Abstract
Salinity stress is a major constraint for crop growth, development, and yield, worldwide. Salinity stress is among the key abiotic stresses that critically impede plant development, causing yield reductions ranging from 15% to 90% in major crops, under moderate-to-high soil salinity levels. Although regarded as climate resilient, very little information is available on pearl millet, regarding its spectrum of physiological to molecular responses, inherent mechanisms exhibited, yield losses, and stress mitigation strategies compared to other cereal crops. High salinity levels in the soil impact the growth and productivity of pearl millet, which is predominantly grown in several arid and semi-arid zones. Therefore, this chapter highlights the differential responses of pearl millet crop to salinity stress, and the need to evaluate for superior genotypic variability with greater stress tolerance mechanisms. This chapter also discusses different approaches that can be employed for crop improvement programs that target salt-tolerant genotypes suitable for varying agro-ecological conditions.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
14.1 Introduction
Pearl millet (Pennisetum glaucum (L.)) is the most extensively cultivated millet crop for grain and forage. It is a staple crop for more than 90 million poor farmers, mainly grown in semi-arid and arid regions of Asia and Africa, where other cereals cannot be cultivated (Srivastava and Kumar 2015). It can survive under extreme environmental conditions and nutrient-deficient soils. These adaptive traits make pearl millet a vital cereal grown in adverse agroclimatic conditions where other crops fail to produce economic yields. Salinity is emerging as a serious threat for pearl millet cultivation, as it is mainly grown under rainfed production systems of arid and semiarid regions.
Salinity stress severely limits crop production and yield potential. Low precipitation, irrigation with saline water, and poor irrigation practices cause salinity stress. Salinization poses a major challenge in both arable and marginal agricultural lands (Litalien and Zeeb 2019). It is estimated that by 2050 the present area of over 833 million hectares of salt-affected soils will expand further fivefold, posing a threat to food security (FAO 2021). The soil is assumed to be saline or influenced by salinity if it has an electrical conductivity of more than 4 dS m−1 at 25 °C, where 4 dS m−1 ≈40 mM NaCl or greater. Cell dehydration, ionic toxicity, nutritional stress, osmotic stress, drought stress, and oxidative stress are some effects of salinity stress (Zhu 2002). The existing and future changes in soil fertility need climate efficient crops for the utilization of marginal lands for cultivation. Millets are considered to be climate resilient that can adapt to poor resource and hardy soils. Thus, understanding the stress tolerance mechanisms and genetic manipulation of millets, especially pearl millet, will aid further in attaining sustainable development efforts to search out enhanced crop performance on marginal and insignificant lands, which still remains undeciphered.
This chapter highlights the spectrum of physiological and molecular responses concerning salt tolerance and mitigation of salt stress in plant salinity stress tolerance, especially pearl millet. Approaches to improve salinity tolerance in pearl millet are limited, including the conventional agronomic, breeding, molecular breeding, and genomic approaches with some success. Various novel approaches have been attempted in pearl millet for enhancing yield under salt-stress environments. These include using adapted germplasm, genetic diversification of adapted landraces through introgression of suitable elite genetic material, and exploitation of heterosis to amalgamate salt tolerance and high yield. Molecular breeding is fast emerging as a supplement approach to enhance drought adaptation at a faster rate with greater precision. Molecular marker-based genetic linkage maps of pearl millet are available and genomic regions determining yield under salinity stress environments have been identified, preparing a road map for marker-assisted selection.
14.2 Physiological Basis of Salt Stress Tolerance in Pearl Millet
Millets are generally climate-contingent crops adapted to marginal and dry lands of arid and semi-arid regions, surviving low rainfall and poor soils. They show exceptional tolerance to various abiotic stresses, including salinity stress. The primary growing areas of the pearl millet crop fall in the arid and semi-arid zones of South Asia and West Africa (Blummel et al. 2003), where the soil salinity stress is one of the major constraints. Pearl millet [Pennisetum glaucum (L.) R. Br] is considered to be fairly tolerant to salinity, having three subtypes of C4 photosynthetic pathway (phosphoenol pyruvate carboxylase (PEP-CK subtype)), NAD-malic enzyme subtype and NADP-malic enzyme subtype) (Wang and Shangguan 2010). Millets benefit greatly from the C4 photosynthetic trait as their Kranz anatomy helps maintain higher CO2 levels in bundle sheath cells, reduces photorespiratory loss, and the carboxylase activity of Rubisco is also enhanced. Many physiological and morphological advantages like lesser CO2 compensation point, higher transpiration and water use efficiency, leaf venation features, and deeper root systems make these crops resilient to harsh climates. The growth and yield remain unaffected to changing CO2 levels or elevated atmospheric CO2 concentration, unlike C3 cereals like rice and wheat. Pearl millet is classified as glycophytes and has an average salt tolerance threshold of 6 (ECe) (dS/m). The availability of fairly high levels of tolerance in Pennisetum species (Ashraf and McNeilly 1987, 1992; Dua 1989; Muscolo et al. 2003) offers a scope to integrate these type of tolerant crop species into appropriate breeding and management programs to improve the productivity of the saline soils. In the current changing climate scenario, millets are promising crops for climate resilience and sustainable productivity. Improved overall plant growth, higher biomass allocation, and partitioning are some additional benefits of C4 photosynthesis alongside WUE and NUE (Sage and Zhu 2011). Higher stomatal conductance, high photo assimilation, and low transpiration benefitted millet crops with increased resource use efficiency. Pearl millet crop adopts various escape mechanisms, avoidance and tolerance to unfavorable stress environments.
The rapid diffusion of photosynthetic metabolites is attributed to leaf vein density apart from their anatomical superiority. These C4 grasses primarily have an efficient water distribution cascade compared to C3 grasses by having a denser leaf venation pattern (higher network of the small longitudinal and transverse veins while keeping a constant density of large longitudinal veins) (Govinda et al. 2012). The leaf veins anatomy described by Altus and Canny (1982) has a hierarchical order and has different structures.
-
The enormous longitudinal veins run from the leaf blade into the sheath. The huge longitudinal veins assist essentially in the longitudinal carriage of photosynthate outside the leaf blade.
-
The little longitudinal veins fundamentally gather photosynthates from adjacent photosynthetic cells.
-
The cross-over (transverse) veins and little longitudinal veins assume an essential part in the sideways carriage of photosynthates from the little to the enormous longitudinal veins.
14.3 Effect of Salinity Stress on Pearl Millet: Morpho-Physiological and Biochemical Changes
Plants grown under saline soil condition experience a significant amount of high osmotic stress, ion toxicities, and nutritional disorder leading to reduced plant productivity. Salinity may inhibit plant growth in two different ways. Primarily presence of excess salts in the soil solution minimizes the ability of the plant to extract water and leads to water deficit stress (Kumar et al. 2016; Yadav and Dagar 2016). Secondly, the entry of excessive amounts of salt in the transpiration stream will injure the cells in the transpiring leaves leading to reduced growth and development (Kumar et al. 2018). This is called the salt-specific or ion excess effect of salinity.
Salt stress affects all the major processes such as germination, growth, photosynthetic pigments and photosynthesis, water relations, and nutrient uptake (Shahzad et al. 2021), as explained below. However, high salinity impacts its growth and productivity in several arid zones (Krishnamurthy et al. 2007a, b). Yield loss under salinity stress in pearl millet grain yield was around 13–22% under salinity levels of 8–12 dS m−1 (Yadav et al. 2020). Studies have also reported an average reduction of 20% in shoot biomass productivity and about 40% in grain yield in pearl millet germplasm (Krishnamurthy et al. 2014; Toderich et al. 2018) under salinity. Under high saline conditions in pearl millet, a significant reduction of about 47–86% in grain yield and 51% in fodder yield was observed (Choudhary et al. 2019; Kulkarni et al. 2006; Ribadiya et al. 2018).
14.3.1 Germination and Seedling Establishment
Seed germination is the primary and vital phase in a plant’s growth cycle determining yield. Salinity stress alters the imbibition of water by seeds by lowering the osmotic potential of surrounding soil water causing toxicity which changes the activities of enzymes of nucleic acid metabolism, alters protein metabolism, disturbs hormonal balance (ABA/GA), and reduces the utilization of seed reserves. Salinity affected sorghum seed germination by inducing dormancy and ion (Na+, Cl−) toxicity (Rajabi Dehnavi et al. 2020). Maiti et al. (2007) and Jain and Dev Sharma (2005) reported that high salinity levels decreased seed germination and seedling growth pearl millet by reducing root length, fresh/dry weights of roots, and coleoptiles. Similar findings were reported in sorghum under salinity stress by Ali et al. (2020).
14.3.2 Growth and Development Changes Affected by Salinity Stress
The first phase of the growth response results from the effect of salt outside the plant. The salt in the soil solution reduces seedling leaf growth and root growth. Neither Na+ nor Cl− builds up in growing tissues at concentrations that inhibit growth because meristematic tissues are primarily fed by the phloem from which salt is effectively excluded and rapidly elongating cells can accommodate the salt that arrives in the xylem within their vacuoles. The second phase of the growth response results from the toxic effect of salt inside the plant. Munns and Tester (2008) reviewed the response of a plant to salinity stress and summarized that reduction in shoot growth occurs in two phases: a rapid response to the increase in external osmotic pressure (Osmotic Stress) and a slower response due to the accumulation of Na+ in leaves (Ionic Stress). In the osmotic phase, which starts immediately after the salt concentration around the roots increases to a threshold level, the rate of shoot growth falls significantly. This is mainly (but not entirely) due to the osmotic effect of the salt outside the roots. The osmotic stress not only has an immediate impact on growth, but also has a more significant effect on growth rates than the ionic stress. Older leaves get concentrated by the salt intake and continue transport into transpiring leaves, leading to leaf death. Excess loading of salts exceeding the vacuole capacity inhibits enzymatic activity. Alternatively, they might build up in the cell walls and dehydrate the cell. The excessive salt concentration decreased the osmotic potential of the soil that restricted the water uptake in sorghum while salinity stress declined yield in pearl millet through membrane rupture, decreased photosynthesis and dry matter partitioning from source to sink (Kumar et al. 2018; Rahim et al. 2020). Genetic differences in tolerance to salinity at both seedling and grain filling stages have been established, and screening techniques standardized. Also, the germplasm and breeding materials with a higher salinity tolerance have been identified to use in breeding programs (Yadav et al. 2012).
14.3.3 Salinity and Ionic Toxicity Effects in Plants
The appropriate ion ratios provide a tool for the physiological response of a plant in relation to its growth and development (Wang et al. 2003). Excessive soluble salts in the soil compete with the uptake and metabolism of essential mineral nutrients. Increased salt uptake induces specific ion toxicities like high Na+, Cl−, or sulfate (SO2−) that decrease the uptake of essential macronutrients (N, P, K, Ca). The imbalance in Na+ and Cl− ion ratios leads to many physiological disorders in plants. Accumulation of Na+ ion interferes with the uptake of potassium (K+) ion and disturbs stomatal regulation causing water loss, and inhibits protein synthesis, photosynthesis and inactivation of enzymes, while the Cl− ion disturbs the chlorophyll production and causes chlorotic toxicity in pearl millet (Hanin et al. 2016; Kumar et al. 2018) and finger millet (Mbinda and Mukami 2021). Excess Na+ accumulation leads to the deterioration of the structural and functional integrity of organelle membranes in pearl millet. Reduced shoot N content and increased K+ and Na+ content are usually associated with an adaptive salinity tolerance mechanism in pearl millet (Dwivedi et al. 2012). According to Krishnamurthy et al. (2007a, b), shoot biomass ratio associated with salt tolerance and shoot Na+ concentration could be used as potential selection criteria for screening of pearl millet germplasm at the vegetative stage for salt stress.
14.3.4 Changes in Plant Water Relations
Determining the water status is an essential physiological parameter while selecting stress-tolerant genotypes (Parida and Das 2005). With the increased concentration of salts in the root medium, leaf water content gets reduced as lower soil water potential interferes with the plant’s ability to extract water from soil and maintain turgor. This leads to water stress in plants, adversely affecting all the physiological and growth mechanisms like cell expansion, cell division, stomatal mechanisms, and key enzymatic activities (Hussain et al. 2010). Radhouane (2013) reported that salinity causes a reduction in the relative water content in pearl millet leaves. However, plants adjust osmotically (accumulate solutes) at low or moderate salt concentration and maintain a potential gradient for the influx of water in finger millet (Hasanuzzaman et al. 2017).
14.3.5 Salinity and Oxidative Stress
Besides the direct impact of salinity on plants, a common consequence of salinity is induction of excessive accumulation of Reactive Oxygen Species (ROS). ROS are products of normal cellular metabolism, but the balance between production and elimination is disturbed under stress conditions in cellular components of plants. Environmental perturbations like salinity stress induce the overproduction of reactive oxygen species (superoxide radical (O2•ˉ), hydroxyl radical (OH•), hydroperoxyl radical (HO2•), hydrogen peroxide (H2O2), alkoxy radical (RO•), peroxy radical (ROO•), singlet oxygen (1O2)), all of which are cytotoxic to plants (Golldack et al. 2014), which results in peroxidation of membrane lipids, oxidation of protein, inactivation of key enzymes, DNA damage, and disruption of membrane integrity (Pooja et al. 2020). Salt stress causes physiological drought leading to stomatal closure, reduces carbon fixation, exposing chloroplasts to excessive excitation energy, which in turn increases the generation of ROS and membrane disruption (Toderich et al. 2018).
14.3.6 Photosynthetic Pigments and Photosynthesis
Reduced water potential is the main reason for reduced photosynthesis under salt stress. Accumulation of high concentrations of Na+ and/or Cl− in the chloroplasts inhibits photosynthesis by injuring chloroplast. Salinity stress is associated with stomatal closure leading to reduced carbon assimilation. Photosystem II (PS II) is a relatively sensitive component concerning abiotic stress, especially salt stress. A considerable decrease in the efficiency of PS II, electron transport chain, and CO2 assimilation rate was reported under the influence of salinity. Salinity stress reduced chlorophyll fluorescence by disrupting the functionality of thylakoids in chloroplasts and oxygen-evolving complex (PS II) reducing the photosynthetic rate in many crops including pearl millet (Kalaji et al. 2011; Yadav et al. 2020). Salinity stress reduces the photosynthetic rate owing to cell membranes’ dehydration, which reduces their permeability to carbon dioxide. Salt toxicity enhanced leaf senescence and changed enzymatic activity regulating photosynthesis, eventually resulting in negative feedback by reduced sink activity in finger millet (Mukami et al. 2020).
14.3.7 Nutrient Imbalance
Salinity induces multiple nutritional disorders affecting crop performance. The dietary disorders may result from the effect of salinity on nutrient availability, competitive uptake, transport, or distribution within the plant. Salinity disturbs nutrient uptake by increasing the soil pH (Zhu 2004) and disrupting the redox potential of soil solution. Decreased nitrogen uptake, for example in saline soils, occurs due to interaction between Na+ and NH4+ or between Cl− and NO3− that ultimately reduces the growth and yield of the crop. This reduction in NO3− uptake is associated with Cl− antagonism. Phosphate concentration in crops decreases with salinity as it affects the activity of PO3− and phosphate concentration in soils is tightly controlled by the sorption process. Salinity increases sodium concentration in plant tissues resulting in ion imbalances of K, Ca, and Mg. Supra-optimal concentrations of sodium ions are toxic to cell metabolism, which can finally lead to growth inhibition.
14.4 Strategies of Adaptation and Tolerance of Pearl Millet to Salt Stress
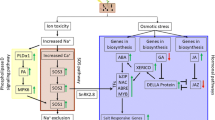
Plants develop various physiological and biochemical mechanisms (as discussed above) to tolerate and survive under salinity stress. Na+ exclusion is often a primary determinant of variability in salinity tolerance within a species, including millets like sorghum and pearl millet (Krishnamurthy et al. 2007a, b). Plant tolerance mechanisms to salinity stress are distinguished into three types: (a) tolerance to osmotic stress, (b) Na+ exclusion, and (c) tissue tolerance. The major form of salt present in the soil is NaCl. A general schema of plant response to salinity stress is depicted in Fig. 14.1.
Maintaining ion homeostasis by ion uptake and compartmentalization is crucial for average plant growth during salt stress. Both glycophytes and halophytes cannot tolerate high salt concentrations in their cytoplasm. Hence, the excess salt is sequestered to the vacuole in older tissues thereby protecting the plant from salinity stress (Munns and Tester 2008). Biological membranes and their associated components play an integral role in maintaining ion concentration within the cytosol during the period of stress by regulating ion uptake and transport (Zhu 2002). Various membrane transporters like carrier proteins, channel proteins, antiporters, and symporters facilitate this phenomenon. The Na+ ion that enters the cytoplasm is then transported to the vacuole via Na+/H+ antiporter (Krishnamurthy et al. 2014). Two types of H+ pumps are present in the vacuolar membrane: vacuolar type H+-ATPase (V-ATPase) and the vacuolar pyrophosphatase (V-PPase). Of these, V-ATPase is the most dominant H+ pump present within the plant cell. Under stressed conditions the survivability of the plant depends upon the activity of V-ATPase. These antiporters are encoded by SOS salt overly sensitive signaling proteins (SOS1, SOS2 and SOS3) helping in ion homeostasis and salt tolerance (Fig. 14.2).
Excess Na+ and high osmolarity are separately sensed by unknown sensors at the plasma membrane level, which then induce an increase in cytosolic (Ca2+). This increase is perceived by SOS3 which activates SOS2. The activated SOS3-SOS2 protein complex phosphorylates SOS1, the plasma membrane Na+/H+ antiporter resulting in the efflux of Na+ ions. SOS2 can regulate NHX1 antiport activity and V-H+ATPase activity independently of SOS3, possibly by SOS3 like Ca2+ binding proteins (SCaBP) that target it to the tonoplast. Salt stress can also induce the accumulation of ABA, which using ABI1 and ABI2, can negatively regulate SOS2 or SOS1 and NHX1. Besides conferring salt tolerance, it regulates pH homeostasis and membrane vesicle trafficking.
During salinity stress, Na+ ion due to its higher concentration in the soil competes with K+ ion for the transporter as they both ions share the same transport mechanism, thereby limiting the uptake of K+. Transporters located on the plasma membrane, belonging to the HKT (histidine kinase transporter) family play an essential role in salt tolerance by regulating transportation of Na+ and K+. A large number of genes and proteins, such as HKT and NHX encoding K+ transporters and Na+, K+/H+ antiporters, protect plants from adverse effects of salinity by maintaining K+ homeostasis, endosomal pH regulation and preventing excess accumulation Na+ in leaves.
14.4.1 Accumulation of Osmolytes or Compatible Solutes
Accumulation of compatible osmolytes reduces the water potential in the plant creating strong water potential gradient which helps in improving water uptake by roots under salinity stress. These compatible solutes include amino acid proline, glycine betaine, sugars, and polyols. As the accumulation of these compounds is proportional to the external osmolarity, they protect cellular structures by maintaining osmotic balance within the cell via continuous water influx (Sorahinobar et al. 2016). Role of compatible osmolytes is not limited to osmotic balance. Compatible solutes also act as low molecular weight chaperones due to their hydrophilic properties. Proline accumulation is a well-known measure adopted for alleviation of salinity stress (Sharma et al. 2019). In pearl millet intracellular proline accumulation during salinity stress not only provided tolerance towards stress but also served as an organic nitrogen reserve during stress recovery (Toderich et al. 2018; Yadav et al. 2020).
Glycine betaine is a non-toxic cellular osmolyte that helps in stress mitigation. Glycine betaine also protects the cell by protein stabilization and reduction in ROS (Veerangamallaiah et al. 2008). Hussain et al.(2010) reported increment in glycine betaine in pearl millet under NaCl treatment. Sugar alcohols are a class of polyols functioning as low molecular weight chaperones and ROS scavenging compounds. Carbohydrates such as sugars (e.g., glucose, fructose, fructans, and trehalose) and starch accumulate under salt stress involving in osmoprotection, carbon storage, and scavenging of reactive oxygen species. Osmotic adjustment is greater in millet races with smaller plants having small organs and cells. Pearl millet varieties well-adapted to saline environments showed good physiological and biochemical responses to increased salinity such as increased proline, total soluble proteins, and epicuticular wax content.
14.4.2 Polyamines
Polyamines (PA) play a crucial role in abiotic stress tolerance including salinity. Increase in the level of polyamines is correlated with stress tolerance in plants. The most common polyamines that are found within the plant system are diamine putrescine (PUT), triamine spermidine (SPD), and tetra-amine spermine (SPM). The positive effects of polyamines have been associated with the maintenance of membrane integrity, regulation of gene expression for the synthesis of osmotically active solutes, active solutes, reduction in ROS production, and controlling accumulation of Na+ and Cl− ion in different organs. When the seedlings of Sorghum bicolour treated with 0.52 mM SPM were subjected to salt stress, improvement in growth and partial increase in the activity of peroxidase and glutathione reductase enzyme was observed with a concomitant decrease in the level of membrane lipid peroxidation (Chai et al. 2010).
14.4.3 Hormonal Regulation
Production of abscisic acid (ABA) is upregulated with salinity induced soil water deficit around the root which helps in partial stomatal closure limiting transpirational water loss. The accumulation of ABA can mitigate the inhibitory effect of salinity on photosynthesis, growth, and translocation of assimilates (Cabot et al. 2009). The positive relationship between ABA accumulation and salinity tolerance is attributed to the accumulation of K+, Ca2+ and compatible solutes in vacuoles of roots, which counteract with the uptake of Na+ and Cl−. ABA is a potential cellular signal that regulates the expression of a number of salt and water deficit responsive genes. ABA treatment in wheat induced the expression of MAPK4-like, TIP 1, and GLP 1 genes under salinity stress (Keskin et al. 2010).
Some other hormones such as salicylic acid (SA) and brassinosteroids (BR) also participate in plant salt stress responses (Yadav et al. 2020). Under salinity stress endogenous level of SA increased along with the increase in the activity of salicylic acid biosynthetic enzyme in rice seedling (Sawada et al. 2006). In an experiment conducted by Jayakannan et al. (2013), SA improved salinity tolerance in Arabidopsis by restoring membrane potential and preventing salt-induced K+ loss via a guard cell outward rectifying K(+) (GORK) channel. Arabidopsis seedling pretreated with SA showed upregulation of H+-ATPase activity, thereby improving K+ retention during salt stress. SA alleviates reduction in photosynthesis under salt stress by enhancing nitrogen and sulfur assimilation and antioxidants.
14.4.4 Antioxidant Regulation
Antioxidant metabolism, including various enzymatic and non-enzymatic antioxidants, plays crucial role in detoxifying ROS induced by salinity stress. Salinity tolerance in pearl millet is positively correlated with the activity of enzymatic antioxidants such as superoxide dismutase (SOD), catalase (CAT) glutathione peroxidase (GPX), glutathione reductase (GR) ascorbate peroxidase (APX), and non-enzymatic antioxidants like ascorbate, anthocyanin, tocopherols, and phenolic compounds (Heidari and Jamshidi 2011; Lakshmi et al. 2017; Khan et al. 2020). Anthocyanin accumulation is largely reported in cereals exposed to salt stress (Mbarki et al. 2018).
14.4.4.1 Case Study on the Response of Pearl Millet to Varying Levels of Salt Stress
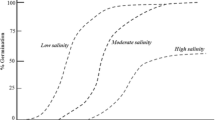
In a field study at ICRISAT, where pearl millet germplasms were screened under different ranges of soil salinity, breeding lines IP 19586 and HHVBC Tall were selected based on good growth and yield productivity (Toderich et al. 2018). While IP 19586 improved pearl millet germplasm line produced an average dry matter production of 27.6–35.0 t ha−1, the HHVBC Tall line was superior since it registered highest value of seed production up to 4.3–4.7 t ha−1. This implies that lines had different salt tolerance strategies at the seed germination stage. The germination rate gradually decreased from 0 to 400 mM NaCl, for the IP 19586 line. The HHVBC Tall line showed tolerance up to a high salinity level (300 mM NaCl), but it was sensitive to extra-high salinity (400 mM NaCl). For other pearl millet varieties, approximately 60% decreases in seed germination rates occurred with 1.5% NaCl treatment (Ali and Idris 2015). The study also revealed that controlled experiments allow for the investigation of plant resistance-related physiological and biochemical characteristics under ionic and osmotic stresses (Toderich et al. 2018). The study revealed the salt tolerance thresholds of pearl millet lines taken for investigation.
Salinity (combined effects of osmotic and ionic stresses) at 200 mM NaCl caused a significant twofold reduction in the growth parameters. Because a PEG-induced osmotic stress with a similar osmotic potential caused no decrease in these growth parameters, we believe the toxic actions of ions resulted in the growth reduction (Munns and Tester 2008). The HHVBC Tall line was more sensitive to ionic stress, having significantly higher proline content. Under extremely high salinity conditions, the most significant differences between the studied genetic lines were revealed. There was significant tissue dehydration in both lines, but especially in HHVBC Tall plants. Furthermore, the pigments significantly decreased in HHVBC Tall plants at 300 mM NaCl. For other P. glaucum varieties, significant reductions in pigment composition occurred at 100–200 mM NaCl (Sneha et al. 2013). The high proline and malondialdehyde (MDA) contents indicate the presence of high stress levels in HHVBC Tall plants under these conditions (Liu et al. 2014). The IP 19586 pearl millet germplasm line, which has a high biomass yield potential, is of interest as a forage resource for livestock in salt-affected agro-landscapes. Since soil salinization is negatively reflected in the grain production, the seed multiplication of the high-grain HHVBC Tall pearl millet line would guarantee stable seed production only in soils with low and medium levels of salinity. The study concluded that IP 19586 line of pearl millet should be used for maximal green biomass production under drought stress (0.6 MPa), while the HHVBC-Tall line should be used to produce high grain yields under low and medium salinity and drought (0.6 MPa) conditions. Depending on the timing and intensity of the salt stress, the harvest index varies from 0.2 to 0.5 (Munns et al. 2006). At the same time, low salinity levels may not reduce grain yield, even though the numbers of leaves, leaf area, and stover biomass are reduced. This may reflect a harvest index that increases with salinity or grain yield that does not decrease until a given (“threshold”) salinity is reached. The findings from the study indicated that the dry biomass yield can be used as a screening and selection criterion for evaluating salt-tolerance behaviors among a large collection of plant accessions. Based on their relative dry biomasses (DMs), it is possible to select the optimum genotypes for each salinity level.
14.5 Molecular Characterization of Salt Stress
Salinity stress triggers various other stresses like osmotic stress, dehydration, and oxidative stress. Disruption of physiological processes such as stomatal movement and reduction in gaseous exchange, chlorophyll content, photosynthetic efficiency etc. are consequences of salinity stress (Chaudhry et al. 2020, 2021). There are manifold changes at the molecular level resulting from salinity stress. Higher salt content in the root environment leads to poor availability of water and nutrients (Ali et al. 2021). High salt environment promotes reactive oxygen species production in plants consequently promoting the production of antioxidant defense components. The signals generated because of the reactive oxygen species are sensed by various molecular components are transmitted through the cell such that plants exhibit transcriptional and post-transcriptional modifications to cope up with the stress. Salt-induced oxidative stress and subsequent damage to cellular organelles and their functions are reported in various crop/plant species.
14.5.1 Molecular Basis of Salt Tolerance in Plants
Plants cope up with cellular changes through sensors and signaling cascades. In plants, the environmental cue to high salinity is first recognized by transmembrane sensors. High sodium concentration causes imbalance in homeostasis of other ions such as K+ and Ca2+ (Julkowska and Testerink 2015). It is reported that Ca2+ levels in cytosol are elevated within seconds of exposure to salinity (Knight et al. 1997). Ca2+ acts as a second messenger in stress signaling and Ca2+ ion concentration acts as one of the earliest steps in salt sensing (Lynch et al. 1989).
Only a few salt sensors have been identified so far in the plant kingdom and they include ROS-based and Na+/Ca2+ membrane transporters (Wu et al. 2021). There are Na+/H+ transporters at both the plasma membrane and tonoplast that enable efflux of Na+ ions out of the cell into the apoplastic space or to sequester them in the vacuole, respectively. The salt overly sensitive (SOS) signaling pathway is well characterized, which converts Ca2+ signaling to salt tolerance. SOS pathway is activated when plants are exposed to high levels of sodium chloride and the pathway helps in regulating osmotic homeostasis under high salinity conditions (Ishitani et al. 2000). When salt concentration in the soil increases, plants grown there tend to accumulate excessive sodium content which adversely affects various metabolic processes leading ultimately to poor plant growth. Optimum sodium levels are maintained in the cell by the coordinated action of several membrane bound transport proteins. Salt overly sensitive mutants were first identified in Arabidopsis thaliana in a genetic screen designed to identify the components of salt sensitivity/tolerance (Zhu et al. 1998). The key components of the SOS signaling pathway involved in Na+ extrusion have been identified as SOS3, SOS2, and SOS1. Arabidopsis plants with a mutation in SOS1 gene were highly sensitive to increased NaCl content in the growing medium and accumulated excessive Na+ in them when compared to the wild-type plants (Shi et al. 2000). Biochemically, SOS1 was identified as a Na+/H+ antiporter located in the plasma membrane (Shi et al. 2000; Qiu et al. 2002) that regulates salt content by mediating Ca2+-dependent microfilament re-organization (Zhao et al. 2011). Na+/H+ exchanger in the vacuolar membrane is also a target for the SOS pathway (Qiu et al. 2004). The expression of SOS1 is regulated by other members of the pathway namely, SOS2 and SOS3. SOS2 is a CBL-interacting protein kinase (CIPK) and SOS3, a (a Ca2+ sensor of the CBL family, Calcineurin B-like). The elevated levels of cytosolic Ca2+ stimulate Ca2+-dependent protein kinase complex (SOS2-SOS3) that phosphorylates and initiates the activity of SOS1. SOS pathway genes are found conserved across species and their presence has been reported in many species such as Brassica juncea, wheat, barley, grapes, and sugarcane. Calcium signal that is initiated as a result of high salinity stress is sensed by SOS3 which is a calcium sensor belonging to calcineurin B-like family. SOS3 along with SOS2 forms a protein complex which activates SOS1 (Na+/H+ antiporter) transcription through protein phosphorylation so that SOS1 regulates efflux of excess sodium ions (Guo et al. 2009). SOS3-SOS2 complex also activates vacuolar Na+/H+ exchanger (NHX) making vacuole accumulate excess Na+ ions.
Ion and nutrient transporters form an integral part of stress signaling. Ion transporters and channel proteins are most often regulated by Ca2+-driven phosphorylation (Kudla et al. 2018). Various receptor kinases are implicated in salinity tolerance. FERONIA (FER) is one such receptor kinase to salt stress identified in Arabidopsis (Feng et al. 2018). Fer is also a member in the chain of Ca2+ signaling and is involved in cell wall integrity restoration and root growth. Increased calcium deposition in the cortex and epidermis of the roots stimulates Ca2+calmodulin-dependent kinases and plasma membrane ATPase along with triggering ROS accumulation. High ROS accumulation and Ca2+ levels regulate the release of the stress hormone abscisic acid and initiate transcriptional changes (Jiang et al. 2012).
K+ is the most abundant cation that is accumulated in cytosol. It is an important nutrient for plant growth, whereas high concentrations of sodium ions are harmful. Plants grown in saline environments will tend to have a high Na+/K+ ratio which is toxic to plants, and they attempt to maintain a high K+/Na+ concentration in the cytosol by regulating Na+, K+, and H+ transporters. High affinity K+ transporters (HKTs) play significant role in maintaining a low Na+/K+ ratio (Hauser and Horie 2010). Na+/K+ transporters, hence, are important candidates in salt signaling and tolerance. Members of this group of genes have been identified from many species and Gmhkt gene from soybean imparted salt tolerance in transgenic tobacco plants (Chen et al. 2011).
One of the immediate effects of high salinity is the production of reactive oxygen species as discussed above. As high levels of them are toxic to the organism in multiple ways, antioxidant mechanism is activated to counter-act the effects. There are various antioxidant enzymes present in plants, some of which had proven roles in tackling salt stress. On overexpressing tobacco ascorbate peroxidase, the transgenic tobacco plants had salinity tolerance (Badawi et al. 2004). Late embryogenesis abundant protein (LEA), that are commonly associated with dehydration stress and cold stress tolerance were also found to have role in salinity stress tolerance (Checker et al. 2012; Chen et al. 2016). Glyoxalase pathway genes (GlyI and GlyII) have substantial, well-characterized roles in salinity tolerance (Singla-Pareek et al. 2003). These genes are involved in de-toxification of ROS and this pathway is ubiquitous in plants and animals. Their effectiveness has been proved in multiple crops through over-expression and transgenic development (Singla-Pareek et al. 2003; Bhomkar et al. 2008; Álvarez Viveros et al. 2013).
14.5.2 Transcription Factors in Salt Tolerance Mechanism
Transcription factors are major players in regulation of gene expression in any kind of stresses in plants. They bind to the cis-acting elements in the promoter regions of stress-responsive genes and regulate their expression. Potentially important transcription factors are generally identified by comparing the transcriptomes of stressed and non-stressed tissues. Various transcription factors responding to salt stress have been identified from many plant species and many have been functionally characterized. bZIP genes were upregulated in wheat genotypes under long-term salinity stress (Johnson et al. 2002). In Arabidopsis, salt stress induced the expression of AtWRKY8, and its target gene was identified as RD29A, a gene highly induced under various types of abiotic stress including salinity (Hu et al. 2013). NAC transcription factor also plays role in imparting salinity tolerance in crops like rice and wheat proved by overexpressing the gene (Song et al. 2011; Nakashima et al. 2007).
As the DREB1A/CBF3 gene from Arabidopsis thaliana was constitutively expressed in citrus and peanut, the transgenic events in both these plants showed tolerance to high salinity (Alvarez-Gerding et al. 2015; Sarkar et al. 2014). Erianthus DREB2 when expressed in sugarcane the resultant plants were better performers at drought and high-salinity conditions (Augustine et al. 2015). OsEREBP2 transcripts were increased after high salinity treatment and hence was a probable candidate for salt tolerance in rice. OsEREBP2 was found to bind the promoter region of a receptor-like kinase, OsRMC which negatively regulated salt stress response in rice (Serra et al. 2013). MYB transcription factors are also found to play roles in salinity tolerance in plants and it was proven by overexpression of OsMYB91 and OsMYB48-1 in rice which resulted in upregulation of salt-stress responsive genes (Zhu et al. 2015; Xiong et al. 2014). To understand the functional contribution of these transcription factors, the common approaches adopted are generation of mutant and overexpression of the gene of interest in both crop species and in model crops like Arabidopsis and tobacco. A transcription factor, SERF1 (Salt-Responsive ERF1) identified in rice showed a root-specific induction under high salt conditions and loss of this gene weakened the MAPK (mitogen-activated Protein Kinase) pathway and salt responsive transcription factors downstream of it (Schmidt et al. 2013). OSBZ8, a bZIP class of ABRE binding transcription factor, was highly expressed in salt tolerant rice genotypes than in susceptible one (Mukherjee et al. 2006). MdMYB46 transcription factor from apple was identified to enhance salt and osmotic tolerance in apple and the RNAi apple lines developed indicated that this factor increased salt tolerance (Chen et al. 2019).
14.5.3 Transgenics for Enhancing Salinity Tolerance
Cis-genic or transgenic expression of salt stress tolerance-related candidate genes characterizes them functionally and is often used to improve the trait in crop plants. SOS pathway genes alone or in combination have imparted salt tolerance in transgenic plants (Shang et al. 2012; Ma et al. 2014). Overexpression of NHX homologues from Arabidopsis thaliana was reported to improve salinity tolerance in major crops (Xue et al. 2004; Asif et al. 2011). Other Na+/H+ antiporters also were good candidates to impart salt tolerance. Vacuolar Na+/H+ antiporter from pokkali rice imparted resistance to salt stress when overexpressed (Amin et al. 2016). Transgenic expression of Na+/H+ antiporters from different species such as barley, Arabidopsis, wheat, and soybean showed promising results (Bayat et al. 2010; Apse et al. 1999; Chen et al. 2008, 2011). Expression of microbial genes like mtlD (mannitol biosynthesis), catalase (ROS neutralization), betA (from E. coli), and hal1 (yeast salt tolerance gene) that are involved in osmo-tolerance in plants have been attempted to develop salinity tolerant genotypes (Abebe et al. 2003; Bhattacharya et al. 2004).
14.5.4 Understanding the Molecular Basis of Salinity Tolerance in Pearl Millet
Development of salinity tolerant genotypes is vital in the given scenario of climate change events, that is occuring globally. Apart from traditional plant breeding approaches, modern molecular biology tools are relevant and are to be used to harness maximum benefits. Whole genome reference for pearl millet that was developed in the recent years has provided momentum to pearl millet genomic research lately (Varshney et al. 2017). Several candidate genes relevant to salinity stress have been identified and cloned from pearl millet and many of them were functionally characterized.
A great extent of diversity towards salinity tolerance is found in various germplasms of pearl millet, at both genotypic and phenotypic levels (Liu et al. 2014). The local indigenous landraces grown in arid regions can survive better in the adverse climatic conditions and are known to be superior to the newly bred cultivars in terms of abiotic stress-tolerance. Therefore, they can represent valuable plant genetic resources and donor germplasm for providing new variations and stress adaptation traits. Some unique genes from pearl millet have already been employed to develop salt-tolerance in rice and groundnut (Santosh Rama Bhadra Rao et al. 2017). It is imperative to analyze the molecular mechanism of salt-tolerance in this critical hardy plant, in order to dig out more such proteins for developing stress-tolerance in the sensitive crop species through tools of biotechnology. Proteomics also represents an essential and complementary approach in the field of abiotic stress. Studies in various germplasm collection of pearl millet have also characterized for salinity tolerance at morphological and physio-biochemical level (Jha 2022) and at the proteomic level where salt susceptible accessions had >25% downregulation of proteins under salinity, while salt tolerant accessions had almost 50% unaltered protein (Jha 2022).
Vacuolar antiporters like Na+/H+ antiporter actively sequester excess Na+ into the vacuoles using the proton motive force developed by ATPases and PPases. These transporters are thereby playing a vital role in regulation of effects of excess salinity. Rajagopal et al. (2007) reported the isolation and characterization of an isoform of vacuolar Na+/H+ antiporter from Pennisetum glaucum (PgNHX1) and through transgenic expression of the gene in Brassica juncea, they proved its role in salinity tolerance (Rajagopal et al. 2007). The same gene had been transformed into rice also and PgNHX1 overexpressing rice plants had high salinity tolerance. The transgenic plants had extensive root system in comparison to the wild type and they set flowers and seeds and completed their life cycle even at high salt concentrations of 150 mM NaCl. The toxicity effects of high ionic concentration during salt stress are alleviated by the combined action of several proteins including ATPases. They are proton pumps that generate the proton motive force required for ion transport, vacuolar ATPases being one among them. A subunit of Vacuolar ATPase gene was cloned from pearl millet along with its promoter and its expression pattern was studied (Tyagi et al. 2005). This gene was highly upregulated under salinity stress, high ABA and calcium and salicylic acid. Its promoter harbored DRE and ABRE elements. More isoforms of the c sub-unit of vacuolar ATPases were cloned and studied later (Tyagi et al. 2006). Bhaskaran and Savithramma (2011) co-expressed a vacuolar H+ pyrophosphatase gene from A. thaliana and PgNHX1, a vacuolar Na+/H+ antiporter gene from pearl millet in tomato plants to enhance their salt tolerance. Transgenics with both the genes had enhanced tolerance to salinity than single gene transformed plants. The dual gene transgenic plants could survive well even at 200 mM NaCl concentrations by enhanced synthesis of chlorophyll and proline contents. These plants could survive in high salt conditions by sequestering the excess Na+ ions into their vacuoles thereby reducing its toxic effects.
A gene encoding voltage-dependent anion channel from Pennisetum glaucum (PgVDAC) was identified from among the genes that are regulated under salt stress and was cloned and characterized (Desai et al. 2006). Expression analysis showed its upregulation under salinity, low temperature, and desiccation, while it did not respond to ABA application. Rice plants that had constitutive expression of PgVDAC could not survive, while mild levels of upregulation imparted adaptiveness to salinity stress. Late embryogenesis abundant (LEA) family members are known to contribute towards adaptation to desiccation and osmotic stress. A cDNA clone coding for group 7 LEA was identified in pearl millet (PgLEA) which when expressed in E. coli showed enhanced tolerance to high temperature and salinity (Reddy et al. 2012). Another late embryogenesis protein, dehydrin (PgDHN) cloned from pearl millet, was induced by high salinity and conferred salt tolerance to pearl millet plants by accumulating in their leaves (Shinde et al. 2018). A NAC transcription factor from pearl millet (PgNAC21) was shown to be induced by salinity stress and ABA treatment (Shinde et al. 2019). NAC proteins are generally highly induced under various abiotic stress conditions. PgNAC21 had ABA responsive elements and MYB factor binding sites in its promoter and MYB binding was proved by a yeast-hybrid assay. Arabidopsis plants overexpressing PgNAC21 had high salinity apart from many other attributes such as overexpression of stress-regulated genes such as GSTF6 (GLUTATHIONE S-TRANSFERASE 6), COR47 (COLD-REGULATED 47), and RD20 (RESPONSIVE TO DEHYDRATION 20).
MicroRNAs are small regulatory RNA sequences of 20–22 bp length having multiple roles in organism development and functions. Shinde et al. (2020) identified 81 conserved and 14 novel miRNAs from a salinity tolerant pearl millet genotype that were salinity responsive. Of these, 30 miRNAs were upregulated and 51 were downregulated under salt stress. Among the target mRNAs of these miRNAs, 25% encoded transcription factors. Majority of the targets identified for these miRNAs were transcription factor and members belonging to ion transporters, protein kinases, detoxification proteins, and heat shock proteins which are involved in salinity tolerance mechanisms.
A comprehensive understanding of the salinity tolerance mechanism in pearl millet was brought out by comparative transcriptome analysis of contrasting genotypes for salinity tolerance, ICMB081 (susceptible) and ICMB 01222 (tolerant) (Shinde et al. 2018). Their study revealed a higher growth rate and a higher leaf sugar accumulation in the tolerant line under salt stress. The most conspicuous differentially expressed genes that were upregulated in tolerant line included genes of ubiquitin-mediated proteolysis and phenyl propanoid pathways and genes from glycolysis/gluconeogenesis pathways. The candidate genes identified in various studies are promising tools for functional characterization of salinity stress tolerance and for employing as donors for transgenics or genome editing.
14.5.5 High-Throughput Approaches for Phenotyping Salt Stress Tolerance in Pearl Millet
Developing quantifiable high-throughput phenotyping (HTP) approaches are the prerequisite to eliminate the bottlenecks in identifying the genetic gains with precision and accelerating the breeding programs. HTPs include employing aerial and ground-based platforms to phenotype the crop behavior (traits) throughout their growth stages under abiotic or biotic stresses. The HTP platforms applies the imaging techniques (satellite imaging, high pixel mobile camera, drone imaging, robot imaging), light sensors, remote sensing, mobile applications that are non-destructive, non-invasive with different levels of automation. These techniques offer wide range of advantages where data can be recorded over a large area with high resolution, are easy to operate and portable, lesser operational cost, and fewer cons like battery capacity, suitability over all areas, high initial costs, weather hindrance, and data interpretation and image quality.
Hence, an in-depth understanding of plant stress is pivotal for improving yield protection for sustainable production systems (Pessarakli 2019). Plant scientists rely on crop phenotyping for precise and reliable trait collection and utilization of genetic resources and tools to accomplish their research goals. Plant phenotyping is defined as the comprehensive assessment of complex traits of plants such as development, growth, resistance, tolerance, physiology, architecture, yield, ecology, and the elementary measurement of individual quantitative parameters that form the foundation for complex trait assessment (Li et al. 2014). Breeding programs generally aim to phenotype large populations for numerous traits throughout the crop cycle (Sandhu et al. 2021a, b). Low-throughput phenotyping has high cost, laborious, destructive, less accurate, low resolution, and environment inference. The development of high-throughput phenotyping (HTP) has largely overcome the problems crop stress phenotyping due to the automation, imaging techniques, and artificial intelligence. HTP has offered great potential for non-destructive and effective field-based plant phenotyping. Under the HTP, manual, semi-autonomous, or autonomous platforms equipped with single or multiple sensors record temporal and spatial data, resulting in storage of large amounts of data and analysis (Kaur et al. 2021; Sandhu et al. 2021b). Several HTP platforms exist and are presently employed to phenotype different abiotic stress-associated traits in various crops (Table 14.1).
These HTP platforms include several imaging platforms, techniques (satellite imagery, mobile camera, Drone imaging, robot imaging), sensors, analytical tools that include indices and formulas (Stress tolerance index, tolerance index, salt tolerance, NDVI, Green leaf index, chlorophyll index), machine and data learning. The outcome of the phenotypic data encompasses a large amount of physiological, morphological, and biochemical data to identify stress tolerant genotypes by assessing their traits under varying environments. The major challenge here is handling the data collected from different formats of platform, interpreting the spectral images, environmental interferences, which have to be effectively optimized for accurate results and cost-effective phenotyping. These approaches also need to consider the performance of crops under multi-locational trials.
14.6 Breeding Strategies for Salt Stress Tolerance in Pearl Millet
Pearl millet and its wild relatives are rated to be fairly tolerant to salinity (Ashraf and McNeilly 1987) and can be more profitably grown in saline soils. Large genotypic variation was reported to exist in pearl millet for salinity response in terms of whole plant response (Ashraf and McNeilly 1987, 1992; Dua 1989). Moreover, availability of high levels of tolerance in other species of Pennisetum (Ashraf and McNeilly 1987, 1992; Muscolo et al. 2003) and within the P. glaucum (Dua 1989) offers a scope for understanding the traits related to tolerance and to integrate these tolerant crop species/genotypes into appropriate management programs to improve the productivity of the saline soils. The differential responses of plants toward salinity stress rely upon their genetic make-up and the environment. Therefore, screening a large number of genotypes is essential to select the superior genotypes with greater stress tolerance. The candidate genes could be identified from those potential genotypes, and transferred to other salt-sensitive crops by plant breeding or transgenic approaches (Jha 2019). Germplasm screening for salinity stress tolerance has been performed in several plant species, namely rice, wheat, maize, and sorghum (Morton et al. 2019), but only a few candidate genes have been identified for stress tolerance, owing to the complex nature of salinity stress (Jha 2018; Lakra et al. 2018). Large genotypic variation has been observed in pearl millet toward salinity stress tolerance. A wide range of pearl millet breeding lines has been evaluated extensively for salt tolerance (Krishnamurthy et al. 2007a, b; Toderich et al. 2018). Typically, the landraces and wild relatives of a crop species exhibit genetic diversity and are known to harbor novel genes for environmental adaptation and other agronomic important traits. Therefore, these genotypes can be used as valuable genetic resources for developing abiotic stress tolerance (Hoang et al. 2016; Quan et al. 2018). Despite having a wide genetic diversity and a large germplasm collection available at the national repositories, limited reports are available for the identification and selection of superior genotypes for abiotic stress tolerance in pearl millet (Shivhare and Lata 2017).
Various traits of the plants’ adaptability under salinity stress conditions have been reported from several studies. Genomic approaches and HTP phenotyping provide new insights and create a pipeline to support breeders to overcome salinity stress for crop improvement. Plants cope with salinity stress using various mechanisms, and these mechanisms can be exploited using strategies as mentioned above. Approaches like mutation breeding, wild relative exploration, Marker assisted breeding (MAB), Double haploid coupled with novel approaches like CRISPRCas9 technology and GWAS (Genome Wide Association Studies) have resulted in developing salinity tolerant crops including millets. In pearl millet, the essential genomic resources are available, although this information is limited in other millets. Pearl millet has a high level of DNA marker polymorphism between elite inbred parental lines of popular hybrids (Vadez et al. 2012). Information on the DNA markers (AFLP, RFLP, RAPD, expressed sequence tag-based (EST) markers, sequence-tagged sites (STSs), simple sequence repeat (SSRs/microsatellites), DArTs, CISP and SNP) for identification of QTLs are already available to develop salt tolerant lines and other abiotic stresses based on the molecular and biochemical bases coupled to higher crop yield. Based on this available marker information, gene linkage map has been constructed which is about 700 cM and map QTLs for salt tolerance (Singh et al. 2016; Sharma et al. 2014). Also, a final genetic map in pearl millet is about 716.7 cM with 23.23/cM overall average density of SNPs and 1.66 unique linkage bins per cM (Vadez et al. 2012).
Screening of pearl millet germplasm has resulted in the development of advanced breeding materials, improved population including OPVs, gene pools and composites, parental lines of potential hybrids, and germplasm accessions with high grain and forage yield presumably with a high degree of salinity tolerance. The available genotypes for salinity stress tolerance in pearl millet documented and reported by Shivhare and Lata (2017) include ICMB 02111, ICMB 94555, ICMB 95333, ICMB 00888, ICMB 01222, ICMP 451, IP 3732, IP 3757, IP8210, and PRLT 2/89-33, 10876 and 10878 (Sudan), 18406 and 18570 (Namibia), and ICMV93753 and ICMV 94474 (India); 863-B, CZI 98-11, CZI 9621, HTP 94/54. These available materials can be released for cultivation after extensive validation of their yield performances at on-farm trials. “HASHAKI I,” a salt tolerant pearl millet variety, has been identified in Uzbekistan in 2012 as a high-forage variety for salt-affected areas. The identified salinity-tolerant pearl millet lines should be utilized in breeding programs to develop salinity-tolerant locally adapted cultivars (both OPVs and hybrids) (Yadav et al. 2012).
With respect to the functional validation of genes and QTLs, voltage-dependent anion channel gene VDAC (Desai et al. 2006), formation of LEA genes (Reddy et al. 2012), PgNHX1 Vacuolar Na+/H+ transporter gene (Verma et al. 2007), and reduced salt uptake DT-QTL at linkage group 2 (Sharma et al. 2011, 2014) are a few of them reported to confer salt tolerance. But till date, only few efforts have been aligned in the area of transcriptome analysis and transgenic approach in pearl millet crop for delineating the mechanism of abiotic stress tolerance (Choudhary et al. 2021). Hence, development of recombinant pearl millet varieties is still in nascent stage despite its economic importance and hence calls for a concerted effort for generation and evaluation of transgenic lines under various stress conditions. Also, in an effort to develop whole genome information, several organizations are taking a consensus effort to sequence the pearl millet genome, to augment breeding programs and generate ample gene resources. In regard to this, Tift 23DB2B1 has been chosen as a global pearl millet reference genotype, to develop its draft genome sequence through whole genome shotgun and bacterial artificial chromosome (BAC) sequence approaches.
14.7 Salinity Management Practices and Recent Advances for Stress Tolerance
Salinity stress affects the physiological and biochemical processes of the plant, which results in reduced seed germination, growth, and yield in many crops including pearl millet. Different management approaches are needed to mitigate the adverse effects of salinity stress to enhance the production of pearl millet. Identification of salt-tolerant genotypes along with appropriate site-specific crop management practices would increase pearl millet productivity in saline soils. Recent approaches in use of plant growth promoting rhizobacteria and advanced genetic and molecular techniques proved effective in reducing the effects of salinity stress in pearl millet.
14.7.1 Agronomic Approaches
Agronomic approaches involving the use of soil amendments (chemical and organic) will reclaim the saline soils and reduce the adverse effects of salinity on crop production. In pearl millet, application of potassium, phosphorus, gypsum, silicon & boron and organic amendments e.g., manure, biochar, compost, and crop residues have been used to improve plant growth under salinity stress. The concentration of sodium is increased in saline conditions resulting in imbalanced uptake and accumulation of essential nutrients. Increase in Na+ content will reduce Ca2+ and K+ content in saline soils. Application of K fertilizer reduces the adverse effects of salinity through its role in stomatal regulation, osmoregulation, protein synthesis, and energy status. Application of potassium in saline soil significantly increased the grain yield and resulted in decreased sodium and increased potassium content in leaves of pearl millet (Heidari and Jamshidi 2011). Application of silicon and potassium humate resulted in better growth parameters and biochemical components under saline conditions (Hassanein et al. 2017). Boron is an important element for many biochemical and physiological reactions of plants (Sezer 2014). Boron application alleviated the negative effect of salinity and improved grain yields in pearl millet (Salem 2020). Boron improved potassium concentration and maintained membrane integrity. Boron (B) application reduces the toxicity of aluminum by stabilizing integrity of the proteins, controlling the activities of antioxidant enzymes and secondary metabolites and lowering reactive oxygen species and Al concentrations (Riaz et al. 2018).
Organic amendments proved as an effective strategy for saline soil amelioration. Organic amendments improve soil chemical and physical properties. Solid waste, vermicompost, and cow dung influence soil salinity and alleviate its adverse effects on the growth of plants by changing the physico-chemical properties of soil. Application of organic amendments improved the growth performance of pearl millet in saline conditions (Diatta 2016; Araújo et al. 2022). Biochar also improved physico-chemical properties of soil, including soil cation exchange capacity, pH, water holding capacity, surface area, and soil structure under abiotic stresses (Bamminger et al. 2016). Biochar application improved potassium availability uptake and decreased sodium availability and uptake under salt stress. It enhanced the soil quality, availability and concentrations of nutrients in plant, and chlorophyll synthesis, on the other hand it reduced Na, Cl, and proline in the leaf tissue (Ding et al. 2022). It is concluded that exogenously applied organic matters such as plant residues, manure, a by-product of municipal or farming activities, etc. are an efficient and feasible way to mitigate the effects of salinity on plant growth and soil health (Meena et al. 2018).
14.7.2 Seed Priming
Seed priming is one of the most economical and easiest technique for successful crop production under salinity stress conditions. It is defined as a pre-sowing technique of treating seeds with various priming agents like water or other chemicals to enhance the germination percentage. It hastens the germination rate and improves seedling establishment in crops like maize, pearl millet, and wheat. To hasten the germination rate, seedling establishment, and crop yields, a number of priming procedures have been developed, including hydropriming (soaking seed in water), osmopriming (soaking seed in nutrients, hormones, or chemicals), and halopriming (soaking seed in salt solution). The positive effect of seed priming arises due to synthesis of certain germination-promoting substances, enhancing pre germination metabolites, early DNA replication, greater ATP availability, enzyme activation, osmotic adjustments, and membrane reorganization through restoring their original structures and reducing leakage of metabolites (Paparella et al. 2015). Primed seeds exhibit reduced photo and thermal dormancy, a wider range of germination temperatures, and a superior ability to deal with weeds and pests in addition to synchronous and quick emergence. Several research findings evidenced the role of seed priming to improve salt stress tolerance in pearl millet. Zida et al. (2017) observed the positive effect of hydro priming on germination rate under saline conditions. Many researchers reported an increase in grain yield to the tune of 13–30% by soaking pearl millet seeds for 8–16 h in water (Jidda and Anaso 2017). Khan et al. (2020) reported that seed priming with silver nanoparticles at 20 mM improved the growth attributes and antioxidant enzyme activities of pearl millet by reducing oxidative damage, and increased the salinity tolerance by reducing Na+ uptake and maintaining the Na+/K+ ratio. Priming pearl millet seeds with salicylic acid @100 μM for 8 h can improve the germination (%) and seedling vigor of the genotypes under higher salinity level (Anju et al. 2019).
14.7.3 Plant Growth-Promoting Rhizobacteria (PGPR) to Ameliorate Salinity Stress
Bacteria used for promoting plant growth are termed as plant growth-promoting rhizobacteria. These are rhizospheric or endophytic bacteria that colonize the rhizosphere and enhance germination, root and shoot length, increase uptake of minerals, and yield, and enhance tolerance against drought and salinity stress (Lugtenberg and Kamilova 2009). A variety of PGPR genera such as Azospirillum, Aeromonas, Acetobacter, Achromobacter, Pseudomonas, Bacillus, Flavobacterium, Chryseobacterium, Sinorhizobium, Bradyrhizobium, etc. have been identified for maintaining the growth of various crop plants grown under high salt environment (Etesami and Maheshwari 2018). PGPR plays a major role in imparting salt tolerance to plants (Paul and Lade 2014; Srivastava and Kumar 2015; Qin et al. 2016). Application of halotolerant bacteria will be advantageous for improving the crop yields in salt affected areas. Mechanisms underlining the ability by which bacteria encourage plant growth and avert damage induced by high salt concentration in the soil include phytohormones production such as indoleacetic acid (IAA), gibberellic acid, cytokinins, and ethylene (Spaepen et al. 2009; Mishra et al. 2010); synthesis of enzyme ACC deaminase to diminish the content of ethylene in the roots of developing plants (Dey et al. 2004); synthesis of osmoprotectants (Berg et al. 2013); solubilization of minerals, like phosphorus and potassium; and modulation of antioxidant enzymes (Kohler et al. 2009). Kushwaha et al. (2020) reported that pearl millet plants treated with bacillus strain resulted in significantly higher plant growth compared to untreated seed grown under saline environment (200 mM NaCl). Kayasth et al. 2014 reported Gordonia sp. a salt tolerant bacterial inoculant for growth promotion of pearl millet under saline soil conditions. Khushdil et al. (2019) reported that pearl millet plants inoculated with Aspergillusterreus improves the NaCl tolerance in pearl millet by ameliorating the physicochemical attributes of the host plants.
14.7.4 Application of Hormones
Plant hormones, namely auxins, gibberellins, cytokinins, abscisic acid, and salicylic acid, are produced within the plant at very low content and have the potential to regulate plant development and ameliorate the effects of various biotic and abiotic stresses including salt stress. Application of hormones in pearl millet elevates osmotic adjustment to maintain turgor, improve nutrient uptake, accumulate antioxidants, and detoxify reactive oxygen species, thereby maintaining membrane and enzyme stability under stress conditions (Kaya et al. 2010). Abscisic acid (ABA) is a signaling molecule that mediates the responses to salt stress (Knight and Knight 2001; Nishiyama et al. 2011), and is considered a vital signal of salt tolerance because of its rapid biosynthesis and significant accumulation in plant cells upon exposure to salt stress conditions. The ameliorations effects on salinity stress were reported in other millet crops. Addition of abscisic acid (ABA) to the induction medium containing 200 mM NaCl improved salt tolerance of finger millet over those without ABA in association with the appearance of several ABA-responsive proteins (Uma et al. 1995). Hussain et al. (2010) and Yadav et al. (2020) reported that application of salicylic acid proved beneficial to mitigate adverse effects of salt stress by significantly improving physiological traits, biochemical traits, and ultimately improved grain yield in pearl millet. Salem (2020) reported that plant height (cm), panicle length (cm), panicles number/m2, grain weight/panicle (g), seed index (g) and grain protein content percentage as well as biological, grain, and protein yields (kg/fed) were significantly increased by salicylic acid (SA) under saline soil conditions.
14.8 Conclusion and Future Perspectives
Though pearl millet is a crop with inbuilt capacity to withstand harsh environments and therefore is capable of cultivation in saline lands for grain and forage production, but still salinity stress acts as a significant abiotic constraint for its cultivation in several areas. There is a significant decline in the growth and yield of pearl millet crop in saline areas, which eventually affects its productivity. As compared to other cereal crops only limited information has been available on response to soil salinity in pearl millet. Efforts should be focused on collection and characterization of pearl millet germplasm so that potential sources of genetic variation for salinity tolerance could be identified and incorporated in crop improvement programs. A concerted effort is also needed to develop and upgrade phenotypic screens for abiotic stress tolerance. Unravelling the whole genome sequence will help in further crop improvement programs and ideotype development.
References
Abebe T, Guenzi AC, Martin B, Cushman JC (2003) Tolerance of mannitol-accumulating transgenic wheat to water stress and salinity. Plant Physiol 131(4):1748–1755. https://doi.org/10.1104/pp.102.003616
Ali S, Idris A (2015) Germination and seedling growth of pearl millet (Pennisetum glaucum L.) cultivars under salinity conditions. Int J Plant Res 1:1–5
Ali AYA, Ibrahim MEH, Zhou G, Nimir NEA, Jiao X, Zhu G, Elsiddig AMI, Suliman MSE, Elradi SBM, Yue W (2020) Exogenous jasmonic acid and humic acid increased salinity tolerance of sorghum. Agron J 112:871–884
Ali M, Afzal S, Parveen A, Kamran M, Javed MR, Abbasi GH, Malik Z, Riaz M, Ahmad S, Chattha MS et al (2021) Silicon mediated improvement in the growth and ion homeostasis by decreasing Na+ uptake in maize (Zea mays L.) cultivars exposed to salinity stress. Plant Physiol Biochem 158:208–218
Altus D, Canny M (1982) Loading of assimilates in wheat leaves. I. The specialization of vein types for separate activities. Funct Plant Biol 9:571
Álvarez Viveros MF, Inostroza-Blancheteau C, Timmermann T et al (2013) Overexpression of GlyI and GlyII genes in transgenic tomato (Solanum lycopersicum Mill.) plants confers salt tolerance by decreasing oxidative stress. Mol Biol Rep 40:3281–3290. https://doi.org/10.1007/s11033-012-2403-4
Alvarez-Gerding X, Espinoza C, Inostroza-Blancheteau C, Arce-Johnson P (2015) Molecular and physiological changes in response to salt stress in Citrus macrophylla W plants overexpressing Arabidopsis CBF3/DREB1A. Plant Physiol Biochem 92:71–80
Amin USM, Sudip B, Elias Sabrina M, Samsad R, Taslima H, Richard M, Seraj Zeba I (2016) Enhanced salt tolerance conferred by the complete 2.3 kb cDNA of the rice vacuolar Na+/H+ antiporter gene compared to 1.9 kb coding region with 5′ UTR in transgenic lines of rice. Front Plant Sci 7:14. https://doi.org/10.3389/fpls.2016.00014
Anju UL, Doddagoudar SR, Pattanashetti SK, Gowda B, Vijaykumar K (2019) Influence of seed priming on seed germination, seedling growth, peroxidase activity, proline and total soluble sugar content of pearl millet (Pennisetum glaucum L.) under salinity stress. IJCS 7(5):508–514
Apse MP et al (1999) Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science 285(5431):1256–1258
Araújo CDA, Lira JBD, Magalhães ALR, Silva TGFD, Gois GC, Andrade APD, Araújo GGLD, Campos FS (2022) Pearl millet cultivation with brackish water and organic fertilizer alters soil properties. Ciência Animal Brasileira 22
Asaari MSM, Mertens S, Dhondt S et al (2019) Analysis of hyperspectral images for detection of drought stress and recovery in maize plants in a high-throughput phenotyping platform. Comput Electron Agric 162:749–758
Ashraf M, McNeilly T (1987) Salinity effects on five cultivars/lines of pearl millet (Pennisetum americanum [L] Leeke). Plant Soil 103:13–19
Ashraf M, McNeilly T (1992) The potential of exploting variations in salinity tolerance in pearl millet (Pennisetum americanum [L] Leeke). Plant Breed 104:234–240
Asif MA, Zafar Y, Iqbal J, Iqbal MM, Rashid U, Ali GM et al (2011) Enhanced expression of AtNHX1, in transgenic groundnut (Arachis hypogaea L.) improves salt and drought tolerance. Mol Biotechnol 49(3):250–256. https://doi.org/10.1007/s12033-011-9399-1
Augustine SM, Narayan JA, Syamaladevi DP, Appunu C, Chakravarthi M, Ravichandran V, Tuteja N, Subramonian N (2015) Overexpression of EaDREB2 and pyramiding of EaDREB2 with the pea DNA helicase gene (PDH45) enhance drought and salinity tolerance in sugarcane (Saccharum spp. hybrid). Plant Cell Rep 34(2):247–263
Badawi GH, Kawano N, Yamauchi Y, Shimada E, Sasaki R, Kubo A, Tanaka K (2004) Over-expression of ascorbate peroxidase in tobacco chloroplasts enhances the tolerance to salt stress and water deficit. Physiol Plant 121(2):231–238
Bamminger C, Poll C, Sixt C, Högy P, Wüst D, Kandeler E et al (2016) Short-term response of soil microorganisms to biochar addition in a temperate agroecosystem under soil warming. Agric Ecosyst Environ 233:308–317
Bayat F, Shiran B, Belyaev DV, Yur’eva NO, Sobol’kova GI, Alizadeh H et al (2010) Potato plants bearing a vacuolar Na+/H+ antiporter HvNHX2 from barley are characterized by improved salt tolerance. Russ J Plant Physiol 57:696–706
Beauchêne K, Leroy F, Fournier A et al (2019) Management and characterization of abiotic stress via PhénoField®, a high-throughput field phenotyping platform. Front Plant Sci 10:904
Berg G, Alavi M, Schmidt CS, Zachow C, Egamberdieva D, Kamilova F, Lugtenberg B (2013) Biocontrol and osmoprotection for plants under salinated conditions. Mol Microb Ecol Rhizosphere 1:561–573
Bhaskaran S, Savithramma DL (2011) Co-expression of Pennisetum glaucum vacuolar Na+/H+ antiporter and Arabidopsis H+-pyrophosphatase enhances salt tolerance in transgenic tomato. J Exp Bot 62(15):5561–5570. https://doi.org/10.1093/jxb/err237
Bhattacharya RC, Maheswari M, Dineshkumar V, Kirti PB, Bhat SR, Chopra VL (2004) Transformation of Brassica oleracea var. capitata with bacterial betA gene enhances tolerance to salt stress. Sci Horticult 100:215–227
Bhomkar P, Upadhyay CP, Saxena M, Muthusamy A, Prakash NS, Poggin K, Hohn T, Sarin NB (2008) Salt stress alleviation in transgenic Vigna mungo L. Hepper (blackgram) by overexpression of the glyoxalase I gene using a novel Cestrum yellow leaf curling virus (CmYLCV) promoter. Mol Breed 22:169–181
Blummel M, Zerbini E, Reddy BVS, Hash CT, Bidinger F, Khan AA (2003) Improving the production and utilization of sorghum and pearl millet as livestock feed: progress towards dual-purpose genotypes. Field Crops Res 84:143–158
Busemeyer L, Mentrup D, Möller K, Wunder E, Alheit K, Hahn V et al (2013) BreedVision—a multi-sensor platform for non-destructive field-based phenotyping in plant breeding. Sensors 13(3):2830–2847
Cabot C, Sibole JV, Barceló J, Poschenrieder C (2009) Abscisic acid decreases leaf Na+ exclusion in salt-treated Phaseolus vulgaris L. J Plant Growth Regul 28(2):187–192
Chai YY, Jiang CD, Shi L, Shi T, Gu W (2010) Effects of exogenous spermine on sweet sorghum during germination under salinity. Biol Plant 54:145–148. https://doi.org/10.1007/s10535-010-0023-1
Chaudhry UK, Gökçe ZN, Gökçe AF (2020) Effects of salinity and drought stresses on the physio-morphological attributes of onion cultivars at bulbification stage. Int J Agric Biol 24(6):1681–1689. https://doi.org/10.17957/IJAB/15.1611
Chaudhry UK, Gökçe ZNÖ, Gökçe AF (2021) Drought and salt stress effects on biochemical changes and gene expression of photosystem II and catalase genes in selected onion cultivars. Biologia 76:1–15. https://doi.org/10.1007/s11756-021-00827-5
Checker VG, Chhibbar AK, Khurana P (2012) Stress-inducible expression of barley Hva1 gene in transgenic mulberry displays enhanced tolerance against drought, salinity and cold stress. Transgenic Res 21:939–957. https://doi.org/10.1007/s11248-011-9577-8
Chen LH, Zhang B, Xu ZQ (2008) Salt tolerance conferred by overexpression of Arabidopsis vacuolar Na+/H+ antiporter gene AtNHX1 in common buckwheat (Fagopyrum esculentum). Transgenic Res 17:121–132
Chen H, He H, Yu D (2011) Overexpression of a novel soybean gene modulating Na+ and K+ transport enhances salt tolerance in transgenic tobacco plants. Physiol Plant 141(1):11–18. https://doi.org/10.1111/j.1399-3054.2010.01412.x
Chen H, Liu L, Wang L, Wang S, Cheng X (2016) VrDREB2A, a DREB-binding transcription factor from Vigna radiata, increased drought and high-salt tolerance in transgenic Arabidopsis thaliana. J Plant Res 129:263–273
Chen K, Song M, Guo Y, Liu L, Xue H, Dai H, Zhang Z (2019) MdMYB46 could enhance salt and osmotic stress tolerance in apple by directly activating stress responsive signals. Plant Biotechnol J 17:2341. https://doi.org/10.1111/pbi.13151
Choudhary S, Vadez V, Tom Hash C et al (2019) Pearl millet mapping population parents: performance and selection under salt stress across environments varying in evaporative demand. Proc Natl Acad Sci India Sect B Biol Sci 89:201–211
Choudhary M, Singh A, Rakshit S (2021) Coping with low moisture stress: remembering and responding. Physiol Plant 172(2):1162–1169
Desai MK, Mishra RN, Verma D, Nair S, Sopory SK, Reddy MK (2006) Structural and functional analysis of a salt stress inducible gene encoding voltage dependent anion channel (VDAC) from pearl millet (Pennisetum glaucum). Plant Physiol Biochem 44(7–9):483–493
Dey RKKP, Pal KK, Bhatt DM, Chauhan SM (2004) Growth promotion and yield enhancement of peanut (Arachis hypogaea L.) by application of plant growth-promoting rhizobacteria. Microbiol Res 159(4):371–394
Diatta S (2016) Improving pearl millet (Pennisetum glaucum (L.) R. Br.) productivity in salt-affected soils in Senegal: a greenhouse and field investigation. Doctoral dissertation, Virginia Tech
Ding Z, Majrashi MA, Ghoneim AM, Ali EF, Eissa MA, Shal RE (2022) Irrigation and biochar effects on pearl millet and kinetics of ammonia volatilization from saline sandy soils. J Soil Sci Plant Nutr 22:1–13
Dua RP (1989) Salinity tolerance in pearl millet. Indian J Agric Res 23:9–14
Dwivedi SL, Upadhyaya HD, Senthilvel S, Tom Hash C, Fukunaga K, Diao X, Santra D, Baltensperge D, Prasad M (2012) Millets genet genomic research. In: Janick J (ed) Plant breeding reviews, vol 35. Wiley, New York, pp 247–375
Etesami H, Maheshwari DK (2018) Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: action mechanisms and future prospects. Ecotoxicol Environ Saf 156:225–246
FAO (2021) Global symposium on salt affected soils. https://www.fao.org/events/global-symposium-on-salt-affected-soils
Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan Q, Liu MC, Maman J, Steinhorst L, Schmitz-Thom I, Yvon R, Kudla J, Wu HM, Cheung AY, Dinneny JR (2018) The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr Biol 28(5):666–675.e5. https://doi.org/10.1016/j.cub.2018.01.023. Epub 2018 Feb 15. PMID: 29456142; PMCID: PMC5894116
Gill T, Gill SK, Saini DK, Chopra Y et al (2022) A comprehensive review of high throughput phenotyping and machine learning for plant stress phenotyping. Phenomics 2:156
Golldack D, Li C, Mohan H, Probst N (2014) Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front Plant Sci 5:151
Govinda R, Shanta K, Vivek T, Jolly C, Coe R, Samart W, Quick W et al (2012) Towards a C4 rice. Asian J Cell Biol 7:13–31
Granier C, Aguirrezabal L, Chenu K et al (2006) PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol 169:623–635
Guo KM, Babourina O, Rengel Z (2009) Na+/H+ antiporter activity of the SOS1 gene: lifetime imaging analysis and electrophysiological studies on Arabidopsis seedlings. Plant Physiol 137:155–165
Hairmansis A, Berger B, Tester M, Roy SJ (2014) Image-based phenotyping for non-destructive screening of diferent salinity tolerance traits in rice. Rice (NY) 7:16
Hanin M, Ebel C, Ngom M, Laplaze L, Masmoudi K (2016) New insights on plant salt tolerance mechanisms and their potential use for breeding. Front Plant Sci 7:1787
Hasanuzzaman M, Davies NW, Shabala L, Zhou M, Brodribb TJ, Shabala S (2017) Residual transpiration as a component of salinity stress tolerance mechanism: a case study for barley. BMC Plant Biol 17:107. https://doi.org/10.1186/s12870-017-1054
Hassanein RA, El Khawas SA, Khafaga HS, Abd El-Nabe AS, Abd Elrady AS (2017) Amelioration of drought stress on physiological performance of pearl millet (Pennisetum americanum) plant grown under saline condition using potassium humate and silicon source. Egypt J Exp Biol 13:57–68
Hauser F, Horie T (2010) A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ 33:552–565
Heidari M, Jamshidi P (2011) Effects of salinity and potassium application on antioxidant enzyme activities and physiological parameters in pearl millet. Agric Sci China 10(2):228–237
Hoang TML, Tran TN, Nguyen TKT, Williams B, Wurm P, Bellairs S, Mundree S (2016) Improvement of salinity stress tolerance in rice: challenges and opportunities. Agronomy 6:54. https://doi.org/10.1007/s43657-022-00048-z
Hu Y, Chen L, Wang H, Zhang L, Wang F, Yu D (2013) Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J 74:730–745
Hussain K, Nawaz K, Majeed A, Khan F, Lin F, Ghani A, Raza G, Afghan S, Zia-ul-Hussain S, Ali K, Shahazad A (2010) Alleviation of salinity effects by exogenous applications of salicylic acid in pearl millet (Pennisetum glaucum (L.) R. Br.) seedlings. Afr J Biotechnol 9(50):8602–8607
Ishitani M, Liu J, Halfter U, Kim CS, Shi W, Zhu JK (2000) SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12:1667–1678. PMID:11006339
Jain A, Dev Sharma A (2005) The effect of phytoharmones (ABA, GA3) on germination, osmoprotectant solution and enzymes of carbohydrates maetabolism in pearl millet. Israel J Plant Sci 12:56–60
Jansen M, Gilmer F, Biskup B et al (2009) Simultaneous phenotyping of leaf growth and chlorophyll fuorescence via GROWSCREEN FLUORO allows detection of stress tolerance in Arabidopsis thaliana and other rosette plants. Funct Plant Biol 36:902–914
Jayakannan M, Bose J, Babourina O (2013) Salycilic acid improves salinity tolerance in Arabidopsis by restoring membrane potential and preventing salt induced K+ loss via GORK channel. J Exp Bot 64(8):2255–2268
Jeudy C, Adrian M, Baussard C et al (2016) RhizoTubes as a new tool for high throughput imaging of plant root development and architecture: test, comparison with pot grown plants and validation. Plant Methods 12:31
Jha S (2018) Proteomics of salinity stress: opportunities and challenges. In: Ramakrishna A, Gill SS (eds) Metabolic adaptations in plants during abiotic stress. CRC Press, Boca Raton
Jha S (2019) Transgenic approaches for enhancement of salinity stress tolerance in plants. In: Singh SP et al (eds) Molecular approaches in plant biology and environmental challenges. Springer Nature, Singapore
Jha S (2022) Proteome responses of pearl millet genotypes under salinity. Plant Gene 29:100347. https://doi.org/10.1016/j.plgene.2021.100347
Jiang C, Belfield EJ, Mithani A, Visscher A, Ragoussis J, Mott R et al (2012) ROS-mediated vascular homeostatic control of root-to-shoot soil Na delivery in Arabidopsis. EMBO J 31:4359–4370. https://doi.org/10.1038/emboj.2012.273
Jidda MB, Anaso AB (2017) Effects of crop improvement technologies on downy mildew of pearl millet [Pennisetum glaucum (L.) R. Br]. J Cereals Oilseeds 8(3):14–20
Johnson RR, Wagner RL, Verhey SD, Walker-Simmons MK (2002) The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol 130(2):837–846
Julkowska MM, Testerink C (2015) Tuning plant signaling and growth to survive salt. Trends Plant Sci 20:586–594
Kalaji HM, Govindjee BK, Kocielniakd J, Zuk-Golaszewska K (2011) Effects of salt stress on photosystem II efficiency and CO2 assimilation of two Syrian barley landraces. Environ Exp Bot 73:64–72
Kaur B, Sandhu KS, Kamal R et al (2021) Omics for the improvement of abiotic, biotic, and agronomic traits in major cereal crops: applications, challenges, and prospects. Plants 10:1989
Kaya C, Tuna AL, Okant AM (2010) Effect of foliar applied kinetin and indole acetic acid on maize plants grown under saline conditions. Turk J Agric For 34(6):529–538
Kayasth M, Kumar V, Gera R (2014) Gordonia sp.: a salt tolerant bacterial inoculant for growth promotion of pearl millet under saline soil conditions. 3 Biotech 4(5):553–557
Keskin BC, Sarikaya AT, Ykel B, Memon AR (2010) Abscicic acid regulated gene expression in bread wheat (Triticum aestivum L.). Aust J Crop Sci 4(8):617–625
Khan I, Raza MA, Awan SA, Shah GA, Rizwan M, Ali B, Tariq R, Hassan MJ, Alyemeni MN, Brestic M, Zhang X (2020) Amelioration of salt induced toxicity in pearl millet by seed priming with silver nanoparticles (AgNPs): the oxidative damage, antioxidant enzymes and ions uptake are major determinants of salt tolerant capacity. Plant Physiol Biochem 156:221–232
Khushdil F, Jan FG, Jan G, Hamayun M, Iqbal A, Hussain A, Bibi N (2019) Salt stress alleviation in Pennisetum glaucum through secondary metabolites modulation by Aspergillus terreus. Plant Physiol Biochem 144:127–134
Knight H, Knight MR (2001) Abiotic stress signalling pathways: specificity and cross-talk. Trends Plant Sci 6:262–267
Knight H, Trewavas AJ, Knight MR (1997) Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J 12:1067–1078
Kohler J, Hernández JA, Caravaca F, Roldán A (2009) Induction of antioxidant enzymes is involved in the greater effectiveness of a PGPR versus AM fungi with respect to increasing the tolerance of lettuce to severe salt stress. Environ Exp Bot 65(2–3):245–252
Krishnamurthy L, Serraj RC, Hash T, Dakheel AJ, Reddy BVS (2007a) Screening sorghum genotypes for salinity tolerant biomass production. Euphytica 156:15–24
Krishnamurthy L, Serraj R, Rai KN, Hash CT and Dakheel AJ (2007b). Identification of pearl millet [Pennisetum glaucum (L.) R. Br.] lines tolerant to soil salinity. Euphytica 158: 179–188
Krishnamurthy L, Upadhyaya HD, Purushothaman R, Gowda CLL, Kashiwagi J, Dwivedi SL, Singh S, Vadez V (2014) The extent of variation in salinity tolerance of the minicore collection of finger millet germplasm. Plant Sci 227(2014):51–59
Kudla J, Becker D, Grill E, Hedrich R, Hippler M, Kummer U, Parniske M, Romeis T, Schumacher K (2018) Advances and current challenges in calcium signaling. New Phytol 218:414. https://doi.org/10.1111/nph.14966
Kulkarni VN, Rai KN, Dakheel AJ, Ibrahim M, Hebbara M, Vadez V (2006) Pearl millet germplasm adapted to saline conditions. SAT eJournal 2(1):1–4
Kumar A, Krishnamurthy SL, Lata C, Kumar P, Devi R, Kulshrestha N, Yadav RK, Sharma SK (2016) Salinity and drought induced changes in gas exchange attributes and chlorophyll fluorescence characteristics of rice (Oryza sativa) varieties. Ind J Agric Sci 86(6):718–726
Kumar A, Sharma SK, Lata C, Devi R, Kulshrestha N, Krishnamurthy SL, Singh K, Yadav RK (2018) Impact of water deficit (salt and drought) stress on physiological, biochemical and yield attributes on wheat (Triticum aestivum) varieties. Ind J Agric Sci 88(10):1624–1632
Kushwaha P, Kashyap PL, Kuppusamy P, Srivastava AK, Tiwari RK (2020) Functional characterization of endophytic bacilli from pearl millet (Pennisetum glaucum) and their possible role in multiple stress tolerance. Plant Biosyst 154(4):503–514
Lakra N, Kaur C, Anwar K, Singla-Pareek SL, Pareek A (2018) Proteomics of contrasting rice genotypes: identification of potential targets for raising crops for saline environment. Plant Cell Environ 41(5):947–969
Lakshmi TV, Varalaxmi Y, Yadav S, Maheswari M (2017) Differential response of antioxidative enzymes to various abiotic stresses in Pennisetum glaucum seedlings. Russ J Plant Physiol 64:889–898
Li L, Zhang Q, Huang D (2014) A review of imaging techniques for plant phenotyping. Sensors 14:20078–20111. https://doi.org/10.3390/s141120078
Litalien A, Zeeb B (2019) Curing the earth: a review of anthropogenic soil salinization and plant-based strategies for sustainable mitigation. Sci Total Environ 698:134235. https://doi.org/10.1016/j.scitotenv.2019.134235
Liu Y, Wang Q, Zhang Y, Cui J, Chen G, Xie B, Wu C, Liu H (2014) Synergistic and antagonistic effects of salinity and pH on germination in switchgrass (Panicum virgatum L.). PLoS One 9:e85282
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting-rhizobacteria. Annu Rev Microbiol 63:541–556. https://doi.org/10.1146/annurev.micro.62.081307.162918
Lynch J, Polito VS, Lauchli A (1989) Salinity stress increases cytoplasmic Ca activity in maize root protoplasts. Plant Physiol 90:1271–1274
Ma DM, Wr WX, Li HW, Jin FX, Guo LN, Wang J et al (2014) Co-expression of the Arabidopsis SOS genes enhances salt tolerance in transgenic tall fescue (Festuca arundinacea Schreb.). Protoplasma 251:219–231. PMID:24022678
Maiti RK, Singh VP, Sarkar NC, Purohit SS, Hernandez-Pinero J (2007) Physical basis of crop growth and productivity. In: Reaearch advances in pearl millet. Springer, Berlin, pp 39–67
Martins SM, de Brito GG, da Gonçalves WC et al (2020) PhenoRoots: an inexpensive non-invasive phenotyping system to assess the variability of the root system architecture. Sci Agric 77. https://doi.org/10.1590/1678-992x-2018-0420
Mbarki S, Sytar O, Zivcak M, Abdelly C, Cerda A, Brestic M (2018) Anthocyanins of coloured wheat genotypes in specific response to salt stress. Molecules 23:1518. https://doi.org/10.3390/molecules23071518
Mbinda W, Mukami A (2021) A review of recent advances and future directions in the management of salinity stress in finger millet. Front Plant Sci 12:734798. https://doi.org/10.3389/fpls.2021.734798
Meena MD, Narjary B, Sheoran P, Jat HS, Joshi PK, Chinchmalatpure AR, Yadav G, Yadav RK, Meena MK (2018) Changes of phosphorus fractions in saline soil amended with municipal solid waste compost and mineral fertilizers in a mustard-pearl millet cropping system. Catena 160:32–40
Mishra M, Kumar U, Mishra PK, Prakash V (2010) Efficiency of plant growth promoting rhizobacteria for the enhancement of Cicer arietinum L. growth and germination under salinity. Adv Biol Res 4(2):92–96
Morton MJL, Awlia M, Al-Tamimi N, Saade S, Pailles Y, Negrão S, Tester M (2019) Salt stress under the scalpel – dissecting the genetics of salt tolerance. Plant J 97:148–163
Mukami A, Ng’etich A, Syombua E, Odour R, Mbinda W (2020) Varietal differences in physiological and biochemical responses to salinity stress in six finger millet plants. Physiol Mol Biol Plants 26:1569–1582. https://doi.org/10.1007/s12298-020-00853-8
Mukherjee K, Choudhury AR, Gupta B et al (2006) An ABRE-binding factor, OSBZ8, is highly expressed in salt tolerant cultivars than in salt sensitive cultivars of indica rice. BMC Plant Biol 6(1):18
Munns R, James RA, Läuchli A (2006) Approaches to increasing the salt tolerance of wheat and other cereals. J Exp Bot 57(5):1025–1043
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Muscolo A, Panuccio MR, Sidari M (2003) Effect of salinity on growth, carbohydrate metabolism and nutritive properties of Kikuyu grass (Pennisetum clandestinum Hochst). Plant Sci 104:1103–1110
Nakashima K, Tran L-SP, Van Nguyen D et al (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51(4):617–630
Nishiyama R, Watanabe Y, Fujita Y, Le DT, Kojima M, Werner T, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Kakimoto T, Sakakibara H, Schmülling T, Tran LSP (2011) Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23:2169–2183
Paparella S, Araujo SS, Rossi G, Wijayasinghe M, Carbonera D, Balestrazzi A (2015) Seed priming: state of the art and new perspectives. Plant Cell Rep 34:1281–1293
Parida AK, Das AB (2005) Salt tolerance and salinity effect on plants: a review. Ecotoxicol Environ Saf 60:324–349
Paul D, Lade H (2014) Plant-growth-promoting rhizobacteria to improve crop growth in saline soils: a review. Agron Sustain Dev 34:737–752
Pérez-Ruiz M, Prior A, Martinez-Guanter J et al (2020) Development and evaluation of a self-propelled electric platform for high-throughput field phenotyping in wheat breeding trials. Comput Electron Agric 169:105237
Pessarakli M (2019) Handbook of plant and crop stress, 4th edn. CRC Press, Taylor & Francis Group, Boca Raton
Pooja, Nandwal AS, Chand M, Kumari A, Rani B, Goel V, Kulshreshtha N (2020) Soil moisture deficit induced changes in antioxidative defense mechanism of four sugarcane varieties differing in their maturity. Ind J Agric Sci 90(3):56–61
Qin Y, Druzhinina IS, Pan X, Yuan Z (2016) Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnol Adv 34(7):1245–1259
Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu J-K (2002) Proc Natl Acad Sci U S A 99:8436–8441
Qiu QS, Guo Y, Quintero FJ, Pardo JM, Schumaker KS, Zhu JK (2004) Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J Biol Chem 279(1):207–215. https://doi.org/10.1074/jbc.M307982200. Epub 2003 Oct 21. PMID: 14570921
Quan R, Wang J, Hui J et al (2018) Improvement of salt tolerance using wild rice genes. Front Plant Sci 8:2269
Radhouane L (2013) Agronomic and physiological responses of pearl millet ecotype (Pennisetum glaucum (L.) R. Br.) to saline irrigation. Emir J Food Agric 25(2):109–116
Rahim Z, Gulnaz P, Gul S, Rehman K (2020) Ameliorating effect of salt stress (KCl, NaCl) on growth and germination parameters of pearl millet (Pennisetum americanum). Pak J Agric Res 33(4):951–956
Rajabi Dehnavi A, Zahedi M, Ludwiczak A, Cardenas Perez S, Piernik A (2020) Effect of salinity on seed germination and seedling development of sorghum (Sorghum bicolor (L.) Moench) Genotypes. Agronomy 10:859. https://doi.org/10.3390/agronomy10060859
Rajagopal D, Agarwal P, Tyagi W et al (2007) Pennisetum glaucum Na+/H+ antiporter confers high level of salinity tolerance in transgenic Brassica juncea. Mol Breed 19:137–151. https://doi.org/10.1007/s11032-006-9052-z
Reddy PS, Reddy GM, Pandey P et al (2012) Cloning and molecular characterization of a gene encoding late embryogenesis abundant protein from Pennisetum glaucum: protection against abiotic stresses. Mol Biol Rep 39:7163–7174. https://doi.org/10.1007/s11033-012-1548-5
Riaz M, Yan L, Wu X, Hussain S, Aziz O, Wang Y, Imran M, Jiang C (2018) Boron alleviates the aluminum toxicity in trifoliate orange by regulating antioxidant defense system and reducing root cell injury. J Environ Manag 208:149–158
Ribadiya TR, Savalia SG, Vadaliya BM, Davara MA (2018) Efect of salinity on yield, yield attributes and quality of pearl millet (Pennisetum glaucum L) varieties. Int J Chem Studies 6(6):878–882
Sadok W, Naudin P, Boussuge B et al (2007) Leaf growth rate per unit thermal time follows QTL-dependent daily patterns in hundreds of maize lines under naturally fluctuating conditions. Plant Cell Environ 30:135–146
Sage RF, Zhu XG (2011) Exploiting the engine of C4 photosynthesis. J Exp Bot 62:2989–3000. https://doi.org/10.1093/jxb/err179
Salem E (2020) Cooperative effect of salicylic acid and boron on the productivity of pearl millet crop under the degraded saline soils conditions. Egypt J Agron 42(2):185–195
Sandhu KS, Patil SS, Pumphrey MO, Carter AH (2021a) Multi-trait machine and deep learning models for genomic selection using spectral information in a wheat breeding program. Plant Genome 14:e20119. https://doi.org/10.1002/TPG2.20119
Sandhu KS, Aoun M, Morris CF, Carter AH (2021b) Genomic selection for end-use quality and processing traits in soft white winter wheat breeding program with machine and deep learning models. Biology 10:689. https://doi.org/10.3390/BIOLOGY10070689
Santosh Rama Bhadra Rao T, Vijaya Naresh J, Sudhakar Reddy P, Reddy MK, Mallikarjuna G (2017) Expression of Pennisetum glaucum eukaryotic translational initiation factor 4A (PgeIF4A) confers improved drought, salinity, and oxidative stress tolerance in groundnut. Front Plant Sci 8:453. https://doi.org/10.3389/fpls.2017.00453
Sarkar T, Thankappan R, Kumar A, Mishra GP, Dobaria JR (2014) Heterologous expression of the AtDREB1A gene in transgenic peanut-conferred tolerance to drought and salinity stresses. PLoS One 9(12):e110507
Sawada H, Shim I-S, Usui K (2006) Induction of benzoic acid 2-hydroxylase and salicylic acid biosynthesis: modulation by salt stress in rice seedlings. Plant Sci 171:263–270. https://doi.org/10.1016/j.plantsci.2006.03.020
Schmidt R, Mieulet D, Hubberten H-M et al (2013) Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 25(6):2115–2131
Serra T, Figueiredo DD, Cordeiro AM et al (2013) OsRMC, a negative regulator of salt stress response in rice, is regulated by two AP2/ERF transcription factors. Plant Mol Biol 82:439–455
Sezer S (2014) Effect of boron fertilizer applications on the growth and B, N uptake of maize (Zea mays L.) under the different soils. J Food Agric Environ 12(2):1323–1327
Shahzad B, Rehman A, Tanveer M, Wang L, Park SK, Ali A (2021) Salt stress in Brassica: effects, tolerance mechanisms, and management. J Plant Growth Regulat 41:781–795. https://doi.org/10.1007/s00344-021-10338-x
Shang G, Li Y, Hong Z, Liu CL, Shao-Zhen HE, Liu QC (2012) Overexpression of SOS genes enhanced salt tolerance in Sweetpotato. J Integr Agric 11:378–386
Sharma PC, Sehgal D, Singh D, Singh G, Yadav RS (2011) A major terminal drought tolerance QTL of pearl millet is also associated with reduced salt uptake and enhanced growth under salt stress. Mol Breed 27(2):207–222
Sharma PC, Singh D, Sehgal D, Singh G, Hash CT, Yadav RS (2014) Further evidence that a terminal drought tolerance QTL of pearl millet is associated with reduced salt uptake. Environ Exp Bot 102:48–57
Sharma A, Shahzad B, Kumar V, Kohli SK, Sidhu GPS, Bali AS, Handa N, Kapoor D, Bhardwaj R, Zheng B (2019) Phytohormones regulate accumulation of osmolytes under abiotic stress. Biomol Ther 9(7):285. https://doi.org/10.3390/biom9070285
Shi H, Ishitani M, Kim C, Zhu J-K (2000) Proc Natl Acad Sci U S A 97:6896–6901
Shinde H, Tanaka K, Dudhate A, Tsugama D, Mine Y, Kamiya T, Gupta SK, Liu S, Takano T (2018) Comparative de novo transcriptomic profiling of the salinity stress responsiveness in contrasting pearl millet lines. Environ Exp Bot 155:619–627., ISSN 0098-8472. https://doi.org/10.1016/j.envexpbot.2018.07.008
Shinde H, Dudhate A, Tsugama D, Gupta SK, Liu S, Takano T (2019) Pearl millet stress-responsive NAC transcription factor PgNAC21 enhances salinity stress tolerance in Arabidopsis. Plant Physiol Biochem 135:546–553. https://doi.org/10.1016/j.plaphy.2018.11.004
Shinde H, Dudhate A, Anand L, Tsugama D, Gupta SK, Liu S, Takano T (2020) Small RNA sequencing reveals the role of pearl millet miRNAs and their targets in salinity stress responses. South African J Bot 132:395–402. https://doi.org/10.1016/j.sajb.2020.06.011. ISSN 0254-6299
Shivhare R, Lata C (2017) Exploration of genetic and genomic resources for abiotic and biotic stress tolerance in pearl millet. Front Plant Sci 7:2069
Singh R, Singh Y, Xalaxo S, Verulkar S, Yadav N, Singh S et al (2016) From QTL to variety-harnessing the benefits of QTLs for drought, flood and salt tolerance in mega rice varieties of India through a multi-institutional network. Plant Sci 242:278–287
Singla-Pareek SL, Reddy MK, Sopory SK (2003) Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc Natl Acad Sci U S A 100:14672–14677
Sneha S, Anirudha R, Amit D, Subhash C (2013) Effect of salinity on seed germination, accumulation of proline and free amino acid in Pennisetum glaucum (L.) R. Br. Pak J Biol Sci 16:877–881. https://doi.org/10.3923/pjbs.2013.877.881
Song S-Y, Chen Y, Chen J, Dai X-Y, Zhang W-H (2011) Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234(2):331–345
Sorahinobar M, Niknam V, Ebrahimzadeh H, Soltanloo H, Behmanesh M, Enferadi ST (2016) Central role of salicylic acid in resistance of wheat against Fusarium graminearum. J Plant Growth Regul 35:477–491. https://doi.org/10.1007/s00344-015-9554-1
Spaepen S, Vanderleyden J, Okon Y (2009) Plant growth-promoting actions of rhizobacteria. Adv Bot Res 51:283–320
Srivastava P, Kumar R (2015) Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J Biol Sci 22(2):123–131
Toderich K, Shuyakaya E, Rakhmankulova Z, Bukerev R, Khujanazarov T, Zhapaev R, Ismail S, Gupta SK, Yamanaka N, Boboev F (2018) Threshold tolerance of new genotypes of Pennisetum glaucum (L.) R. Br. to salinity and drought. Agronomy 8:230
Tyagi W, Rajagopal D, Singla-Pareek SL, Reddy MK, Sopory SK (2005) Cloning and regulation of a stress-regulated Pennisetum glaucum vacuolar ATPase c gene and characterization of its promoter that is expressed in shoot hairs and floral organs. Plant Cell Physiol 46(8):1411–1422. https://doi.org/10.1093/pcp/pci154
Tyagi W, Singla-Pareek S, Nair S et al (2006) A novel isoform of ATPase c subunit from pearl millet that is differentially regulated in response to salinity and calcium. Plant Cell Rep 25:156–163. https://doi.org/10.1007/s00299-005-0055-8
Uma S, Prasad TG, Udaya Kumar M (1995) Genetic variability in recovery growth and synthesis of stress proteins in response to polyethylene glycol and salt stress in finger millet. Ann Bot 76(1):43–49
Vadez V, Hash T, Kholova J (2012) Phenotyping pearl millet for adaptation to drought. Front Physiol 3:386
Varshney R, Shi C, Thudi M et al (2017) Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat Biotechnol 35:969–976. https://doi.org/10.1038/nbt.3943
Veerangamallaiah G, Jyothsnakumari G, Thippaswamy M, Reddy PCO, Surabhi GK, Sriranganayakulu G, Mahesh Y, Rajasekhar B, Madhurarekha C, Sudhakar C (2008) Proteomic analysis of salt stress responses in foxtail millet (Setaria italic L. cv. Prasad). Plant Sci 175:631
Verma D, Singla-Pareek SL, Rajagopal D, Reddy MK, Sopory SK (2007) Functional validation of a novel isoform of Na+/H+ antiporter from Pennisetum glaucum for enhancing salinity tolerance in rice. J Biosci 32(6):21–628
Virlet N, Sabermanesh K, Sadeghi-Tehran P, Hawkesford MJ (2016) Field Scanalyzer: an automated robotic feld phenotyping platform for detailed crop monitoring. Funct Plant Biol 44:143–153
Wang K, Shangguan Z (2010) Photosynthetic characteristics and resource utilization efficiency of maize (Zea mays L.) and millet (Setaria italica L.) in a semi-arid hilly loess region in China. N Z J Crop Hortic Sci 38:247–254. https://doi.org/10.1080/01140671.2010.503987
Wang W, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218:1–14
Wu H, Hill CB, Stefano G, Bose J (2021) New insights into salinity sensing, signaling and adaptation in plants. Front Plant Sci 11:1843
Xiong H, Li J, Liu P, Duan J, Zhao Y, Guo X, Li Y, Zhang H, Ali J, Li Z (2014) Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS One 9(3):e92913
Xue ZY, Zhi DY, Xue GP, Zhang H, Zhao YX et al (2004) Enhanced salt tolerance of transgenic wheat (Tritivum aestivum L.) expressing a vacuolar Na+/H+ antiporter gene with improved grain yields in saline soils in the field and a reduced level of leaf Na+. Plant Sci 167:849–859. https://doi.org/10.1016/j.plantsci.2004.05.034
Yadav RK, Dagar JC (2016) Innovations in utilization of poor-quality water for sustainable agricultural production. In: Dagar et al (eds) Innovative saline agriculture. Springer, Berlin, pp 219–261. ISBN 978-81-322-2770-0
Yadav OP, Rai KN, Gupta SK (2012) Pearl millet: genetic improvement for tolerance to abiotic stresses. In: Improving crop productivity in sustainable agriculture. Wiley Blackwell, Hoboken, pp 261–268. ISBN 978-3-527-33242-7
Yadav T, Kumar A, Yadav RK, Yadav G, Kumar R, Kushwaha M (2020) Salicylic acid and thiourea mitigate the salinity and drought stress on physiological traits governing yield in pearl millet-wheat. Saudi J Biol Sci 27(8):2010–2017
Zhao YK, Wang T, Zhang WS, Li X (2011) SOS3 mediates lateral root development under low salt stress through regulation of auxin redistribution and maxima in Arabidopsis. New Phytol 189:1122–1134. PMID: 21087263
Zhou J, Reynolds D, Websdale D et al (2017) CropQuant: an automated and scalable feld phenotyping platform for crop monitoring and trait measurements to facilitate breeding and digital agriculture. BioRxiv. https://doi.org/10.1101/161547
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Zhu JK (2004) Plant salt tolerance and the SOS pathway. In: Proceedings of Italian Society of Agricultural Genetics. ISBN 88-900622-5-8
Zhu JK, Liu J, Xiong L (1998) Genetic analysis of salt tolerance in Arabidopsis: evidence for a critical role of potassium nutrition. Plant Cell 10:1181–1191
Zhu N, Cheng S, Liu X, Du H, Dai M, Zhou D-X, Yang W, Zhao Y (2015) The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci 236:146–156
Zhu F, Saluja M, Dharni JS et al (2021) PhenoImage: an open-source graphical user interface for plant image analysis. Plant Phenome J 4:e20015. https://doi.org/10.1002/ppj2.20015
Zida PE, Soalla WR, Paco SÃ (2017) Hydropriming of pearl millet (Pennisetum glaucum L.) in northern and Central Burkina Faso applying six hours of soaking and overnight drying of seeds. Afr J Agric Res 12(49):3441–3446
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
S, S., Swarna, R., Jinu, J., Dheeraj, C., Talwar, H.S. (2024). Salinity Stress in Pearl Millet: From Physiological to Molecular Responses. In: Tonapi, V.A., Thirunavukkarasu, N., Gupta, S., Gangashetty, P.I., Yadav, O. (eds) Pearl Millet in the 21st Century. Springer, Singapore. https://doi.org/10.1007/978-981-99-5890-0_14
Download citation
DOI: https://doi.org/10.1007/978-981-99-5890-0_14
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-5889-4
Online ISBN: 978-981-99-5890-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)