Abstract
The development of bacterial resistance against current antibiotic drugs necessitates a continuous renewal of the arsenal of efficacious drugs. This imperative has not been met by the output of antibiotic research and development of the past decades for various reasons, including the declining efforts of large pharma companies in this area. Moreover, the majority of novel antibiotics are chemical derivatives of existing structures that represent mostly step innovations, implying that the available chemical space may be exhausted. This review negates this impression by showcasing recent achievements in lead finding and optimization of antibiotics that have novel or unexplored chemical structures. Not surprisingly, many of the novel structural templates like teixobactins, lysocin, griselimycin, or the albicidin/cystobactamid pair were discovered from natural sources. Additional compounds were obtained from the screening of synthetic libraries and chemical synthesis, including the gyrase-inhibiting NTBI’s and spiropyrimidinetrione, the tarocin and targocil inhibitors of wall teichoic acid synthesis, or the boronates and diazabicyclo[3.2.1]octane as novel β-lactamase inhibitors. A motif that is common to most clinically validated antibiotics is that they address hotspots in complex biosynthetic machineries, whose functioning is essential for the bacterial cell. Therefore, an introduction to the biological targets—cell wall synthesis, topoisomerases, the DNA sliding clamp, and membrane-bound electron transport—is given for each of the leads presented here.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
The “golden era of antibiotic drug discovery” (Davies 2006) commenced with the breakthrough discovery of the sulfonamides and β-lactams in the 1930s and lasted o the mid 1960s, as the majority of the antibiotics used today had been discovered in that period. Since then, the speed of novel antibiotic discovery has become slower. This is expressed by a declining absolute number of launched antibiotics, and by the fact that only few of those addressed a new mode of action (Policy 2010; Boucher et al. 2013; O’Connell et al. 2013; Butler et al. 2013b). Indeed, most antibiotics launched in the past 30 years are chemical analogs of well-established drug classes. It has to be clearly stated that most of such analogs (so-called ‘step-innovations’) confer a medical benefit (Wright et al. 2014), e.g., through enlarged safety windows, improved ADMET properties, circumvented bacterial efflux or degradation systems (Brötz-Oesterhelt and Brunner 2008), and sometimes impressive shifts in antibacterial activity from a distinct activity against Gram-positive or Gram-negative bacteria to a broad-spectrum antibiotic activity, as demonstrated by the fourth and fifth generation of cephalosporins, advanced tetracycline, or fluoroquinolone analogs (Wright et al. 2014). However, the evolution of bacterial resistance against these analogs is often faster due to cross resistances (Coates et al. 2011).
Since 2000 five new classes of antibiotics have entered the market (Scheme 1): linezolid (2000), daptomycin (2003), retapamulin (2007), fidaxomicin (2010), and bedaquiline (2012) (Butler et al. 2013b).
These successes are a contribution to improve the innovation gap in antibiotic research and development, but they are clearly insufficient to cover the increasing medical need of a sustainable supply with effective antibiotics (O’Neill 2015). All of these five antibiotics only address infections with Gram-positive pathogens. Furthermore, retapamulin is confined to topical administrations, and fidaxomicin and bedaquiline are narrow spectrum agents that are only approved for treatments of C. difficile and MDR-TB, respectively. There remains a particularly strong, unmet need for novel antibiotics effective against Gram-negative pathogens. Because the lack of innovation (Policy 2010; Boucher et al. 2013; O’Connell et al. 2013) in antibiotic drug research coincides with an increasing occurrence of infections with multi-drug-resistant pathogens (Boucher et al. 2009; Peterson 2009) associated with high mortality and morbidity (Pendleton et al. 2013), there are concerns that the increasing lack of effective therapeutics could develop into a serious threat for public health, or even into a fallback to a so-called pre-antibiotic era (Piddock 2012; Ventola 2015; Wright 2015b).
The anthropological reasons for this situation are manifold; they include the inadequate clinical use of existing antibiotics (Gilbert 2015; Sanchez and Demain 2015; Shiva 2015), extended misuse of antibiotics in intensive animal husbandry for food production (Bengtsson and Greko 2014; Littmann et al. 2015), and the economically driven exodus of big pharma companies from the antibiotics research field that contributed to the innovation gap mentioned above (Lowther 1979; Powers 2003; Projan 2003; Spellberg et al. 2004; Taubes 2008; Torres 2010; O’Connell et al. 2013).
Beyond these anthropological acceleration forces (Breu et al. 2001; Gillings 2013), we have to accept that bacterial resistance to antibiotics is not a side effect of modern drug therapy, but an inherent part of bacterial evolution to fight for their evolutionary niche with other bacteria and further organisms (Wright 2012; Wright and Poinar 2012; Rodríguez-Rojas et al. 2013).
It has been estimated that bacteria-producing antibacterial metabolites originated at least hundreds of millions of years ago (Baltz 2008; Wright and Poinar 2012). During the evolution of antibacterial metabolites, these bacteria had to intrinsically coevolve resistance mechanisms for self-preservation (Wright 2012; Wright and Poinar 2012; Perry et al. 2014), since in most cases they probably possess the same biological target for the drug. Therefore, it can be assumed that resistance mechanisms have existed for just as long as the corresponding antibacterial metabolites (O’Connell et al. 2013). This hypothesis is supported by a metagenomic analysis of DNA found in permafrost sediments, which was determined to be 30,000 years old and led to the discovery of genes encoding for resistance to β-lactams, tetracyclines, and glycopeptide antibacterials (D’Costa et al. 2011). This is most probably also true for purely synthetic antibiotics, as nature provides natural product antibiotics directed to nearly every known druggable target in bacteria (Lin et al. 1997; Keller et al. 2007; Johnston et al. 2016). Furthermore, we have to consider that every time an antibiotic is administered there is a significant influence on the resistome (Gillings 2013), which is defined as the collection of all genes in pathogenic and non-pathogenic bacteria that could contribute to a phenotype of antibiotic resistance (Frankel et al. 2006; Wright 2007, 2010; Forsberg et al. 2012; Gillings 2013; Nesme and Simonet 2015). This influence can lead to the evolution of resistance also in non-pathogenic bacteria in the patient or after clearance of the drug into the general environment, i.e., in organisms which have not been originally targeted by the treatment. The resistance genes can easily spread via horizontal gene transfer (HGT) (Syvanen 2012; Baltrus 2013; Polz et al. 2013) by mobile plasmids (Smillie et al. 2010; Harrison and Brockhurst 2012; Carattoli 2013), transposons (Casacuberta and Gonzalez 2013), or outer membrane vesicle (OMV) (Berleman and Auer 2013; Brown et al. 2015; Perez-Cruz et al. 2015; Schwechheimer and Kuehn 2015) transport through the whole pan-genome (Medini et al. 2005; Tettelin et al. 2008; Lapierre and Gogarten 2009) of the global microbiome (Whitman et al. 1998).
These facts imply that in pronounced contrast to other medical indications, the efficacy of antibacterial drugs deteriorates over time. Therefore, the identification of novel antimicrobials, especially with new modes of action (Fischbach and Walsh 2009; Wattal and Goel 2011), is a continuous, necessary task to keep a life-saving headway in the permanent race between bacterial evolution and the protection of human health (Rodríguez-Rojas et al. 2013). In addition, the way antibiotics are handled today should be seriously revised, since studies have shown that smart policies for the prudent use of antibiotics in the clinic and throughout agriculture can make a significant difference in the occurrence and the level of resistance (European Centre for Disease Prevention and Control: Annual Report of the European Antimicrobial Resistance Surveillance Network, EARS-Net 2012).
The threat of multi-drug-resistant pathogens has already been recognized and gained international political attention, leading to several national and international programs and initiatives for the research and development of novel antibiotics (Policy 2010; Boucher et al. 2013; Rex 2014; Bush 2015; Eichberg 2015). At this stage, most of the compounds that have recently been launched or that are currently undergoing phase II/III clinical trials are analogs of existing classes (Pucci and Bush 2013; Hesterkamp 2016).
As the marketed classes of antibiotics have been extensively reviewed before (Butler et al. 2013b; Zetts 2014; Paris 2015), we would like to focus this review on the feasibility to discover novel structural templates with antibiotic activity, and to advance such compounds to late-stage pre-clinical as well as clinical development (Boucher et al. 2013; O’Connell et al. 2013; Butler et al. 2013b; Brown et al. 2014; Xu et al. 2014a; Bush 2015; Paris 2015). For this purpose, a brief overview of existing druggable targets and common features among them is given, followed by a review of promising novel scaffolds that address existing as well as novel targets. The presented scaffolds result from a personal and non-comprehensive selection of compounds addressing essential bacterial machineries, i.e., DNA replication, cell wall synthesis, and membrane components.
2 Lessons Learned from Druggable Targets in Bacteria
In the over one hundred years of antibiotic research, numerous different bacterial targets and their corresponding interaction with antibiotics have been investigated (Tommasi et al. 2015). As in other indications, the majority of drug-like, optimized compounds did not reach the clinics, as they addressed non-valid targets hampered by an insufficient conservation in the microbial spectrum, unexpected metabolic bypass mechanisms, non-essentiality under in vivo conditions or unexpectedly rapid resistance formations (Payne et al. 2007). A special characteristic of antibiotic R&D is the dominance of natural products as the source of approved drugs (Koehn and Carter 2005; Bérdy 2012; Newman and Cragg 2012; Kirst 2013; Bauer and Brönstrup 2014; Butler et al. 2014; Schaefer 2014; Harvey et al. 2015). In fact, the far majority of antibiotics on the market are derived from secondary metabolites of bacteria (Gerth et al. 2003; Clardy et al. 2006; Diez et al. 2012; Schäberle et al. 2014; Elshahawi et al. 2015), marine microorganisms (Blunt et al. 2011), fungi (Schueffler and Anke 2014; Stadler and Hoffmeister 2015), and (in rare cases) plants (Gibbons 2004; Savoia 2012; Upadhyay et al. 2014). This is no surprise, considering that nature provides a huge diversity of structures, which have been evolutionary optimized for binding their biological targets and used by their producers to fight for their ecological niche against competing or harmful microbes. Even though most of the antibacterial natural products need further semi-synthetic optimization to fulfill the ADME (Absorption, Distribution, Metabolism, and Excretion) properties required for a drug applied to humans, nature is still the most effective source for novel antibacterial lead structures (Cragg and Newman 2013; Brown et al. 2014). Natural products have also often disclosed novel, relevant biological targets, which were subsequently addressed by synthetic compounds.
In Table 1, the most important approved antibiotic classes are summarized according to the metabolic pathway they address, their molecular targets, and their distinct mechanism of action. The targets they address share important common features, i.e., essentiality for the survival of the pathogen, an evolutionary conservation in a variety of pathogens, and a high evolutionary distance to the mammalian counterparts (Brötz-Oesterhelt and Brunner 2008). Antibiotics mainly applied in monotherapies such as β-lactams, glycopeptides, or fluoroquinolones show additional common characteristics: The most successful antibiotics all interact with large biological structures consisting of multi-enzyme complexes like the protein or cell wall biosynthesis machinery, whose impairment has drastic consequences for the bacterial cell that are hard to repair or compensate (Table 1) (Silver 2007; Brötz-Oesterhelt and Brunner 2008). In contrast, the few antibiotics targeting single enzymes such as sulfonamides and benzyl diaminopyrimidines have to be applied in combination therapies to avoid rapid resistance.
Furthermore, it is noticeable that there are fewer antibiotics available for the treatment of infections caused by Gram-negative pathogens compared to Gram-positive ones. The main reason for that issue is the difficulty for antibiotics to cross the outer membrane of Gram-negative bacteria (Zgurskaya et al. 2015). The surface of the outer membrane of the Gram-negative bilayer is covered with lipopolysaccharides (LPS), lipidic structures embedded and anchored into the bilayer that consist of three, covalently connected structural elements: A proximal hydrophobic lipid A, a core oligosaccharide region, and a distal O-antigen polysaccharide (Cohen 2011). It has been shown that LPS molecules interact with each other on the cell surface through divalent cations, thereby forming a permeability barrier (Nikaido 2003; Zhang et al. 2013). Due to the embedded cations, hydrophobic molecules have been shown to partition poorly into LPS and to permeate across the outer membrane bilayer with extremely low rates. Therefore, the LPS-containing bilayer serves as an efficient barrier for diffusion-mediated uptake of many chemical scaffolds known to be active against Gram-positive bacteria (Zhang et al. 2013). The uptake of most antibiotics into Gram-negative bacteria is mediated by porins, β-barrel-shaped proteins that act as a pore through which small, polar molecules can diffuse. Because the physicochemical constraints for porin diffusion are hardly met by existing compound collections designed for high affinity binding of canonical eukaryotic targets and for assuring oral bioavailability, attempts to discover novel lead structures active against Gram-negative pathogens have been by-and-large unsuccessful (Payne et al. 2007; Tommasi et al. 2015). Thus, the searches for relevant chemical matter (e.g., natural products preoptimized by microorganisms) or for alternative strategies [e.g., active transport machineries of Gram-negative pathogens (Ji et al. 2012; Górska et al. 2014; Mislin and Schalk 2014; Saha et al. 2013; Wang et al. 2014; Johnstone and Nolan 2015; Page 2013; Górska et al. 2014; Mislin and Schalk 2014; Xu et al. 2014b; Zgurskaya et al. 2015] need to be intensified significantly.
3 Lipid II and Lipid III—The Bottlenecks in Peptidoglycan and Wall Teichoic Acid Synthesis
Known, clinically validated targets represent the most promising choice for the development of novel antibiotics, in particular when novel chemical scaffolds that address different binding sites compared to existing drugs break resistance.
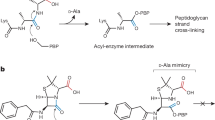
An excellent example for such a target is lipid II 6 (Scheme 2) (Breukink and de Kruijff 2006; de Kruijff et al. 2008), an amphiphilic, membrane-anchored peptidoglycan precursor molecule that is essential for cell membrane functionality.
The cell walls of all bacteria contain a layer of peptidoglycan, a biopolymer of the alternating amino sugars N-acetylglucosamine (D-NAc-Glu, NAG), and N-acetylmuramic acid (NAc-Mur, NAM), which is modified with pentapeptides of the sequence L-alanyl-γ-D-glutamyl-L-lysyl-D-alanyl-D-alanine (in the case of S. aureus) that is attached to the 3-hydroxy group of the NAc-Mur sugar (Schwartz et al. 2001; Breukink and de Kruijff 2006; de Kruijff et al. 2008). The cross linkage of the peptides by penicillin-binding proteins (PBPs) to macromolecules confers a structural rigidity and mechanical strength that prevents the bacterial protoplast to burst under the osmotic internal pressure.
The synthesis of lipid II as the precursor monomer of the peptidoglycan layer begins on the cytoplasmic site of the plasma membrane (Scheme 3, right side). N-Acetyl-muramic-acid-pentapeptide uridinyl pyrophosphate (UDP-NAc-Mur) is attached by the enzyme phosphor-MurNAc-pentapetide translocase (MraY) (Chung et al. 2013) to bactoprenol phosphate, which is embedded in the phospholipid membrane. The resulting membrane-anchored lipid I is then further glycosylated by the peripherally membrane-associated enzyme MurG (Mohammadi et al. 2007) leading to the formation of lipid II. Next, the FemXAB transpeptidases catalyze the coupling of a pentaglycine-moiety to the D-lysine of the pentapeptide unit and furthermore, the amidation of the γ-D-glutamate of the pentapeptide unit by the MurT/GatD two-enzyme complex leading to the corresponding D-isoglutamin (Münch et al. 2012; Zapun et al. 2013). This modified lipid II now contains the complete peptidoglycan monomer linked via a pyrophosphate to the bactoprenol membrane anchor, which is translocated through the phospholipid membrane by the MurJ flippase and presented on the extracellular membrane surface (Mohammadi et al. 2011; Butler et al. 2013a; Sham et al. 2014; Meeske et al. 2015).
On the extracellular membrane surface the peptidoglycan precursor is released, incorporated into the growing peptidoglycan layer and crosslinked (Zapun et al. 2013) with each other catalyzed by the PBPs (Sauvage et al. 2008).
The peptidoglycan layer in Gram-positive bacteria is generally around 20–80 nm thick and contains up to 20 sublayers with vast amounts of peptidoglycan subunits, which all have to be translocated through the membrane in the form of the membrane-anchored lipid II after assembly on the cytosolic membrane surface. Since supply with bactoprenol phosphate in bacteria is restricted (Storm and Strominger 1974; Kramer et al. 2004), the amount of lipid II that can be generated is limited. Thus, the lipid II cycle is a highly dynamic process that represents the bottleneck in bacterial cell wall synthesis. Therefore, the lipid II cycle is an ideal target for antibiotics (Breukink and de Kruijff 2006).
The first approved antibiotic targeting lipid II was the glycopeptide vancomycin 7 (Scheme 4). Glycopeptide antibiotics are active against a broad variety of Gram-positive pathogens and act by binding to the D-Ala-D-Ala-dipeptide terminus in lipid II on the extracellular membrane surface via several hydrogen bonds and ionic interactions (Jia et al. 2013; Münch et al. 2015).
Vancomycin has been for a long time the antibiotic of last resort against Gram-positive pathogens (Williams and Bardsley 1999). Due to the difficulty for bacteria to circumvent binding to an essential metabolite (rather than a protein target), the first significant levels of resistance to vancomycin took almost three decades to occur (Boneca and Chiosis 2003). However, nowadays there is an increasing prevalence of resistance against vancomycin (Williams and Bardsley 1999), which is caused in most cases by the exchange of the terminal D-alanine of the lipid II structure by a D-lactate (Williams and Bardsley 1999; Kahne et al. 2005), which results in the loss of a single hydrogen bond to the D-Ala residue, and a 100-fold drop in binding affinity (Mccomas et al. 2003).
Another well-investigated antibiotic interacting with lipid II is the peptide l antibiotic nisin 8 (Scheme 5). Nisin, obtained by fermentation of Lactobacillus lactis on natural substrates such as milk or dextrose, is widely used as a safe food preservation additive (Cleveland et al. 2001), since orally administered nisin is completely degraded by digesting enzymes in the stomach.
Nisin binds to the pyrophosphate moiety of lipid II (Scheme 5) forming a phosphate cage that prevents the release of the peptidoglycan subunits and leads to the formation of a pore in the membrane via inter-membrane assembly of a nisin–lipid II 8:4 complex (Hsu et al. 2004; Bauer and Dicks 2005; Martin and Breukink 2007).
The first isoprene unit and the pyrophosphate in lipid II and the N-terminal part of nisin have been identified as determinants for the nisin–lipid II interaction (Chan et al. 1989; Kuipers et al. 1993; Brötz et al. 1998; Wiedemann et al. 2001). Additionally, several other lipid II interacting compounds are currently under preclinical and clinical investigation such as ramoplanin, mannopeptimycin, lysobactin (Lee et al. 2016b), and plusbacin A3 (Breukink and de Kruijff 2006).
Recently, a novel lipid II interacting cyclodepsipeptide has been isolated from the Gram-negative β-proteobacterium Eleftheria terrae (Ling et al. 2015). Teixobactin 9 (Scheme 6) is an undecapeptide bearing four R-amino acids and seven S-amino acids including the unusual S-allo-enduracididine. The N-terminal R-phenylalanine is mono methylated, and the C-terminus of the molecule is cyclized to the hydroxyl function of a R-threonine side chain in the sequence, forming a 13-membered depsipeptide ring.
Teixobactin was discovered within a screen of so far uncultured soil bacteria by in situ cultivation in diffusion chambers using the so-called iChip technology (Nichols et al. 2010; Piddock 2015). The compound showed highly potent antibacterial activity against a broad variety of Gram-positive pathogens including several multi-drug-resistant strains such as methicillin-resistant S. aureus (MRSA) and vancomycin-resistant E. faecalis (VRE). Teixobactin was also highly active against C. difficile (MIC = 0.005 µg/mL) and M. tuberculosis (MIC = 0.125 µg/mL). The compound was ineffective against Gram-negative bacteria with the exception of E. coli (asmB1), a mutant having a defective outer membrane barrier.
In vitro experiments have shown that teixobactin bound under formation of stable 2:1 drug:target complexes to lipid I and lipid II by interaction with the pyrophosphate sugar moiety (Schemes 2 and 3) (Ling et al. 2015). Furthermore, at higher concentrations teixobactin was able to completely inhibit the YbjG-catalyzed mono-dephosphorylation of bactoprenol pyrophosphate, which is an essential step of the bactoprenol phosphate recycling with in the lipid II cycle of peptidoglycan biosynthesis (Scheme 3).
In vivo the binding to lipid II is believed to be the primary effect of teixobactin, since lipid I is not exposed to the extracellular site. Nevertheless, the exact nature of the sugar in the binding motif seems to be less important, since it has been demonstrated that teixobactin efficiently bound lipid III 10 (Scheme 7), bearing a NAc-Glu instead of NAc-Mur attached to the pyrophosphate.
Lipid III is a precursor molecule in the synthesis of wall teichoic acids (WTA). Wall teichoic acids are glycol-phosphopolyol biopolymers that are attached to the NAc-Mur moieties on the surface of the peptidoglycan layer of Gram-positive bacteria (Pereira et al. 2008; Brown et al. 2013), rendering the bacterial surface strongly anionic (Scheme 3).
WTA biosynthesis inhibition is not per se lethal for Gram-positive bacteria. Early-stage WTA inhibition can lead to cells showing enhanced sensitivity to certain drugs (see next chapter) (Sewell and Brown 2014; Lee et al. 2016a). In contrast, late-stage inhibition of membrane-bound WTA precursors is lethal due to accumulation of toxic intermediates and the depletion of cellular pools of bactoprenol phosphate (D’Elia et al. 2006b; Swoboda et al. 2011; Brown et al. 2013; Wang et al. 2013), which is essential for peptidoglycan biosynthesis as discussed above. Furthermore, it is known that teichoic acids anchor autolysins to prevent uncontrolled hydrolysis of peptidoglycan (Bierbaum and Sahl 1985; Dubée et al. 2011). Thus, inhibition of WTA synthesis leads to enhanced concentrations of free autolysins, which significantly contributes to the outstanding antibacterial activity of teixobactin. With its binding to lipid II and lipid III, teixobactin is targeting two non-protein lipid structures that are bottlenecks of cell wall synthesis in Gram-positive bacteria (Wright 2015a). It is therefore no surprise that the escape strategies for the pathogens are limited. Whether a resistance mechanism already exists is not known, but so far it was not possible to obtain teixobactin-resistant mutants of S. aureus or M. tuberculosis at 4x MIC or in serial passaging experiments (Ling et al. 2015). As the teixobactin producer is a Gram-negative bacterium, it is intrinsically resistant against re-entry of the drug due to the impermeability of the outer membrane, and therefore does not encode a resistance mechanism that could be horizontally transferred to Gram-positive species (Kåhrström 2015).
Preliminary pharmacokinetic and pharmacodynamics data from in vivo studies in mice were promising (100 % survival rate in a mouse efficacy study). The compound retained its potency in blood serum, showed overall good stability and low toxicity to mammalian cells. Therefore, teixobactin could become a valuable candidate to fight back the antibiotic crisis (Scheme 8).
Not surprisingly, the compound has already motivated medicinal chemists to synthesize the first analogs (Jad et al. 2015; Parmar et al. 2016). Two research groups independently published the simple solid-supported synthesis of the teixobactin analog 11 (Scheme 7), where the unusual S-allo-enduracididine is substituted by the structurally related, simpler arginine. In a second analog (12), the four R-amino acids were additionally substituted by S-amino acids. Antibacterial tests demonstrated that substitution of S-allo-enduracididine leads to a 10-fold drop in activity for analog 11. Furthermore, the R-configuration of several amino acids in the teixobactin sequence seems to be essential for the activity, since analog 12 leads to a 640-fold drop in activity compared to natural teixobactin. Nevertheless, more detailed structure–activity relationship (SAR) studies are needed to determine the exact binding interactions and to further optimize this emerging natural lead structure.
4 Early and Late-Stage WTA Biosynthesis Inhibition—Restoring the Efficacy of β-Lactams Against Gram-Positive Pathogens
The predominant resistance mechanisms toward β-lactams in Gram-negative and Gram-positive pathogens are different. Resistance in Gram-negative pathogens normally is achieved via two general mechanisms. One escape strategy is the production of degradation enzymes such as β-lactamases, rendering the antibiotic inactive due to hydrolysis of the β-lactam pharmacophore (Bush 2010; Rawat and Nair 2010). Alternatively, several Gram-negative pathogens overexpress membrane-embedded efflux pumps, which actively transport drugs out of the bacteria (Amaral et al. 2014; Blair et al. 2014). Therefore, considerable efforts have been made to restore the antibacterial activity of β-lactams against Gram-negative pathogens through inhibition of β-lactamases or of efflux pumps. While marketed β-lactamase inhibitors utilize a β-lactam motif themselves for suicide inhibition, chemists have recently identified and optimized novel structural templates: 1,6-diazabicyclo[3.2.1]octane-2-carboxamide-based, reversible β-lactamase inhibitors such as avibactam 13, relebactam 14, and OPO595 15 (Scheme 9) have been advanced to clinical trials as additive in combination therapies with different β-lactam antibiotics (Hirsch et al. 2012; Livermore et al. 2013; Lapuebla et al. 2015a; Livermore et al. 2015; Paris 2015; Toussaint and Gallagher 2015). Avibactam has already been approved for combination therapy with ceftazidime and is in clinical trials for combination therapy with several other β-lactams (Zetts 2014; Paris 2015). Furthermore, RPX7009 16, a novel boronic acid-based β-lactamase inhibitor, is currently in phase III clinical trials in combination therapy with meropenem. Notably, some congeners of the boronic acid-based class of compounds also cover metallo-β-lactamases like NDM-1 in addition to serine-β-lactamases (Hecker et al. 2015; Paris 2015; Lapuebla et al. 2015b). The significant progress achieved on this target class contributes to alleviate the upcoming antibiotic crisis in short to medium term.
Also, several promising efflux pump inhibitors for the AcrAB-TolC and MexAB-OrpM efflux pumps have been discovered (Scheme 9) (Askoura et al. 2011; Tegos et al. 2011; Opperman and Nguyen 2015; Venter et al. 2015), but so far none of them has entered the clinic.
The predominant resistance mechanism against β-lactams in clinically important Gram-positive pathogens is different, thus requiring a different strategy: It has been shown that resistance in MRSA is achieved by the horizontally acquired penicillin binding protein PBP2A, which has a considerably lower affinity for β-lactam antibiotics compared to the original, bifunctional transglycosylase/transpeptidase PBP2 (Pinho and Errington 2005; Lim and Strynadka 2002). β-Lactams acylate the active site for transpeptidation on PBP2 and thereby inhibit the autonomous localization to the division septum. In MRSA, PBP2A can take over the transpeptidase activity and additionally, act as a structural scaffold to recruit acylated PBP2 to the division septum to just exert its essential transglycosylation function (Pinho et al. 2001; Pinho and Errington 2005). This PBP2/PBP2A cooperation leads to the formation of peptidoglycan strands with low level of crosslinking (Leski and Tomasz 2005; Atilano et al. 2010), which are substrates for PBP4, a secondary transpeptidase that catalyzes the formation of highly crosslinked peptidoglycan (Wyke et al. 1981; Atilano et al. 2010).
It has been demonstrated that the correct localization of PBP4 at the division septum, which is crucial for the resistance mechanism, depends on the presence of wall teichoic acid (WTA) polymers (Scheme 10) (Atilano et al. 2010; Brown et al. 2012). As WTAs are also responsible for the anchoring of autolysins of the peptidoglycan as discussed above, simultaneous inhibition of WTA during the presence of β-lactams leads to depleted levels of crosslinked peptidoglycan and to non-viable bacteria, thus circumventing the resistance mechanism toward β-lactams and restoring their antibacterial activity against Gram-positive bacteria (Maki et al. 1994; Swoboda et al. 2011; Farha et al. 2013; Wang et al. 2013; Sewell and Brown 2014; Lee et al. 2016a).
Model of β-lactam resistance in MRSA and role of WTA. Adapted from Sewell and Brown (2014)
WTA biosynthesis, as summarized earlier in Scheme 3, is categorized into two distinct groups: The non-essential, WTA early-stage genes tarO, tarA, and mnaA, responsible for attachment of N-acetylglucosamine (NAG, D-NAc-Glu) and N-acetylmuramic acid (NAc-Mur, NAM) to membrane-anchored bactoprenol phosphate, and the conditionally essential WTA late-stage genes tarB, tarD, tarF, tarI, tarJ, tarL, tarG, and tarH, responsible for the further assembly of the WTA biopolymers and finally for translocation through the phospholipid membrane to the extracellular environment (Lazarevic and Karamata 1995; Meredith et al. 2008; Pereira et al. 2008; Brown et al. 2013).
As mentioned earlier, the inhibition of late-stage wall teichoic acid biosynthesis steps or deletions of the encoding genes leads to antibacterial effects due to accumulation of toxic intermediates and the depletion of cellular pools of bactoprenol phosphate (D’Elia et al. 2006b; Swoboda et al. 2011; Brown et al. 2013; Wang et al. 2013). In contrast, the inhibition of early-stage biosynthesis steps catalyzed by TarO and TarA or deletion of their corresponding genes leads to viable bacteria.
Paradoxically, it was demonstrated that the essentiality of late-stage WTA biosynthesis steps can be suppressed by the concomitant inactivation of early-stage WTA biosynthesis steps. This phenomenon has been referred to as the essential gene paradox (Scheme 11) (D’Elia et al. 2006a, b, 2009a, b; Farha et al. 2013; Sewell and Brown 2014; Lee et al. 2016a). Probably, concomitant inhibition of early-stage steps avoids depletion of the bactoprenol phosphate pool, which seems to be the lethal factor in late-stage WTA inhibition due to its significant effect on peptidoglycan biosynthesis. Bacteria under such early- and late-stage inhibitory pressure are synthetic-viable, but show significantly higher drug sensitivity. The concomitant early-stage inhibition of WTA synthesis, which represents also the major resistance mechanism toward late-stage WTA inhibitors, results in decreased fitness of the bacteria.
Schematic presentation of the essential gene paradox in WTA biosynthesis. Adapted from Sewell and Brown (2014)
The first reported late-stage WTA biosynthesis inhibitor is targocil 21 (Scheme 12), which is a selective TarG inhibitor identified by the high throughput screening (HTS) of a library of 55000 small molecules against wild-type S. aureus and a tarO-deletion mutant (Lee et al. 2010; Swoboda et al. 2010). Targocil has been proven to induce significant cell wall stress (Campbell et al. 2012) and displayed a submicromolar MIC against MRSA. Targocil treatment came with a high frequency of resistance selection (FOR 7 × 10−7 at 8 × MIC (Lee et al. 2010)), which was substantially reduced in co-application with the β-lactam oxacillin, suggesting that the mechanism of resistance is an early-stage WTA biosynthesis gene mutation.
In another screening of a focused library of 20000 small molecules bioactive against S. aureus performed by researchers from Merck, six additional TarG inhibitors were identified (Wang et al. 2013). Interestingly, none of these inhibitors was structurally related to targocil. Four of them, L275 22, L640 23, L541 24, and L638 25, contain a tricyclic 1,2,3,4-tetrahydrocyclopenta[b]indole core. Compound L555 26 is an active member of a large group of screened 4 N-aryl-4H-1,2,4-triazoles all depicting two chiral centers. Compound L524 27 is a 3C-aryl-4H-1,2,4-triazol bearing a bicycle[2.2.2]octane motif. All these hits had submicromolar antibacterial activity against MRSA. Furthermore, combination experiments with the β-lactam imipenem showed clear synergistic effects. In contrast, concomitant inhibition of the early-stage WTA biosynthesis enzyme TarO by addition of the specific inhibitor tunicamycin A 28 (see Scheme 13) led to suppression of TarG inhibition and increased MICs. Since the TarG inhibitory effect of L275, L638, L640, and L555 could not be fully suppressed, it is likely that these compounds display an off-target associated non-specific cellular toxicity or even address secondary targets (Wang et al. 2013). The frequencies of resistance development for compounds L275 22 (FOR <3.6 × 10−8 at 8 × MIC) and L638 25 (FOR 1.9 × 10−10 at 8 × MIC) were significantly lower than for targocil.
Beyond tunicamycin A 28, which displayed selective inhibitory activity for TarO in bacteria, but was cytotoxic for eukaryotic cells due to a promiscuous inhibition of UDP-HexNAc:polyprenol-P HexNAc-1-P family of enzymes including GlcNAc phosphotransferase (Swoboda et al. 2011), the first early-stage WTA biosynthesis inhibitor reported was ticlopidine 29. Ticlopidine is an adenosine diphosphate receptor inhibitor approved as the antiplatelet drug Tilicid®, which has been identified as a TarO inhibitor in a screening of a library of 2080 previously approved drugs (Farha et al. 2013). Ticlopidine was demonstrated to have strong synergistic antibacterial activity against MRSA in combination with the β-lactam cefuroxime both in vitro and in vivo, while displaying no antibacterial activity when applied alone. In addition, ticlopidine as an approved drug has already optimized ADMET properties, a known side effect profile and established manufacturing processes. Nevertheless, it remains to be clarified whether the adenosine diphosphate receptor inhibition, the primary effect of ticlopidine, can be tolerated under the conditions of antibiotic therapy regimens. Recently, researchers at Merck reported a screen of 2.8 million synthetic small molecules for inhibitors of early-stage WTA biosynthesis in MRSA. Tarocin A 30 and tarocin B 31 (Scheme 13) were identified as compounds that could suppress TarG inhibition with IC50’s of 26 and 6 µM, respectively (Lee et al. 2016a). The compounds were subsequently proven to selectively inhibit TarO. Interestingly, none of them is structurally related to ticlopidine: While tarocin A has an oxazolidin-2-one core, tarocin B consists of a benzimidazol scaffold bearing a bicyclo[2.2.1]heptane side chain.
Both compounds lack any intrinsic growth inhibitory activity against MRSA, MRSE, MSSA, or several other pathogens. In contrast to tunicamycin A, tarocin A and B show no cytotoxic activity against HeLa cells.
Furthermore, it could be demonstrated that both compounds as well as derivatives with improved water solubility (tarocin A1, tarocinA2, structures not shown) were able to restore β-lactam efficacy against MRSA and MRSE, as strong synergistic effects with imipenem and dicloxacillin in vitro and in vivo were observed. For example, panels of 108 different dicloxacillin-resistant MRSA strains and 66 dicloxacillin-resistant MRSE strains had significantly higher susceptibilities against a combination of dicloxacillin and tarocin A2 versus dicloxacillin alone (Lee et al. 2016a). In summary, the inhibition of WTA synthesis represents a novel and innovative strategy to restore the efficacy of β-lactam antibiotics in Gram-positive pathogens; the clinical proof of this concept has yet to be established, though.
5 DNA Gyrase Inhibition—Blocking the Relaxation of Supercoiled Bacterial DNA
Bacterial topoisomerases constitute a target class that has been successfully addressed in antibiotic therapy. Topoisomerases are enzymes controlling the topological state of DNA within bacterial cells and are therefore an integral part of essential processes such as DNA replication and transcription (Collin et al. 2011; Ehmann and Lahiri 2014). Topoisomerases in general are found in eukaryotic and prokaryotic cells and can be divided into two main types, type I and type II (Schoeffler and Berger 2008). Type I topoisomerases catalyze the relaxation of supercoiled DNA through the transient break of one DNA strand, while type II topoisomerases induce the relaxation through a transient double-strand breakage in an ATP hydrolysis-depending sequence (Liu et al. 1980). All topoisomerases are able to relax supercoiled DNA, but only the DNA gyrase (a type II topoisomerase in bacteria) can also introduce negative supercoils (Bates and Maxwell 2007; Nöllmann et al. 2007; Schoeffler and Berger 2008). There are significant differences between eukaryotic and prokaryotic type II topoisomerases. While eukaryotic type II topoisomerases are homodimers of large single-subunit enzymes, the prokaryotic type II topoisomerases, such as DNA gyrase (or topoisomerase II) and the structurally closely related topoisomerase IV, are A2B2 complexes of the subunits GyrA and GyrB (Scheme 14) (Champoux 2001; Wang 2002).
The DNA gyrase has three interfaces that exist in a closed or open conformation: The N-terminal domain of GyrB (N-gate), the GyrA-GyrB-DNA interface presenting tyrosine residues in the active cleaving site (DNA gate) and the C-terminal area of coiled coils (C-gate). The details of the complex mechanism of supercoiling by DNA gyrase are still under investigation (Gubaev and Klostermeier 2014). Nevertheless, the so-called two-gate model (Roca and Wang 1992, 1994; Roca et al. 1996) (Scheme 14c) is strongly supported by biochemical and structural data.
The DNA binds with the G-segment (gate segment) to the gyrase at the interface of the N-terminus of the GyrA dimer and the TOPRIM domain of GyrB. Upon binding of two molecules adenosin triphosphate (ATP) the GyrB subunits dimerize, which closes the N-gate and traps the so-called T-segment (transport segment) of DNA. Then the DNA gyrase transiently cleaves the G-segment via a double-strand breakage of the DNA by the formation of covalent DNA-phosphotyrosyl bonds. The hydrolysis of one ATP molecule leads to the rotation of GyrB, opening of the DNA gate, and the T-segment is transported through the cleaved DNA site. The subsequent religation of the G-segment introduces two negative supercoils into the DNA. Finally, the release of the T-segment and the hydrolysis of the second ATP molecule sets the gyrase into its starting position (Costenaro et al. 2007; Collin et al. 2011).
In general, inhibitors of the DNA gyrase can be divided into two groups: Those blocking the ATP binding site and those interfering with the DNA binding or the DNA strand passage within the gyrase. The latter are called catalytic site inhibitors or gyrase poisons. Due to the close structural similarity of DNA gyrase and topoisomerase IV, inhibitors possessing these mechanisms of action usually show activity against both targets. This dual-targeting phenomenon often results in low mutation frequencies for drug resistance and has probably significantly contributed to the success of antibacterial drugs targeting topoisomerases.
The first ATP-site inhibitor discovered was novobiocin 32 (Scheme 15), a bacterial metabolite produced by Streptomyces niveus, introducing the class of aminocoumarins as gyrase-inhibiting compounds.
The compounds bind to the GyrB subunit near the ATP binding site with very high affinity and thereby block the functionality of the gyrase. Although the aminocoumarins show potent antibacterial activity against Gram-positive bacteria, their therapeutic application is limited due to poor activity against most Gram-negative pathogens and in particular due to poor water solubility and in vivo toxicities (Maxwell 1993; Maxwell and Lawson 2003). Therefore, novel ATP-site inhibitors possessing more drug-like scaffolds have been investigated in recent years, with some of them already demonstrating their efficacy in vivo (Scheme 15) (Ehmann and Lahiri 2014).
Structures of the aminocoumarins novobiocin and chlorobiocin as well as advanced ATP-site inhibitors with novel scaffolds (name-giving structural motifs are highlighted in red) and their antibacterial activities, adapted from Ehmann and Lahiri (2014)
Tari et al. presented the pyrrolopyrimidine 34 showing broad-spectrum antibacterial activity with impressive MIC90 values of 0.008 µg/mL against S. aureus (Tari et al. 2013). Compound 34 displays also good activity against Gram-negative pathogens representing a major advancement for ATP-site inhibitors. Furthermore, the 2-pyridinyl urea 35 (Basarab et al. 2013), the 2-aminopyrimidine 36 (Uria-Nickelsen et al. 2013), aminobenzimidazols 37 (Finn 2013) and 38 (Chopra et al. 2012), the pyrrolamide 39 (Shahul et al. 2014) and the thiazolopyridone 40 (Kale et al. 2013, 2014) were reported as efficient ATP-site inhibitors against Gram-positive bacteria, and, for some of them (38, 39 and 40) also moderate activities against M. tuberculosis were revealed.
The first catalytic site inhibitor of the DNA gyrase was nalidixic acid 41 (Scheme 16), introducing the antibacterial class of quinolones. The first generation of quinolones had relatively weak antimicrobial activity, but with the introduction of the fluoroquinolones such as ciprofloxacin 42 or levofloxacin 43 very potent antimicrobials were available, displaying broad-spectrum activity against Gram-positive and Gram-negative pathogens (Collin et al. 2011). Such catalytic site inhibitors stabilize the DNA cleavage complex of the gyrase or topoisomerase IV and thereby poison the enzyme. This has been shown to be a very effective mode of inhibition of type II topoisomerases, since very low concentration of the inhibiting compounds bound to their target can lead to sufficient protein-stabilized DNA breaks, initiating a cascade which cumulates in cell death (Anderson and Osheroff 2001; Kohanski et al. 2010). Nevertheless, significant occurrence of resistance toward fluoroquinolones has been reported (Jacoby 2005), mostly associated with mutations in the quinolone resistance-determining regions in GyrA and GyrB (or ParC and ParE, the equivalents in topoisomerase IV) (Yoshida et al. 1990, 1991; Jacoby 2005).
Structures of nalidixic acid and the fluoroquinolones ciprofloxacin and moxifloxacin as well as advanced catalytic site inhibitors with novel scaffolds and their antibacterial activities (adapted from Ehmann et al.) (Ehmann and Lahiri 2014)
In the recent years, intensive research led to the discovery of a group of several non-fluoroquinolone inhibitors of the gyrase that bind near the catalytic site, but are mechanistically and microbiologically distinct from the fluoroquinolones (Ehmann and Lahiri 2014).
These compounds are simply called novel bacterial topoisomerase inhibitors (NBTIs). Most NTBIs possess a 4-aminopiperidine moiety bridging two aromatic motifs. Investigations on the mechanism of action have been shown that NBTIs also stabilize the DNA–protein complex, but in contrast to the fluoroquinolones they bind to the gyrase in the presence of intact, unbroken DNA strands (Bax et al. 2010).
The first frontrunner advanced to clinical trials was GSK966587 44 developed by researchers at GlaxoSmithKline (Miles et al. 2013). The compound bears a tricyclic 3-fluoro-4,5-dihydro-7H-pyrrolo[3,2,1-de][1,5]naphthyridin-7-one core and showed potent activity against Gram-positive bacteria (Ehmann and Lahiri 2014). A follower of the same series, gepotidacin 45 (Mayer and Janin 2014), is in several clinical phase II trials investigating the treatment of respiratory tract infections caused by S. pneumoniae, acute bacterial skin and skin structure infections caused by S. aureus and uncomplicated urogenital gonorrhea (Paris 2015). Additionally, researchers at AstraZeneca reported further NTBIs showing potent activity against Gram-positive bacteria such as AZD9742 46 (Reck et al. 2012) and NTBI 5463 47 (Dougherty et al. 2014). The latter presents a 4-aminocyclohexyl bridge between the two aromatic regions with promising activity against Gram-negative pathogens. Noteworthy, all of these compounds show activity against fluoroquinolone-resistant bacterial strains.
Recently, several catalytic site inhibitors of the DNA gyrase with novel unique scaffolds different from the early NBTIs (Scheme 16) have been discovered. Researchers at AstraZeneca reported compound ETX0914 48 (Scheme 17) with an unusual spiropyrimidinetrione scaffold that showed potent broad-spectrum activity against Gram-positive as well as Gram-negative bacteria (Jacobsson et al. 2014; Alm et al. 2015; Kern et al. 2015; Su et al. 2016). The compound was active against fluoroquinolone-resistant strains, suggesting that the exact mode of inhibition is also different from the fluoroquinolones (Basarab et al. 2015). Genetic analysis of resistant mutants showed that mutations in the conserved GyrB TOPRIM domain are correlated to decreased susceptibilities to ETX0914 (Alm et al. 2015). Currently, the compound is in clinical phase II trials for the treatment of uncomplicated gonorrhea (Paris 2015). Another compound, which has recently been shown to have extremely potent antibacterial activities (up to MIC = 0.002 µg/mL) against Gram-positive pathogens including several multi-drug-resistant and also fluoroquinolone-resistant strains, is 2,4-diiodoemodin 49 (Chen et al. 2014), a semi-synthetic derivative of the secondary metabolite emodin 50, which naturally occurs in plants and fungi. Interestingly, 49 has been shown to be an inhibitor of DNA gyrase and bacterial topoisomerase I, which is a unique dual-targeting mode of this compound. Additionally, unlike emodin, which caused cytotoxic effects through strong inhibition of the human topoisomerase IIα, 2,4-diiodoemodin showed only very little inhibitory effect against this enzyme. Nevertheless, a further understanding of this new class of antibacterial haloemodins, for example with respect to target promiscuity and the associated side effects, is needed to advance this structural template.
A novel structural class of gyrase inhibitors of proteobacterial origin, consisting of oligomeric pseudopeptides formed by chains of coupled para-aminobenzoic acids, has recently been discovered by two research groups from two different Gram-negative producer strains.
Albicidin 51 (Scheme 18) has been isolated from Xanthomonas albilineans (Cociancich et al. 2015), a xylem-invading plant pathogen that causes leaf scald disease in sugarcane (Royer et al. 2004). The compound is a phytotoxin that blocks the DNA gyrase in sugarcane chloroplasts (Birch and Patil 1987a, b). Additionally, albicidin has been demonstrated to be a nanomolar inhibitor of bacterial DNA gyrase with a broad-spectrum antibacterial activity against Gram-positive and Gram-negative pathogens (Hashimi et al. 2007; Cociancich et al. 2015; Kretz et al. 2015). Albicidin stabilizes the DNA gyrase cleavage complex with relaxed or supercoiled DNA in the presence of ATP via binding to the GyrA subunit, leading to a similar DNA fragmentation pattern as ciprofloxacin. Nevertheless, only low levels of cross resistance to albicidin in some fluoroquinolone- and couvermycin-resistant strains have been observed (Hashimi et al. 2007). The existence of a potent gyrase inhibitor produced by X. albilineans has been known for decades, but since the bacterium produces only very small amounts of the compound, the compound’s structure could be elucidated only recently, when Süssmuth and coworkers expressed heterologously in X. axonopodis vp. vesicatoria and established an extended purification protocol (Cociancich et al. 2015).
The structure of albicidin consists of the non-proteinogenic α-amino acid (S)-β-cyanoalanine, the two aromatic δ-amino acids para-aminobenzoic acid (PABA) and 2-hydroxy-3-methoxy-para-aminobenzoic acid (PABA-D1) as well as 2-methyl-para-coumaric acid (MPCA) that represents the N-terminus of this oligomeric pseudopeptide. Biosynthetically albicidin is a product of the polyketide synthase-nonribosomal peptide synthase (PKS-NRPS) machinery (Huang et al. 2001; Vivien et al. 2007; Piel 2010). Albicidin has been successfully targeted by a total synthesis by Süssmuth and coworkers, thus confirming the unusual structure (Kretz et al. 2015).
In parallel, the structure of the cystobactamid 919-2 52 (Scheme 18) was published by Müller and coworkers (Baumann et al. 2014; Herrmann et al. 2016).
The compound was isolated together with the truncated homolog cystobactamid 507 53 (Moreno et al. 2015) and the isomer cystobactamid 919-1 54 from cultivations of myxobacterium Cystobacter sp. Cbv34. Cystobactamid 919-2 shares the general PABA-derived oligomeric, pseudopeptidic structure of albicidin, but the central amino acid is a non-proteinogenic (S,S)-β-methoxy asparagine, and it possesses a different oxidation pattern on the right-hand site of the molecule. Additionally, there is a para-nitrobenzoic acid (PNBA) unit at the N-terminus. In accordance with the structural similarity of cystobactamids and albicidin, also cystobactamid 919-2 displayed very potent broad-spectrum antibacterial activity against Gram-positive and Gram-negative pathogens due to inhibition of the DNA gyrase at the GyrA subunit with low levels of cross resistance to fluoroquinolone-resistant strains (Baumann et al. 2014). Both compounds were regarded as “natural quinolones” representing the first natural products inhibiting the GyrA subunit of DNA gyrase (Baumann et al. 2014; Johnston and Magarvey 2015). Currently, chemical optimization studies of albicidin and cystobactamid 919-2 to improve their properties as antibacterial drugs are ongoing (personal communication, M. Brönstrup).
6 Bacterial DnaN Polymerase Sliding Clamp—A Highly Conserved, Multi-enzyme Interacting Core Architecture of the Bacterial Replisome
As mentioned earlier, multi-enzyme complexes or structures synthesized by multi-enzymatic pathways are less prone to bacterial resistance development, since their malfunctioning is hard to bypass.
In this context the bacterial replisome machinery, which consists of at least twelve interacting enzymes that are highly conserved in bacteria, is a good structure to target (Robinson et al. 2012). Next to the DNA gyrase, which also belongs to the bacterial replisome and has already been discussed, the bacterial DnaN polymerase sliding clamp has recently attracted attention as an antibacterial target (Georgescu et al. 2008b; Kjelstrup et al. 2013; Wolff et al. 2014; Yin et al. 2014a, b; Holzgrabe 2015; Kling et al. 2015; Yin et al. 2015).
The bacterial DnaN or β-clamp is a ring-shaped homodimer, with each monomer composed of three globular domains, which functions as a crucial subunit of the DNA polymerase III holoenzyme (Scheme 19). The general protein–DNA interactions between polymerases and DNA templates are weak (Mizrahi et al. 1985). Therefore, the association of the polymerase to the DNA strand is one of the rate-limiting steps of the replication. The assembly of the two β-subunits of the bacterial β-clamp around the DNA, mediated by ATP hydrolysis, leads to strong and specific protein–protein interactions between the β-clamp and the polymerase III (Pol III), thereby preventing polymerase dissociation and dramatically increasing the polymerase activity (Stukenberg et al. 1991; Argiriadi et al. 2006; Georgescu et al. 2008a; Donnell et al. 2013; Cho et al. 2014). Therefore, the β-clamp serves as a processivity-promoting factor in DNA replication (Stukenberg et al. 1991).
Furthermore, as the β-clamp shows direct protein–protein interactions with a multitude of other enzymes involved in the replication process such as the α-polymerase, the ε-proofreading subunits (Ozawa et al. 2013), or the clamp loader γ/τ complex (Stukenberg et al. 1991), it can be seen as the linchpin of the replisome (Robinson et al. 2012). The important functionality within the replication, its moderate copy number per cell (330–600 homodimers) (Leu et al. 2000), its highly conserved structure across the bacterial species (Burnouf et al. 2004; Argiriadi et al. 2006; Georgescu et al. 2008a; Gui et al. 2011; Wolff et al. 2014; Niiranen et al. 2015), and its significant structural divergence to the mammalian counterpart (proliferating cell nuclear antigen, PCNA) (Bloom 2009; Robinson et al. 2012; Donnell et al. 2013) make the β-clamp a particularly attractive antibacterial target.
In 2001, Jennings and coworkers discovered that the general protein–protein interaction between the β-clamp and several β-clamp-interacting proteins is based on a consensus sequence of amino acids in a linear pentapeptidic-binding motif, given as QL[S/D]LF and QLxLx[L/F][S/D] (Scheme 20) (Dalrymple et al. 2001).
Jennings and coworkers were able to identify the first synthetic nonapeptides containing these linear binding motifs that inhibited either the binding of the α-subunit or of the δ-subunit of E. coli Pol III to the corresponding E. coli β-clamp, displaying IC50 values of 1.33–14.6 µM or 8.8–12.9 µM, respectively.
It was demonstrated in further SAR studies in 2004 (Wijffels et al. 2004) and 2011 (Wijffels et al. 2011) that lipophilic N-terminal extensions of the pentapeptidic linear binding motif as well as substitutions with more lipophilic phenylalanine derivatives led to tremendous increases of the inhibitory effect of the binding of the α-subunit of E. coli Pol III to the corresponding E. coli β-clamp (IC50 up to 20 nM). These linear motifs (LM) bind to the LM-binding pocket, which is highly conserved across Gram-negative and Gram-positive bacteria (Argiriadi et al. 2006) and consists of two subsites (I and II) at the interface of the two domains on each monomer of the β-clamp homodimer (Dalrymple et al. 2001; Bunting et al. 2003; Burnouf et al. 2004; Georgescu et al. 2008a, b).
In 2008, O’Donnell and coworkers published the first non-peptidic small molecule inhibitor of the β-clamp, RU-7 58 (Scheme 21) (Georgescu et al. 2008b). The compound was identified by screening a library of 30000 polar small molecules for their inhibitory effect on α-β2-binding by a fluorescence anisotropy titration assay. RU-7 inhibited the E. coli relication system with a Ki of 10 µM by binding to the subunit I of the LM-binding pocket, but showed no inhibitory effect on the PCNA-dependent eukaryotic system in the yeast S. cereviase.
An other small molecule, the oxim ether 59, was identified by an in silico screen of 32000 compounds followed by an α-β2-binding assay (Wijffels et al. 2011). The compound, which also bound to the subunit I of the LM-binding pocket, displayed an IC50 value for inhibition of α-β2-binding of 40 µM.
In 2014, Oakley and coworkers reported that the weak antibacterial effects of non-steroidal anti-inflammatory drugs (NSAIDs) such as (R)-vedaprofen 60, bromfenac 61, or (S)-carprofen 62 (Scheme 22) are due to their binding to the subunit I of the LM-binding pocket of the β-clamp (Yin et al. 2014a).
Recently, a fragment-based screening of 352 compounds utilizing X-ray crystallography led to the discovery of tetrahydrocarbazol 63 (Scheme 22) as a β-clamp binding small molecule, which is structurally closely related to carprofen and was also shown to bind into the subunit I of the β-clamp (Yin et al. 2014b). Further, SAR studies led to the identification of N-acetamido-THC 64 with an 10-fold decreased IC50 for the inhibition of the β-clamp (Yin et al. 2015).
Whereas all compounds discussed so far were binding to the LM-binding pocket of the β-clamp, Lobner-Olesen and coworkers recently reported cyclic octapeptides (65–67, Scheme 23) inhibiting the DnaN–DnaN monomer interaction of the homodimer (Kjelstrup et al. 2013).
The compounds were identified using the SICLOPPS technology (Scott et al. 1999) for the intracellular in vivo production of cyclic 21-mer peptides. A six-amino-acid-long randomized sequence was used to build a library of over 900000 combinations, which were subsequently screened for their ability to inhibit DnaN–DnaN interactions in the S. aureus replisome. The initial six-amino-acid-long hit sequences were incorporated in an octapeptide and synthesized utilizing the Impact Twin System (New England Biolabs) and evaluated for their antibacterial activity. Interestingly, while cyclic octapeptides displayed MIC values of 50 µg/mL and 21 µg/mL, respectively, the corresponding linear peptides were totally inactive.
An issue with all of the synthetic DnaN binders described above is that their antibacterial potency is mostly weak. This again illustrates that the ability to penetrate into the bacterial cell is a crucial hurdle for new antibiotic templates.
This is fundamentally different for griselimycin (GM) 68 (Scheme 24), a long known natural product from bacterial ferments that attracted renewed interest as an anti-tuberculosis drug (Holzgrabe 2015; Kling et al. 2015). Griselimycin is a cyclic decadepsipeptide containing two unusual L-(4R)-4-methylproline and one unsubstituted L-proline in the linear sequence, which is cyclized with its C-terminal glycine onto the hydroxyl group of an internal threonine side chain. Additionally, the compound possesses several posttranslational N-methylations, and the N-terminus is acetylated. Griselimycin was isolated from two strains of Streptomyces in the 1960s (Terlain and Thomas 1969, 1971a, b), and was found to exhibit antibacterial activity specifically against Corynebacteria including M. tuberculosis, though the exact mode of action and target were unknown. The pharmaceutical company Rhône-Poulenc started with early investigations for the development of griselimycin as an anti-tuberculosis drug. The first studies in humans were promising, despite poor pharmacokinetics of the compound (Bénazet et al. 1966; Noufflard-Guy-Loé and Berteaux 1965). But first derivatization programs to find analogs with improved pharmacokinetics (Bouchaudon 1964; Jolles 1971) were soon terminated in the 1970s after rifampin became available for tuberculosis treatment. Since novel tuberculosis drugs are desperately needed and GM was reported to be highly active against multi-drug-resistant M. tuberculosis, researchers from Sanofi-Aventis, Müller, and coworkers re-investigated the use of griselimycin as anti-tuberculosis drug (Kling et al. 2015).
First, a metabolic stability profiling of natural, less-abundant analogs of GM led to the identification of Pro8 as main site of metabolic degradation responsible for the poor pharmacokinetic parameters of griselimycin. This result was supported by the finding that the natural derivative methylgriselimycin (MGM) 69 (Scheme 24) showed a remarkably increased stability toward degradation compared to griselimycin. Since MGM is produced in only small amounts, a solid-supported total synthesis for griselimycins was established to provide access to several GM analogs bearing Pro8 modifications, supposed to improve potency and metabolic stability of the lead structure. From biological evaluation of these analogs cyclohexylgriselimycine (CGM) 70 (Scheme 24) was identified, displaying MIC values of 0.06 µg/mL and 0.2 µg/mL in the drug susceptible M. tuberculosis strain H37Rv and within macrophage-like RAW264.7 cells, respectively. CGM revealed a time-dependent in vitro activity, and although the unbound fraction in plasma was very low, the MIC dropped only fivefold in the presence of human plasma. CGM showed high-level activity against a panel of M. tuberculosis strains covering a broad geographical and evolutionary diversity, including strains mono-resistant to first- and second-line anti-tuberculosis drugs. The overall ADMET properties of CGM also improved; in particular CGM displayed a high-level oral bioavailability of 89 %, a moderate clearance (1.1 L/h/kg), and an expanded half-life and drug exposure, enabling a once daily administration.
In vivo activity studies in a mouse model of tuberculosis revealed a minimum-effective dose (MED) of 50 mg/kg, and mice treated with 600 mg/kg daily dose were proven culture-negative after 4 weeks of treatment. Giving the promising result of CGM administered in monotherapy, a combination therapy with first-line anti-tuberculosis drugs such as rifampin (RIF), pyrazinamide (PZA), and isoniazid (INH) was investigated. In an in vivo mouse model of TB, CGM was administered at MED of 100 mg/kg alone and in combination with other TB drugs in a model of chronic TB. CGM alone was demonstrated to be as active as INH, the most bactericidal first-line anti-tuberculosis drug, and showed significantly improved activity in combination with RIF compared to the standard combination therapy (INH/RIF/PZA).
The analysis of the genes of a GM- and MGM-producing Streptomyces strain, which is naturally resistant to GM, led to the identification of griR, a homolog of the dnaN gene (51 % of identity). Introduction of the griR gene to Streptomyces coelicolor, a strain susceptible to GM, allowed the strain to survive in the presence of GM, suggesting that overexpression of GriR mediates GM resistance and that the griselimycins interact with DnaN polymerase sliding clamp (β-clamp) of the replisome. Further, genomic-based investigations on the resistance mechanisms in vitro and in vivo in M. smegmatis and M. tuberculosis revealed that resistance in these species is achieved by overexpression of DnaN. However, this process occurred with very low rate and was accompanied by considerable loss of fitness, expressed in a negative correlation of growth to increasing level of resistance.
Finally, the interaction between GM, MGM, and CGM with DnaN from M. smegmatis, M. tuberculosis, and E. coli as well as human sliding clamp (PCNA) was analyzed and characterized by surface plasmon resonance (SPR) and by a crystal structure analysis. SPR analysis demonstrated a high affinity binding of GM, MGM, and CGM to mycobacterial sliding clamps in the picomolar range, whereas significantly lower binding to the sliding clamp of E. coli and no binding between PCNA and griselimycins was detected. The analysis of co-crystals of GM and CGM with DnaN from M. smegmatis and M. tuberculosis revealed that the griselimycins bind to the LM-binding pocket, filling both subsite I and II. The linear N-terminal segment and the adjacent half of the macrocycle of GM and CGM superimpose very well with the natural linear peptidic-binding motif of the LM-binding pocket.
Since there is no preexisting resistance of griselimycins due to the different modes of action compared to the approved anti-tuberculosis drugs, and since resistance occurs at an extremely low frequency associated with fitness loss of the pathogens, the griselimycins are attractive candidates for the treatment of drug-sensitive and multi-drug-resistant tuberculosis. Clinical trials with CGM have not been reported yet, though.
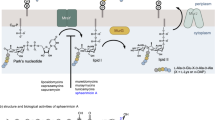
7 Menaquinone as a Small Molecule Antibacterial Target
Isoprenoid quinones such as phylloquinone (vitamin K1) 71, ubiquinone (coenzyme Q10) 72, and menaquinone 73 (Scheme 25) are present in almost all living organisms as plasma membrane-anchored electron carriers or antioxidants (Kawamukai 2002; Aguilaniu et al. 2005; Kurosu and Begari 2010; Nowicka and Kruk 2010; Cervellati and Greco 2016). These compounds are involved in several essential, live-sustaining metabolic processes such as the generation of adenosine triphosphate (ATP) in the respiratory chain. The general structure of these compounds consists of a polar, quinone-containing head and an unpolar, hydrophobic side chain, which allows the molecules to anchor in the phospholipid bilayer of cell membranes.
While phylloquinone and menaquinone show a methylnaphtoquinone core, the polar head group of ubiquinone consists of a dimethoxy-benzoquinone. The length and saturation of the side chain can differ in the particular species. The quinone moiety in these compounds is crucial for their biological activity as electron carriers in cellular redox processes, as it is reduced by a two-step reversible reduction process via the intermediate semiquinone to the corresponding hydroquinone. While ubiquinone is utilized as coenzyme for the ATP generation in the mammalian respiratory chain, in most Gram-positive and anaerobic Gram-negative bacteria menaquinone is the major isoprenoid quinone, responsible for the electron transfer in the bacterial respiratory chain and a multitude of other metabolic redox processes (Kurosu and Begari 2010; Fujimoto et al. 2012).
The biosynthesis of menaquinone in E. coli is catalyzed by seven menaquinone-specific enzymes, MenA-G, which are encoded in two gene clusters (Young 1975; Schoepp-Cothenet et al. 2009). It starts from chorismate 74 (Scheme 26), an intermediate of the shikimate pathway. First, MenF isomerizes chorismate 74 to isochorismate 75. MenD, a thiamin diphosphate-dependent enzyme, then mediates the Stetter-type 1,4-conjugate addition of α-ketoglutarate to isochorismate, leading to the formation of 76. The elimination of the pyruvate moiety is then catalyzed by MenH, yielding 2-succinyl-6-hydroxy-3-cyclohexadiene-1-carboxylate 77, and subsequently MenC forces the elimination of water providing O-succinylbenzoate 78. The O-succinyl-CoA ligase MenE attaches 78 to coenzyme A and a Diekmann-type cyclization catalyzed by MenB and subsequent thioester hydrolysis by yfbB leads to formation of 1,4-dihydroxy-2-naphthoate 81. Finally, prenylation by MenA and methylation mediated by MenG gives menaquinone 73 (Kurosu and Begari 2010). The most common menaquinones contain 7, 8, or 9 isoprene units; however, menaquinones with 4, 5, 6, 10, 11, 12, or 13 isoprene units have been isolated from bacteria.
Since it has been demonstrated that menaquinone is crucial for bacterial growth (Dhiman et al. 2009; Fujimoto et al. 2012) and colony development (Pelchovich et al. 2013), it is not a surprise that menaquinone biosynthesis has already been investigated as a target for the development of novel antibiotics (Kurosu and Begari 2010).
In the last years, a few inhibitors of individual enzymes of menaquinone biosynthesis have been reported (Kurosu and Begari 2010). For example, allylaminomethanone A 84 and phenylethylaminomethanone A 85 (Scheme 27) are inhibitors of MenA, which mimic the structure of the product of MenA catalysis, demethylmenaquinone, and display MIC values of 1.5 and 12.5 µg/mL, respectively, against M. tuberculosis (Kurosu et al. 2007; Debnath et al. 2012). Furthermore, the vinylsulfonamide 86, which was designed as mimic of O-succinylbenzoate-CoA ester, was reported to efficiently inhibit MenE at low nanomolar concentrations (Lu et al. 2012).
Recently, the novel natural cyclodepsipeptide lysocin E 87 (Scheme 28) was reported to target the redox metabolism in bacteria by directly binding to menaquinone (Hamamoto et al. 2014). Since the biosynthesis of menaquinone is a multi-enzymatic pathway as described above and the role of menaquinone is crucial for viability of bacteria, menaquinone seems to be a much smarter choice for an antibacterial target than the individual enzymes of the biosynthesis, even more when considering that an alternative biosynthetic pathway for menaquinones starting from futalosine exists in several Gram-positive bacteria (Hiratsuka et al. 2008).
The cyclic structure of 87 displays a head-to-tail lactonization of the hydroxyl function of the N-terminal threonine to the carboxylic acid of the C-terminal threonine. Furthermore, the compound contains five D-amino acids, (3R)-3-hydroxy-5-methyl-hexanoic acid capping the N-terminal amino group and a N-methylation at the present phenylalanine.
Lysocin E was discovered by an extensive screening campaign in invertebrates, where culture supernatants of soil bacteria were tested in a silkworm infection model. The compound has shown good antibacterial activity against several Gram-positive pathogens including MRSA.
Lysocin E induced potassium leakage through the bacterial membrane, which led to a rapid loss of membrane potential and bacteriolysis. The hemolytic activity against mammalian cells was very low, and further investigations demonstrated that the MIC value of 87 against S. aureus increased in a dose-dependent manner upon addition of menaquinone. The direct binding of menaquinone in a 1:1 complex was further indicated by micro-calorimetric measurements, detecting an exothermic response upon addition of lysocin to menaquinone. Interestingly, no binding of the compound to the structurally similar ubiquinone was observed, suggesting that the naphthoquinone core in menaquinone is crucial for the interaction.
Furthermore, the first in vivo studies in S. aureus infected mice showed a promising ED50 value of 0.5 mg/kg body weight, even smaller than that of vancomycin. This is remarkable in view of the moderate–good MIC value in vitro. Even a dose of 400 mg/kg body weight administered by intraperitoneal injection was tolerated, and no increase of biochemical markers for tissue damage of liver and kidneys was observed. Lysocin E has already been successfully addressed in a solid-supported total synthesis by Inoue and coworkers, and the first structure–activity relationship studies have revealed that the N-methylation at the phenylalanine in lysocin E is crucial for its activity (Murai et al. 2015).
In summary, lysocin E demonstrates that the depletion of essential small molecule mediators of energy metabolism is a viable, but yet underexplored antibiotics strategy. It also highlights that in vivo phenotypic assays, a highly successful discovery strategy of pharmaceutical research before the advent of in vitro assay systems, are still a powerful way to disclose urgently needed, novel antibiotic lead compounds.
8 Conclusions
Antibiotic lead finding suffers from insufficient volume and scale since the pharmaceutical industry has considerably reduced its efforts in this indication, inter alia due to an unsatisfying outcome of a target-centric, high throughput screening-based antibiotic research strategy (Payne et al. 2007; Tommasi et al. 2015), and due to uncertain economic revenues. But it does not suffer from a shortage of validated biological mechanisms, or from an exhaustion of chemical matter, as demonstrated by the showcases of this review. Microbial natural products continue to play a dominant role as sources of novel antibiotics, and their discovery is facilitated by multiple advances in natural product research (Harvey et al. 2015): Classical strategies like the screening of extracts in relevant phenotypic assays (e.g., lysocin), or a re-investigation of neglected compounds of the past with modern chemical and genetic methods (as with griselimycin), are complemented by the cultivation of underexplored sources (e.g., teixobactin, cystobactamid), the heterologous expression of (almost) silent gene clusters (e.g., albicidin) or genome mining techniques (Doroghazi et al. 2014). Apart from being promising leads and drug candidates, natural products also teach important lessons and question existing ‘rules’ of drug discovery. Griselimycin, for example, demonstrates that peptides of elevated (>1000 Da) molecular weight can not only disrupt protein–protein interactions at picomolar concentrations, but at the same time exhibit high oral bioavailability, and high stability in plasma. Teixobactin, on the other hand, confirmed the rule that a dual molecular mode of action is advantageous with respect to suppression of resistance. It also underlines the common principle that natural antibiotics preferably target hotspots in complex biosynthetic machineries, as their malfunctioning is hard to circumvent by bacterial mutations.
New structural templates also arise from synthetic libraries that are screened against innovative targets, as exemplified by the inhibitors of WTA biosynthesis, the DNA sliding clamp, the DNA gyrase, or the β-lactamases. While many synthetic hits from the past suffered from insufficient translocation or permeation across bacterial membranes, the examples above demonstrate how an early incorporation of whole cell assays in the optimization cascade (or directly in the primary screening assay) assured the successful optimization of synthetic structures to antibiotics.
Our selection of case studies has been subjective and non-comprehensive, as it did not discuss the numerous additional innovative approaches to fight bacterial infections that include antibiotics (e.g., translation inhibitors, antimicrobial peptides, or actively transported conjugates), non-antibiotic pathogenicity-blocking small molecules [e.g., secretion system inhibitors, quorum sensing inhibitors, immune stimulants (Rasko and Sperandio 2010)], biomolecules [e.g., lysins, toxin binders (Morrison 2015)], or even larger entities [e.g., bacteriophages, probiotics] (Czaplewski et al. 2016). However, the few examples provide sufficient arguments that from a scientific point of view, the discovery and development of novel antibiotics is highly rewarding.
References
Aguilaniu H, Durieux J, Dillin A (2005) Metabolism, ubiquinone synthesis, and longevity. Genes Dev 19:2399–2406. doi:10.1101/gad.1366505
Aldred KJ, Kerns RJ, Osheroff N (2014) Mechanism of quinolone action and resistance. Biochemistry 53:1565–1574. doi:10.1021/bi5000564
Alm RA, Lahiri SD, Kutschke A et al (2015) Characterization of the novel DNA gyrase inhibitor AZD0914: low resistance potential and lack of cross-resistance in Neisseria gonorrhoeae. Antimicrob Agents Chemother 59:1478–1486. doi:10.1128/AAC.04456-14
Amaral L, Martins A, Spengler G, Molnar J (2014) Efflux pumps of Gram-negative bacteria: what they do, how they do it, with what and how to deal with them. Front Pharmacol 4:Article168. doi:10.3389/fphar.2013.00168
Anderson VE, Osheroff N (2001) Type II topoisomerases as targets for quinolone antibacterials: turning Dr. Jekyll into Mr. Hyde. Curr Pharm Des 7:337–353. doi:10.2174/1381612013398013
Anderson RJ, Groundwater PW, Todd A, Worsley AJ (2012) Antibacterial agents: chemistry, mode of action, mechanisms of resistance and clinical applications. Wiley, New York
Argiriadi MA, Goedken ER, Bruck I, et al (2006) Crystal structure of a DNA polymerase sliding clamp from a Gram-positive bacterium. BMC Struct Biol 6:2. doi:10.1186/1472-6807-6-2
Askoura M, Mottawea W, Abujamel T, Taher I (2011) Efflux pump inhibitors (EPIs) as new antimicrobial agents against Pseudomonas aeruginosa. Libyan J Med 6:1–8. doi:10.3402/ljm.v6i0.5870
Atilano ML, Pereira PM, Yates J et al (2010) Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc Natl Acad Sci USA 107:18991–18996. doi:10.1073/pnas.1004304107
Baltrus DA (2013) Exploring the costs of horizontal gene transfer. Trends Ecol Evol 28:489–495. doi:10.1016/j.tree.2013.04.002
Baltz RH (2008) Renaissance in antibacterial discovery from actinomycetes. Curr Opin Pharmacol 8:557–563. doi:10.1016/j.coph.2008.04.008
Basarab GS, Manchester JI, Bist S et al (2013) Fragment-to-hit-to-lead discovery of a novel pyridylurea scaffold of ATP competitive dual targeting type II topoisomerase inhibiting antibacterial agents. J Med Chem 56:8712–8735. doi:10.1021/jm401208b
Basarab GS, Kern GH, McNulty J et al (2015) Responding to the challenge of untreatable gonorrhea: ETX0914, a first-in-class agent with a distinct mechanism-of-action against bacterial Type II topoisomerases. Sci Rep 5:11827. doi:10.1038/srep11827
Bates AD, Maxwell A (2007) Energy coupling type II topoisomerase why do they hydrolyze ATP? Biochemistry 46:7929–7941. doi:10.1021/bi700789g
Bauer A, Brönstrup M (2014) Industrial natural product chemistry for drug discovery and development. Nat Prod Rep 31:35–60. doi:10.1039/c3np70058e
Bauer R, Dicks LMT (2005) Mode of action of lipid II-targeting lantibiotics. Int J Food Microbiol 101:201–216. doi:10.1016/j.ijfoodmicro.2004.11.007
Baumann S, Herrmann J, Raju R et al (2014) Cystobactamids: myxobacterial topoisomerase inhibitors exhibiting potent antibacterial activity. Angew Chem Int Ed 53:14605–14609. doi:10.1002/anie.201409964
Bax BD, Chan PF, Eggleston DS et al (2010) Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature 466:935–940. doi:10.1038/nature09197
Bénazet et al (1966) Antibiotics—advances in research, production and clinical use. In: Herold M, Gabriel Z (eds) Proceedings of the congress on antibiotics held in Prague, 15–19 June, 1964. Butterworths, London, pp 262–264
Bengtsson B, Greko C (2014) Antibiotic resistance–consequences for animal health, welfare, and food production. Ups J Med Sci 119:96–102. doi:10.3109/03009734.2014.901445
Bérdy J (2012) Thoughts and facts about antibiotics: where we are now and where we are heading. J Antibiot 65:441. doi:10.1038/ja.2012.54
Berleman J, Auer M (2013) The role of bacterial outer membrane vesicles for intra- and interspecies delivery. Environ Microbiol 15:347–354. doi:10.1111/1462-2920.12048
Bierbaum G, Sahl HG (1985) Induction of autolysis of staphylococci by the basic peptide antibiotics Pep 5 and nisin and their influence on the activity of autolytic enzymes. Arch Microbiol 141:249–254. doi:10.1007/BF00408067
Birch RG, Patil SS (1987a) Correlation between albicidin production and chlorosis induction by Xanthomonas albilineans, the sugarcane leaf scald pathogen. Physiol Mol Plant Pathol 30:199–206. doi:10.1016/0885-5765(87)90033-6
Birch RG, Patil SS (1987b) Evidence that an albicidin-like phytotoxin induces chlorosis in sugarcane leaf scald disease by blocking plastid DNA replication. Physiol Mol Plant Pathol 30:207–214. doi:10.1016/0885-5765(87)90034-8
Biswas S, Brunel J-M, Dubus J-C et al (2012) Colistin: an update on the antibiotic of the 21st century. Expert Rev Anti Infect Ther 10:917–934. doi:10.1586/eri.12.78
Blair JMA, Richmond GE, Piddock LJV (2014) Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol 9:1165–1177. doi:10.2217/fmb.14.66
Bloom LB (2009) Loaing clamps for DNA replication and repair. DNA Repair 8:570–578. doi:10.1016/j.dnarep.2008.12.014.Loading
Blunt JW, Copp BR, Munro MHG et al (2011) Marine natural products. Nat Prod Rep 28:196–268. doi:10.1039/c005001f
Boneca IG, Chiosis G (2003) Vancomycin resistance: occurrence, mechanisms and strategies to combat it. Expert Opin Ther Targets 7:311–328. doi:10.1517/14728222.7.3.311
Bouchaudon J (1964) Nouveau cyclopeptide, sa préparation et les médicaments qui le contiennent
Boucher HW, Talbot GH, Bradley JS et al (2009) Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi:10.1086/595011
Boucher HW, Talbot GH, Benjamin DK et al (2013) 10 x ′20 Progress–development of new drugs active against gram-negative bacilli: an update from the Infectious diseases society of America. Clin Infect Dis 56:1685–1694. doi:10.1093/cid/cit152
Bozdogan B, Appelbaum PC (2004) Oxazolidinones: activity, mode of action, and mechanism of resistance. Int J Antimicrob Agents 23:113–119. doi:10.1016/j.ijantimicag.2003.11.003
Breu F, Guggenbichler S, Wollmann J (2001) Human as the world’s greatest evolutionary force. Science 293:1786–1790
Breukink E, de Kruijff B (2006) Lipid II as a target for antibiotics. Nat Rev Drug Discov 5:321–332. doi:10.1038/nrd2004
Brötz H, Josten M, Wiedemann I et al (1998) Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol Microbiol 30:317–327. doi:10.1046/j.1365-2958.1998.01065.x
Brötz-Oesterhelt H, Brunner NA (2008) How many modes of action should an antibiotic have? Curr Opin Pharmacol 8:564–573. doi:10.1016/j.coph.2008.06.008
Brown S, Xia G, Luhachack LG et al (2012) Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proc Natl Acad Sci USA 109:18909–18914. doi:10.1073/pnas.1209126109
Brown S, Santa Maria JP, Walker S (2013) Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol 67:313–336. doi:10.1146/annurev-micro-092412-155620
Brown DG, Lister T, May-Dracka TL (2014) New natural products as new leads for antibacterial drug discovery. Bioorg Med Chem Lett 24:413–418. doi:10.1016/j.bmcl.2013.12.059
Brown L, Wolf JM, Prados-Rosales R, Casadevall A (2015) Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat Rev Microbiol 13:620–630. doi:10.1038/nrmicro3480
Bunting KA, Roe SM, Pearl LH (2003) Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the β-clamp. EMBO J 22:5883–5892. doi:10.1093/emboj/cdg568
Burnouf DY, Olieric V, Wagner J et al (2004) Structural and biochemical analysis of sliding clamp/ligand interactions suggest a competition between replicative and translesion DNA polymerases. J Mol Biol 335:1187–1197. doi:10.1016/j.jmb.2003.11.049
Bush K (2010) Bench-to-bedside review: the role of beta-lactamases in antibiotic-resistant Gram-negative infections. Crit Care 14:224. doi:10.1186/cc8892
Bush K (2015) Investigational agents for the treatment of Gram-negative bacterial infections: a reality check. ACS Infect Dis 1:509–511. doi:10.1021/acsinfecdis.5b00100
Butler EK, Davis RM, Bari V et al (2013a) Structure-function analysis of MurJ reveals a solvent-exposed cavity containing residues essential for peptidoglycan biogenesis in Escherichia coli. J Bacteriol 195:4639–4649. doi:10.1128/JB.00731-13
Butler MS, Blaskovich MA, Cooper MA (2013b) Antibiotics in the clinical pipeline in 2013. J Antibiot 66:571–591. doi:10.1038/ja.2013.86
Butler MS, Robertson AAB, Cooper MA (2014) Natural product and natural product derived drugs in clinical trials. Nat Prod Rep 31:1612–1661. doi:10.1039/c4np00064a
Campbell J, Singh AK, Swoboda JG et al (2012) An antibiotic that inhibits a late step in wall teichoic acid biosynthesis induces the cell wall stress stimulon in Staphylococcus aureus. Antimicrob Agents Chemother 56:1810–1820. doi:10.1128/AAC.05938-11
Carattoli A (2013) Plasmids and the spread of resistance. Int J Med Microbiol 303:298–304. doi:10.1016/j.ijmm.2013.02.001
Casacuberta E, Gonzalez J (2013) The impact of transposable elements in environmental adaptation. Mol Ecol 22:1503–1517. doi:10.1111/mec.12170
Cervellati R, Greco E (2016) In vitro antioxidant activity of ubiquinone and ubiquinol, compared to vitamin E. Helv Chim Acta 99:41–45. doi:10.1002/hlca.201500124
Champoux JJ (2001) DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem 70:369–413. doi:10.1161/01.RES.0000255691.76142.4a
Chan WC, Bycroft BW, Lian L-Y, Roberts GCK (1989) Isolation and characterisation of two degradation products derived from the peptide antibiotic nisin. FEBS Lett 252:29–36. doi:10.1016/0014-5793(89)80884-1
Chen Q, Duan F, Li X et al (2014) Haloemodin as novel antibacterial agent inhibiting DNA gyrase and bacterial topoisomerase i. J Med Chem 57:3707–3714. doi:10.1021/jm401685f
Cho WK, Jergic S, Kim D et al (2014) Loading dynamics of a sliding DNA clamp. Angew Chem Int Ed 53:6768–6771. doi:10.1002/anie.201403063
Chopra I, Roberts M (2001) Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 65:232–260. doi:10.1128/MMBR.65.2.232
Chopra S, Matsuyama K, Tran T et al (2012) Evaluation of gyrase B as a drug target in Mycobacterium tuberculosis. J Antimicrob Chemother 67:415–421. doi:10.1093/jac/dkr449
Chung BC, Zhao J, Gillespie RA et al (2013) Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science 341:1012–1016. doi:10.1126/science.1236501
Clardy J, Fischbach MA, Walsh CT (2006) New antibiotics from bacterial natural products. Nat Biotechnol 24:1541–1550. doi:10.1038/nbt1266
Cleveland J, Montville TJ, Nes IF, Chikindas ML (2001) Bacteriocins: Safe, natural antimicrobials for food preservation. Int J Food Microbiol 71:1–20. doi:10.1016/S0168-1605(01)00560-8
Coates AR, Halls G, Hu Y (2011) Novel classes of antibiotics or more of the same? Br J Pharmacol 163:184–194. doi:10.1111/j.1476-5381.2011.01250.x
Cociancich S, Pesic A, Petras D et al (2015) The gyrase inhibitor albicidin consists of p-aminobenzoic acids and cyanoalanine. Nat Chem Biol 11:195–197. doi:10.1038/nchembio.1734
Cohen GN (2011) Microbial biochemistry, 2nd edn. Springer, Heidelberg
Cole ST, Riccardi G (2011) New tuberculosis drugs on the horizon. Curr Opin Microbiol 14:570–576. doi:10.1016/j.mib.2011.07.022
Collin F, Karkare S, Maxwell A (2011) Exploiting bacterial DNA gyrase as a drug target: Current state and perspectives. Appl Microbiol Biotechnol 92:479–497. doi:10.1007/s00253-011-3557-z
Connor EE (1998) Sulfonamide antibiotics. Prim Care Update Ob Gyns 5:32–35
Costenaro L, Grossmann JG, Ebel C, Maxwell A (2007) Modular structure of the full-length DNA gyrase B subunit revealed by small-angle X-ray scattering. Structure 15:329–339. doi:10.1016/j.str.2007.01.013
Cragg GM, Newman DJ (2013) Natural products: a continuing source of novel drug leads. Biochim Biophys Acta-Gen Subj 1830:3670–3695. doi:10.1016/j.bbagen.2013.02.008
Czaplewski L, Bax R, Clokie M et al (2016) Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect Dis 16:239–251. doi:10.1016/S1473-3099(15)00466-1
D’Costa VM, King CE, Kalan L et al (2011) Antibiotic resistance is ancient. Nature 477:457–461. doi:10.1038/nature10388
D’Elia MA, Millar KE, Beveridge TJ, Brown ED (2006a) Wall teichoic acid polymers are dispensable for cell viability in Bacillus subtilis. J Bacteriol 188:8313–8316. doi:10.1128/JB.01336-06
D’Elia MA, Pereira MP, Chung YS et al (2006b) Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J Bacteriol 188:4183–4189. doi:10.1128/JB.00197-06
D’Elia MA, Henderson JA, Beveridge TJ et al (2009a) The N-acetylmannosamine transferase catalyzes the first committed step of teichoic acid assembly in Bacillus subtilis and Staphylococcus aureus. J Bacteriol 191:4030–4034. doi:10.1128/JB.00611-08
D’Elia MA, Pereira MP, Brown ED (2009b) Are essential genes really essential? Trends Microbiol 17:433–438. doi:10.1016/j.tim.2009.08.005
Dalrymple BP, Kongsuwan K, Wijffels G et al (2001) A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc Natl Acad Sci U S A 98:11627–11632. doi:10.1073/pnas.191384398
Davies J (2006) Where have all the antibiotics gone? Can J Infect Dis Med Microbiol 17:287–290
Davis BD (1987) Mechanism of bactericidal action of aminoglycosides. Microbiol Rev 51:341–350 doi:0146-0749/87/030341-10$02.00/0
de Kruijff B, van Dam V, Breukink E (2008) Lipid II: a central component in bacterial cell wall synthesis and a target for antibiotics. Prostaglandins Leukot Essent Fat Acids 79:117–121. doi:10.1016/j.plefa.2008.09.020
Debnath J, Siricilla S, Wan B et al (2012) Discovery of selective menaquinone biosynthesis inhibitors against Mycobacterium tuberculosis. J Med Chem 55:3739–3755. doi:10.1021/jm201608g
Dhiman RK, Mahapatra S, Slayden RA et al (2009) Menaquinone synthesis is critical for maintaining mycobacterial viability during exponential growth and recovery from non-replicating persistence. Mol Microbiol 72:85–97. doi:10.1111/j.1365-2958.2009.06625.x
Diez J, Martinez JP, Mestres J et al (2012) Myxobacteria: natural pharmaceutical factories. Microb Cell Fact 11:52–54. doi:10.1186/1475-2859-11-52
Donnell MO, Langston L, Stillman B et al (2013) Principles and concepts of DNA replication in bacteria, archea, and eukarya. Cold Spring Harb Perspect Biol 5:a010180. doi:10.1101/cshperspect.a010108
Doroghazi JR, Albright JC, Goering AW et al (2014) A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat Chem Biol 10:963–968. doi:10.1038/nchembio.1659
Dougherty TJ, Nayar A, Newman JV et al (2014) NBTI 5463 is a novel bacterial type II topoisomerase inhibitor with Gram-negative antibacterial activity and in vivo efficacy. Antimicrob Agents Chemother. doi:10.1128/aac.02778-13
Dubée V, Chau F, Arthur M et al (2011) The in vitro contribution of autolysins to bacterial killing elicited by amoxicillin increases with inoculum size in Enterococcus faecalis. Antimicrob Agents Chemother 55:910–912. doi:10.1128/AAC.01230-10
Edwards DI (1993) Nitroimidazole drugs–action and resistance mechanisms. I. Mechanisms of action. J Antimicrob Chemother 31:9–20. doi:10.1093/jac/31.1.9
Ehmann DE, Lahiri SD (2014) Novel compounds targeting bacterial DNA topoisomerase/DNA gyrase. Curr Opin Pharmacol 18:76–83. doi:10.1016/j.coph.2014.09.007
Eichberg MJ (2015) Public funding of clinical-stage antibiotic development in the United States and European Union. Heal Secur 13:156–165. doi:10.1089/hs.2014.0081
Elshahawi SI, Shaaban KA, Kharel MK, Thorson JS (2015) A comprehensive review of glycosylated bacterial natural products. Chem Soc Rev 44:7591–7697. doi:10.1039/C4CS00426D
Epand RM, Walker C, Epand RF, Magarvey NA (2016) Molecular mechanisms of membrane targeting antibiotics. BBA Biomembranes 1858:980–987. doi:10.1016/j.bbamem.2015.10.018
European Centre for Disease Prevention and Control (2012) Annual report of the European antimicrobial resistance surveilance network (EARS-Net). ECDC, Stockholm
Farha MA, Leung A, Sewell EW et al (2013) Inhibition of WTA synthesis blocks the cooperative action of pbps and sensitizes MRSA to β-lactams. ACS Chem Biol 8:226–233. doi:10.1021/cb300413m
Finn J (2013) Evaluation of WO2012177707 and WO2012097269: Vertex’s phosphate prodrugs of gyrase and topoisomerase antibacterial agents. Expert Opin Ther Pat 23:1233–1237. doi:10.1517/13543776.2013.820707
Fischbach MA, Walsh CT (2009) Antibiotics for emerging pathogens. Science 325:1089–1093. doi:10.1126/science.1176667
Floss HG, Yu T-W (2005) Rifamycin-mode of action, resistance, and biosynthesis. Chem Rev 105:621–632. doi:10.1021/cr030112j
Forsberg KJ, Reyes A, Wang B et al (2012) The shared antibiotic resistome of soil bacteria and human pathogens. Science 337:1107–1111. doi:10.1126/science.1220761
Frankel RB, Kalmijn AJ, Amann R et al (2006) Sampling the antibiotic resistome. Science 311:374–378. doi:10.1126/science.1120800
Fujimoto N, Kosaka T, Yam M (2012) Menaquinone as well as ubiquinone as a crucial component in the Escherichia coli respiratory chain. In: Ekinci D (ed) Chemical biology, pp 187–208
Georgescu RE, Kim SS, Yurieva O et al (2008a) Structure of a sliding clamp on DNA. Cell 132:43–54. doi:10.1016/j.cell.2007.11.045
Georgescu RE, Yurieva O, Kim S-S et al (2008b) Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp. Proc Natl Acad Sci USA 105:11116–11121. doi:10.1073/pnas.0804754105
Gerth K, Pradella S, Perlova O et al (2003) Myxobacteria: Proficient producers of novel natural products with various biological activities—past and future biotechnological aspects with the focus on the genus Sorangium. J Biotechnol 106:233–253. doi:10.1016/j.jbiotec.2003.07.015
Gibbons S (2004) Anti-staphylococcal plant natural products. Nat Prod Rep 21:263–277. doi:10.1039/b212695h
Giguère S (2013) Lincosamides, pleuromutilins, and streptogramins. Antimicrob Ther Vet Med 199–210. doi:10.1016/0007-1935(89)90107-3
Gilbert GL (2015) Knowing when to stop antibiotic therapy. Med J Aust 202:121–123. doi:10.5694/mja14.01201
Gillings MR (2013) Evolutionary consequences of antibiotic use for the resistome, mobilome, and microbial pangenome. Front Microbiol 4:1–10. doi:10.3389/fmicb.2013.00004
Godtfredsen WO, Jahnsen S, Lorck H et al (1962) Fusidic acid: a new antibiotic. Nature 193:987
Górska A, Sloderbach A, Marszałł MP (2014) Siderophore-drug complexes: Potential medicinal applications of the “Trojan horse” strategy. Trends Pharmacol Sci 1–8. doi:10.1016/j.tips.2014.06.007
Gubaev A, Klostermeier D (2014) Reprint of “the mechanism of negative DNA supercoiling: a cascade of DNA-induced conformational changes prepares gyrase for strand passage”. DNA Repair (Amst) 20:130–141. doi:10.1016/j.dnarep.2014.06.006
Gui W-J, Lin S-Q, Chen Y-Y et al (2011) Crystal structure of DNA polymerase III β sliding clamp from Mycobacterium tuberculosis. Biochem Biophys Res Commun 405:272–277. doi:10.1016/j.bbrc.2011.01.027
Hamamoto H, Urai M, Ishii K et al (2014) Lysocin E is a new antibiotic that targets menaquinone in the bacterial membrane. Nat Chem Biol 11:127–133. doi:10.1038/nchembio.1710
Hammes WP, Neuhaus FC (1974) On the mechanism of action of vancomycin: inhibition of peptidoglycan synthesis in Gaffkya homari. Antimicrob Agents Chemother 6:722–728. doi:10.1128/AAC.6.6.722
Harrison E, Brockhurst MA (2012) Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol 20:262–267. doi:10.1016/j.tim.2012.04.003
Harvey AL, Edrada-Ebel R, Quinn RJ (2015) The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov 14:111–129. doi:10.1038/nrd4510
Hashimi SM, Wall MK, Smith AB et al (2007) The phytotoxin albicidin is a novel inhibitor of DNA gyrase. Antimicrob Agents Chemother 51:181–187. doi:10.1128/AAC.00918-06
Hawkey PM, Livermore DM (2012) Carbapenem antibiotics for serious infections. BMJ 344:1–7. doi:10.1136/bmj.e3236
Hawser S, Lociuro S, Islam K (2006) Dihydrofolate reductase inhibitors as antibacterial agents. Biochem Pharmacol 71:941–948. doi:10.1016/j.bcp.2005.10.052
Hecker SJ, Reddy KR, Totrov M et al (2015) Discovery of a cyclic boronic acid β-lactamase inhibitor (RPX7009) with utility vs class A serine carbapenemases. J Med Chem 58:3682–3692. doi:10.1021/acs.jmedchem.5b00127
Heide L (2014) New aminocoumarin antibiotics as gyrase inhibitors. Int J Med Microbiol 304:31–36. doi:10.1016/j.ijmm.2013.08.013
Henry RJ (1943) The mode of action of sulfonamides. Bacteriol Rev 7:175–262. doi:10.1128/AEM.02766-10
Herrmann J, Lukežič T, Kling A, et al (2016) Strategies for the discovery and development of new antibiotics from natural products: three case studies. Curr Top Microbiol Immunol
Hesterkamp T (2016) Antibiotics clinical development and pipeline. Curr Top Microbiol Immunol. doi:10.1007/82_2015_451
Hiratsuka T, Furihata K, Ishikawa J et al (2008) An alternative menaquinone biosynthetic pathway operating in microorganisms. Science 321:1670–1673. doi:10.1126/science.1160446
Hirsch EB, Ledesma KR, Chang KT et al (2012) In vitro activity of MK-7655, a novel beta-lactamase inhibitor, in combination with imipenem against carbapenem-resistant Gram-negative bacteria. Antimicrob Agents Chemother 56:3753–3757. doi:10.1128/AAC.05927-11
Holzgrabe U (2015) New griselimycins for treatment of tuberculosis. Chem Biol 22:981–982. doi:10.1016/j.chembiol.2015.08.002
Hooper DC, Wolfson JS, McHugh GL et al (1982) Effects of novobiocin, coumermycin-a1, clorobiocin, and their analogs on Escherichia-coli Dna gyrase and bacterial-growth. Antimicrob Agents Chemother 22:662–671
Hsu S-TD, Breukink E, Tischenko E et al (2004) The nisin-lipid II complex reveals a pyrophosphate cage that provides a blueprint for novel antibiotics. Nat Struct Mol Biol 11:963–967. doi:10.1038/nsmb830
Huang G, Zhang L, Birch RG (2001) A multifunctional polyketide-peptide synthetase essential for albicidin biosynthesis in Xanthomonas albilineans. Microbiology 147:631–642
Jacobsson S, Golparian D, Alm RA et al (2014) High in vitro activity of the novel spiropyrimidinetrione AZD0914, a DNA gyrase inhibitor, against multidrug-resistant Neisseria gonorrhoeae isolates suggests a new effective option for oral treatment of gonorrhea. Antimicrob Agents Chemother 58:5585–5588. doi:10.1128/AAC.03090-14
Jacoby GA (2005) Mechanisms of bacterial resistance to quinolones. 41:S120–S126
Jad YE, Acosta GA, Naicker T et al (2015) Synthesis and biological evaluation of a teixobactin analogue. Org Lett 17:6182–6185. doi:10.1021/acs.orglett.5b03176
Ji C, Juárez-Hernández RE, Miller MJ (2012) Exploiting bacterial iron acquisition: siderophore conjugates. Future Med Chem 4:297–313. doi:10.4155/fmc.11.191
Jia Z, O’Mara ML, Zuegg J et al (2013) Vancomycin: ligand recognition, dimerization and super-complex formation. FEBS J 280:1294–1307. doi:10.1111/febs.12121
Johnston CW, Magarvey NA (2015) Natural products: untwisting the antibiotic’ome. Nat Chem Biol 11:177–178. doi:10.1038/nchembio.1757
Johnston CW, Skinnider MA, Dejong CA et al (2016) Assembly and clustering of natural antibiotics guides target identification. Nat Chem Biol 12:233–239. doi:10.1038/nchembio.2018
Johnstone TC, Nolan EM (2015) Beyond iron: non-classical biological functions of bacterial siderophores. Dalton Trans 44:6320–6339. doi:10.1039/c4dt03559c
Jolles G (1971) New cyclopeptides, GB Patent 1,252,553
Kahne D, Leimkuhler C, Lu W, Walsh C (2005) Glycopeptide and lipoglycopeptide antibiotics. Chem Rev 105:425–448. doi:10.1021/cr030103a
Kåhrström CT (2015) Antimicrobials: a new drug for resistant bugs. Nat Rev Microbiol 13:126–127. doi:10.1038/nrmicro3429
Kale MG, Raichurkar A, Shahul Hameed P et al (2013) Thiazolopyridine ureas as novel antitubercular agents acting through inhibition of DNA gyrase B. J Med Chem 56:8834–8848. doi:10.1021/jm401268f
Kale RR, Kale MG, Waterson D et al (2014) Thiazolopyridone ureas as DNA gyrase B inhibitors: optimization of antitubercular activity and efficacy. Bioorg Med Chem Lett 24:870–879. doi:10.1016/j.bmcl.2013.12.080
Kalman D, Barriere S (1990) Review of the pharmacology, pharmacokinetics, and clinical use of cephalosporins. Tex Heart Inst J 17:203–215
Kannan K, Mankin AS (2011) Macrolide antibiotics in the ribosome exit tunnel: species-specific binding and action. Ann N Y Acad Sci 1241:33–47. doi:10.1111/j.1749-6632.2011.06315.x
Kawamukai M (2002) Biosynthesis, Bioproduction and Novel Roles of Ubiquinone. J Biosci Bioeng 94:511–517
Keller S, Schadt HS, Ortel I, Süssmuth RD (2007) Action of atrop-abyssomicin C as an inhibitor of 4-amino-4-deoxychorismate synthase PabB. Angew Chem Int Ed 46:8284–8286. doi:10.1002/anie.200701836
Kern G, Palmer T, Ehmann DE, et al (2015) Inhibition of Neisseria gonorrhoeae type II Topoisomerases by the Novel Spiropyrimidinetrione AZD0914. J Biol Chem M115.663534. doi:10.1074/jbc.M115.663534
Kirst HA (2013) Developing new antibacterials through natural product research. Expert Opin Drug Discov 8:479–493. doi:10.1517/17460441.2013.779666
Kjelstrup S, Hansen PMP, Thomsen LE et al (2013) Cyclic Peptide Inhibitors of the β-Sliding Clamp in Staphylococcus aureus. PLoS ONE. doi:10.1371/journal.pone.0072273
Kling A, Lukat P, Almeida DV et al (2015) Targeting DnaN for tuberculosis therapy using novel griselimycins. Science 348:1106–1112. doi:10.1126/science.aaa4690
Koehn FE, Carter GT (2005) The evolving role of natural products in drug discovery. Nat Rev Drug Discov 4:206–220. doi:10.1038/nrd1657
Kohanski MA, Dwyer DJ, Collins JJ (2010) How antibiotics kill bacteria: from targets to networks. Nat Rev Microbiol 8:423–435. doi:10.1038/nrmicro2333
Kotra LP, Haddad J, Mobashery S (2000) Aminoglycosides: perspectives on mechanisms of action and resistance and strategies to counter resistance. Antimicrob Agents Chemother 44:3249–3256. doi:10.1128/AAC.44.12.3249-3256.2000.Updated
Kramer NE, Smid EJ, Kok J et al (2004) Resistance of Gram-positive bacteria to nisin is not determined by Lipid II levels. FEMS Microbiol Lett 239:157–161. doi:10.1016/j.femsle.2004.08.033
Kretz J, Kerwat D, Schubert V et al (2015) Total synthesis of albicidin: A lead structure from xanthomonas albilineans for potent antibacterial gyrase inhibitors. Angew Chem Int Ed 54:1969–1973. doi:10.1002/anie.201409584
Kuipers OP, Rollema HS, de Vos WM, Siezen RJ (1993) Biosynthesis and secretion of a precursor of nisin Z by Lactococcus lactis, directed by the leader peptide of the homologous lantibiotic subtilin from Bacillus subtilis. FEBS Lett 330:23–27. doi:10.1016/0014-5793(93)80911-D
Kurosu M, Begari E (2010) Vitamin K2 in electron transport system: are enzymes involved in vitamin K2 biosynthesis promising drug targets? Molecules 15:1531–1553. doi:10.3390/molecules15031531
Kurosu M, Narayanasamy P, Biswas K et al (2007) Discovery of 1,4-didydroxy-2-naphthoate prenyltransferase inhibitors: new drug leads for multidrug-resistant gram-positive pathogens. J Med Chem 50:3973–3975. doi:10.1021/jm070638m
Lapierre P, Gogarten JP (2009) Estimating the size of the bacterial pan-genome. Trends Genet 25:107–110. doi:10.1016/j.tig.2008.12.004
Lapuebla A, Abdallah M, Olafisoye O et al (2015a) Activity of imipenem with relebactam against gram-negative pathogens from New York City: Table 1. Antimicrob Agents Chemother 59:5029–5031. doi:10.1128/AAC.00830-15
Lapuebla A, Abdallah M, Olafisoye O, et al (2015b) Activity of meropenem combined with RPX7009, a novel β-lactamase inhibitor, against Gram-negative clinical isolates in New York City. Antimicrob Agents Chemother 59:AAC.00843–15. doi:10.1128/AAC.00843-15
Lazarevic V, Karamata D (1995) The tagGH operon of Bacillus subtilis 168 encodes a two-component ABC transporter involved in the metabolism of two wall teichoic acids. Mol Microbiol 16:345–355. doi:10.1111/j.1365-2958.1995.tb02306.x
Lee K, Campbell J, Swoboda JG et al (2010) Development of improved inhibitors of wall teichoic acid biosynthesis with potent activity against Staphylococcus aureus. Bioorg Med Chem Lett 20:1767–1770. doi:10.1016/j.bmcl.2010.01.036
Lee SH, Wang H, Labroli M, et al (2016a) TarO-specific inhibitors of wall teichoic acid biosynthesis restore b -lactam efficacy against methicillin-resistant staphylococci. Sci Transl Med 8:329ra32. doi:10.1126/scitranslmed.aad7364
Lee W, Schaefer K, Qiao Y et al (2016b) The mechanism of action of lysobactin. J Am Chem Soc 138:100–103. doi:10.1021/jacs.5b11807
Leski TA, Tomasz A (2005) Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus. J Bacteriol 2:1815–1824. doi:10.1128/JB.187.5.1815
Leu FP, Hingorani MM, Turner J, O’Donnell M (2000) The δ subunit of DNA polymerase III holoenzyme serves as a sliding clamp unloader in Escherichia coli. J Biol Chem 275:34609–34618. doi:10.1074/jbc.M005495200
Lewis K (2013) Platforms for antibiotic discovery. Nat Rev Drug Discov 12:371–387. doi:10.1038/nrd3975
Lim D, Strynadka NCJ (2002) Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat Struct Biol 9:870–876. doi:10.1038/nsb858
Lin AH, Murray RW, Vidmar TJ, Marotti KR (1997) The oxazolidinone eperezolid binds to the 50S ribosomal subunit and competes with binding of chloramphenicol and lincomycin. Antimicrob Agents Chemother 41:2127–2131
Ling LL, Schneider T, Peoples AJ et al (2015) A new antibiotic kills pathogens without detectable resistance. Nature 517:455–459. doi:10.1038/nature14098
Lipsky BA, Baker CA (1999) Fluoroquinolone toxicity profiles: a review focusing on newer agents. Clin Infect Dis 28:352–364. doi:10.1086/515104
Littmann J, Buyx A, Cars O (2015) Antibiotic resistance: An ethical challenge. Int J Antimicrob Agents 46:359–361. doi:10.1016/j.ijantimicag.2015.06.010
Liu LF, Liu CC, Alberts BM (1980) Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell 19:697–707. doi:10.1016/S0092-8674(80)80046-8
Livermore DM, Warner M, Mushtaq S (2013) Activity of MK-7655 combined with imipenem against enterobacteriaceae and Pseudomonas aeruginosa. J Antimicrob Chemother 68:2286–2290. doi:10.1093/jac/dkt178
Livermore DM, Mushtaq S, Warner M, Woodford N (2015) Activity of OP0595/β-lactam combinations against Gram-negative bacteria with extended-spectrum, AmpC and carbapenem-hydrolysing β-lactamases. J Antimicrob Chemother 70:3032–3041. doi:10.1093/jac/dkv239
Lowther GE (1979) The united states food and drug administration and the practice of optometry. J Am Optom Assoc 50:579–582
Lu X, Zhou R, Sharma I et al (2012) Stable analogues of OSB-AMP: potent Inhibitors of MenE, the o-succinylbenzoate-CoA synthetase from bacterial menaquinone biosynthesis. ChemBioChem 13:129–136. doi:10.1002/cbic.201100585
Maki H, Yamaguchi T, Murakami K (1994) Cloning and characterization of a gene affecting the methicillin resistance level and the autolysis rate in Staphylococcus aureus. J Bacteriol 176:4993–5000
Maren TH (1976) Relatons between structure and biological activity of sulfonamides. Annu Rev Pharmacol Toxicol 16:309–327. doi:10.1146/annurev.pa.16.040176.001521
Martin NI, Breukink E (2007) Expanding role of lipid II as a target for lantibiotics. Future Microbiol 2:513–525. doi:10.2217/17460913.2.5.513
Mast Y, Wohlleben W (2014) International Journal of Medical Microbiology Streptogramins – Two are better than one ! 304:44–50
Maxwell A (1993) The interaction between coumarin drugs and DNA gyrase. Mol Microbiol 9:681–686. doi:10.1111/j.1365-2958.1993.tb01728.x
Maxwell A, Lawson DM (2003) The ATP-binding site of type II topoisomerases as a target for antibacterial drugs. Curr Top Med Chem 3:283–303. doi:10.2174/1568026033452500
Mayer C, Janin YL (2014) Non-quinolone inhibitors of bacterial type IIA topoisomerases: a feat of bioisosterism. Chem Rev 114:2313–2342
Mccomas CC, Mccomas CC, Crowley BM et al (2003) Partitioning the loss in vancomycin binding affinity for D-Ala-D-Lac into lost H-bond and repulsive lone pair contributions. J Am Chem Soc 125:9314–9315
Medini D, Donati C, Tettelin H et al (2005) The microbial pan-genome. Curr Opin Genet Dev 15:589–594. doi:10.1016/j.gde.2005.09.006
Meeske AJ, Sham L-T, Kimsey H et al (2015) MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc Natl Acad Sci U S A 112:6437–6442. doi:10.1073/pnas.1504967112
Meredith TC, Swoboda JG, Walker S (2008) Late-stage polyribitol phosphate wall teichoic acid biosynthesis in Staphylococcus aureus. J Bacteriol 190:3046–3056. doi:10.1128/JB.01880-07
Michalopoulos AS, Livaditis IG, Gougoutas V (2011) The revival of fosfomycin. Int J Infect Dis 15:e732–e739. doi:10.1016/j.ijid.2011.07.007
Miles TJ, Hennessy AJ, Bax B et al (2013) Novel hydroxyl tricyclics (e.g., GSK966587) as potent inhibitors of bacterial type IIA topoisomerases. Bioorg Med Chem Lett 23:5437–5441. doi:10.1016/j.bmcl.2013.07.013
Miller EL (2002) The penicillins: a review and update. J Midwifery Women’s Heal 47:426–434. doi:10.1016/S1526-9523(02)00330-6
Mislin GL, Schalk IJ (2014) Siderophore-dependent iron uptake systems as gates for antibiotic Trojan horse strategies against Pseudomonas aeruginosa. Metallomics 6:408–420. doi:10.1039/c3mt00359k
Mital A (2009) Synthetic nitroimidazoles: biological activities and mutagenicity relationships. Sci Pharm 77:497–520. doi:10.3797/scipharm.0907-14
Mizrahi V, Henrie RN, Marlier JF et al (1985) Rate-limiting steps in the DNA polymerase I reaction pathway. Biochemistry 24:4010–4018. doi:10.1021/bi00336a031
Mohammadi T, Karczmarek A, Crouvoisier M et al (2007) The essential peptidoglycan glycosyltransferase MurG forms a complex with proteins involved in lateral envelope growth as well as with proteins involved in cell division in Escherichia coli. Mol Microbiol 65:1106–1121. doi:10.1111/j.1365-2958.2007.05851.x
Mohammadi T, van Dam V, Sijbrandi R et al (2011) Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J 30:1425–1432. doi:10.1038/emboj.2011.61
Moreno M, Elgaher WAM, Herrmann J et al (2015) Synthesis and biological evaluation of cystobactamid 507: a bacterial topoisomerase inhibitor from Cystobacter sp. Synlett 26:1175–1178. doi:10.1055/s-0034-1380509
Morrison C (2015) Antibacterial antibodies gain traction. Nat Rev Drug Discov 14:737–738. doi:10.1038/nrd4770
Mullane KM, Gorbach S (2011) Fidaxomicin: first-in-class macrocyclic antibiotic. Expert Rev Anti Infect Ther 9:767–777. doi:10.1586/eri.11.53
Münch D, Roemer T, Lee SH et al (2012) Identification and in vitro analysis of the GatD/MurT enzyme-complex catalyzing lipid II amidation in Staphylococcus aureus. PLoS Pathog 8:1–11. doi:10.1371/journal.ppat.1002509
Münch D, Engels I, Müller A et al (2015) Structural variations of the cell wall precursor lipid II and their influence on binding and activity of the lipoglycopeptide antibiotic oritavancin. Antimicrob Agents Chemother 59:772–781. doi:10.1128/AAC.02663-14
Murai M, Kaji T, Kuranaga T et al (2015) Total synthesis and biological evaluation of the antibiotic lysocin E and its enantiomeric, epimeric, and N-demethylated analogues. Angew Chem Int Ed 54:1556–1560. doi:10.1002/anie.201410270
Nesme J, Simonet P (2015) The soil resistome: a critical review on antibiotic resistance origins, ecology and dissemination potential in telluric bacteria. Environ Microbiol 17:913–930. doi:10.1111/1462-2920.12631
Newman DJ, Cragg GM (2012) Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod 75:311–335. doi:10.1021/np200906s
Nichols D, Cahoon N, Trakhtenberg EM et al (2010) Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl Environ Microbiol 76:2445–2450. doi:10.1128/AEM.01754-09
Nicolau DP (2008) Carbapenems: a potent class of antibiotics. Expert Opin Pharmacother 9:23–37. doi:10.1517/14656566.9.1.23
Niiranen L, Lian K, Johnson KA, Moe E (2015) Crystal structure of the DNA polymerase III β subunit (β-clamp) from the extremophile Deinococcus radiodurans. BMC Struct Biol. doi:10.1186/s12900-015-0032-6
Nikaido H (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. doi:10.1128/MMBR.67.4.593
Nöllmann M, Crisona NJ, Arimondo PB (2007) Thirty years of Escherichia coli DNA gyrase: From in vivo function to single-molecule mechanism. Biochimie 89:490–499. doi:10.1016/j.biochi.2007.02.012
Noufflard-Guy-Loé H, Berteaux S (1965) Rev Tuberc Pneumol 29:301–326
Novak R (2011) Are pleuromutilin antibiotics finally fit for human use? Ann N Y Acad Sci 1241:71–81. doi:10.1111/j.1749-6632.2011.06219.x
Nowicka B, Kruk J (2010) Occurrence, biosynthesis and function of isoprenoid quinones. Biochim Biophys Acta Bioenerg 1797:1587–1605. doi:10.1016/j.bbabio.2010.06.007
O’Connell KMG, Hodgkinson JT, Sore HF et al (2013) Combating multidrug-resistant bacteria: Current strategies for the discovery of novel antibacterials. Angew Chem Int Ed 52:10706–10733. doi:10.1002/anie.201209979
O’Neill J (2015) Securing new drugs for future generations : the pipeline of antibiotics. Wellcome Trust, London
Oakley AJ, Prosselkov P, Wijffels G et al (2003) Flexibility revealed by the 1.85 Å crystal structure of the $β$ sliding-clamp subunit of Escherichia coli DNA polymerase III. Acta Crystallogr Sect D 59:1192–1199. doi:10.1107/S0907444903009958
Oliphant CM, Green GM (2002) Quinolones: a comprehensive review. Am Fam Physician 65:455–464
Opperman TJ, Nguyen ST (2015) Recent advances toward a molecular mechanism of efflux pump inhibition. Front Microbiol 6:1–16. doi:10.3389/fmicb.2015.00421
Ozawa K, Horan NP, Robinson A et al (2013) Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits α, ε, θ and β reveals a highly flexible arrangement of the proofreading domain. Nucleic Acids Res 41:5354–5367. doi:10.1093/nar/gkt162
Page MGP (2013) Siderophore conjugates. Ann N Y Acad Sci 1277:115–126. doi:10.1111/nyas.12024
Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA (2011) Carbapenems: past, present, and future. Antimicrob Agents Chemother 55:4943–4960. doi:10.1128/AAC.00296-11
Paris L (2015) Report of the Antibiotics resistance project: antibiotics currently in clinical development. http://www.pewtrusts.org/en/multimedia/data-visualizations/2014/antibiotics-currently-in-clinical-development
Parmar A, Iyer A, Vincent CS et al (2016) Efficient total syntheses and biological activities of two teixobactin analogues. Chem Commun 52:6060–6063. doi:10.1039/C5CC10249A
Payne DJ, Gwynn MN, Holms DJ, Pompliano DL (2007) Drugs for bad bugs: confronting the challanges of antibacterial discovery. Nat Rev Drug Discov 6:29–40
Pelchovich G, Omer-Bendori S, Gophna U (2013) Menaquinone and iron are essential for complex colony development in Bacillus subtilis. PLoS ONE 8:1–14. doi:10.1371/journal.pone.0079488
Pendleton JN, Gorman SP, Gilmore BF (2013) Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther 11:297–308. doi:10.1586/eri.13.12
Pereira MP, Schertzer JW, D’Elia MA et al (2008) The wall teichoic acid polymerase TagF efficiently synthesizes poly(glycerol phosphate) on the TagB product lipid III. ChemBioChem 9:1385–1390. doi:10.1002/cbic.200800026
Perez-Cruz C, Delgado L, Lopez-Iglesias C, Mercade E (2015) Outer-inner membrane vesicles naturally secreted by gram-negative pathogenic bacteria. PLoS ONE 10:1–18. doi:10.1371/journal.pone.0116896
Perry JA, Westman EL, Wright GD (2014) The antibiotic resistome: what’ s new? Curr Opin Microbiol 21:45–50. doi:10.1016/j.mib.2014.09.002
Peterson LR (2009) Bad bugs, no drugs: no ESCAPE revisited. Clin Infect Dis 49:992–993. doi:10.1086/605540
Piddock LJV (2012) The crisis of no new antibiotics-what is the way forward? Lancet Infect Dis 12:249–253. doi:10.1016/S1473-3099(11)70316-4
Piddock LJV (2015) Teixobactin, the first of a new class of antibiotics discovered by ichip technology? J Antimicrob Chemother 70:2679–2680. doi:10.1093/jac/dkv175
Piel J (2010) Biosynthesis of polyketides by trans-AT polyketide synthases. Nat Prod Rep 27:996–1047. doi:10.1039/b816430b
Pinho MG, Errington J (2005) Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol Microbiol 55:799–807. doi:10.1111/j.1365-2958.2004.04420.x
Pinho MG, de Lencastre H, Tomasz A (2001) An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc Natl Acad Sci USA 98:10886–10891. doi:10.1073/pnas.191260798
Policy IP (2010) The 10 × ‘20 initiative: pursuing a global commitment to develop 10 new antibacterial drugs by 2020. Clin Infect Dis 50:1081–1083. doi:10.1086/652237
Polz MF, Alm EJ, Hanage WP (2013) Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet 29:170–175. doi:10.1016/j.tig.2012.12.006
Powers JH (2003) Development of drugs for antimicrobial-resistant pathogens. Curr Opin Infect Dis 16:547–551. doi:10.1097/01.qco.0000104294.87920.b4
Projan SJ (2003) Why is big Pharma getting out of antibacterial drug discovery? Curr Opin Microbiol 6:427–430. doi:10.1016/j.mib.2003.08.003
Pucci MJ, Bush K (2013) Investigational antimicrobial agents of 2013. Clin Microbiol Rev 26:792–821. doi:10.1128/CMR.00033-13
Rasko DA, Sperandio V (2010) Anti-virulence strategies to combat bacteria-mediated disease. Nat Rev Drug Discov 9:117–128. doi:10.1038/nrd3013
Rawat D, Nair D (2010) Extended-spectrum beta-lactamases in gram negative bacteria. J Glob Infect Dis 2:263–274. doi:10.4103/0974-777X.68531
Reck F, Alm RA, Brassil P et al (2012) Novel N-linked aminopiperidine inhibitors of bacterial topoisomerase type II with reduced p K a: antibacterial agents with an improved safety profile. J Med Chem 55:6916–6933. doi:10.1021/jm300690s
Redgrave LS, Sutton SB, Webber MA, Piddock LJV (2014) Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol 22:438–445. doi:10.1016/j.tim.2014.04.007
Rex JH (2014) ND4BB: addressing the antimicrobial resistance crisis. Nat Rev Microbiol 12:231–232. doi:10.1038/nrmicro3245
Robinson A, Causer RJ, Dixon NE (2012) Architecture and conservation of the bacterial DNA replication machinery, an underexploited drug target. Curr Drug Targets 13:352–372. doi:10.2174/138945012799424598
Roca J, Wang JC (1992) The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell 71:833–840. doi:10.1016/0092-8674(92)90558-T
Roca J, Wang JC (1994) DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell 77:609–616. doi:10.1016/0092-8674(94)90222-4
Roca J, Berger JM, Harrison SC, Wang JC (1996) DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc Natl Acad Sci U S A 93:4057–4062. doi:10.1073/pnas.93.9.4057
Rodríguez-Rojas A, Rodríguez-Beltrán J, Couce A, Blázquez J (2013) Antibiotics and antibiotic resistance: A bitter fight against evolution. Int J Med Microbiol 303:293–297. doi:10.1016/j.ijmm.2013.02.004
Royer M, Costet L, Vivien E et al (2004) Albicidin pathotoxin produced by Xanthomonas albilineans is encoded by three large PKS and NRPS genes present in a gene cluster also containing several putative modifying, regulatory, and resistance genes. Mol Plant Microbe Interact 17:414–427
Saha R, Saha N, Donofrio RS, Bestervelt LL (2013) Microbial siderophores: a mini review. J Basic Microbiol 53:303–317. doi:10.1002/jobm.201100552
Sanchez S, Demain AL (eds) (2015) Antibiotics: current innovations and future trends. Caister Academic Press, Norfolk
Sauvage E, Kerff F, Terrak M et al (2008) The penicillin-binding proteins: Structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev 32:234–258. doi:10.1111/j.1574-6976.2008.00105.x
Savoia D (2012) Plant-derived antimicrobial compounds: alternatives to antibiotics. Future Microbiol 7:979–990. doi:10.2217/fmb.12.68
Schäberle TF, Lohr F, Schmitz A, König GM (2014) Antibiotics from myxobacteria. Nat Prod Rep 31:953. doi:10.1039/c4np00011k
Schaefer B (2014) Natural Products in the Chemical Industry. Springer, Berlin & Heidelberg
Schoeffler AJ, Berger JM (2008) DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys 41:41–101. doi:10.1017/S003358350800468X
Schoepp-Cothenet B, Lieutaud C, Baymann F et al (2009) Menaquinone as pool quinone in a purple bacterium. Proc Natl Acad Sci U S A 106:8549–8554. doi:10.1073/pnas.0813173106
Schueffler A, Anke T (2014) Fungal natural products in research and development. Nat Prod Rep 1425–1448. doi:10.1039/C4NP00060A
Schwartz B, Markwalder JA, Wang Y (2001) Lipid II: total synthesis of the bacterial cell wall precursor and utilization as a substrate for glycosyltransfer and transpeptidation by penicillin binding protein (PBP) 1b of Eschericia coli. J Am Chem Soc 123:11638–11643. doi:10.1021/ja0166848
Schwechheimer C, Kuehn MJ (2015) Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 13:605–619. doi:10.1038/nrmicro3525
Scott LJ (2013) Fidaxomicin: a review of its use in patients with clostridium difficile infection. Drugs 73:1733–1747. doi:10.1007/s40265-013-0134-z
Scott CP, Abel-Santos E, Wall M et al (1999) Production of cyclic peptides and proteins in vivo. Proc Natl Acad Sci U S A 96:13638–13643. doi:10.1073/pnas.96.24.13638
Sewell EWC, Brown ED (2014) Taking aim at wall teichoic acid synthesis: new biology and new leads for antibiotics. J Antibiot 67:43–51. doi:10.1038/ja.2013.100
Shahul HP, Solapure S, Mukherjee K et al (2014) Optimization of pyrrolamides as mycobacterial gyrb atpase inhibitors: structure-activity relationship and in vivo efficacy in a mouse model of tuberculosis. Antimicrob Agents Chemother 58:61–70. doi:10.1128/AAC.01751-13
Sham L-T, Butler EK, Lebar MD et al (2014) MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345:220–222
Sharma M, Chauhan PM (2012) Dihydrofolate reductase as a therapeutic target for infectious diseases: opportunities and challenges. Future Med Chem 4:1335–1365. doi:10.4155/fmc.12.68
Shiva F (2015) Antibiotic prescription and bacterial resistance. Arch Pediatr Infect Dis 3:e21540. doi:10.5812/pedinfect.21540
Silver LL (2007) Multi-targeting by monotherapeutic antibacterials. Nat Rev Drug Discov 6:41–55. doi:10.1038/nrd2202
Singh GS (2004) β-Lactams in the new millennium. Part-I : monobactams and carbapenems. Med Chem (Los Angeles) 4:62–92
Smillie C, Garcillan-Barcia MP, Francia MV et al (2010) Mobility of plasmids. Microbiol Mol Biol Rev 74:434–452. doi:10.1128/MMBR.00020-10
Spellberg B, Powers JH, Brass EP et al (2004) Trends in antimicrobial drug development: implications for the future 90502:1279–1286
Stadler M, Hoffmeister D (2015) Fungal natural products-the mushroom perspective. Front Microbiol 6:1–4. doi:10.3389/fmicb.2015.00127
Storm DR, Strominger JL (1974) Binding of bacitracin to cells and protoplasts of Micrococcus lysodeikticus. J Biol Chem 249:1823–1827
Stukenberg PT, Studwell-Vaughan PS, O’Donnell M, O’Donnell M (1991) Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem 266:11328–11334. doi:10.1074/jbc.M111.338145
Su X-H, Wang B-X, Le W-J et al (2016) Multidrug-resistant neisseria gonorrhoeae Isolates from Nanjing, China, are sensitive to killing by a novel DNA gyrase inhibitor, ETX0914 (AZD0914). Antimicrob Agents Chemother 60:621–623. doi:10.1128/AAC.01211-15
Swoboda JG, Meredith TC, Cambell J et al (2010) Discovery of a small molecule that blocks wall teichoic acid biosynthesis in Stphylococcus aureus. ACS Chem Biol 5:839–849
Swoboda JG, Mylonakis E, Wilkinson BJ, Walker S (2011) Synthetic lethal compound combinations reveal a fundamental connection between wall teichoic acid and peptidoglycan biosynthesis in Staphylococcus aureus. ACS Chem Biol 6:106–116
Syvanen M (2012) Evolutionary implications of horizontal gene transfer. Annu Rev Genet 46:341–358. doi:10.1146/annurev-genet-110711-155529
Tari LW, Li X, Trzoss M et al (2013) Tricyclic GyrB/ParE (TriBE) inhibitors: a new class of broad-spectrum dual-targeting antibacterial agents. PLoS ONE 8:e84409. doi:10.1371/journal.pone.0084409
Taubes G (2008) The bacterial fight back. Science 321:356–361
Tegos GP, Haynes M, Strouse JJ et al (2011) Microbial efflux pump inhibition: tactics and strategies. Curr Pharm Des 17:1291–1302. doi:10.1038/nature11130.Reduced
Terlain B, Thomas JP (1969) C R Hebd Seances Acad Sci Ser C 269:1546–1549
Terlain B, Thomas JP (1971a) Bull Chim Soc Fr 6:2349–2356
Terlain B, Thomas JP (1971b) Bull Chim Soc Fr 6:2357–2362
Tettelin H, Riley D, Cattuto C, Medini D (2008) Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol 11:472–477. doi:10.1016/j.mib.2008.09.006
‘t Hart P, Oppedijk SF, Breukink E, Martin NI (2016) New insights into nisin’s antibacterial mechanism revealed by binding studies with synthetic lipid II analogues. Biochemistry 55:232–237. doi:10.1021/acs.biochem.5b01173
Thaker M, Spanogiannopoulos P, Wright GD (2010) The tetracycline resistome. Cell Mol Life Sci 67:419–431. doi:10.1007/s00018-009-0172-6
Tommasi R, Brown DG, Walkup GK et al (2015) ESKAPEing the labyrinth of antibacterial discovery. Nat Rev Drug Discov 14:529–542. doi:10.1038/nrd4572
Torres C (2010) Up against the wall. Nat Med 16:628–631. doi:10.1038/nm0610-628
Toussaint KA, Gallagher JC (2015) β-Lactam/β-lactamase inhibitor combinations: from then to now. Ann Pharmacother 49:86–98. doi:10.1177/1060028014556652
Upadhyay A, Upadhyaya I, Kollanoor-Johny A, Venkitanarayanan K (2014) Combating pathogenic microorganisms using plant-derived antimicrobials: a minireview of the mechanistic basis. Biomed Res Int Article ID 761741. doi:10.1155/2014/761741
Uria-Nickelsen M, Neckermann G, Sriram S et al (2013) Novel topoisomerase inhibitors: microbiological characterisation and in vivo efficacy of pyrimidines. Int J Antimicrob Agents 41:363–371. doi:10.1016/j.ijantimicag.2012.12.001
Venter H, Mowla R, Ohene-Agyei T, Ma S (2015) RND-type drug efflux pumps from Gram-negative bacteria: molecular mechanism and inhibition. Front Microbiol 6:377. doi:10.3389/fmicb.2015.00377
Ventola CL (2015) The antibiotic resistance crisis: part 1: causes and threats. P & T 40:277–283
Vivien E, Pitorre D, Cociancich S et al (2007) Heterologous production of albicidin: a promising approach to overproducing and characterizing this potent inhibitor of DNA gyrase. Antimicrob Agents Chemother 51:1549–1552. doi:10.1128/AAC.01450-06
Wang JC (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol 3:430–440. doi:10.1038/nrm831
Wang H, Gill CJ, Lee SH et al (2013) Discovery of wall teichoic acid inhibitors as potential anti-MRSA beta-lactam combination agents. Chem Biol 20:272–284. doi:10.1016/j.chembiol.2012.11.013
Wang W, Qiu Z, Tan H, Cao L (2014) Siderophore production by actinobacteria. Biometals 27:623–631. doi:10.1007/s10534-014-9739-2
Watanakunakorn C (1984) Mode of action and in-vitro activity of vancomycin. J Antimicrob Chemother 14:7–18. doi:10.1093/jac/14.suppl_D.7
Wattal C, Goel N (2011) Tackling antibiotic resistance. Nat Rev Microbiol 9:894–896. doi:10.1038/nrmicro2693
Whitman WB, Coleman DC, Wiebe WJ (1998) Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 95:6578–6583. doi:10.1073/pnas.95.12.6578
Wiedemann I, Breukink E, Van Kraaij C et al (2001) Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J Biol Chem 276:1772–1779. doi:10.1074/jbc.M006770200
Wijffels G, Dalrymple BP, Prosselkov P et al (2004) Inhibition of protein interactions with the beta 2 sliding clamp of Escherichia coli DNA polymerase III by peptides from beta 2-binding proteins. Biochemistry 43:5661–5671. doi:10.1021/bi036229j
Wijffels G, Johnson WM, Oakley AJ et al (2011) Binding inhibitors of the bacterial sliding clamp by design. J Med Chem 54:4831–4838. doi:10.1021/jm2004333
Williams DH, Bardsley B (1999) The vancomycin group of antibiotics and the fight against resistant bacteria. Angew Chem Int Ed 38:1172–1193. doi:10.1002/(SICI)1521-3773(19990503)38:9<1172:AID-ANIE1172>3.0.CO;2-C
Wolff P, Amal I, Oliéric V et al (2014) Differential modes of peptide binding onto replicative sliding clamps from various bacterial origins. J Med Chem 57:7565–7576. doi:10.1021/jm500467a
Wright GD (2007) The antibiotic resistome: the nexus of chemical and genetic diversity. NatRevMicrobiol 5:175–186. doi:10.1038/nrmicro1614
Wright GD (2010) The antibiotic resistome. Exp Opin Drug Disc 5:779–788. doi:10.1517/17460441.2010.497535
Wright GD (2012) The origins of antibiotic resistance. In: Coates ARM (ed) Handbook of experimental pharmacology, pp 45–65
Wright G (2015a) An irresistible newcomer. Nature 517:442–443. doi:10.1038/nature14193
Wright GD (2015b) Solving the antibiotic crisis. ACS Infect Dis 1:80–84. doi:10.1021/id500052s
Wright GD, Poinar H (2012) Antibiotic resistance is ancient: implications for drug discovery. Trends Microbiol 20:157–159. doi:10.1016/j.tim.2012.01.002
Wright PM, Seiple IB, Myers AG (2014) The evolving role of chemical synthesis in antibacterial drug discovery. Angew Chem Int Ed 53:8840–8869. doi:10.1002/anie.201310843
Wyke AW, Ward JB, Hayes MV, Curtis NAC (1981) A role in vivo for penicillin-binding protein-4 of Staphylococcus aureus. Eur J Biochem 119:389–393. doi:10.1111/j.1432-1033.1981.tb05620.x
Xu Z-Q, Flavin MT, Flavin J (2014a) Combating multidrug-resistant Gram-negative bacterial infections. Expert Opin Investig Drugs 23:163–182. doi:10.1517/13543784.2014.848853
Xu Z-Q, Flavin MT, Flavin J (2014b) Combating multidrug-resistant Gram-negative bacterial infections. Expert Opin Investig Drugs 23:163–182. doi:10.1517/13543784.2014.848853
Yahav D, Farbman L, Leibovici L, Paul M (2011) Colistin: new lessons on an old antibiotic: EBSCOhost. Clin Microbiol Infect 18:18–29
Yin Z, Wang Y, Whittell LR et al (2014a) DNA replication is the target for the antibacterial effects of nonsteroidal anti-inflammatory drugs. Chem Biol 21:481–487. doi:10.1016/j.chembiol.2014.02.009
Yin Z, Whittell LR, Wang Y et al (2014b) Discovery of lead compounds targeting the bacterial sliding clamp using a fragment-based approach. J Med Chem 57:2799–2806. doi:10.1021/jm500122r
Yin Z, Whittell LR, Wang Y et al (2015) Bacterial sliding clamp inhibitors that mimic the sequential binding mechanism of endogenous linear motifs. J Med Chem 58:4693–4702. doi:10.1021/acs.jmedchem.5b00232
Yoshida H, Bogaki M, Nakamura M, Nakamura S (1990) Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother 34:1271–1272. doi:10.1128/aac.34.6.1271
Yoshida H, Bogaki M, Nakamura M et al (1991) Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob Agents Chemother 35:1647–1650. doi:10.1128/AAC.35.8.1647
Young IG (1975) Biosynthesis of bacterial menaquinones. Menaquinone mutants of Escherichia coli. Biochemistry 14:399–406. doi:10.1021/bi00673a029
Yunis AA (1988) Chloramphenicol: relation of structure to activity and toxicity. Annu Rev Pharmacol Toxicol 28:83–100 doi:10.1146/annurev.pa.28.040188.000503
Zaffiri L, Gardner J, Toledo-Pereyra LH (2012) History of antibiotics. From salvarsan to cephalosporins. J Invest Surg 25:67–77. doi:10.3109/08941939.2012.664099
Zapun A, Philippe J, Abrahams KA et al (2013) In vitro reconstitution of peptidoglycan assembly from the gram-positive pathogen streptococcus pneumoniae. ACS Chem Biol 8:2688–2696. doi:10.1021/cb400575t
Zetts R (2014) Report of the Antibiotics resistance project: antibiotics currently in clinical development. http://www.pewtrusts.org/en/multimedia/data-visualizations/2014/antibiotics-currently-in-clinical-development
Zgurskaya HI, López CA, Gnanakaran S (2015) Permeability barrier of Gram-negative cell envelopes and approaches to bypass it. ACS Infect Dis 1:512–522. doi:10.1021/acsinfecdis.5b00097
Zhanel GG, Walkty AJ, Karlowsky JA (2015) Fidaxomicin: a novel agent for the treatment of clostridium difficile infection. Can J Infect Dis Med Microbiol 26:305–313
Zhang G, Meredith TC, Kahne D (2013) On the essentiality of lipopolysaccharide to Gram-negative bacteria. Curr Opin Microbiol 16:779–785. doi:10.1016/j.mib.2013.09.007
Zhao M, Goedecke T, Gunn J, et al (2013) Protostane and fusidane triterpenes: a mini-review. Molecules 18:4054–4080. doi:10.3390/molecules18044054
Acknowledgments
We thank Dr. Dr. Werner Tegge, Giambattista Testolin, Isabell Schneider and Dr. Verena Fetz for fruitful discussions and for proofreading of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this chapter
Cite this chapter
Klahn, P., Brönstrup, M. (2016). New Structural Templates for Clinically Validated and Novel Targets in Antimicrobial Drug Research and Development. In: Stadler, M., Dersch, P. (eds) How to Overcome the Antibiotic Crisis . Current Topics in Microbiology and Immunology, vol 398. Springer, Cham. https://doi.org/10.1007/82_2016_501
Download citation
DOI: https://doi.org/10.1007/82_2016_501
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-49282-7
Online ISBN: 978-3-319-49284-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)