Abstract
Ubiquitination is one of the most diverse forms of protein post-translational modification that changes the function of the landscape of substrate proteins in response to stimuli, without the need for “de novo” protein synthesis. Ubiquitination is involved in almost all aspects of eukaryotic cell biology, from the best-studied role in promoting the removal of faulty or unnecessary proteins by the way of the ubiquitin proteasome system and autophagy-lysosome pathway to the recruitment of proteins in specific non-proteolytic signaling pathways, as emerged by the more recent discoveries about the protein signature with peculiar types of ubiquitin chains. Spermatogenesis, on its own, is a complex cellular developmental process in which mitosis, meiosis, and cell differentiation coexist so to result in the continuous formation of haploid spermatozoa. Successful spermatogenesis is thus at the same time a mixed result of the precise expression and correct intracellular destination of structural proteins and enzymes, from one hand, and the fine removal by targeted degradation of unfolded or damaged proteins as well as of obsolete, outlived proteins, from the other hand. In this minireview, I will focus on the importance of the ubiquitin system all over the spermatogenic process, discussing both proteolytic and non-proteolytic functions of protein ubiquitination. Alterations in the ubiquitin system have been in fact implicated in pathologies leading to male infertility. Notwithstanding several aspects of the multifaceted world of the ubiquitin system have been clarified, the physiological meaning of the so-called ubiquitin code remains still partially elusive. The studies reviewed in this chapter provide information that could aid the investigators to pursue new promising discoveries in the understanding of human and animal reproductive potential.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Ubiquitin code
- Ubiquitin-proteasome system
- Autophagy-lysosome pathway
- USP8
- Meiotic-sex-chromosome inactivation
- Mitophagy
- Acrosome biogenesis
- Centrosome inheritance
Background
Any cells of an organism have developed mechanisms to sense and respond to their environment. Post-translational protein modifications have evolved as the universal tool that cells have developed to switch on/off dynamic processes. The post-translational protein modification known as protein ubiquitination determines the half-life, stabilization, refolding, translocation and subcellular sorting of proteins crucial for cell physiology [1,2,3,4,5,6,7]. Moreover, modification by ubiquitination is critical for the down-regulation of steroid hormone receptors, plasma membrane receptors and transporters, and ion channels [7,8,9,10,11,12,13,14]. Cumulatively, the ubiquitin system functions as the key regulator of proteostasis, i.e., the maintenance of a healthy proteome essential for cell metabolism, cell proliferation and differentiation, organelle biogenesis, and stress adaptation [15].
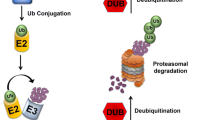
The ubiquitin (Ub) system is complex existing different hierarchical relationships among its various components [16, 17]. The signaling molecule resides in the 76-amino acid, 8.5 kDa Ub moiety covalently ligated to the selected protein through a series of actions that require the involvement of ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin protein ligases (E3s) [18,19,20]. E1s, E2s, and E3s work cooperatively to hallmark the protein in question by catalyzing the formation of an isopeptide bond between the C-terminal glycine of Ub and the amino group on the side chain of a lysine residue in the tagged protein (Fig. 1). Recent evidence has shown that Ub can also be attached to other residues, including cysteines, serines, threonines, and the N-terminus of the polypeptide backbone [21,22,23,24,25]. Further, the Ub tag in the substrate protein could consist in mono-, multi- and poly-ubiquitination. The lysine residues present in Ub are seven, namely, Lys6, Lys11, Lys27, Lys29, Lys33, Lys48 and Lys63 (Figs. 1 and 2). It follows that in polyubiquitination, additional Ub moieties can be added through conjugation to the same internal Lys residue as in the previously bound Ub moiety; this topology gives rise to homotypic Ub chains (Fig. 2). It has to be highlighted, however, that in addition to homotypic chains: (a) mixed Ub chains can be generated if different linkages alternate at succeeding positions of the chain (Fig. 2); (b) certain E3 ligases can generate branched Ub structures via Ub lysines 6, 27, and 48 through autoubiquitination [26] (Fig. 2). The different Ub-chain topologies thus resulting dictate different physiological consequences [23,24,25, 27] (Fig. 2). For instance, it was proposed that Lys48-based polyubiquitination serves as a signal for degradation by the proteasome [16], while monoubiquitination is rather considered a signal for non-proteolytic functions such as histone modification involved in epigenetic control of gene expression [28], budding of viruses [29], and cytoskeleton arrangement [30]. To add to the complexity, recent studies, however, have shown that mono- and multi-monoubiquitination could serve as degradation signal for some tagged proteins [27].
Patterns of protein ubiquitination. Upper half of the cartoon, The addition of Ub unities at specific lysine residues on the target substrate occurs through a series of actions that require the involvement of ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin protein ligase (E3); Lower half, Different topologies of ubiquitination: mono-ubiquitination occurs at a defined lysine residue while there is multi-mono-ubiquitination when multiple lysine residues are modified with one ubiquitin each. Poly-ubiquitination occurs when a polymeric chain of a substrate-attached Ub is just added to the substrate. These chains can be short and contain only two Ub molecules or long and incorporate more than ten Ub moieties. Here, it is illustrated the chain of four Ub molecules, i.e., the canonical tag for proteasomal degradation. In the boxed area, structure of Ub showing the seven lysine (K) residues and the methionine (Met) at the amino-terminus
Complexity of the Ub code. Upper half of the cartoon, The amino-acid sequence of Ub is reported; the lysine residues (K), evidenced in blue, are numbered starting from the N-terminus methionine 1 (M, red). Below, schematic representation of the increasing complexity of the poly-ubiquitin modification. Mono-ubiquitin can be extended on K-residues or the N-terminal M, thus giving rise to eight homotypic poly-ubiquitin chains. In addition to homotypic chains, there are heterotypic chains that are generated with more than one type of linkage in mixed or branched polymers. Further complexity rises from the cross talk between Ub and Ub-like proteins, such as SUMO (S) and Nedd8 (N8), or other post-translational modifications like acetylation (Ac) and phosphorylation (P). Lower half, Physiological roles associated with different types of Ub tags. (A). Proteasome-mediated degradation has been traditionally associated to poly-ubiquitination via K48; (B). Receptor-mediated endocytosis is often associated to multi-mono-ubiquitination as well as mono-ubiquitination; (C). K6- and K63-linked Ub chains are related to mitophagy while K27-linked poly-ubiquitin is involved in autophagy; (D). Mono-ubiquitination is associated to DNA damage while K63-linked chains are related to nuclear protein interactions as well as sumoylation is related to chromatin/heterochromatin modifications; (E). Intracellular protein trafficking and signaling (earmarked by yellow star) are associated individually to specific types of poly-ubiquitination (K63-linked, M1-linked, K11/K63 mixed and defined branched chains) as well as to mono-ubiquitination
Eukaryotic cells dispose of two major degradation systems, the ubiquitin–proteasome system (UPS) and the autophagy-lysosome pathway (ALP). The UPS is the canonical proteolytic route for cytosolic damaged, misfolded, and/or short-lived proteins [15, 16, 31]. The ALP functions predominantly in the degradation of biomacromolecules delivered by way of endocytosis, phagocytosis, autophagy, or biosynthetic transport [15, 31, 32], including the removal of dysfunctional or superfluous cellular organelles. Albeit apparently functionally separated, the two systems intersect and communicate; the turnover of proteasomes (proteaphagy) is, in fact, mediated by autophagy [33] and how the UPS and ALP collaborate has been recently dissected and elucidated by studying the control of ribosome recycling and turnover [34]. At the present state of knowledge with canonical certainties put in discussion (for example, no essentials of Lys48-based poly-ubiquitin chains for proteasome targeting), a not yet clarified enigma remains, i.e., which is/are the typology/ies of Ub signature, if any, necessary for substrate targeting to the either UPS or ALP systems. UPS and ALP, more recently, have been the object of extensive studies and documentation. For this reason, from a general point of view, I remind to refer to the excellent reviews, some of which reported here, already available in the literature to avoid redundant repetitions.
Protein ubiquitination, however, regulates also non-proteolytic events, including membrane protein trafficking, vesicular-based transport, synaptic plasticity, protein kinase activation, DNA repair, and chromatin dynamic [5, 11, 17, 20, 35,36,37]. To date, several reports have shown how ubiquitination is critical for proper protein localization and recognition by signaling and regulatory complexes, thus affecting from a general point of view cellular signaling and/or homeostasis [6, 24, 25, 38, 39]. In such a context, Lys63-linked Ub chains were assumed to have a non-degradative role in cellular signaling and intracellular trafficking [24, 40] (Fig. 2).
Ubiquitination of target proteins is a reversible modification. The enzymes that oppose the function of E3 ligases are known as deubiquitinases (DUBs) [41,42,43,44,45,46,47]. Protein deubiquitination is important for several reasons. When it occurs before the commitment of a substrate to either UPS or ALP proteolysis, it negatively regulates protein degradation. To this regards, it has been suggested a kind of proofreading mechanism wherein ubiquitin is removed from proteins inappropriately targeted to the proteolysis [41]. On the other hand, surely some DUBs work as parts of the proteasome itself by removing Ub moieties from proteins committed to degradation; thereby these DUBs allows the recycling of free Ub within the cell and keep the proteasome free of unanchored Ub chains that can compete with ubiquitinated substrates for Ub-binding sites [43]. Evidence has also accumulated about the different ways through which DUBs recruit their protein substrates. There are DUBs that bind directly to an Ub signal that they just then cleave without any direct relation with the protein substrate (a kind of aspecific housekeeping enzyme) [41,42,43], and there are DUBs that target either selected types of Ub chains [46,47,48] or selected proteins [47], thus influencing specifically peculiar cellular pathways and/or processes. These DUBs, besides the canonical catalytic domain, possess additional protein-protein interaction domains. In brief, DUBs are far from being uniform in structure and function and this feature may explain because progress in understanding DUB function has lagged behind that of the Ub conjugation machinery. The human genome encodes about 100 DUBs while about 20 DUBs exist in Saccharomyces cerevisiae. At the present, six structurally distinct DUB families have been described: Ub-C-terminal hydrolases (UCHs; 4 members in humans), Ub-specific proteases (USPs; 54 human members), ovarian tumour proteases (OTUs; 16 human members), the Josephin family (4 human members), the motif interacting with ubiquitin (MIU)- containing novel DUB family (MINDYs; 4 human members) and a family of Zn-dependent JAB1/MPN/MOV34 metalloenzymes (16 human members) [49].
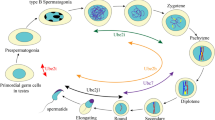
Spermatogenesis is the process that leads to the production, starting from a pool of diploid stem cells named spermatogonia, of highly differentiated haploid spermatozoa. The complexity of spermatogenesis relies collectively on the coexistence of mitosis, meiosis, and cell differentiation in a unique process. The Ub system is essential for the proteostasis of each eukaryotic cell; there are however a number of properties that appear to be peculiarly related to the processes that articulate spermatogenesis. Albeit not intended to be an exhaustive review, some recently emerged aspects of Ub system in spermatogenesis that are discussed here can serve as a guide for tackling future studies on this topic.
Proteolytic Functions of the Ub System along Spermatogenesis
Spermatogonial Development
To date, the role of the UPS in protein turnover in spermatogonial stem cells (SSCs)/spermatogonia (SPGs) has been rather unexplored. In the testis while some SSCs self-renewal, other SSCs differentiate; this occurs for the entire adult life of the male. In vitro, SSCs undergo asymmetric divisions; thereby, it was proposed that SSC asymmetric divisions contribute to their self-renewal and differentiation [50]. One of the first members of the Ub system discovered to be present in the testis was the deubiquitinating enzyme Uchl1, otherwise known as PGP 9.5, [51]. SSCs/SPGs express Uchl1 that regenerates monoubiquitin from ubiquitinated proteins targeted to the proteasome. Interestingly, Uchl1 was found to segregate asymmetrically to the two daughter cells from asymmetric division and, more specifically, the daughter cell with a high Uchl1 level resulted to be positive to the undifferentiated spermatogonial marker Plzf; conversely, the one with a low Uchl1 level expressed the differentiated spermatogonial marker c-Kit [52]. Remarkably, the authors succeeded to show asymmetric segregation of Uchl1 and Plzf not only with cultured SSCs, but also in situ in seminiferous tubules [52]. This suggests that a full functionality of Uchl1 is consistent with maintaining the undifferentiated state of SSCs. It is however fair to remember that there is no general agreement about the possibility of asymmetric division of SSCs in vivo in mammals [53]. A recent, sophisticated study suggests a different and generic mechanism underlying the fate of SSCs: these stem cells appear not to rely on asymmetric division, but SSCs with higher level of SHISA6, a cell-autonomous Wnt inhibitor, remain in the undifferentiated cell pool, while those with lower levels of the inhibitor are inclined to differentiate [54].
The HR23B gene encodes a mammalian homolog of Saccharomyces cerevisiae RAD23, a nuclear protein containing an ubiquitin-like domain involved in nucleotide excision repair (NER). Defective NER is associated with three clinically and genetically heterogeneous human syndromes: xeroderma pigmentosum (XP), Cockayne syndrome (CS), and trichothiodystrophy (TTD) [55]. HR23B gene has been validated as a sensitivity determinant for histone deacetylase inhibitor (HDACI)-induced apoptosis so that HR23B−/−mice were generated to assay its biological relevance in vivo [56]. Unexpectedly, HR23B deficiency did not result in a NER defect, but HR23B−/− mice showed impaired embryonic development and, if survived, in addition to a retarded growth, male mice resulted to be sterile. This sterility is consequence of an early failure of spermatogenesis since HR23B knockout yields a phenotype like that known as Sertoli cell-only syndrome [56], with the total absence of male germ cells. Recently, HR23B/RAD23 have been shown to interact with the proteasome to which it delivers multi-ubiquitinated proteins and to play a proteolytic role involved not only in NER, but also in stress response, transcription and ER-associated protein degradation [57].
By gene expression analysis using microarrays, numerous genes codifying for gene products related to the Ub system have been identified in SSCs/SPGs. Among these there are the E1 enzymes Uba1 and Uba6, E2 enzyme Ube2, E3 enzymes Huwe1, Trim47, and Rnf149 [58]. A part the evidence of their expression, to date these UPS genes are still waiting for elucidation of their specific functions in SSC/SPG biology. An exception could be Huwe1. Despite the fact that Huwe1 has a broad range of tissue expression with a cytoplasmic protein distribution, in neurons and spermatogonia/pachytene spermatocytes the E3 ubiquitin ligase is present in the nucleus [59]. This aspect, together with the fact that in vitro experiments have shown that Huwe1 binds to the E2 enzyme UBC4 thus ubiquitinating the histones H2A and H2B [60], has led to propose that Huwe1 plays an important role in the proliferation and differentiation of spermatogonia, as well as in the regulation of sex chromosomes inactivation during meiosis. Collectively, these roles have been experimentally confirmed by inactivation of the Hewe1 gene at key stages during spermatogenesis [61].
Meiosis
The importance of the Ub system during meiosis (and mitosis) emerged outstanding since the discovery (late 1980s/early 1990s), through studies of cell division in frog and clam oocytes [62, 63], that it is the cyclin-Cdk complex to drive cells into M phase and that the concentration of cyclin fluctuates, rising gradually during interphase to then falling rapidly to zero at the end of M prophase.
Meiosis is a highly regulated cell division process subdivided in sequential phases such as meiotic entry, genetic recombination of homologous chromosomes, meiotic sex chromosome inactivation (MSCI), double cell division, and meiotic exit. Despite temporal differences in their meiotic programs, both sexes require a fully functional Ub system, like various studies exploiting gene-targeting technologies have shown. Given the complexity of the epigenetic regulation, including that relies on the ubiquitin code, of meiosis in mammals, I suggest here to refer to published reviews [64,65,66]. Below, a few representative examples related to UPS in spermatogenesis are considered.
Meiotic Entry
Fizzy-related 1 (FZR1) is an activator of the Anaphase promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase that promotes the metaphase-anaphase transition in mitosis [67]. Recently, it is emerging a role for APC/CFZR1 activity in oocyte meiosis [68] while loss of APC/CFZR1 activity in the male germline leads to infertility due to the absence of mature spermatozoa [69]. As to the male infertility, spermatogonia of APC/CFZR1−/− undergo abnormal proliferation and show delayed entry into meiosis. Although early recombination events could initiate, the developing spermatocytes fail to progress beyond zygotene and undergo apoptosis. The authors [69] proposed that the requirement for APC/CFZR1-mediated cyclin B1 degradation is crucial in early meiosis because cyclin B1 must be kept in check during early prophase I so to allow the completion of recombination events.
Meiotic Progression
There are a number of UPS genes that have been described to be potentially involved at various time-points of meiotic progression, like the components of APC/C complex cited above, (for a review, see ref. [58]); for most of them, however, the effective function of their gene products during spermatogenesis has yet to be elucidated. Proteolysis is rightly thought to be essential in regulating the cell cycle also in meiosis; checkpoint protein requirements, however, are emerged to be different between somatic cell mitosis and germ cell meiosis. Accurate chromosome segregation during spermatogenesis, for instance, is highly dependent on BubR1, but not Mad2, Bub3, Rae1 and Nup98 that are all components of the mitotic spindle checkpoint, the stability of which is down-regulated through the UPS [70]. Therefore it may be that the control of meiotic cell cycle is, at least partially, under the surveillance of non-conventional and germ cell signal-responsive variants of the Ub system. This topic deserves to be better defined in the future.
Spermiogenesis
Spermiogenesis is an extraordinary differentiation process through which haploid round spermatids metamorphose to become streamlined spermatozoa with en extreme cell polarization and a flagellum conferring the capacity to move. In higher eukaryotes spermatozoa are in fact the only cells that leave their organism to exert out of the confines of the body their basic function, fertilization. The differentiation steps include the morphological changes occurring as consequence of the nuclear chromatin compaction, acrosome formation, axoneme development, mitochondria reorganization, head elongation, tail assembling and, last but not least, removal of most cytoplasm and cellular structures/organelles now acting only as a rubbish.
Chromatin Condensation
Chromatin remodeling is a gradual process continuing from spermatogonia proliferation, through meiosis, to the formation of mature sperm. To acquire its stream-like shape and simultaneously protect the genetic material during the long trip to the egg, sperm have evolved a peculiar chromatin organization. Briefly, nucleosomes are disassembled, transition proteins replace somatic and testis-specific histones and, in turn, protamines replace transition proteins. As a consequence, sperm acquires a highly condensed and inaccessible organization of chromatin with haploid DNA efficiently packaged into peculiar, small donut-shaped, organizational units known as toroids [71]. Interest in studying the regulation of sperm chromatin organization is multiple, because any alterations in protein composition or integrity of the chromatin may contribute to male infertility.
Moreover, many proteins that function only temporarily are formed during replacement of nucleoproteins [72, 73]. An example is the H2A histone variant H2A.Lap1. H2A.Lap1 dynamically loads onto the inactive X chromosome in later stages of round spermatid, thus enabling the transcriptional activation of genes that were previously repressed, i.e., the MSCI (meiotic sex chromosome inactivation) silenced genes [74]. As parenthetic clause, it is here noticed that the activation of sex-linked genes required for male reproduction is briefly resumed successively in the section of this minireview devoted to non-proteolytic functions of the Ub system along spermatogenesis. The histone-to-protamine transition is however essential to produce fertile spermatozoa; consequently, histones as well as the temporarily functioning/transition proteins must be eliminated. The presence of efficient degradation machinery, possibly residing within the nucleus of spermatids themselves, is therefore mandatory. The UPS provides, at least in part, this machinery. Histones are in fact targets of multiple dynamic post-translational modifications, particularly on their C-terminal tails. The major ones are acetylation, methylation, phosphorylation, sumoylation, and, last but not least, ubiquitination [73, 75]. A testis-specific isoform of the E2 enzyme UBC4, namely UBC4-testis, is induced in round spermatids and early elongated spermatids [76]. UBC4-testis interacts with the testis-specific E3 ligase E3Histone/LASU1 in ubiquitinating histones H1, H2A, H2B, H3 and H4 at early spermiogenesis [60]. The poly-ubiquitinated histones are then targeted to the proteasome for degradation. Evidence of the presence of poly-ubiquitinated proteins as well as of proteasomes in the nuclei of both rodent and human spermatids has been provided [77]. Lastly, it could be worth of mention to remember that studies in Drosophila melanogaster have shown that 12 of the 33 subunits of the 26 S proteasome are represented by paralogous genes [78] and, in each case, one of the paralogos is testis-specific [79]. Knock out of one of these paralogs, namely a6T, is resulted in abnormal nuclear morphology and maturation yielding a male sterile phenotype [80] suggesting a role of sperm proteasome in chromatin remodeling.
Acrosome Biogenesis and Axoneme Development
Acrosome formation has been described to be coupled to an increase in anti-ubiquitin labeling [81]; further, proteasomes have been found to mark the surface of the developing acrosome [82] and an E3 ligase, UBR7, has been identified inside the acrosome [83]. A direct involvement of the proteolytic function of UPS in the biogenesis of the vacuole, however, has still to be found. Parallelly, E3 Ub ligases like MARCH10 [84] and MARCH7 [85] as well as proteasomes [82] have been detected in the mammalian sperm flagellum. Interestingly, spermatozoa from male mice deficient for the Ub ligase HERC4 are resulted to be affected with a 50% reduction in tail motility, although HERC4 substrate targets remain elusive [86]. To date it is, however, not still defined a role of the UPS-mediated degradation in the axoneme formation.
Mitochondria Reorganization
During spermatogenesis the number of mitochondria undergoes spatiotemporal variation. Mammalian spermatogonia have about 2000–3000 mitochondria/cell whereas a mammalian sperm has about 70–80 mitochondria, all confined in the mitochondrial sheath that is the final element of the sperm tail to be formed. Remarkably, the number of mitochondria decreases dramatically, concomitantly with their peculiar redistribution, in the haploid phase of spermatogenesis. Thereby, spermiogenesis implies a massive mitochondrial reorganization that requires, from one hand, removal/degradation of most organelles and, from the other hand, the microtubule-mediated transport of mitochondria to the forming tail midpiece [87]. It is to decipher and clarify on the basis of which mechanism most mitochondria are lost but a number survives going to relocate at the tail mitochondrial sheath. This aspect has so far received minimal attention. There could be a signal that hallmarks the mitochondria that have to be eliminated or, vice versa, that have to be saved. Alternatively, the two different fates, i.e., survival or elimination, could reside in a merely casual passive selection. Undoubtedly, elongating spermatids to transform into elongated spermatids loss most their cytoplasm, where the majority of mitochondria reside, and this occurs in the form of the cytoplasmic lobe/residual body that is then resorbed mostly by Sertoli cells [88]. Such phenomenon exhibits the feature and fate of a phagosome englobed in the host cell; in other words, sperm mitochondria are not eliminated through endogenous mitophagy, i.e., the autophagy-mediated destruction of damaged mitochondria. On the other hand, mitochondria in both spermatids and mature sperm are ubiquitinated [89]. Although the exact mechanism that supervises such (global or partial?) ubiquitination is still vague, this indicates that the mitochondria destined to the tail mid-piece, the only to be present in mature sperm, are ubiquitinated. This argument will be further considered in the section “Passing of the baton between Non-Proteolytic functions and Proteolytic functions of the Ub system in sperm biology”.
The other crucial aspect, in addition to their massive removal, of mitochondria reorganization during spermiogenesis is the formation of the tail mitochondrial sheath, a sperm-specific structure wrapping both the axonemal complex and nine outer dense fibers in a left-handed double helical array. It is evident that becoming part of an ‘ex novo’ developed organelle that highly polarizes the differentiated sperm, the flagellum, requires unique structural-remodeling capacities and a well-functioning cytoskeleton with its ability to transport protein complexes, vesicles, and organelle over long distances. As to this topic I suggest to read the review [87], albeit the written ‘codes’ that determine which cargos bind to a protein adaptor or motor protein, differently from somatic cells, remain virtually unknown.
Non-proteolytic Functions of the Ub System along Spermatogenesis
Ubiquitination could also regulate non-proteolityc events such as protein activity, protein interactions and subcellular localization [25], transport of newly synthesized mature proteins to the membranes [3, 13], DNA damage repair and DNA replication mechanisms [90], and cellular signaling by recruiting proteins to concur to particular signaling pathways like inflammatory signaling and apoptotic cell death [91]. As already said, the complexity of spermatogenesis relies on the coexistence of mitosis, meiosis, and cell differentiation in a unique process where the Ub system plays direct role also in non-proteolytic (NP) control mechanisms. For instance, modifications in chromatin organization are related not only to the histone-to-protamine transition with related histone degradation, but also to peculiar phenomena like the meiotic synapsis and desynapsis of chromosomes, homologous recombination, and meiotic sex chromosome inactivation (MSCI). Parallelly, the metamorphosis of haploid spermatids into spermatozoa is accomplished also through non-proteolytic functions by components of the Ub system. For space constraints and convenience of clarity for the readers, here I highlight only few aspects as representative of the topic that heads this paragraph.
NP-Ub System and Meiotic DNA
Histone ubiquitination can both stimulate and repress various cellular processes [92]. For example, ubiquitination of H2A at gene promoter regions suppresses gene transcription [93,94,95], while intragenic ubiquitination of H2B facilitates transcription elongation [96,97,98,99]. Histone ubiquitination is also associated with DNA damage responses where H2A and H2B ubiquitination are enriched at sites of DNA damage [100,101,102,103]. These post-translation chromatin modifications can occur both in somatic and germ cells and they are not further discussed here. Below, events characterized to be spermatogenic-specific are reported.
During mammalian male meiosis, the heteromorphic sex chromosomes (X and Y) condense in a separate chromatin domain known as the XY or sex body [104]. X and Ychromosomes are largely heterologous and show homologous synapsis only at the small pseudoautosomal region (PAR). BRCA1, a multifunctional DNA repair protein that possesses transcriptional, ubiquitin ligase (E3), and heterochromatin-related gene silencing activity [105], detects the non-synapsis regions of chromosomal axial elements and promotes the localization of ATR, a member of the PI3-like kinase family, to the X, Ychromosomes; this will lead to the phosphorylation of histone H2AX at serine 139 [106]. Hence, i.e., from its formation on, the XY body is positive for phosphorylated H2AX [107]. Phosphorylation of H2AX initiates repression of genes on the sex chromosomes [108], a process called ‘meiotic sex chromosome inactivation’ (MSCI). Remarkably, because all this process can occur, H2AX needs to be previously ubiquitinated; the polycomb repressive complexes PRC1 and PRC2 catalyse the mono-ubiquitination of H2AX at lysine 119 [75], giving thus the start to the process of MSCI. Moreover, the E2 enzyme HR6B, which is enriched in the XY body of pachytene spermatocytes, functions in the maintenance of X chromosome silencing in both spermatocytes and spermatids. Conversely, Hrb6-KO mice show derepression of X-chromosomal gene activity that leads to abnormal global upregulation of gene transcription from the X chromosome [109].
RNF8 possesses a RING domain at its C-terminus and is counted among the E3 Ub ligases. RNF8 also participates in the DNA damage response and ubiquitinates histones, promoting the recruitment of downstream DNA damage response factors, such as 53BP1, BRCA1 and Rad51 [100, 110, 111]. It was suggested that RNF8 could play an important role in the chromatin remodeling occurring during MSCI. A study carried out to check such a hypothesis led to the generation of RNF8-deficient mice uncovering, however, a surprising result. Absence of H2A ubiquitination by RNF8 in spermatocytes of RNF8-KO mice did not affect XY body formation, MSCI, or meiotic progression, but its absence in elongating spermatids invalidated nucleosome removal at the histone-to-protamine transition; consequently, this results in a defective spermiogenesis and infertility [95]. Moreover, a further study [112] has shown that ubiquitination-deficient mutations in MIWI protein, a Piwi family member - Piwi proteins are essentials for gametogenesis in animals [113] - cause male infertility. The Authors [112] have identified the culprit; MIWI binds, sequestering it, RNF8 in the cytoplasm of early spermatids and MIWI degradation by APC/C in late spermatids is required for nuclear translocation of RNF8. However, ubiquitination-deficient mutations in MIWI prevent MIWI from degradation so that RNF8 cannot translocate into the nucleus to catalyze histone ubiquitination and trigger histone removal.
As last mention to illustrate the ubiquitin regulatory network that is at the head of the meiotic sex-genes silencing/postmeiotic sex-genes activation, I report the intriguing model proposed by Adams and co-workers [114]. According to the authors “regulation of ubiquitin leads to the organization of poised enhancers and promoters during meiosis, which induce subsequent gene activation from the otherwise silent sex chromosomes in postmeiotic spermatids”. Due to MSCI, silenced sex-genes must escape silencing for activation in spermatids thereby ensuring their function for male reproduction. Adams et al. [114] succeded in demostrating that RNF8 and SCML2, a germ-line specific Polycomb protein, cooperate to regulate histone ubiquitination during meiosis to establish active histone modifications for subsequent gene activation. For example, the authors show that SCML2 deubiquitinates RNF8-independent H2AK119ub but does not deubiquitinate RNF8-dependent polyubiquitination. RNF8-dependent polyubiquitination is required for the establishment of H3K27 acetylation, a marker of active enhancers, while persistent H2AK119ub inhibits establishment of H3K27 acetylation.
As concluding remark, I make a general recall to the epigenetic of the male gamete. Major epigenetic signatures include DNA methylation, histone modifications/localizations, and expression profiles of non-coding RNAs. These marks drive gene expression patterns in the cell and have a profound impact in the early phases of embryo development. Fertile mammalian sperm have epigenetic modifications consistent with gene ‘poising’ at the promoters of genes involved in development; these epigenetic signatures include the localization of retained histones that are not removed at the histone-to protamine transition and are signed by ubiquitin modifications. The field of epigenetics is burgeoning, thus I invite the readers to give a glance at this topic [114,115,116,117].
NP-Ub System and Spermiogenesis
The post-genomic era has provided insight into the complexity of the Ub system [7]; comprehensive proteomics studies have identified tens-of-thousands of ubiquitination sites on thousands of proteins [118]. As reported above, proteins can be mono-, multi-, poly-ubiquitinated while Ub molecule has seven Lys residues all of which can be ubiquitinated; moreover, new emerging findings reveal that Ub can be not only ubiquitinated, but also modified by other modifications. These last include SUMOylation, i.e., the addition of the Ub-like modifier protein SUMO that is conceptually similar to polyubiquitin chain formation [119], phosphorylation [120, 121] and acetylation [122]. Altogether, these complex patterns constitute a ‘ubiquitin code’, which is read by hundreds of proteins that incorporate ubiquitin-binding domains. Thus the more recent experimental evidence has shaken the long-standing dogma according to which Ub constitutes a targeting signal for protein degradation only. As to the Ub system and spermiogenesis, however, it remains a consistent part of work to be still done; most studies in the literature, in fact, have been devoted to sperm ubiquitination as ‘degradative’ signal only.
NP-Ub System and Activation of Silenced Genes
As to this topic, I send back to the works just discussed above dealing with the male epigenome, in particular to the silencing/activation of sex-linked genes.
NP-Ub System and Acrosome Biogenesis
Protein ubiquitination functions in protein trafficking in both endocytic and secretory pathways [32]. One of the cytomorphogenic events that hallmark spermiogenesis is the biogenesis of the acrosome. The acrosome is a unique membranous organelle located over the anterior part of the sperm nucleus, rich of hydrolytic enzymes and considered to be indispensable for fertilization [123]. Originally described as a modified lysosome, it was then proposed as a direct Golgi-derived secretory vesicle and, more recently, as a lysosome-related organelle (LRO) [124, 125]. According to the experimental evidence that both the endocytic and biosynthetic machineries concur to acrosome biogenesis [123,124,125,126,127,128] and that acrosome functionality reflects the modular LRO-like structural organization [123, 128], the notion that the acrosome is a LRO is now currently accepted. The endosomal system constitutes a network of progressively maturing vesicles characterized by modular organization, high spatial regulation and interconnectivity with the biosynthetic route. The ubiquitin-specific protease USP8, originally named UBPy [129], participates in the endosomal sorting of transmembrane proteins both in somatic and male germ cells [127, 128, 130,131,132,133] where it interacts with the proline-rich SH3 domain of the signal-transducing adaptor Hbp/STAM2, a component of the endosomal sorting complex ESCRT-0 [124, 132]. USP8 deubiquitinates both cargo proteins, typically signaling molecules as transmembrane tyrosine kinase receptors [129,130,131, 133], and ESCRT-0 proteins [132] thus modulating both the function of the signaling molecules and the stability of components of the endosomal trafficking machinery (for a recent review see [134]). The proteolytic cleavage of USP8 increases the DUB enzymatic activity whereas USP8 phosphorylation-dependent association with 14–3-3 proteins inhibits the deubiquitinating activity [135]. Somatic mutations of human USP8 cause Cushing’s disease [136, 137] and defects in the down-regulation of USP8 are found more and more to be related to tumorigenesis [134, 138]. As to germ cells, human USP8 has been identified as candidate gene for male fertility traits [139, 140]. Consistently with the above findings, USP8 is resulted to cooperate in acrosome biogenesis by regulating the trafficking of vesicular cargoes destined to the forming LRO; one of such cargos has been identified in a molecular variant of the tyrosine kinase membrane receptor MET [127]. As further remark, it is to remember that USP8 possesses a MIT (microtubule interacting and trafficking/transport) domain at its aminus-terminus, which could provide a direct link between the sorted vesicular cargo and microtubules [124]. Spermatids are characterized to exhibit peculiar microtubule arrays, such as the cortical microtubule network (early spermatids), the manchette (elongating spermatids), the axonemal microtubules (elongated spermatids); all these cytoskeletal structures function as tracks during spermiogenesis [87, 124, 127]. Like the cortical microtubule array supplies the tracks along which USP8-signed cargo is trafficked to the acrosome, it might be that USP8 is involved also in the manchette-mediated transport (USP8 locates on the manchette both at light [124] and electron microscopy [128] level) and/or intra-flagellar transport. These two hypotheses are still to be explored.
In conclusion, acrosomogenesis is a clear example of a non-proteolytic involvement of the Ub system. It remains, however, to decipher which are the Ub marks (Lys63-, Lys11-, Lys27-, Lys33-, Lys48- linked or other types of linkage) and ubiquitin chain architectures that sign the targeted cargos and how these signatures are “translated” by the spermatid protein transport systems to build the acrosome.
Passing of the Baton Between Non-Proteolytic Functions and Proteolytic Functions of the Ub System in Sperm Biology
As already noticed, the number of mitochondria decreases notably during spermiogenesis and the mitochondria located at the sperm tail mid-piece are ubiquitinated. It remains as an unsolved question to understand the physiological meaning of such ubiquitination (signal for degradation or signal for regulation of organelle localization or both? In other words, may be that in order to be successively degraded these organelles must be recognized and, therefore, signaled?). More E3 Ub ligases have been coupled to spermatid/sperm tail suggesting a potential involvement for each of these in the construction of the flagellum [83,84,85]. To date, however, these assertions remain generic.
In Drosophila spermiogenesis the pink1-parkin pathway has been shown to play a critical role in regulating mitochondrial morphology and function [141]. PINK1 encodes a putative serine/threonine kinase with a mitochondrial targeting sequence, while PARKIN is an E3 Ub ligase responsible for directing the autophagic clearance of defective mitochondria [142]. In human PARKIN mutations are responsible for a familiar form of autosomal recessive juvenile parkinsonism [143]. At present, it is not known if there is a direct involvement of PARKIN in mammalian spermiogenesis/tail morphogenesis. It has been, however, demonstrated that the murine PACRG (Parkin co-regulated gene) protein interacts with MEIG1 (meiosis-expressed gene 1) that migrates to the manchette in elongating spermatids; together, PACRG/MEIG1 form a complex in the manchette that is necessary to transport cargos, such as SPAG16L, to build the sperm tail [144]. As said, USP8 too locates at the spermatid manchette and, to complete the localization of USP8 in haploid mouse male germ cells, USP8 localizes also at the centrosome and principal piece of the flagellum [82]. In somatic cells USP8 is known to play a critical role in the control of mitochondrial quality; it is, in fact, required for the efficient recruitment of PARKIN to depolarized mitochondria so to trigger subsequent mitophagy [145]. Such a direct interaction between a DUB and E3 ligase is not surprising; E3 ligases are often regulated by DUBs [24] and in the case of PARKIN, USP8 deubiquitinates directly the ligase by acting on non-canonical Lys6-linked Ub chains [145]. Thus, when levels of USP8 are reduced or its activity is inhibited, an accumulation of Lys6-linked Ub conjugates on PARKIN delays its overall activity in mitochondrial quality control. Consequently, USP8 activity is crucial for Parkinson’s pathogenesis [145] as indicated also by further proof. Another USP8 protein target is αlpha-synuclein. USP8 removes Lys63-linked Ub chains on αlpha-synuclein, thus contributing to the accumulation of misfolded αlpha-synuclein into Lewy bodies, cellular inclusions characteristic of the Parkinson disease [146]. Upon the experimental evidence on USP8 capacity of hydrolyzing different Lys-linked Ub chains [147] with repercussions on mitochondrial quality, it could be worthy of consideration to investigate about a potential involvement of USP8 in tailoring the ubiquitin signature (and fate) of mitochondria during sperm tail development. A recall to the wobbler mice might be useful. The Wobbler mouse is a spontaneous mutant used as model of motor neuron degeneration [148] associated to male infertility (wobbler sperm are acrosomeless and immotile, with a tail mid-piece characterized by a disorganized and mislocalized mitochondrial sheath, see [128, 149]). Relevant USP8 upregulation has been found in wobbler cells (in particular, spinal cord oligodendrocytes and spermatids) selectively affected by the wobbler mutation [150, 151]. This suggests that increased levels of USP8 could reflect the induction of USP8-mediated (rescuing or maladaptive) responses to the disorder.
A statement that is widely accepted among reproductive/developmental biologists and geneticists is that almost all eukaryotic animals inherit their mitochondria from the maternal parent. Until some decades ago, the prevailing explanation for such a phenomenon has been a passive model of simple dilution of the few paternal mitochondria by an excess copy number of the oocyte mitochondria [152]. Recent studies in Caenorhabditis elegans, which produces non-flagellated amoeboid sperm with mitochondria of canonical morphology, have brought to the light the involvement of autophagy in degradation of paternal mitochondria after fertilization [153]. Further research towards such a direction in the mouse has shown the presence of the autophagy receptor p62 and the ubiquitin-like modifier of autophagy LC3 in the sperm tail [154]. A successive study carried out always on mouse yields contrasting results [155]. The Authors employed embryos obtained by transgenic oocytes, expressing GFP-tagged autophagosome LC3, which were fertilized with transgenic spermatozoa bearing red fluorescent protein (RFP) labeled-mitochondria. It was thus provided evidence against sperm mitophagy; the authors stated in fact that maternal inheritance of mtDNA is not an active process of sperm mitochondria elimination achieved through autophagy, but may be a passive process as a result of pre-fertilization sperm mtDNA elimination [155]. Successively Politi and coworkers [156], by investigating the fate of Drosophila paternal mitochondria after fertilization, have found that paternal mitochondrial destruction is mediated by a common endocytic and autophagic pathway that implies a divergence from the classic autophagic pathway of damaged mitochondria. Still more recently, a study from Sutovsky’s group [157] has established that sperm mitophagy, at least in higher mammals, occurs post-fertilization and relies on a combined action of both ALP and UPS systems employing an unconventional, ubiquitin-recognizing autophagic pathway independent of canonical autophagy receptors such as LC3. Since propagation of paternal mitochondrial genes results in heteroplasmy, which could be potentially detrimental for embryo development [158], it would be desirable that this topic could be clarified, at least in human, in consideration of the diffused employment of assisted reproductive technologies.
As conclusive mention, I want to reserve a hint for another sperm organelle that, at a first glance, could appear to be out of place in such a context, the centrosome. Generally speaking, centrosome is a non-membrane bound organelle composed of two centrioles surrounded by pericentriolar material (PCM) where γ-tubulin nucleates microtubules as a part of the centrosomal γ-tubulin ring complex (γ-TuRC) [159]. Centrosomes are in fact major microtubule-organizing center of the cell (MTOC). Contrary to mitochondria, at fertilization sperm centrosome is inherited as first embryonic MTOC indispensable for early embryogenesis [160]. As known the oocyte’s centrosome is reduced and casts off into the first and second polar body during oogenesis in mammals [161]; so, once a sperm enters the oocyte’s cytoplasm at fertilization, the male gamete provides the essential centrosome in the form of the distal centriole that previously functioned as basal body for the development of the sperm tail. Centrosomes, however, contain also proteasomes [82, 162] that are thought to regulate the degradation of local ubiquitin-conjugates. Indeed, more molecular species referable to components of the Ub system have been found to reside at the centrosome and results have been obtained indicating that these selected components participate in the ubiquitin-dependent regulation of centrosome architecture [163]. Going back to the assembly of the first centrosome upon fertilization in mammals and, more generally, in vertebrates, it is now widely shared, with the exception of mice and some other murine animals, that the zygote forms its centrosome from the paternal distal centriole and from a pool of maternal factors that have been characterized to some extent but non been comprehensively identified [164]. Clearly, the enrichment in the research on centrioles and PCM during fertilization and embryonic development could improve our understanding of fertilization and aid in more efficient diagnoses of human infertility.
Concluding Remark
In this review, I have attempted to provide an overview of how the Ub system in its integrity is not only involved, but dictates the biological progressing of spermatogenesis. In comparison to other post-translational modifications like phosphorylation, the temporal, spatial, and substrate context of ubiquitination is extremely intricate. The complexity of the Ub signal has recently become even more evident with the discovery that Ub itself can be post-translationally modified, including its phosphorylation [25]. Here, I have deliberately avoided dealing this last topic because it is emerging at present and no research in this connection has been so far addressed to spermatogenesis. Being not this an exhaustive review, my point of view has been to supply sufficient information to the readers to recognize that Ub system is not only as a blunt tool used during spermatogenesis to degrade proteins via the proteasome. This view has been, generally speaking, superseded in the last years with the impressive acquisition of data on the plethora of cellular functions that the Ub system governs; this has to be applied to spermatogenesis too. What may become the next frontier in ‘Ub system and male germ cell differentiation/function’ can be the NP-roles of Ub system during spermatogenesis. The idea that ‘signaling’ pools of ubiquitinated proteins could compete with the ‘proteasome-targeted’ pools provides a challenge to identifying and studying the roles of the Ub chain types that characterize such signaling pools. Here, some proposals for further investigation are put forward. To recall only some examples: (a) Which Ub modification does include the localization of retained histones that are not removed at the histone-to protamine transition?; (b) Which are the Ub sorting signals that, like topogenic sequences, address selected proteins to an unconventional organelle such as the acrosome is?; (c) Given that Ub chains are formed as structurally distinct polymers via different linkages, which is/are the Ub chain/s that promote/s protein trafficking at the manchette and centrosome/basal body?; (d) Through which sorting signal, if any, are spermatid mitochondria selected for their removal by Sertoli cell’s phagocytosis and/or destination to the tail mitochondrial sheath? Which Ub chain topology does mark the sperm tail mitochondria?; and still other, going on, unsolved questions.
The improved understanding of Ub-system biology during spermatogenesis could be hopefully employed in the exploration of human reproduction/fertility and, given the strong increase in the use of Assisted Reproductive Technologies, pre-implanted ART offspring.
References
Hershko, A., Ciechanover, A., Heller, H., Haas, A. L., et al. (1980). Proposed role of ATP in protein breakdown: Conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proceedings of the National Academy of Sciences of the United States of America, 77, 1783–1786.
Hochstrasser, M. (1992). Ubiquitin and intracellular protein degradation. Current Opinion in Cell Biology, 4, 1024–1031.
Hicke, L., & Dunn, R. (2003). Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annual Review of Cell and Developmental Biology, 19, 141–172.
von Mikecz, A. (2006). The nuclear ubiquitin-proteasome system. Journal of Cell Science, 119, 1977–1984.
Kirkin, V., & Dikic, I. (2007). Role of ubiquitin- and Ubl-binding proteins in cell signaling. Current Opinion in Cell Biology, 19, 199–205.
Acconcia, F., Sigismund, S., & Polo, S. (2009). Ubiquitin in trafficking: the network at work. Experimental Cell Research, 315, 1610–1618.
Clague, M. J., Heride, C., & Urbé, S. (2015). The demographics of the ubiquitin system. Trends in Cell Biology, 25, 417–426.
Abdel-Hafiz, H. A., & Horwitz, K. B. (2014). Post-translational modifications of the progesterone receptors. The Journal of Steroid Biochemistry and Molecular Biology, 140, 80–89. https://doi.org/10.1016/j.j
Zhu, J., Zhao, C., Kharman-Biz, A., Zhuang, T., Jonsson, P., Liang, N., Williams, C., Lin, C. Y., Qiao, Y., Zendehdel, K., Strömblad, S., Treuter, E., & Dahlman-Wright, K. (2014). The atypical ubiquitin ligase RNF31 stabilizes estrogen receptor α and modulates estrogen-stimulated breast cancer cell proliferation. Oncogene, 33, 4340–4351. https://doi.org/10.1038/onc.2013.573
Marmor, M. D., & Yarden, Y. (2004). Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene, 23, 2057–2070.
Parks, E. E., & Ceresa, B. P. (2014). Cell surface epidermal growth factor receptors increase Src and c-Cbl activity and receptor ubiquitylation. The Journal of Biological Chemistry, 289, 25537–25545. https://doi.org/10.1074/jbc.M114.579581
Fatehchand, K., Ren, L., Elavazhagan, S., Fang, H., Mo, X., Vasilakos, J. P., Dietsch, G. N., Hershberg, R. M., Tridandapani, S., & Butchar, J. P. (2016). Toll-like receptor 4 ligands down-regulate Fcγ Receptor IIb (FcγRIIb) via MARCH3 protein-mediated ubiquitination. Journal of Biological Chemistry, 291, 3895–3904. https://doi.org/10.1074/jbc.M115.701151
Foot, N., Henshall, T., & Kumar, S. (2017). Ubiquitination and the regulation of membrane proteins. Physiological Reviews, 97, 253–281.
Ramachandran, S., Osterhaus, S. R., Parekh, K. R., Jacobi, A. M., Behlke, M. A., & McCray, P. B., Jr. (2016). SYVN1, NEDD8, and FBXO2 proteins regulate ΔF508 cystic fibrosis transmembrane conductance regulator (CFTR) ubiquitin-mediated proteasomal degradation. The Journal of Biological Chemistry, 291, 25489–25504.
Dikic, I. (2017). Proteasomal and Autophagic degradation systems. Annual Review of Biochemistry, 86, 193–224. https://doi.org/10.1146/annurev-biochem-061516-044908
Hershko, A., & Ciechanover, A. (1998). The ubiquitin system. Annual Review of Biochemistry, 67, 425–479.
Berruti, G. (2014). Male germ cell differentiation signaling events; Role of Phosphorylation and Ubiquitination. Reference Mod Biomedicine Science, 11, 2144–2156. https://doi.org/10.1016/B978-0-12-801238-3.04014-9
O'Connor, H. F., & Huibregtse, J. M. (2017). Enzyme-substrate relationships in the ubiquitin system: approaches for identifying substrates of ubiquitin ligases. Cellular and Molecular Life Sciences, 74, 3363–3375. https://doi.org/10.1007/s00018-017-2529-6
Zheng, N., & Shabek, N. (2017). Ubiquitin ligases: Structure, function, and regulation. Annual Review of Biochemistry, 86, 129–157.
Witting, K. F., Mulder, M. P. C., & Ovaa, H. (2017). Advancing our understanding of ubiquitination using the Ub-toolkit. Journal of Molecular Biology, S0022-2836(17), 30165–30161. https://doi.org/10.1016/j.jmb.2017.04.002
Petroski, M. D., & Deshaies, R. J. (2005). Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin–RING ubiquitin-ligase complex SCF-Cdc34. Cell, 123, 1107–1120.
Petroski M.D., Zhou X., Dong G., Daniel-Issakani S., Payan D.G, Huang J. Substrate modification with lysine 63-linked ubiquitin chains through the UBC13-UEV1A ubiquitin-conjugating enzyme. The Journal of Biological Chemistry 2007; 282: 29936–29945.
Komander, D. (2009). The emerging complexity of protein ubiquitination. Biochemical Society Transactions, 37, 937–953.
Komander, D., & Rape, M. (2012). The ubiquitin code. Annual Review of Biochemistry, 81, 203–239.
Swatek, K. N., & Komander, D. (2016). Ubiquitin modifications. Cell Research, 26, 399–422.
Ben-Saadon, R., Zaaroor, D., Ziv, T., & Ciechanover, A. (2006). The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Molecular Cell, 24, 701–715.
Livneh, I., Kravtsova-Ivantsiv, Y., Braten, O., Kwon, Y. T., & Ciechanover, A. (2017). Monoubiquitination joins polyubiquitination as an esteemed proteasomal targeting signal. BioEssays, 39. https://doi.org/10.1002/bies.201700027
Robzyk, K., Recht, J., & Osley, M. (2000). Rad6-dependent ubiquitination of histone H2B in yeast. Science, 287, 501–504.
Patnaik, A., Chau, V., & Wills, J. W. (2000). Ubiquitin is part of the retrovirus budding machinery. Proceedings of the National Academy of Sciences of the United States of America, 97, 13069–13074.
Lin, S., Lu, S., Mulaj, M., Fang, B., et al. (2016). Monoubiquitination inhibits the actin bundling activity of fascin. The Journal of Biological Chemistry, 291, 27323–27333.
Cohen-Kaplan, V., Livneh, I., Avni, N., Cohen-Rosenzweig, C., & Ciechanover, A. (2016). The ubiquitin-proteasome system and autophagy: Coordinated and independent activities. The International Journal of Biochemistry & Cell Biology, 79, 403–418.
Saftig, P., & Klumperman, J. (2009). Lysosome biogenesis and lysosomal membrane proteins: Trafficking meets function. Nature Reviews. Molecular Cell Biology, 10, 623–635. https://doi.org/10.1038/nrm2745
Marshall, R. S., & Vierstra, R. D. (2015). Eat or be eaten: The autophagic plight of inactive 26S proteasomes. Autophagy, 11, 1927–1928. https://doi.org/10.1080/15548627.2015.1078961
An, H., & Harper, J. W. (2020). Ribosome abundance control via the ubiquitin-proteasome system and autophagy. Journal of Molecular Biology, 432, 170–184.
Ulrich, H. D., & Walden, H. (2010 Jul). Ubiquitin signalling in DNA replication and repair. Nature Reviews. Molecular Cell Biology, 11(7), 479–489. https://doi.org/10.1038/nrm2921
Zhu, X., Xing, R., Tan, R., Dai, R., & Tao, Q. (2017). The RNF146 E3 ubiquitin ligase is required for the control of Wnt signaling and body pattern formation in Xenopus. Mechanisms of Development, 147, 28–36. https://doi.org/10.1016/j.mod.2017.08.001
Smith GA, Fearnley GW, Abdul-Zani I, Wheatcroft SB, Tomlinson DC, Harrison MA, Ponnambalam S. Ubiquitination of basal VEGFR2 regulates signal transduction and endothelial function. Biology Open 2017; pii: bio.027896. doi: https://doi.org/10.1242/bio.027896.
Rana, A. S. J. B., Ge, Y., & Strieter, E. R. (2017). Ubiquitin chain enrichment middle-down mass spectrometry (UbiChEM-MS) reveals cell-cycle dependent formation of Lys11/Lys48 branched ubiquitin chains. Journal of Proteome Research, 16, 3363–3369. https://doi.org/10.1021/acs.jproteome.7b00381
Dwane, L., Gallagher, W. M., Ní Chonghaile, T., & O'Connor, D. P. (2017). The emerging role of non-traditional ubiquitination in oncogenic pathways. Cell Death and Differentiation, 24, 903–916. https://doi.org/10.1038/cdd.2017.42
Wu, X., & Karin, M. (2015). Emerging roles of Lys63-linked polyubiquitylation in immune responses. Immunological Reviews, 266, 161–174. https://doi.org/10.1111/imr.12310
Amerik, A. Y., & Hochstrasser, M. (1695). Mechanism and function of deubiquitinating enzymes. Biochimica et Biophysica Acta, 2004, 189–207.
Love, K. R., Catic, A., Schlieker, C., & Ploegh, H. L. (2007). Mechanisms, biology and inhibitors of deubiquitinating enzymes. Nature Chemical Biology, 3, 697–705.
Reyes-Turcu, F. E., Ventii, K. H., & Wilkinson, K. D. (2009). Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annual Review of Biochemistry, 78, 363–397. https://doi.org/10.1146/annurev.biochem.78.082307.091526
Suresh, B., Lee, J., Hong, S. H., Kim, K. S., & Ramakrishna, S. (2015). The role of deubiquitinating enzymes in spermatogenesis. Cellular and Molecular Life Sciences, 72, 4711–4720. https://doi.org/10.1007/s00018-015-2030-z
Lim, K. H., Song, M. H., & Baek, K. H. (2016). Decision for cell fate: deubiquitinating enzymes in cell cycle checkpoint. Cellular and Molecular Life Sciences, 73, 1439–1455. https://doi.org/10.1007/s00018-015-2129-2
McCann, A. P., Scott, C. J., Van Schaeybroeck, S., & Burrows, J. F. (2016). Deubiquitylating enzymes in receptor endocytosis and trafficking. The Biochemical Journal, 473, 4507–4525.
Mevissen, T. E. T., & Komander, D. (2017). Mechanisms of Deubiquitnase specificity and regulation. Annual Review of Biochemistry, 86, 34.1–34.33.
Keusekotten, K., Elliott, P. R., Glockner, L., Fiil, B. K., Damgaard, R. B., et al. (2013). OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell, 153, 1312–1326.
Leznicki, P., & Kulathu, Y. (2017). Mechanisms of regulation and diversification of deubiquitylating enzyme function. Journal of Cell Science, 130, 1997–2006. https://doi.org/10.1242/jcs.201855
Lin, H. (1997). The tao of stem cells in the germline. Annual Review of Genetics, 31, 455–491.
Bradbury and Thompson. (1985). 1985 Immunoassay of the neuronal and neuroendocrine marker PGP 9.5 in human tissues. Journal of Neurochemistry, 44, 651–653.
Luo, J., Megee, S., & Dobrinski, I. (2009). Asymmetric distribution of UCH-L1 in spermatogonia is asso- ciated with maintenance and differentiation of spermatogonial stem cells. Journal of Cellular Physiology, 220, 460–468.
de Rooij, D. G., & Griswold, M. D. (2012). Questions about spermatogonia posed and answered since 2000. Journal of Andrology., 33, 1085–1095. https://doi.org/10.2164/jandrol.112.016832
Tokue, M., Ikami, K., Mizuno, S., Takagi, C., Miyagi, A., Takada, R., et al. (2017). SHISA6 confers resistance to differentiation-promoting Wnt/β-catenin signaling in mouse Spermatogenic stem cells. Stem Cell Reports, 8, 561–575. https://doi.org/10.1016/j.stemcr.2017.01.006
Bootsma, D., Kraemer, K. H., Cleaver, J. E., & Hoeijmakers, J. H. J. (2001). Nucleotide excision repair syndromes: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. In C. R. Scriver, A. L. Beaudet, W. S. Sly, D. Valle, B. Vogelstein, & K. W. Kinzler (Eds.), The metabolic and molecular bases of inherited disease (1st ed., pp. 677–703). McGraw-Hill.
Ng, J. M., Vrieling, H., Sugasawa, K., Ooms, M. P., Grootegoed, J. A., Vreeburg, J. T., et al. (2002). Developmental defects and male sterility in mice lacking the ubiquitin-like DNA repair gene mHR23B. Molecular and Cellular Biology, 22, 1233–1245.
Liang, R. Y., Chen, L., Ko, B. T., Shen, Y. H., Li, Y. T., Chen, B. R., Lin, K. T., Madura, K., & Chuang, S. M. (2014). Rad23 interaction with the proteasome is regulated by phosphorylation of its ubiquitin-like (UbL) domain. Journal of Molecular Biology, 426, 4049–4060. https://doi.org/10.1016/j.jmb.2014.10.004
Bose, R., Manku, G., Culty, M., & Wing, S. S. (2014). Ubiquitin-proteasome system in spermatogenesis. Advances in Experimental Medicine and Biology, 759, 181–213. https://doi.org/10.1007/978-1-4939-0817-2_9
Sheng, K., Liang, X., Huang, S., & Xu, W. (2014). The role of histone ubiquitination during spermatogenesis. BioMed Research International, 2014, 870695. https://doi.org/10.1155/2014/870695
Liu, Z., Oughtred, R., & Wing, S. S. (2005). Characterization of E3Histone, a novel testis ubiquitin protein ligase which ubiquitinates histones. Molec Cell Biol., 25, 2819–2831.
Bose, R., Sheng, K., Moawad, A. R., Manku, G., O'Flaherty, C., Taketo, T., Culty, M., Fok, K. L., & Wing, S. S. (2017). Ubiquitin ligase Huwe1 modulates spermatogenesis by regulating spermatogonial differentiation and entry into meiosis. Scientific Reports, 7(1), 17759. https://doi.org/10.1038/s41598-017-17902-0
Blow, J. J., & Nurse, P. (1990). A cdc2-like protein is involved in the initiation of DNA replication in Xenopus egg extracts. Cell, 62, 855–862.
Draetta G, Luca F. Westendorf J., Brizuela L, Ruderman J, Beach D. Cdc2 kinase is complexed with both cyclin a and B, evidence for proteolytic inactivation of MPF. Cell 1989; 56: 829–838.
Wang, L., Cao, C., Wang, F., Zhao, J., & Li, W. (2017). H2B ubiquitination: Conserved molecular mechanism, diverse physiologic functions of the E3 ligase during meiosis. Nucleus, 8, 461–468.
Menon DU, Shibata Y, Mu W, Magnuson T. Mammalian SWI/SNF collaborates with a polycomb-associated protein to regulate male germline transcription in the mouse. Development 2019; 146, dev174094. doi:https://doi.org/10.1242/dev.174094.
Serrano-Quilez, J., Roig-Soucase, S., & Rodriguez-Navarro, S. (2020). Sharing Marks: H3K4 methylation and H2B ubiquitination as features of meiotic recombination and transcription. International Journal of Molecular Sciences, 21, 4510. https://doi.org/10.3390/ijms21124510
Peters, J. M. (2006). The anaphase promoting complex/cyclosome: A machine designed to destroy. Nature Reviews. Molecular Cell Biology, 7, 644–656.
Holt, J. E., Tran, S. M., Stewart, J. L., Minahan, K., García-Higuera, I., Moreno, S., & Jones, K. T. (2011). The APC/C activator FZR1 coordinates the timing of meiotic resumption during prophase I arrest in mammalian oocytes. Development, 138, 905–913.
Holt, J. E., Pye, V., Boon, E., Stewart, J. L., García-Higuera, I., Moreno, S., Rodríguez, R., Jones, K. T., & McLaughlin, E. A. (2014). The APC/C activator FZR1 is essential for meiotic prophase I in mice. Development, 141, 1354–1365. https://doi.org/10.1242/dev.104828
Jeganathan, K. B., & van Deursen, J. M. (2006). Differential mitotic checkpoint protein requirements in somatic and germ cells. Biochemical Society Transactions, 34, 583–586.
McCarthy, S., & Ward, W. S. (1999). Functional aspects of mammalian sperm chromatin. Human Fertility, 2, 56–60.
Sassone-Corsi, P. (2002). Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science, 296, 2176–2178.
Montellier, E., Boussouar, F., Rousseaux, S., et al. (2013). Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes & Development, 27, 1680–1692.
Soboleva, T. A., Nekrasov, M., Pahwa, A., Williams, R., Huttley, G. A., & Tremethick, D. J. (2011). A unique H2A histone variant occupies the transcriptional start site of active genes. Nature Structural & Molecular Biology, 19(1), 25–30. https://doi.org/10.1038/nsmb.2161
Crichton, J. H., Playfoot, C. J., & Adams, I. R. (2014). The role of chromatin modifications in progression through mouse meiotic prophase. Journal of Genetics and Genomics, 41, 97–106. https://doi.org/10.1016/j.jgg.2014.01.003
Wing, S. S., Bedard, N., Morales, C., Hingamp, P., & Trasler, J. (1996). A novel rat homolog of the Saccharomyces cerevisiae ubiquitin-conjugating enzymes UBC4 and UBC5 with distinct biochemical features is induced during spermatogenesis. Molecular and Cellular Biology, 16, 4064–4072.
Haraguchi, C. M., Mabuchi, T., Hirata, S., Shoda, T., Tokumoto, T., Hoshi, K., & Yokota, S. (2007). Possible function of caudal nuclear pocket: Degradation of nucleoproteins by ubiquitin-proteasome system in rat spermatids and human sperm. The Journal of Histochemistry and Cytochemistry, 55, 585–595.
Yuan, X., Miller, M., & Belote, J. M. (1996). Duplicated proteasome subunit genes in Drosophila melanogaster encoding testes-specific isoforms. Genetics, 144, 147–157.
Belote, J. M., & Zhong, L. (2009). Duplicated proteasome subunit genes in Drosophila and their roles in spermatogenesis. Heredity, 103, 23–31.
Zhong L, Belote JM. The testis-specific proteasome subunit Prosalpha6T of D. melanogaster is required for individualization and nuclear maturation during spermatogenesis. Development 2007; 134: 3517–3525.
Haraguchi, C. M., Mabuchi, T., Hirata, S., Shoda, T., Hoshi, K., & Yokota, S. (2004). Ubiquitin signals in the developing acrosome during spermatogenesis of rat testis: An immunoelectron microscopic study. The Journal of Histochemistry and Cytochemistry, 52, 1393–1403.
Berruti, G., & Martegani, E. (2005). The deubiquitinating enzyme mUBPy interacts with the sperm-specific molecular chaperone MSJ-1: The relation with the proteasome, acrosome, and centrosome in mouse male germ cells. Biology of Reproduction, 72, 14–21.
Zimmerman, S. W., Yi, Y. J., Sutovsky, M., van Leeuwen, F. W., Conant, G., & Sutovsky, P. (2014). Identification and characterization of RING- finger ubiquitin ligase UBR7 in mammalian spermatozoa. Cell and Tissue Research, 356, 261–278.
Iyengar PV1, Hirota T, Hirose S, Nakamura N. Membrane-associated RING-CH 10 (MARCH10 protein) is a microtubule-associated E3 ubiquitin ligase of the spermatid flagella. J Biol Chem. 2011; 286: 39082–90. doi: https://doi.org/10.1074/jbc.M111.256875.
Zhao B1, Ito K, Iyengar PV, Hirose S, Nakamura N. MARCH7 E3 ubiquitin ligase is highly expressed in developing spermatids of rats and its possible involvement in head and tail formation. Histochem Cell Biol. 2013; 139: 447–460. doi: https://doi.org/10.1007/s00418-012-1043-z.
Rodriguez, C. I., & Stewart, C. L. (2007). Disruption of the ubiquitin ligase HERC4 causes defects in spermatozoon maturation and impaired fertility. Developmental Biology, 312, 501–508.
Pleuger C, Lehti MS, Dunleavy JE, Fietz D, O'Bryan MK. Haploid male germ cells-the Grand Central Station of protein transport. Hum Reprod Update. 2020 18; 26: 474–500. doi: https://doi.org/10.1093/humupd/dmaa004.
Oko, R., & Clermont, Y. (1998). Spermiogenesis. In E. Knobil & J. D. Neil (Eds.), Encyclopedia of reproduction (pp. 602–609). Academic Press.
Sutovsky, P., Moreno, R. D., Ramalho-Santos, J., Dominko, T., Simerly, C., & Schatten, G. (2000). Ubiquitin tag for sperm mitochondria. International Review of Cytology, 195, 1–65.
Nakagawa, T., & Nakayama, K. (2015). Protein monoubiquitylation: Targets and diverse functions. Genes to Cells, 20, 543–562.
Rittinger, K., & Ikeda, F. (2017). Linear ubiquitin chains: enzymes, mechanisms and biology. Open Biology, 7, 170026. https://doi.org/10.1098/rsob.170026
Weake, V. M., & Workman, J. L. (2008). Histone ubiquitination: Triggering gene activity. Molecular Cell, 29, 653–663. https://doi.org/10.1016/j.molcel.2008.02.014
Cao, R., Tsukada, Y., & Zhang, Y. (2005). Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Molecular Cell, 20, 845–854.
de Napoles, M., Mermoud, J. E., Wakao, R., Tang, Y. A., Endoh, M., Appanah, R., Nesterova, T. B., et al. (2004). Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Developmental Cell, 7, 663–676.
Lu, L.-Y., Wu, J., Ye, L., Gavrilina, G. B., Saunders, T. L., & Yu, X. (2010). RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Developmental Cell, 18, 371–384.
Fleming A.B, Kao C.F, Hillyer C., Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Molecular Cell 2008; 31: 57–66.
Minsky, N., Shema, E., Field, Y., Schuster, M., Segal, E., & Oren, M. (2008). Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nature Cell Biology, 10, 483–488.
Pavri, R., Zhu, B., Li, G., Trojer, P., Mandal, S., Shilatifard, A., & Reinberg, D. (2006). Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell, 125, 703–717.
Zhu B, Zheng Y, Pham A.D, Mandal S.S, Erdjument-Bromage H., Tempst P, Reinberg D. Monoubiquitination of human histone H2B: The factors involved and their roles in HOX gene regulation. Molecular Cell 2005; 20: 601–611.
Huen, M. S., Grant, R., Manke, I., Minn, K., Yu, X., Yaffe, M. B., & Chen, J. (2007). RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell, 131, 901–914.
Zhao G.Y, Sonoda E, Barber L.J, Oka H, Murakawa Y, Yamada K. Ikura T et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Molecular Cell 2007; 25: 663–675.
Doil, C., Mailand, N., Bekker-Jensen, S., Menard, P., Larsen, D. H., Pepperkok, R., Ellenberg, J., Panier, S., Durocher, D., Bartek, J., et al. (2009). RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell, 136, 435–446.
Stewart, G. S., Panier, S., Townsend, K., Al-Hakim, A. K., Kolas, N. K., Miller, E. S., Nakada, S., Ylanko, J., Olivarius, S., Mendez, M., et al. (2009). The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell, 136, 420–434.
Solari, A. J. (1974). The behavior of the XY pair in mammals. International Review of Cytology, 38, 273–317.
Starita, L. M., & Parvin, J. D. (2003). The multiple nuclear functions of BRCA1: Transcription, ubiquitination and DNA repair. Current Opinion in Cell Biology, 15, 345–350.
Turner, J. M., Aprelikova, O., Xu, X., Wang, R., Kim, S., et al. (2004). BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Current Biology, 14, 2135–2142.
Mahadevaiah, S. K., Turner, J. M., Baudat, F., Rogakou, E. P., de Boer, P., et al. (2001). Recombinational DNA double-strand breaks in mice precede synapsis. Nature Genetics, 27, 271–276.
Fernandez-Capetillo, O., Mahadevaiah, S. K., Celeste, A., Romanienko, P. J., Camerini-Otero, R. D., et al. (2003). H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Developmental Cell, 4, 497–508.
Mulugeta Achame, E., Wassenaar, E., Hoogerbrugge, J. W., Sleddens-Linkels, E., Ooms, M., Sun, Z. W., & van IJcken WF, Grootegoed JA, Baarends WM. (2010). The ubiquitin-conjugating enzyme HR6B is required for maintenance of X chromosome silencing in mouse spermatocytes and spermatids. BMC Genomics, 11, 367. https://doi.org/10.1186/1471-2164-11-367
Kolas, N. K., Chapman, J. R., Nakada, S., Ylanko, J., Chahwan, R., Sweeney, F. D., Panier, S., Mendez, M., Wildenhain, J., Thomson, T. M., et al. (2007). Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science, 318, 1637–1640.
Mailand, N., Bekker-Jensen, S., Faustrup, H., Melander, F., Bartek, J., Lukas, C., & Lukas, J. (2007). RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell, 131, 887–900.
Gou, L. T., Kang, J. Y., Dai, P., Wang, X., Li, F., Zhao, S., Zhang, M., Hua, M. M., Lu, Y., Zhu, Y., Li, Z., Chen, H., Wu, L. G., Li, D., Fu, X. D., Li, J., Shi, H. J., & Liu, M. F. (2017). Ubiquitination-deficient mutations in human Piwi cause male infertility by impairing histone-to-protamine exchange during spermiogenesis. Cell, 169, 1090–1104. https://doi.org/10.1016/j.cell.2017.04.034
Carmell, M. A., Girard, A., van de Kant, H. J., Bourc’his, D., Bestor, T. H., de Rooij, D. G., & Hannon, G. J. (2007). MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Developmental Cell, 12, 503–514.
Adams, S. R., Maezawa, S., Alavattam, K. G., Abe, H., Sakashita, A., Shroder, M., Broering, T. J., Sroga Rios, J., Thomas, M. A., Lin, X., Price, C. M., Barski, A., Andreassen, P. R., & Namekawa, S. H. (2018). RNF8 and SCML2 cooperate to regulate ubiquitination and H3K27 acetylation for escape gene activation on the sex chromosomes. PLoS Genetics, 14(2), e1007233. https://doi.org/10.1371/journal.pgen.1007233
Gannon, J. R., Emery, B. R., Jenkins, T. G., & Carrell, D. T. (2014). The sperm epigenome: Implications for the embryo. Advances in Experimental Medicine and Biology, 791, 53–66.
Sin, H. S., Barski, A., Zhang, F., Kartashov, A. V., Nussenzweig, A., Chen, J., Andreassen, P. R., & Namekawa, S. H. (2012 Dec 15). RNF8 regulates active epigenetic modifications and escape gene activation from inactive sex chromosomes in post-meiotic spermatids. Genes & Development, 26(24), 2737–2748. https://doi.org/10.1101/gad.202713.112
Luo, M., Zhou, J., Leu, N. A., Abreu, C. M., Wang, J., Anguera, M. C., de Rooij, D. G., Jasin, M., & Wang, P. J. (2015). Polycomb protein SCML2 associates with USP7 and counteracts histone H2A ubiquitination in the XY chromatin during male meiosis. PLoS Genetics, 11(1), e1004954. https://doi.org/10.1371/journal.pgen.1004954
Kim, W., Bennett, E. J., Huttlin, E. L., et al. (2011). Systematic and quantitative assessment of the ubiquitin-modified proteome. Molecular Cell, 44, 325–340.
Galisson, F., Mahrouche, L., Courcelles, M., et al. (2011). A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Molecular & Cellular Proteomics, 10, M110.004796–M110.004796.
Peng, J., Schwartz, D., Elias, J. E., et al. (2003). A proteomics approach to understanding protein ubiquitination. Nature Biotechnology, 21, 921–926.
Swaney, D. L., Beltrao, P., Starita, L., et al. (2013). Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nature Methods, 10, 676–682.
Ohtake, F., Saeki, Y., Sakamoto, K., et al. (2015). Ubiquitin acetylation inhibits polyubiquitin chain elongation. EMBO Reports, 16, 192–201.
Berruti, G. (2016). Towards defining an 'origin'-the case for the mammalian acrosome. Seminars in Cell & Developmental Biology, 59, 46–53. https://doi.org/10.1016/j.semcdb.2016.01.013
Berruti, G., Ripolone, M., & Ceriani, M. (2010). USP8, a regulator of endosomal sorting, is involved in mouse acrosome biogenesis through interaction with the spermatid ESCRT-0 complex and microtubules. Biology of Reproduction, 82, 930–939. https://doi.org/10.1095/biolreprod.109.081679
Berruti, G., & Paiardi, C. (2011). Acrosome biogenesis: Revisiting old questions to yield new insights. Spermatogenesis., 1, 95–98.
Wang, H., Wan, H., Li, X., Liu, W., Chen, Q., Wang, Y., Yang, L., Tang, H., Zhang, X., et al. (2014). Atg7 is required for acrosome biogenesis during spermatogenesis in mice. Cell Research, 24, 852–869.
Berruti, G., & Paiardi, C. (2015). USP8/UBPy-regulated sorting and the development of sperm acrosome: The recruitment of MET. Reproduction, 149, 633–644.
Gioria, M., Pasini, M. E., & Berruti, G. (2017). Dynamic of contribution of UBPy-sorted cargo to acrosome biogenesis: effects of its derailment in a mouse model of globozoospermia, the infertile Vps54 (L967Q) mutant. Cell Tissue Research, 369, 413–427. https://doi.org/10.1007/s00441-017-2592-1
Gnesutta, N., Ceriani, M., Innocenti, M., Mauri, I., Zippel, R., Sturani, E., Borgonovo, B., Berruti, G., & Martegani, E. (2001). Cloning and characterization of mouse UBPY, a deubiquitinating enzyme that interacts with the ras guanine nucleotide exchange factor CDC25(mm)/Ras-GRF1. The Journal of Biological Chemistry, 276, 39448–39454.
Wright, M. H., Berlin, I., & Nash, P. D. (2011). Regulation of endocytic sorting by ESCRT- DUB-mediated deubiquitination. Cell Biochemistry and Biophysics, 60, 39–46.
Berruti, G., & Martegani, E. (2013). Ubiquitin Specific Peptidase 8. In N. D. Rawlings & G. S. Salvesen (Eds.), Handbook of proteolytic enzymes (3rd ed., pp. 2070–2075). Academic.
Kato, M., Miyazawa, K., & Kitamura, N. (2000). A deubiquitinating enzyme UBPY interacts with the Src homology 3 domain of Hrs-binding protein via a novel binding motif PX(V/I)(D/N)RXXKP. The Journal of Biological Chemistry, 275, 37481–37487.
Ceriani, M., Amigoni, L., D’Aloia, A., Berruti, G., & Martegani, E. (2015). The deubiquitinating enzyme UBPy/USP8 interacts with TrkA and in- hibits neuronal differentiation in PC12 cells. Experimental Cell Research, 333, 49–59.
Dufner, A., & Knobeloch, K. P. (2019). Ubiquitin-specific protease 8 (USP8/UBPy): A prototypic multidomain deubiquitinating enzyme with pleiotropic functions. Biochemical Society Transactions, 47(6), 1867–1879. https://doi.org/10.1042/BST20190527
Mizuno, E., Kitamura, N., & Komada, M. (2007). 14-3-3-dependent inhibition of the deubiquitinating activity of UBPY and its cancellation in the M phase. Experimental Cell Research, 313, 3624–3634.
Reincke, M., Sbiera, S., Hayakawa, A., Theodoropoulou, M., Osswald, A., Beuschlein, F., Meitinger, T., Mizuno-Yamasaki, E., Kawaguchi, K., Saeki, Y., et al. (2015). Mutations in the deubiquitinase gene USP8 cause Cushing's disease. Nature Genetics, 47, 31–38. https://doi.org/10.1038/ng.3166
Wanichi, I. Q., de Paula Mariani, B. M., Frassetto, F. P., Siqueira, S. A. C., de Castro Musolino, N. R., Cunha-Neto, M. B. C., Ochman, G., Cescato, V. A. S., Machado, M. C., Trarbach, E. B., Bronstein, M. D., & Fragoso, M. C. B. V. (2019). Cushing’s disease due to somatic USP8 mutations: A systematic review and meta-analysis. Pituitary, 22(4), 435–442. https://doi.org/10.1007/s11102-019-00973-9
Sun, J., Shen, D., Gao, Y., Zheng, Y., Zhao, L., Maa, M., Liu, H., & Chen, X. (2020). Dow-regulation of USP8 suppresses HER-3 positive gastric cancer cells proliferation. Oncotargets and Therapy, 13, 7973–7984. https://doi.org/10.2147/OTT.S264108
Kosova, G., Scott, N. M., Niederberger, C., Prins, G. S., & Ober, C. (2012). Genome-wide association study identifies candidate genes for male fertility traits in humans. American Journal of Human Genetics, 90, 950–961.
Cerván-Martín, M., Bossini-Castillo, L., Rivera-Egea, R., Garrido, N., Luján, S., Romeu, G., Santos-Ribeiro, S., Ivirma Group, Lisbon Clinical Group, Castilla, J. A., Gonzalvo, M. C., Clavero, A., Vicente, F. J., et al. (2020). Evaluation of male fertility-associated loci in a European population of patients with severe spermatogenic impairment. Journal of Personalized Medicine, 11(1), 22. https://doi.org/10.3390/jpm11010022
Clark, I. E., Dodson, M. W., Jiang, C., Cao, J. H., Huh, J. R., Seol, J. H., Yoo, S. J., Hay, B. A., & Guo, M. (2006). Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature, 441, 1162–1166.
Narendra, D., Tanaka, A., Suen, D. F., & Youle, R. J. (2008). Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. The Journal of Cell Biology, 183, 795–803.
Durcan, T. M., & Fon, E. A. (2015). The three 'P's of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes & Development, 29, 989–999. https://doi.org/10.1101/gad.262758
Li, W., Tang, W., Teves, M. E., Zhang, Z., Zhang, L., Li, H., Archer, K. J., Peterson, D. L., Williams, D. C., Jr., Strauss, J. F., 3rd, & Zhang, Z. (2015). A MEIG1/PACRG complex in the manchette is essential for building the sperm flagella. Development, 142, 921–930.
Durcan, T. M., Tang, M. Y., Pérusse, J. R., Dashti, E. A., Aguileta, M. A., McLelland, G. L., Gros, P., Shaler, T. A., Faubert, D., Coulombe, B., & Fon, E. A. (2014). USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO Journal, 33, 2473–2491. https://doi.org/10.15252/embj.201489729
Alexopoulou, Z., Lang, J., Perrett, R. M., Elschami, M., Hurry, M. E., Kim, H. T., Mazaraki, D., Szabo, A., Kessler, B. M., Goldberg, A. L., Ansorge, O., Fulga, T. A., & Tofaris, G. K. (2016). Deubiquitinase Usp8 regulates α-synuclein clearance and modifies its toxicity in Lewy body disease. Proceedings of the National Academy of Sciences of the United States of America, 113, E4688–E4697. https://doi.org/10.1073/pnas.1523597113
Oh, Y. M., Lee, S. B., Choi, J., Suh, H. Y., Shim, S., Song, Y. J., Kim, B., Lee, J. M., Oh, S. J., Jeong, Y., Cheong, K. H., Song, P. H., & Kim, K. A. (2014). USP8 modulates ubiquitination of LRIG1 for Met degradation. Scientific Reports, 4, 4980. https://doi.org/10.1038/srep04980
Boillée, S., Berruti, G., Meccariello, R., Grannec, G., Razan, F., Pierantoni, R., Fasano, S., & Junier, M. P. (2002). Early defect in the expression of mouse sperm DNAJ 1, a member of the DNAJ/heat shock protein 40 chaperone protein family, in the spinal cord of the wobbler mouse, a murine model of motoneuronal degeneration. Neuroscience, 113, 825–835.
Paiardi, C., Pasini, M. E., Gioria, M., & Berruti, G. (2011). Failure of acrosome formation and globozoospermia in the wobbler mouse, a Vps54 spontaneous recessive mutant. Spermatogenesis, 1, 52–62.
Paiardi, C., Pasini, M. E., Amadeo, A., Gioria, M., & Berruti, G. (2014). The ESCRT-deubiquitinating enzyme USP8 in the cervical spinal cord of wild-type and Vps54 recessive (wobbler) mutant mice. Histochemistry and Cell Biology, 141, 57–73.
Chianese, R., Scarpa, D., Berruti, G., Cobellis, G., Pierantoni, R., Fasano, S., & Meccariello, R. (2010). Expression and localization of the deubiquitinating enzyme mUBPy in wobbler mouse testis during spermiogenesis. General and Comparative Endocrinology, 166, 289–295. https://doi.org/10.1016/j.ygcen.2009.09.014
Gyllensten, U., Wharton, D., Josefsson, A., & Wilson, A. C. (1991). Paternal inheritance of mitochondrial DNA in mice. Nature, 352, 255–257.
Sato, M., and Sato, K. (2011). Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 2011; 334: 1141–1144.
Al Rawi, S., Louvet-Vallée, S., Djeddi, A., Sachse, M., Culetto, E., Hajjar, C., Boyd, L., Legouis, R., & Galy, V. (2011). Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science, 334, 1144–1147.
Luo, S. M., Ge, Z. J., Wang, Z. W., Jiang, Z. Z., Wang, Z. B., Ouyang, Y. C., Hou, Y., Schatten, H., & Sun, Q. Y. (2013). Unique insights into maternal mitochondrial inheritance in mice. Proceedings of the National Academy of Sciences of the United States of America, 110, 13038–13043.
Politi, Y., Gal, L., Kalifa, Y., Ravid, L., Elazar, Z., & Arama, E. (2014). Paternal mitochondrial destruction after fertilization is mediated by a common endocytic and autophagic pathway in Drosophila. Developmental Cell, 29, 305–320.
Song, W. H., Yi, Y. J., Sutovsky, M., Meyers, S., & Sutovsky, P. (2016). Autophagy and ubiquitin-proteasome system contribute to sperm mitophagy after mammalian fertilization. Proceedings of the National Academy of Sciences of the United States of America, 113, E5261–E5270. https://doi.org/10.1073/pnas.1605844113
Sharpley, M. S., Marciniak, C., Eckel-Mahan, K., McManus, M., Crimi, M., Waymire, K., et al. (2012). Heteroplasmy of mouse mtDNA is genetically unstable and results in altered behavior and cognition. Cell, 151, 333–343.
Berruti, G., & Aivatiadou, E. (2006). mUBPy is a novel centrosome-associated protein and interacts with gamma-tubulin. Journal of Submicroscopic Cytology and Pathology, 38, 77–83.
Simerly, C., Wu, G. J., Zoran, S., Ord, T., Rawlins, R., Jones, J., Navara, C., Gerrity, M., Rinehart, J., Binor, Z., Asch, R., & Schatten, R. (1995). The paternal inheritance of the centrosome, the cell’s microtubule-organizing center, in humans, and the implications for infertility. Nature Medicine, 1, 47–52.
Krioutchkova, M. M., & Onishchenko, G. E. (1999). Structural and functional characteristics of the centrosome in gametogenesis and early embryogenesis of animals. Intern Rev Cytol, 185, 107–156.
Puram, S. V., Kim, A. H., Park, H. Y., Anckar, J., & Bonni, A. (2013). The ubiquitin receptor S5a/Rpn10 links centrosomal proteasomes with dendrite development in the mammalian brain. Cell Reports, 4, 19–30. https://doi.org/10.1016/j.celrep.2013.06.006
Al-Hakim, A. K., Bashkurov, M., Gingras, A. C., Durocher, D., & Pelletier, L. (2012). Interaction proteomics identify NEURL4 and the HECT E3 ligase HERC2 as novel modulators of centrosome architecture. Molecular & Cellular Proteomics, 11, M111.014233. https://doi.org/10.1074/mcp.M111.014233
Inoue, D., Stemmer, M., Thumberger, T., Ruppert, T., Barenz, F., Wittbrodt, J., & Gruss, O. J. (2017). Expression of the novel maternal centrosome assembly factor Wdr8 is required for vertebrate embryonic mitoses. Nature Communication, 8, 14090. https://doi.org/10.1038/ncomms14090
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Berruti, G. (2021). Destruction or Reconstruction: A Subtle Liaison between the Proteolytic and Signaling Role of Protein Ubiquitination in Spermatogenesis. In: Cheng, C., Sun, F. (eds) Molecular Mechanisms in Spermatogenesis. Advances in Experimental Medicine and Biology, vol 1381. Springer, Cham. https://doi.org/10.1007/978-3-030-77779-1_11
Download citation
DOI: https://doi.org/10.1007/978-3-030-77779-1_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-77778-4
Online ISBN: 978-3-030-77779-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)