Abstract
Cells and therefore tissues ordinarily experience some form of mechanical stimulation as they are often in mechanically diverse and dynamic environments. As a result we have learned that cells have the astounding ability to sense and respond to their environment. This seemingly innate behavior of cells has intrigued many researchers in the field of cell mechanics for decades and compelled efforts aimed at characterizing its behaviors and underlying mechanisms. While many techniques exist, in the context of this chapter, novel techniques we have developed and implemented will be examined as well as new emergent behaviors we have discovered. The behaviors that will be discussed have relevance in various areas of pathology and physiology including collective cell migration and cancer metastasis.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Emergent Behavior
- Human Airway Smooth Muscle
- Glassy System
- Human Airway Smooth Muscle Cell
- Collective Migration
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
3.1 Introduction
Professor Yoram Lanir has performed pioneering works in the field of biomechanics. His efforts helped lay the groundwork for theoretical and experimental studies aimed at elucidating the basic functions of cells, tissues, and organs and the constitutive mechanical laws that govern them. More importantly, he has contributed significantly to our understanding of how lessons learned from structures at the cellular level influence physiologic and pathophysiologic function at the tissue level. Complementing his work, here we present new methodologies in cell mechanics developed by our group and describe the emergent cellular behaviors we have discovered using those approaches.
Mechanics are part of our everyday life. For the father teaching his son to catch a ball as for the commuter running to catch a train, cells cooperate to allow bones, tissues, and other structures to exert and experience mechanical force. Throughout their lifetime, cells and tissues experience mechanical stimulation, and depending on the cell’s location and physiology such stimuli may be in the form of tension, compression, or shear, and may be static or cyclic (Garanich et al. 2007; Butcher et al. 2004; Cheng et al. 2009; Richard et al. 2007; Owan et al. 1997). As a result, cells have evolved the ability to sense and respond to their local microenvironment (Kung 2005; Eastwood et al. 1998; Hu et al. 2004; Smith et al. 2003; Thompson 1961; Steward et al. 2009). Such behavior has intrigued scientists from the time of D’Arcy Thompson (Thompson 1961) and inspired efforts aimed at characterizing these behaviors and their underlying mechanisms. While a plethora of approaches exists (Cheng et al. 2009; Steward et al. 2009; Bao and Suresh 2003; Zhu et al. 2000; Chien 2007; Puig-De-Morales et al. 2001; Bellin et al. 2009), we will examine techniques used to define the physical forces a cell exerts on its substrate and upon neighboring cells, and the techniques used to characterize cellular material properties.
3.2 Contractile Forces and Traction Microscopy
Since the pioneering work of Cyril Harris, cellular contractility has been of interest to many groups (Harris 1988; Harris et al. 1980; Oliver et al. 1995, 1998, 1999; Dembo and Wang 1999; Dembo et al. 1996). For the adherent cell in isolation, cellular contractility provides insights into the mechanism of force transmission between the cell and its underlying substrate. Force transmission involves a feedback loop in which integrins connected to the extracellular matrix (ECM) sense and transmit mechanical information through focal adhesions, which contain proteins including vinculin, talin, paxillin, tensin, and zyxin (Zhu et al. 2000; Ridley et al. 2003; Wang et al. 1993; Yoshigi et al. 2005; Vogel and Sheetz 2006). Focal adhesions transmit these mechanical signals to the cytoskeleton (Ridley et al. 2003; Yoshigi et al. 2005; Revenu et al. 2004; DeMali et al. 2003; Cao et al. 1993). Drawing from this information, the cytoskeleton adjusts its physical or chemical activity until intracellular homeostasis is achieved (Ridley et al. 2003; Vogel and Sheetz 2006; DeMali et al. 2003; Norman et al. 1998; Insall and Machesky 2009; Rodriguez et al. 2003). Force transmission and therefore contractility is known to differ between cell types as well as between single cells and integrated monolayers of the same cell type. Contractility is also important in numerous cellular processes including migration, embryogenesis, morphogenesis, metastasis, and wound healing (Ridley et al. 2003; Larsen et al. 2006; Poukkula et al. 2011; Tambe et al. 2011; Friedl et al. 2004; Rorth 2007, 2011; Inaki et al. 2012; Trepat and Fredberg 2011; Trepat et al. 2009). To measure contractile forces or tractions at the cellular level we use traction force microscopy (Dembo and Wang 1999; Dembo et al. 1996; Trepat et al. 2007).
Dembo and Wang were the first to quantify the spatial distributions of the tractions exerted by single cells (Dembo and Wang 1999). Tractions were obtained by measuring displacement fields generated by a single cell adherent upon a flexible, polyacrylamide gel. Embedded within that gel were fluorescent markers located beneath the gel surface. Although the approach described by Dembo et al. is computationally intensive, this method provides detailed maps of the traction forces exerted by a single cell. An alternative to this approach consists of using microfabricated, elastomeric microposts (Maruthamuthu et al. 2011; Sniadecki and Chen 2007). Using this method, the deflection caused by an adherent cell on vertically aligned microposts can be directly measured and used to deduce tractions (Maruthamuthu et al. 2011; Sniadecki and Chen 2007).
We extended the approach of Dembo by developing Fourier Transform Traction Microscopy (FTTM) (Trepat et al. 2009; Butler et al. 2002). Compared with Dembo’s method, FTTM is computationally efficient. Efficiency arises from its mathematical foundation in Fourier analysis, and can be applied in either of two subcases, constrained and unconstrained FTTM. In the former tractions are deduced without prior specification of cellular boundaries, while in the latter tractions are deduced including prespecified cellular boundaries.
Experimentation and implementation are as follows. A single cell is cultured on a ligand-coated, flexible substrate and allowed to spread. Phase contrast images of cells on top of the gel and fluorescent images of beads embedded within the gel are obtained. In addition, another image is taken after the cells have been detached from the gel, representing an unstressed traction-free state referred to as the reference image (Butler et al. 2002). After image acquisition, post-processing begins by initially compensating for artifactual translational shifts between the two pairs of fluorescent images, which are ordinarily attributable to drift of the microscope stage. A fast Fourier transform algorithm is then used to compute a two-dimensional cross-correlation function between the two images. Cross-correlation is used to generate a uniform local displacement from which one image is translated with respect to another, yielding shift-corrected images. The shift-corrected images are divided into smaller windows or pixel areas from which the correlation function is once again used to calculate the local displacement between the reference window and its corresponding window in the experimental image. The displacement, which is calculated for each pixel within the image, produces a discretized gel displacement field. One advantage of FTTM worth noting is its insensitivity to explicit bead identification and bead density due to the utilization of the cross-correlation approach. Alternatively, a potential disadvantage is that this method provides an estimated displacement field while others provide a direct measurement (Dembo and Wang 1999; Butler et al. 2002). The estimated displacement field represents the “raw data” from which unconstrained and constrained tractions are calculated; these tradeoffs were analyzed by the group of Gardel (Maruthamuthu et al. 2011). The Boussinesq solution in Fourier space provides the direct solution of the tractions as a function of the two-dimensional inverse Fourier transform (Butler et al. 2002). This method yields unconstrained tractions as it does not impose traction boundary conditions upon the solution, contrary to the Dembo approach (Butler et al. 2002). Calculating constrained tractions is slightly more complex as this presents a mixed boundary value problem, and is subject to serious potential artifacts caused by improperly identifying the cell boundary. Computing constrained tractions is an iterative process that requires the initial calculation of the traction field as an unconstrained traction and calculation of new traction field by specifying all tractions outside of a user-specified boundary to zero. We initially developed this method for single cells and later expanded this to monolayers (Tambe et al. 2011; Trepat et al. 2009), as shown in Fig. 3.1.
3.3 Monolayer Stress Microscopy and Intercellular Stresses
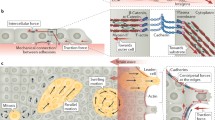
Having introduced FTTM we now extend the discussion to a new technique we recently developed, monolayer stress microscopy (MSM) (Tambe et al. 2011; Trepat and Fredberg 2011). The traction forces that the individual cell exerts on its substrate have been well established and useful in elucidating various cellular processes (Oliver et al. 1995, 1998; Ridley et al. 2003). However, extending our analysis to a monolayer of cells, we know that there are also intercellular forces and therefore intercellular stresses between each cell that they exert on their nearest neighbors. Newton’s Laws demand that the traction forces produced at the cell–substrate interface across the monolayer be balanced by intercellular stresses between the cells (Tambe et al. 2011; Trepat and Fredberg 2011). Therefore combining FTTM with MSM we are now able to measure previously elusive intercellular stresses within a monolayer. The intercellular stress within a monolayer is defined as the local intercellular force per unit area of cell–cell contact (Tambe et al. 2011) and is composed of two components: a normal stress defined as the stress acting perpendicular to the local intercellular junction, and shear stress defined as the stress acting parallel to the local intercellular junction (Fig. 3.2). By assuming the monolayer to be a thin elastic sheet, a computationally less complex, yet rigorous two-dimensional force balance can be done (Tambe et al. 2011). The two-dimensional force balance from Newton’s laws yields the distribution of line forces (force per unit length) everywhere within the monolayer, which are converted to stresses (force per unit area) using the average monolayer height (Tambe et al. 2011). This method is almost model-independent and is built upon the key assumption that the monolayer be treated as a continuum in which all forces are in balance.
Arrows depicting intercellular stresses at the cell junction (a) and intercellular stress maps of migrating monolayers of RPMEC (b). Adapted from Tambe et al. (2011)
3.4 Optical Magnetic Twisting Cytometry and Cell MaterialProperties
Optical magnetic twisting cytometry (OMTC) is used to measure cell material properties including cell stiffness and has helped elucidate the physical mechanisms of force transmission across the cell membrane (Wang et al. 1993). OMTC was first introduced by Crick (Crick and Hughes 1950; Crick 1950), describing a novel method that used phagocytosed small, magnetic particles to be manipulated by three principal motions; twisting, dragging, or prodding. Other groups, including our own (Hu et al. 2004; Puig-De-Morales et al. 2001; Maksym et al. 2000; Fabry et al. 2001a), later improved upon this technique. While slightly more complex, the basic operation remains similar to what was introduced some time ago. In general an OMTC system contains the following components: high-voltage generator for generating current in coils to magnetize magnetic particles, separate current sources and computer to manipulate and control the particles, and a microscope and camera with complementary image acquisition software to observe and acquire images of particle movement (Hu et al. 2004; Maksym et al. 2000). Cell preparation and OMTC measurement protocols used by us as well as others begin by incubating sparsely plated or confluent cells with ligand-coated ferromagnetic beads for a period of 15–20 min in an incubator. In contrast with the Crick method, this brief period allows beads to bind to integrin receptors on the cell surface, linking them to the cytoskeleton through focal adhesions, instead of being phagocytosed (Wang et al. 1993; Crick and Hughes 1950; Crick 1950). After incubation, beads are initially magnetized with a strong magnetic field. Subsequently, a set of twisting coils create a twisting field perpendicular to the initial magnetization and provide a twisting torque at frequencies ranging from 0.5 Hz to frequencies ranging in excess of 5 Hz, Fig. 3.3. During twisting, image acquisition occurs at a specific temporal resolution synchronized with the twisting cycle. A bead-tracking program is then used to compute the bead motion signal. This software is designed to search for beads with specific size, contrast, and shape properties (Hu et al. 2004) (Fig. 3.3). Once, identified, an intensity-based weighted center of mass of the bead is performed in which the respective x and y coordinates of the bead center is calculated, defining the bead motion signal. A Fourier transformation of the bead motion signal (x∼) and torque (T) allow the calculation of the complex modulus (G) (in dimensions of Pascals per nanometer), which can be related to the stiffness (G ′), friction (G ″), and hysteresivity (η) (3.1). Therefore OMTC can be a useful tool in determining the effects of various agonist and antagonist on cell material properties.
Top (a) and side (b) schematic of OMTC device. Image of magnetic beads on single cells marked with bead-tracking program (c). Adapted from Fabry et al. (2001a)
3.5 Three Emergent Behaviors
Many cellular behaviors have been elegantly explained by detailed biochemical mechanisms, while explanation from a physical perspective remains limited and poorly understood (Richard et al. 2007; Cao et al. 1993; Norman et al. 1998; Huang et al. 2004; Lauffenburger 2000; Zheng et al. 1995; Hall and Nobes 2000). This lack of explanation of cellular phenomena from a physical perspective served as a motivation for our group, as a complete physical and chemical explanation is needed to better understand cellular behavior. Here, we present three emergent cellular behaviors that raise exciting questions and controversies.
3.5.1 Fluidization/Reinforcement
Cells have the ability to sense and respond to mechanical forces. For example, pulmonary vascular smooth muscle cells exhibit stretch-induced VEGF and FGF-2 expression (Quinn et al. 2001) and endothelial cells exhibit fluid shear stress-induced polarization (Chien 2007). Stretch in particular is important in various cellular processes including proliferation, differentiation, and gene expression (Eastwood et al. 1998; Wang and Thampatty 2006). An important response to stretch is reinforcement (Choquet et al. 1997; Matthews et al. 2006; von Wichert et al. 2003). Reinforcement describes a phenomenon in which the cytoskeleton increases stiffness and recruits actin stress fibers as stretch is experienced by the cell, exhibiting an active strain-stiffening behavior along with associated structural changes (Krishnan et al. 2009; Chen et al. 2010). Contrary to stretch-induced reinforcement, earlier studies suggested the cytoskeleton fluidized in response to stretch (Choquet et al. 1997; Matthews et al. 2006). These paradoxical findings were baffling as no agreed-upon consensus existed on a simple question: How does the cell respond to stretch? To address this paradox we developed a novel system that allows us to stretch cells while simultaneously using OMTC (Trepat et al. 2007). We used our system on multiple cell types including human airway smooth muscle (HASM) cells, human lung fibroblasts, Madin–Darby canine kidney epithelial (MDCK) cells, and human bronchial epithelial cells to (1) determine if the cell fluidizes or resolidifies in response to stretch and (2) determine if this response is shared among mammalian cell types of diverse mesenchymal lineages. Following stretch, cell stiffness immediately decreased significantly and then gradually returned to baseline over time, suggesting that in response to stretch, cells initially fluidize and gradually resolidify (Trepat et al. 2007). Fluidization in response to stretch was also observed to occur despite chemical perturbation, but with slight change in magnitude and temporal scales (Trepat et al. 2007). Contrary to fluidization, chemical perturbation revealed resolidification to be ATP-dependent (Trepat et al. 2007). Our findings revealed that among mammalian cells resolidification and fluidization are universal responses (Trepat et al. 2007). While fluidization remains relatively new within the field it raises many new and exciting questions: What are the molecular components involved in fluidization and resolidification? How do these two responses stay in balance? The latter is exactly what we next sought out to answer.
Fluidization was further probed as a function of cell contractility using a method we developed called Cell Mapping Rheometry (CMR) (Krishnan et al. 2009). CMR uses a novel punch-indentation system to induce biaxial or uniaxial homogeneous deformations on cells cultured on a flexible substrate (Krishnan et al. 2009). The displacements of the substrate are calculated by tracking fluorescent markers embedded within the substrate using FTTM. HASM cells were subjected to biaxial and uniaxial stretch to probe whether fluidization was dependent on stretch isotropy. In response to stretch, cells immediately decreased contractility followed by a gradual recovery. This stretch-induced fluidization was independent of stress isotropy and agreed quite well with results found previously using OMTC (Trepat et al. 2007; Krishnan et al. 2009). However the question of whether fluidization or resolidification dominated in a mechanical physiological loading condition remained unanswered. To address this, we extended our system to induce non-homogeneous stretch on cells by applying force at a localized area on the substrate (Krishnan et al. 2009). Cells that underwent non-homogeneous stretch responded by reinforcement, while cells experiencing homogenous stretch fluidized. Being that we as well as others believe that homogenous stretch is more physiologically relevant, we concluded that mammalian cells primarily fluidize in response to stretch (Pirentis et al. 2011). While fluidization is believed to be a critical determinate of cell response to mechanical stimulation we should not disregard the fact that in certain physiological conditions reinforcement does prevail. This raises the question of how, if at all, does the interplay between fluidization and reinforcement change in response to other forces such as fluid shear, compression, or even a combination of both? All of this has yet to be elucidated.
3.5.2 Plithotaxis
Shifting our attention from single cells to cell monolayers, we now focus on a recent emergent phenomena describing collective cellular migration. Cellular migration is an essential step for many physiological processes including morphogenesis, wound healing, and regeneration (Ridley et al. 2003; Friedl et al. 2004). For example, organs including the kidney, lung, breast, and salivary glands form through branching morphogenesis that requires the coordinated collective migration of mesenchymal and epithelial cells to form sprouting vessels and ducts (Rorth 2007; Ewald et al. 2008; Vasilyev et al. 2009). Collective migration has also recently been shown to be important in cancer, suggesting cellular migration to be ubiquitous in not only physiology, but pathology as well (Friedl et al. 2004; Scotton et al. 2001). A suggested and widely accepted mechanism proposes this process to be solely dependent on biochemical signaling (Ridley et al. 2003; Rorth 2007). Forexample the EGF receptor has been shown to guide oogenesis (Duchek and Rorth 2001) and FGF has been shown to guide tracheal branching morphogenesis of the Drosophila (Ribeiro et al. 2002). Other studies report cell migration to be driven by internal biochemical mechanisms in which cells within a monolayer relay their chemical state to each other through intercellular connections (Rorth 2007), further illustrating a common theme whereby cells moving as a collective do so by sensing and responding to endogenous and exogenous chemical cues. While the role of biochemistry in cell migration is clearly evident, the role of physics is less so. Cells within a monolayer are physically linked to their underlying substrate through integrins and other transmembrane proteins including syndecans (Bellin et al. 2009; Poukkula et al. 2011). These transmembrane proteins are linked to focal adhesions, which are coupled to the cytoskeleton, but this is not the full story. In addition to being physically linked to their underlying substrate, the cells themselves are physically linked to each other through cell–cell junctions (Gomez et al. 2011; Weber et al. 2012; Borghi et al. 2010). While the mechanics of cell–cell junctions remain poorly understood, it is agreed upon that they sense and exert forces of theirown.
Liu et al. initially examined intercellular forces between a pair of geometrically constrained cells and reported intercellular force magnitude to regulate cell–cell contact size (Liu et al. 2010). We later expanded this finding to geometrically constrained endothelial monolayers and determined intercellular forces to be dependent on substrate stiffness and Rho kinase activity (Krishnan et al. 2011). Complementary to our work the Gardel group later concluded intercellular forces to be independent of cell size and morphology (Maruthamuthu et al. 2011). These studies demonstrated that groups of cells exert intercellular forces at their cell–cell junctions, opening the door for many more exciting questions to be answered. For example: How do these forces differ for larger groups of cells (>1000) as would be seen physiologically? What if any are the spatial and temporal fluctuations observed by these forces? How, if at all, does this affect collective cellular migration?
To answer these questions we used traction microscopy and MSM to observe the physical dynamics of migrating cell monolayers (Tambe et al. 2011). We cultured monolayers of rat pulmonary microvascular endothelial cells and MDCK cells on elastic gels and observed their migration. Using MSM we reported for the first time high-resolution normal and shear intercellular stress maps, revealing stress distributions that were extremely heterogeneous. Also the velocity vector of each migrating cell was found to correlate with the maximum normal stress (or minimal shear stress), implying that cells collectively migrating do so such as to minimize shear stress on its junctions, a phenomenon we have termed plithotaxis (Tambe et al. 2011; Trepat and Fredberg 2011). Plithotaxis represents the first physical explanation of collective cellular migration. This mechanically guided behavior was inhibited by calcium chelation and treatment of anti-cadherin antibodies in MCF10A breast cancer cells, suggesting cells must be physically linked to each other in order to migrate via plithotaxis (Tambe et al. 2011). Whether plithotaxis works cooperatively with other well-known cell guidance mechanisms such as chemotaxis, mechanotaxis, and durotaxis or if plithotaxis exists in vivo remains unanswered. There is also the possibility that plithotaxis is integral in cellular processes beyond collective cell migration where the boundary conditions of cells are different.
3.5.3 Viewing the Cell as a Soft Glassy Material
Stretch-induced fluidization, reinforcement, and plithotaxis all imply an emerging yet surprising concept of cells being comparable to a class of materials in physics known as soft glassy materials (SGMs). SGM are quite ubiquitous in nature and include colloidal suspensions, pastes, foams, and slurries (Sollich 1998; Sollich et al. 1997). While materials classified as SGM may vary in chemical properties, they share similar mechanical properties. All materials classified as SGM share these three common mechanical characteristics: (1) they are soft (young’s modulus = <1 kPA), (2) their dynamics are “scale free,” and (3) the frictional stress is proportional to the elastic stress with a constant of proportionality known as the hysteresivity, η (where η is on the order of 0.1) (Sollich 1998; Sollich et al. 1997; Krishnan et al. 2008). In accordance, SGM also have the ability to selectively phase transition between solid-like and liquid-like states (Sollich 1998). An initial suggestion that cells are analogous to SGM was provided by Fabry (Fabry et al. 2001b) when OMTC was used to demonstrate that multiple cell types exhibited a scaling law behavior that governed their elastic and frictional properties over a wide range of temporal scales and biological conditions. Trepat later combined OMTC with stretch on single cells, revealing the cell’s elastic and frictional properties to not only exhibit a scaling law behavior, but to be scale-free as well (Trepat et al. 2007). Complementary to our stretch studies, single cells subjected to osmotically induced compressive stress were observed to become much more solid-like (Zhou et al. 2009). Cell material properties and their similarities to SGM have been documented for single cells, but the question of whether this behavior remains true for groups of cells such as monolayers needed to be investigated.
To address this we used FTTM and MSM to observe cooperativity of intercellular stresses between cells within monolayers over relatively long distances (10–15 cell diameters) (Tambe et al. 2011). Intercellular stress cooperativity was observed to become enhanced over greater distances when comparing intercellular stress transmission to increasing cell density over time (Tambe et al. 2011), reflecting an increase in the dynamic heterogeneity of intercellular stress as cell density increased. Complementary to cooperativity of intercellular forces, Angelini (Angelini et al. 2011) recently reported cell migration velocity to decrease as cell density increased, a behavior remarkably similar to a glassy system transitioning from a liquid to solid phase (Krishnan et al. 2008).
This glassy-like behavior has led us as well as others to propose that cellular migration be viewed in terms of a glassy system. It should be cautioned that while these cellular systems have been observed to possess similar qualities of glassy systems, exhaustive studies still need to be done to prove beyond a doubt that these living, complex systems are in fact comparable to the inert glassy systems we see and use everyday.
3.6 Concluding Remarks
We have presented emergent behaviors on the cellular level that illustrate physical mechanisms which dictate biological responses and have forced researchers to rethink how they view the cell and its processes. We now know that mammalian cells primarily fluidize in response to stretch and epithelial and endothelial monolayers are mechanically guided as they collectively migrate through their environment. Finally, the ability of cells to fluidize, reinforce, and migrate by plithotaxis are all properties similar to SGMs. Supplementary to our findings is evidence linking these phenomena to systems beyond our fields of research, including cancer and morphogenesis. As there still exist many unknowns, these newly defined concepts represent only the tip of the iceberg of the underlying physical phenomena that influence cellular behavior.
References
Angelini TE, Hannezo E, Trepat X, Marquez M, Fredberg JJ, Weitz DA. Glass-like dynamics of collective cell migration. Proc Natl Acad Sci U S A. 2011;108(12):4714–9. doi:1010059108 [pii] 10.1073/pnas.1010059108.
Bao G, Suresh S. Cell and molecular mechanics of biological materials. Nat Mater. 2003;2(11):715–25. doi:10.1038/nmat1001 nmat1001 [pii].
Bellin RM, Kubicek JD, Frigault MJ, Kamien AJ, Steward Jr RL, Barnes HM, Digiacomo MB, Duncan LJ, Edgerly CK, Morse EM, Park CY, Fredberg JJ, Cheng CM, LeDuc PR. Defining the role of syndecan-4 in mechanotransduction using surface-modification approaches. Proc Natl Acad Sci U S A. 2009;106(52):22102–7. doi:0902639106 [pii] 10.1073/pnas.0902639106.
Borghi N, Lowndes M, Maruthamuthu V, Gardel ML, Nelson WJ. Regulation of cell motile behavior by crosstalk between cadherin- and integrin-mediated adhesions. Proc Natl Acad Sci U S A. 2010;107(30):13324–9. doi:1002662107 [pii] 10.1073/pnas.1002662107.
Butcher JT, Penrod AM, Garcia AJ, Nerem RM. Unique morphology and focal adhesion development of valvular endothelial cells in static and fluid flow environments. Arterioscler Thromb Vasc Biol. 2004;24(8):1429–34. doi:10.1161/01.ATV.0000130462.50769.5a01.ATV.0000130462.50769.5a [pii].
Butler JP, Tolic-Norrelykke IM, Fabry B, Fredberg JJ. Traction fields, moments, and strain energy that cells exert on their surroundings. Am J Physiol Cell Physiol. 2002;282(3):C595–605. doi:10.1152/ajpcell.00270.2001.
Cao LG, Fishkind DJ, Wang YL. Localization and dynamics of nonfilamentous actin in cultured cells. J Cell Biol. 1993;123(1):173–81.
Chen C, Krishnan R, Zhou E, Ramachandran A, Tambe D, Rajendran K, Adam RM, Deng L, Fredberg JJ. Fluidization and resolidification of the human bladder smooth muscle cell in response to transient stretch. PLoS One. 2010;5(8):e12035. doi:10.1371/journal.pone.0012035.
Cheng CM, Steward Jr RL, LeDuc PR. Probing cell structure by controlling the mechanical environment with cell-substrate interactions. J Biomech. 2009;42(2):187–92. doi:S0021-9290(08)00520-4 [pii]10.1016/j.jbiomech.2008.10.014.
Chien S. Mechanotransduction and endothelial cell homeostasis: the wisdom of the cell. Am J Physiol Heart Circ Physiol. 2007;292(3):H1209–24. doi:01047.2006 [pii] 10.1152/ajpheart.01047.2006.
Choquet D, Felsenfeld DP, Sheetz MP. Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell. 1997;88(1):39–48. doi:S0092-8674(00)81856-5 [pii].
Crick FC. The physical properties of the cytoplasm. A study by means of the magnetic particle method. Part 2. Theoretical treatment. Exp Cell Res. 1950;1:505–33.
Crick FC, Hughes AW. The physical properties of the cytoplasm. A study by means of the magnetic particle method. Part 1. Exp Cell Res. 1950;1:37–80.
DeMali KA, Wennerberg K, Burridge K. Integrin signaling to the actin cytoskeleton. Curr Opin Cell Biol. 2003;15(5):572–82. doi:S0955067403001091 [pii].
Dembo M, Wang YL. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J. 1999;76(4):2307–16. doi:S0006-3495(99)77386-8 [pii] 10.1016/S0006-3495(99)77386-8.
Dembo M, Oliver T, Ishihara A, Jacobson K. Imaging the traction stresses exerted by locomoting cells with the elastic substratum method. Biophys J. 1996;70(4):2008–22. doi:S0006-3495(96)79767-9 [pii] 10.1016/S0006-3495(96)79767-9.
Duchek P, Rorth P. Guidance of cell migration by EGF receptor signaling during Drosophila oogenesis. Science. 2001;291(5501):131–3. doi:10.1126/science.291.5501.131291/5501/131 [pii].
Eastwood M, McGrouther DA, Brown RA. Fibroblast responses to mechanical forces. Proc Inst Mech Eng H. 1998;212(2):85–92.
Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev Cell. 2008;14(4):570–81. doi:S1534-5807(08)00111-1 [pii] 10.1016/j.devcel.2008.03.003.
Fabry B, Maksym GN, Shore SA, Moore PE, Panettieri Jr RA, Butler JP, Fredberg JJ. Selected contribution: time course and heterogeneity of contractile responses in cultured human airway smooth muscle cells. J Appl Physiol. 2001a;91(2):986–94.
Fabry B, Maksym GN, Butler JP, Glogauer M, Navajas D, Fredberg JJ. Scaling the microrheology of living cells. Phys Rev Lett. 2001b;87(14):148102.
Friedl P, Hegerfeldt Y, Tusch M. Collective cell migration in morphogenesis and cancer. Int J Dev Biol. 2004;48(5–6):441–9. doi:10.1387/ijdb.041821 041821 [pii].
Garanich JS, Mathura RA, Shi ZD, Tarbell JM. Effects of fluid shear stress on adventitial fibroblast migration: implications for flow-mediated mechanisms of arterialization and intimal hyperplasia. Am J Physiol Heart Circ Physiol. 2007;292(6):H3128–3135. doi:00578.2006 [pii]10.1152/ajpheart.00578.2006.
Gomez GA, McLachlan RW, Yap AS. Productive tension: force-sensing and homeostasis of cell-cell junctions. Trends Cell Biol. 2011;21(9):499–505. doi:S0962-8924(11)00105-X [pii] 10.1016/j.tcb.2011.05.006.
Hall A, Nobes CD. Rho GTPases: molecular switches that control the organization and dynamics of the actin cytoskeleton. Philos Trans R Soc Lond B Biol Sci. 2000;355(1399):965–70. doi:10.1098/rstb.2000.0632.
Harris AK. Fibroblasts and myofibroblasts. Methods Enzymol. 1988;163:623–42.
Harris AK, Wild P, Stopak D. Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science. 1980;208(4440):177–9.
Hu S, Eberhard L, Chen J, Love JC, Butler JP, Fredberg JJ, Whitesides GM, Wang N. Mechanical anisotropy of adherent cells probed by a three-dimensional magnetic twisting device. Am J Physiol Cell Physiol. 2004;287(5):C1184–91. doi:10.1152/ajpcell.00224.2004 00224.2004 [pii].
Huang H, Kamm RD, Lee RT. Cell mechanics and mechanotransduction: pathways, probes, and physiology. Am J Physiol Cell Physiol. 2004;287(1):C1–11. doi:10.1152/ajpcell.00559.2003 287/1/C1 [pii].
Inaki M, Vishnu S, Cliffe A, Rorth P. Effective guidance of collective migration based on differences in cell states. Proc Natl Acad Sci U S A. 2012;109(6):2027–32. doi:1115260109 [pii] 10.1073/pnas.1115260109.
Insall RH, Machesky LM. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev Cell. 2009;17(3):310–22. doi:S1534-5807(09)00348-7 [pii] 10.1016/j.devcel.2009.08.012.
Krishnan R, Trepat X, Nguyen TT, Lenormand G, Oliver M, Fredberg JJ. Airway smooth muscle and bronchospasm: fluctuating, fluidizing, freezing. Respir Physiol Neurobiol. 2008; 163(1–3):17–24. doi:S1569-9048(08)00099-2 [pii] 10.1016/j.resp.2008.04.006.
Krishnan R, Park CY, Lin YC, Mead J, Jaspers RT, Trepat X, Lenormand G, Tambe D, Smolensky AV, Knoll AH, Butler JP, Fredberg JJ. Reinforcement versus fluidization in cytoskelet al mechanoresponsiveness. PLoS One. 2009;4(5):e5486. doi:10.1371/journal.pone.0005486.
Krishnan R, Klumpers DD, Park CY, Rajendran K, Trepat X, van Bezu J, van Hinsbergh VW, Carman CV, Brain JD, Fredberg JJ, Butler JP, van Nieuw Amerongen GP. Substrate stiffening promotes endothelial monolayer disruption through enhanced physical forces. Am J Physiol Cell Physiol. 2011;300(1):C146–54. doi:ajpcell.00195.2010 [pii] 10.1152/ajpcell.00195.2010.
Kung C. A possible unifying principle for mechanosensation. Nature. 2005;436(7051):647–54. doi:nature03896 [pii]10.1038/nature03896.
Larsen M, Wei C, Yamada KM. Cell and fibronectin dynamics during branching morphogenesis. JCell Sci. 2006;119(Pt 16):3376–84. doi:jcs.03079 [pii] 10.1242/jcs.03079.
Lauffenburger DA. Cell signaling pathways as control modules: complexity for simplicity? Proc Natl Acad Sci U S A. 2000;97(10):5031–3. doi:97/10/5031 [pii].
Liu Z, Tan JL, Cohen DM, Yang MT, Sniadecki NJ, Ruiz SA, Nelson CM, Chen CS. Mechanical tugging force regulates the size of cell-cell junctions. Proc Natl Acad Sci U S A. 2010;107(22):9944–9. doi:0914547107 [pii] 10.1073/pnas.0914547107.
Maksym GN, Fabry B, Butler JP, Navajas D, Tschumperlin DJ, Laporte JD, Fredberg JJ. Mechanical properties of cultured human airway smooth muscle cells from 0.05 to 0.4 Hz. J Appl Physiol. 2000;89(4):1619–32.
Maruthamuthu V, Sabass B, Schwarz US, Gardel ML. Cell-ECM traction force modulates endogenous tension at cell-cell contacts. Proc Natl Acad Sci U S A. 2011;108(12):4708–13. doi:1011123108 [pii]10.1073/pnas.1011123108.
Matthews BD, Overby DR, Mannix R, Ingber DE. Cellular adaptation to mechanical stress: role of integrins, Rho, cytoskelet al tension and mechanosensitive ion channels. J Cell Sci. 2006;119(Pt 3):508–18. doi:119/3/508 [pii] 10.1242/jcs.02760.
Norman JC, Jones D, Barry ST, Holt MR, Cockcroft S, Critchley DR. ARF1 mediates paxillin recruitment to focal adhesions and potentiates Rho-stimulated stress fiber formation in intact and permeabilized Swiss 3T3 fibroblasts. J Cell Biol. 1998;143(7):1981–95.
Oliver T, Dembo M, Jacobson K. Traction forces in locomoting cells. Cell Motil Cytoskeleton. 1995;31(3):225–40. doi:10.1002/cm.970310306.
Oliver T, Jacobson K, Dembo M. Design and use of substrata to measure traction forces exerted by cultured cells. Methods Enzymol. 1998;298:497–521.
Oliver T, Dembo M, Jacobson K. Separation of propulsive and adhesive traction stresses in locomoting keratocytes. J Cell Biol. 1999;145(3):589–604.
Owan I, Burr DB, Turner CH, Qiu J, Tu Y, Onyia JE, Duncan RL. Mechanotransduction in bone: osteoblasts are more responsive to fluid forces than mechanical strain. Am J Physiol. 1997;273(3 Pt 1):C810–5.
Pirentis AP, Peruski E, Iordan AL, Stamenovic D. A model for stress fiber realignment caused by cytoskelet al fluidization during cyclic stretching. Cell Mol Bioeng. 2011;4(1):67–80. doi:10.1007/s12195-010-0152-9.
Poukkula M, Cliffe A, Changede R, Rorth P. Cell behaviors regulated by guidance cues in collective migration of border cells. J Cell Biol. 2011;192(3):513–24. doi:jcb.201010003 [pii] 10.1083/jcb.201010003.
Puig-De-Morales M, Grabulosa M, Alcaraz J, Mullol J, Maksym GN, Fredberg JJ, Navajas D. Measurement of cell microrheology by magnetic twisting cytometry with frequency domain demodulation. J Appl Physiol. 2001;91(3):1152–9.
Quinn TP, Schlueter M, Soifer SJ, Gutierrez JA. Cyclic mechanical stretch induces VEGF and FGF-2 expression in pulmonary vascular smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2001;282(5):L897–903.
Revenu C, Athman R, Robine S, Louvard D. The co-workers of actin filaments: from cell structures to signals. Nat Rev Mol Cell Biol. 2004;5(8):635–46. doi:10.1038/nrm1437.
Ribeiro C, Ebner A, Affolter M. In vivo imaging reveals different cellular functions for FGF and Dpp signaling in tracheal branching morphogenesis. Dev Cell. 2002;2(5):677–83. doi:S1534580702001715 [pii].
Richard MN, Deniset JF, Kneesh AL, Blackwood D, Pierce GN. Mechanical stretching stimulates smooth muscle cell growth, nuclear protein import, and nuclear pore expression through mitogen-activated protein kinase activation. J Biol Chem. 2007;282(32):23081–8. doi:M703602200 [pii]10.1074/jbc.M703602200.
Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302(5651):1704–9. doi:10.1126/science.1092053302/5651/1704 [pii].
Rodriguez OC, Schaefer AW, Mandato CA, Forscher P, Bement WM, Waterman-Storer CM. Conserved microtubule-actin interactions in cell movement and morphogenesis. Nat Cell Biol. 2003;5(7):599–609. doi:10.1038/ncb0703-599 ncb0703-599 [pii].
Rorth P. Collective guidance of collective cell migration. Trends Cell Biol. 2007;17(12):575–9. doi:S0962-8924(07)00246-2 [pii] 10.1016/j.tcb.2007.09.007.
Rorth P. Whence directionality: guidance mechanisms in solitary and collective cell migration. Dev Cell. 2011;20(1):9–18. doi:S1534-5807(10)00595-2 [pii] 10.1016/j.devcel.2010.12.014.
Scotton CJ, Wilson JL, Milliken D, Stamp G, Balkwill FR. Epithelial cancer cell migration: a role for chemokine receptors? Cancer Res. 2001;61(13):4961–5.
Smith PG, Deng L, Fredberg JJ, Maksym GN. Mechanical strain increases cell stiffness through cytoskelet al filament reorganization. Am J Physiol Lung Cell Mol Physiol. 2003;285(2): L456–63. doi:10.1152/ajplung.00329.2002 00329.2002 [pii].
Sniadecki NJ, Chen CS. Microfabricated silicone elastomeric post arrays for measuring traction forces of adherent cells. Methods Cell Biol. 2007;83:313–28. doi:S0091-679X(07)83013-5 [pii]10.1016/S0091-679X(07)83013-5.
Sollich P. Rheological constitutive equation for a model of soft glassy materials. Phys Rev E. 1998;58:738–59.
Sollich P, Lequeneux F, Hebraud P, Cates ME. Rheology of soft glassy materials. Phys Rev Lett. 1997;78:2020–3.
Steward Jr RL, Cheng CM, Wang DL, Leduc PR. Probing cell structure responses through a shear and stretching mechanical stimulation technique. Cell Biochem Biophys. 2009;56(2–3): 115–24. doi:10.1007/s12013-009-9075-2.
Tambe DT, Hardin CC, Angelini TE, Rajendran K, Park CY, Serra-Picamal X, Zhou EH, Zaman MH, Butler JP, Weitz DA, Fredberg JJ, Trepat X. Collective cell guidance by cooperative intercellular forces. Nat Mater. 2011;10(6):469–75. doi:nmat3025 [pii] 10.1038/nmat3025.
Thompson DAW. On growth and form. Abridged ed. Cambridge: Cambridge University Press; 1961.
Trepat X, Fredberg JJ. Plithotaxis and emergent dynamics in collective cellular migration. Trends Cell Biol. 2011;21(11):638–46. doi:S0962-8924(11)00127-9 [pii] 10.1016/j.tcb.2011.06.006.
Trepat X, Deng L, An SS, Navajas D, Tschumperlin DJ, Gerthoffer WT, Butler JP, FredbergJJ. Universal physical responses to stretch in the living cell. Nature. 2007;447(7144):592–5. doi:nature05824 [pii]10.1038/nature05824.
Trepat X, Wasserman MR, Angelini TE, Millet E, Weitz DA, Butler JP, Fredberg JJ. Physical forces during collective migration. Nat Phys. 2009;5:426–30.
Vasilyev A, Liu Y, Mudumana S, Mangos S, Lam PY, Majumdar A, Zhao J, Poon KL, Kondrychyn I, Korzh V, Drummond IA. Collective cell migration drives morphogenesis of the kidney nephron. PLoS Biol. 2009;7(1):e9. doi:08-PLBI-RA-2396 [pii] 10.1371/journal.pbio.1000009.
Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol. 2006;7(4):265–75. doi:nrm1890 [pii]10.1038/nrm1890.
von Wichert G, Haimovich B, Feng GS, Sheetz MP. Force-dependent integrin-cytoskeleton linkage formation requires downregulation of focal complex dynamics by Shp2. EMBO J. 2003;22(19):5023–35. doi:10.1093/emboj/cdg492.
Wang JH, Thampatty BP. An introductory review of cell mechanobiology. Biomech Model Mechanobiol. 2006;5(1):1–16. doi:10.1007/s10237-005-0012-z.
Wang N, Butler JP, Ingber DE. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260(5111):1124–7.
Weber GF, Bjerke MA, DeSimone DW. A mechanoresponsive cadherin-keratin complex directs polarized protrusive behavior and collective cell migration. Dev Cell. 2012;22(1):104–15. doi:S1534-5807(11)00465-5 [pii] 10.1016/j.devcel.2011.10.013.
Yoshigi M, Hoffman LM, Jensen CC, Yost HJ, Beckerle MC. Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskelet al reinforcement. J Cell Biol. 2005;171(2):209–15. doi:jcb.200505018 [pii]10.1083/jcb.200505018.
Zheng Y, Olson MF, Hall A, Cerione RA, Toksoz D. Direct involvement of the small GTP-binding protein Rho in lbc oncogene function. J Biol Chem. 1995;270(16):9031–4.
Zhou EH, Trepat X, Park CY, Lenormand G, Oliver MN, Mijailovich SM, Hardin C, Weitz DA, Butler JP, Fredberg JJ. Universal behavior of the osmotically compressed cell and its analogy to the colloidal glass transition. Proc Natl Acad Sci U S A. 2009;106(26):10632–7. doi:0901462106 [pii] 10.1073/pnas.0901462106.
Zhu C, Bao G, Wang N. Cell mechanics: mechanical response, cell adhesion, and molecular deformation. Annu Rev Biomed Eng. 2000;2:189–226. doi:2/1/189 [pii] 10.1146/annurev.bioeng.2.1.189.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media, LLC
About this chapter
Cite this chapter
Steward, R.L., Rosner, S.R., Fredberg, J.J. (2016). Emergent Behaviors in Cell Mechanics. In: Kassab, G., Sacks, M. (eds) Structure-Based Mechanics of Tissues and Organs. Springer, Boston, MA. https://doi.org/10.1007/978-1-4899-7630-7_3
Download citation
DOI: https://doi.org/10.1007/978-1-4899-7630-7_3
Publisher Name: Springer, Boston, MA
Print ISBN: 978-1-4899-7629-1
Online ISBN: 978-1-4899-7630-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)