermatogenesis. Heredity. 2009;103(1):23–31.
Kornberg RD, Thomas JO. Chromatin structure; oligomers of the histones. Science. 1974;184(4139):865–8.
Meistrich ML, Mohapatra B, Shirley CR, Zhao M. Roles of transition nuclear proteins in spermiogenesis. Chromosoma. 2003;111(8):483–8.
Oliva R. Protamines and male infertility. Hum Reprod Update. 2006;12(4):417–35.
Shirley CR, Hayashi S, Mounsey S, Yanagimachi R, Meistrich ML. Abnormalities and reduced reproductive potential of sperm from Tnp1- and Tnp2-null double mutant mice. Biol Reprod. 2004;71(4):1220–9.
Zhao M, Shirley CR, Hayashi S, Marcon L, Mohapatra B, Suganuma R, et al. Transition nuclear proteins are required for normal chromatin condensation and functional sperm development. Genesis. 2004;38(4):200–13.
Cho C, Willis WD, Goulding EH, Jung-Ha H, Choi YC, Hecht NB, et al. Haploinsufficiency of protamine-1 or -2 causes infertility in mice. Nat Genet. 2001;28(1):82–6.
Cho C, Jung-Ha H, Willis WD, Goulding EH, Stein P, Xu Z, et al. Protamine 2 deficiency leads to sperm DNA damage and embryo death in mice. Biol Reprod. 2003;69(1):211–7.
Chen HY, Sun JM, Zhang Y, Davie JR, Meistrich ML. Ubiquitination of histone H3 in elongating spermatids of rat testes. J Biol Chem. 1998;273(21):13165–9.
Rathke C, Baarends WM, Jayaramaiah-Raja S, Bartkuhn M, Renkawitz R, Renkawitz-Pohl R. Transition from a nucleosome-based to a protamine-based chromatin configuration during spermiogenesis in Drosophila. J Cell Sci. 2007;120(Pt 9):1689–700.
Qian MX, Pang Y, Liu CH, Haratake K, Du BY, Ji DY, et al. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell. 2013;153(5):1012–24.
Haraguchi CM, Mabuchi T, Hirata S, Shoda T, Hoshi K, Yokota S. Ubiquitin signals in the developing acrosome during spermatogenesis of rat testis: an immunoelectron microscopic study. J Histochem Cytochem. 2004;52(11):1393–403.
Lerer-Goldshtein T, Bel S, Shpungin S, Pery E, Motro B, Goldstein RS, et al. TMF/ARA160: a key regulator of sperm development. Dev Biol. 2010;348(1):12–21.
Berruti G, Martegani E. The deubiquitinating enzyme mUBPy interacts with the sperm-specific molecular chaperone MSJ-1: the relation with the proteasome, acrosome, and centrosome in mouse male germ cells. Biol Reprod. 2005;72(1):14–21.
Berruti G, Ripolone M, Ceriani M. USP8, a regulator of endosomal sorting, is involved in mouse acrosome biogenesis through interaction with the spermatid ESCRT-0 complex and microtubules. Biol Reprod. 2010;82(5):930–9.
Moreno RD, Palomino J, Schatten G. Assembly of spermatid acrosome depends on microtubule organization during mammalian spermiogenesis. Dev Biol. 2006;293(1):218–27.
Rodriguez CI, Stewart CL. Disruption of the ubiquitin ligase HERC4 causes defects in spermatozoon maturation and impaired fertility. Dev Biol. 2007;312(2):501–8.
Fischer KA, Van Leyen K, Lovercamp KW, Manandhar G, Sutovsky M, Feng D, et al. 15-Lipoxygenase is a component of the mammalian sperm cytoplasmic droplet. Reproduction. 2005;130(2):213–22.
Crimmins S, Sutovsky M, Chen PC, Huffman A, Wheeler C, Swing DA, et al. Transgenic rescue of ataxia mice reveals a male-specific sterility defect. Dev Biol. 2009;325(1):33–42.
Iyengar PV, Hirota T, Hirose S, Nakamura N. Membrane-associated RING-CH 10 (MARCH10 protein) is a microtubule-associated E3 ubiquitin ligase of the spermatid flagella. J Biol Chem. 2011;286(45):39082–90.
Zhao B, Ito K, Iyengar PV, Hirose S, Nakamura N. MARCH7 E3 ubiquitin ligase is highly expressed in developing spermatids of rats and its possible involvement in head and tail formation. Histochem Cell Biol. 2013;139(3):447–60.
Liu YQ, Bai G, Zhang H, Su D, Tao DC, Yang Y, et al. Human RING finger protein ZNF645 is a novel testis-specific E3 ubiquitin ligase. Asian J Androl. 2010;12(5):658–66.
Dwyer JL, Richburg JH. Age-dependent alterations in spermatogenesis in itchy mice. Spermatogenesis. 2012;2(2):104–16.
Roest HP, van Klaveren J, de Wit J, van Gurp CG, Koken MH, Vermey M, et al. Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell. 1996;86(5):799–810.
Escalier D, Bai XY, Silvius D, Xu PX, Xu X. Spermatid nuclear and sperm periaxonemal anomalies in the mouse Ube2b null mutant. Mol Reprod Dev. 2003;65(3):298–308.
Escalier D. New insights into the assembly of the periaxonemal structures in mammalian spermatozoa. Biol Reprod. 2003;69(2):373–8.
Wright A, Reiley WW, Chang M, Jin W, Lee AJ, Zhang M, et al. Regulation of early wave of germ cell apoptosis and spermatogenesis by deubiquitinating enzyme CYLD. Dev Cell. 2007;13(5):705–16.
Rivkin E, Kierszenbaum AL, Gil M, Tres LL. Rnf19a, a ubiquitin protein ligase, and Psmc3, a component of the 26S proteasome, tether to the acrosome membranes and the head–tail coupling apparatus during rat spermatid development. Dev Dyn. 2009;238(7):1851–61.
Morokuma Y, Nakamura N, Kato A, Notoya M, Yamamoto Y, Sakai Y, et al. MARCH-XI, a novel transmembrane ubiquitin ligase implicated in ubiquitin-dependent protein sorting in developing spermatids. J Biol Chem. 2007;282(34):24806–15.
Manandhar G, Sutovsky P, Joshi HC, Stearns T, Schatten G. Centrosome reduction during mouse spermiogenesis. Dev Biol. 1998;203(2):424–34.
Manandhar G, Schatten H, Sutovsky P. Centrosome reduction during gametogenesis and its significance. Biol Reprod. 2005;72(1):2–13.
Wojcik C, Benchaib M, Lornage J, Czyba JC, Guerin JF. Proteasomes in human spermatozoa. Int J Androl. 2000;23(3):169–77.
Bialy LP, Ziemba HT, Marianowski P, Fracki S, Bury M, Wojcik C. Localization of a proteasomal antigen in human spermatozoa: immunohistochemical electron microscopic study. Folia Histochem Cytobiol. 2001;39(2):129–30.
Berruti G, Aivatiadou E. MUBPy is a novel centrosome-associated protein and interacts with gamma-tubulin. J Submicrosc Cytol Pathol. 2006;38(1):77–83.
Taylor EB, Rutter J. Mitochondrial quality control by the ubiquitin–proteasome system. Biochem Soc Trans. 2011;39(5):1509–13.
Alukal JP, Lamb DJ. Intracytoplasmic sperm injection (ICSI)—what are the risks? Urol Clin North Am. 2008;35(2):277–88. ix–x.
Zhu H, Zhou ZM, Huo R, Huang XY, Lu L, Lin M, et al. Identification and characteristics of a novel E1 like gene nUBE1L in human testis. Acta Biochim Biophys Sin. 2004;36(3): 227–34.
Du Y, Liu ML, Jia MC. Identification and characterization of a spermatogenesis-related gene Ube1 in rat testis. Sheng Li Xue Bao. 2008;60(3):382–90.
Odorisio T, Mahadevaiah SK, McCarrey JR, Burgoyne PS. Transcriptional analysis of the candidate spermatogenesis gene Ube1y and of the closely related Ube1x shows that they are coexpressed in spermatogonia and spermatids but are repressed in pachytene spermatocytes. Dev Biol. 1996;180(1):336–43.
Koken MH, Hoogerbrugge JW, Jasper-Dekker I, de Wit J, Willemsen R, Roest HP, et al. Expression of the ubiquitin-conjugating DNA repair enzymes HHR6A and B suggests a role in spermatogenesis and chromatin modification. Dev Biol. 1996;173(1):119–32.
Wing SS, Jain P. Molecular cloning, expression and characterization of a ubiquitin conjugation enzyme (E2(17)kB) highly expressed in rat testis. Biochem J. 1995;305(Pt 1):125–32.
Wing SS, Bedard N, Morales C, Hingamp P, Trasler J. A novel rat homolog of the Saccharomyces cerevisiae ubiquitin-conjugating enzymes UBC4 and UBC5 with distinct biochemical features is induced during spermatogenesis. Mol Cell Biol. 1996;16(8):4064–72.
Singh RK, Kabbaj MH, Paik J, Gunjan A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol. 2009;11(8):925–33.
Bedard N, Hingamp P, Pang Z, Karaplis A, Morales C, Trasler J, et al. Mice lacking the UBC4-testis gene have a delay in postnatal testis development but normal spermatogenesis and fertility. Mol Cell Biol. 2005;25(15):6346–54.
Pertceva JA, Dorogova NV, Bolobolova EU, Nerusheva OO, Fedorova SA, Omelyanchuk LV. The role of Drosophila hyperplastic discs gene in spermatogenesis. Cell Biol Int. 2010;34(10):991–6.
van der Laan R, Uringa EJ, Wassenaar E, Hoogerbrugge JW, Sleddens E, Odijk H, et al. Ubiquitin ligase Rad18Sc localizes to the XY body and to other chromosomal regions that are unpaired and transcriptionally silenced during male meiotic prophase. J Cell Sci. 2004;117(Pt 21):5023–33.
van der Laan R, Roest HP, Hoogerbrugge JW, Smit EM, Slater R, Baarends WM, et al. Characterization of mRAD18Sc, a mouse homolog of the yeast postreplication repair gene RAD18. Genomics. 2000;69(1):86–94.
Tezel G, Nagasaka T, Iwahashi N, Asai N, Iwashita T, Sakata K, et al. Different nuclear/cytoplasmic distributions of RET finger protein in different cell types. Pathol Int. 1999;49(10):881–6.
Kopanja D, Roy N, Stoyanova T, Hess RA, Bagchi S, Raychaudhuri P. Cul4A is essential for spermatogenesis and male fertility. Dev Biol. 2011;352(2):278–87.
Frew IJ, Hammond VE, Dickins RA, Quinn JM, Walkley CR, Sims NA, et al. Generation and analysis of Siah2 mutant mice. Mol Cell Biol. 2003;23(24):9150–61.
Riparbelli MG, Callaini G. The Drosophila parkin homologue is required for normal mitochondrial dynamics during spermiogenesis. Dev Biol. 2007;303(1):108–20.
Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A. 2003;100(7):4078–83.
Smith CL, DeVera DG, Lamb DJ, Nawaz Z, Jiang YH, Beaudet al, et al. Genetic ablation of the steroid receptor coactivator–ubiquitin ligase, E6-AP, results in tissue-selective steroid hormone resistance and defects in reproduction. Mol Cell Biol. 2002;22(2):525–35.
Li MW, Lee WM, Lui WY. Expression of Itch in Sertoli cells is controlled via the interaction of E2F1/DP1 complex with E2F and GATA motifs. Spermatogenesis. 2011;1(2):152–8.
Lui WY, Lee WM. cAMP perturbs inter-Sertoli tight junction permeability barrier in vitro via its effect on proteasome-sensitive ubiquitination of occludin. J Cell Physiol. 2005;203(3):564–72.
Ma T, Keller JA, Yu X. RNF8-dependent histone ubiquitination during DNA damage response and spermatogenesis. Acta Biochim Biophys Sin. 2011;43(5):339–45.
Nian H, Zhang W, Shi H, Zhao Q, Xie Q, Liao S, et al. Mouse RING finger protein Rnf133 is a testis-specific endoplasmic reticulum-associated E3 ubiquitin ligase. Cell Res. 2008;18(7):800–2.
Nian H, Fan C, Liao S, Shi Y, Zhang K, Liu Y, et al. RNF151, a testis-specific RING finger protein, interacts with dysbindin. Arch Biochem Biophys. 2007;465(1):157–63.
Bohgaki T, Bohgaki M, Cardoso R, Panier S, Zeegers D, Li L, et al. Genomic instability, defective spermatogenesis, immunodeficiency, and cancer in a mouse model of the RIDDLE syndrome. PLoS Genet. 2011;7(4):e1001381.
Zhao S, Gou LT, Zhang M, Zu LD, Hua MM, Hua Y, et al. piRNA-triggered MIWI ubiquitination and removal by APC/C in late spermatogenesis. Dev Cell. 2013;24(1):13–25.
Lin H, Keriel A, Morales CR, Bedard N, Zhao Q, Hingamp P, et al. Divergent N-terminal sequences target an inducible testis deubiquitinating enzyme to distinct subcellular structures. Mol Cell Biol. 2000;20(17):6568–78.
Bedard N, Yang Y, Gregory M, Cyr DG, Suzuki J, Yu X, et al. Mice lacking the USP2 deubiquitinating enzyme have severe male subfertility associated with defects in fertilization and sperm motility. Biol Reprod. 2011;85(3):594–604.
Meccariello R, Chianese R, Scarpa D, Berruti G, Cobellis G, Pierantoni R, et al. UBPy/MSJ-1 system during male germ cell progression in the frog, Rana esculenta. Gen Comp Endocrinol. 2007;153(1–3):275–9.
Chianese R, Scarpa D, Berruti G, Cobellis G, Pierantoni R, Fasano S, et al. Expression and localization of the deubiquitinating enzyme mUBPy in wobbler mouse testis during spermiogenesis. Gen Comp Endocrinol. 2010;166(2):289–95.
Berruti G, Martegani E. mUBPy and MSJ-1, a deubiquitinating enzyme and a molecular chaperone specifically expressed in testis, associate with the acrosome and centrosome in mouse germ cells. Ann N Y Acad Sci. 2002;973:5–7.
Lee KH, Song GJ, Kang IS, Kim SW, Paick JS, Chung CH, et al. Ubiquitin-specific protease activity of USP9Y, a male infertility gene on the Y chromosome. Reprod Fertil Dev. 2003;15(1–2):129–33.
Krausz C, Degl’Innocenti S, Nuti F, Morelli A, Felici F, Sansone M, et al. Natural transmission of USP9Y gene mutations: a new perspective on the role of AZFa genes in male fertility. Hum Mol Genet. 2006;15(18):2673–81.
Sun C, Skaletsky H, Birren B, Devon K, Tang Z, Silber S, et al. An azoospermic man with a de novo point mutation in the Y-chromosomal gene USP9Y. Nat Genet. 1999;23(4):429–32.
Luddi A, Margollicci M, Gambera L, Serafini F, Cioni M, De Leo V, et al. Spermatogenesis in a man with complete deletion of USP9Y. N Engl J Med. 2009;360(9):881–5.
Zhang J, Shao XG, Shi YB, Yan L, Wang L, Tian H, et al. Polymorphism of Usp26 correlates with idiopathic male infertility. Zhonghua Nan Ke Xue. 2012;18(2):105–8.
Zhang J, Qiu SD, Li SB, Zhou DX, Tian H, Huo YW, et al. Novel mutations in ubiquitin-specific protease 26 gene might cause spermatogenesis impairment and male infertility. Asian J Androl. 2007;9(6):809–14.
Wei L, Shi YC, Cui YX, Huang YF. Mutation of the USP26 gene in spermatogenesis dysfunction. Zhonghua Nan Ke Xue. 2010;16(1):65–7.
Stouffs K, Lissens W, Tournaye H, Van Steirteghem A, Liebaers I. Alterations of the USP26 gene in Caucasian men. Int J Androl. 2006;29(6):614–7.
Stouffs K, Lissens W, Tournaye H, Van Steirteghem A, Liebaers I. Possible role of USP26 in patients with severely impaired spermatogenesis. Eur J Hum Genet. 2005;13(3):336–40.
Shi YC, Wei L, Cui YX, Shang XJ, Wang HY, Xia XY, et al. Association between ubiquitin-specific protease USP26 polymorphism and male infertility in Chinese men. Clin Chim Acta. 2011;412(7–8):545–9.
Kim YK, Kim YS, Yoo KJ, Lee HJ, Lee DR, Yeo CY, et al. The expression of Usp42 during embryogenesis and spermatogenesis in mouse. Gene Expr Patterns. 2007;7(1–2):143–8.
Kwon J, Kikuchi T, Setsuie R, Ishii Y, Kyuwa S, Yoshikawa Y. Characterization of the testis in congenitally ubiquitin carboxy-terminal hydrolase-1 (Uch-L1) defective (gad) mice. Exp Anim. 2003;52(1):1–9.
Kwon J. The new function of two ubiquitin C-terminal hydrolase isozymes as reciprocal modulators of germ cell apoptosis. Exp Anim. 2007;56(2):71–7.
Kwon J, Wang YL, Setsuie R, Sekiguchi S, Sakurai M, Sato Y, et al. Developmental regulation of ubiquitin C-terminal hydrolase isozyme expression during spermatogenesis in mice. Biol Reprod. 2004;71(2):515–21.
Osawa Y, Wang YL, Osaka H, Aoki S, Wada K. Cloning, expression, and mapping of a mouse gene, Uchl4, highly homologous to human and mouse Uchl3. Biochem Biophys Res Commun. 2001;283(3):627–33.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media New York
About this chapter
Cite this chapter
Bose, R., Manku, G., Culty, M., Wing, S.S. (2014). Ubiquitin–Proteasome System in Spermatogenesis. In: Sutovsky, P. (eds) Posttranslational Protein Modifications in the Reproductive System. Advances in Experimental Medicine and Biology, vol 759. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-0817-2_9
Download citation
DOI: https://doi.org/10.1007/978-1-4939-0817-2_9
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4939-0816-5
Online ISBN: 978-1-4939-0817-2
eBook Packages: MedicineMedicine (R0)