Abstract
Telomeres are specialized nucleoprotein structures localized at the ends of eukaryotic chromosomes. Telomere biology is frequently associated with human cancers where dysfunctional telomeres have been proved to participate in genetic instability. Due to end replication problem, there is a loss of nucleotides from telomere after every cell division. Thus, after limited divisions, normal somatic cells cease to divide and undergo senescence and/or apoptosis. This inability of normal cells to continually proliferate is bypassed in immortal or cancer cells via reactivation of telomerase. Telomerase is a highly specialized ribonucleoprotein molecule that promotes addition of telomeric repeats at the 3′ end of the chromosome. Multiple disease-specific studies have unveiled involvement of various genetic and epigenetic mechanisms in regulation of telomerase expression. Further, selective expression of telomerase in cancer cells in comparison with normal somatic cells makes telomerase a suitable therapeutic target. Thus, large numbers of natural and artificial compounds targeting telomerase have been screened in multiple cancers. This chapter focuses on the telomerase structure, function, and its therapeutic implications in cancer.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
In 1930s, first ever evidence of specialized structure presented at the chromosomal ends which prevented end-to-end fusion of chromosome was unveiled. Hermann Muller, a geneticist, termed this structure as “telomere” which was obtained from Greek words meaning “end” (telos) and “part” (meros). Muller in 1938 in a lecture entitled “The Remaking of Chromosomes” proposed that ....the terminal gene must have a special function, that of sealing the ends of the chromosome, so to speak, and that for some reason, a chromosome cannot persist indefinitely without having its ends thusly “sealed.” This gene may accordingly be distinguished by a special term, the “telomere” (Muller 1938).
Simultaneously, Barbara McClintock while performing experiments on maize indicated that these naturally occurring chromosome ends or telomeres demonstrate a very special property which was necessary for chromosomal stability thus supported Muller’s work (McClintock 1941). Later on, in 1970s, studies performed in Tetrahymena and yeast established that structure protecting the chromosome ends is tandem repeat of hexanucleotide blocks that could function across the diverse species and is structurally and functionally conserved (Blackburn and Gall 1978; Klobutcher et al. 1981). Then, in mid-1980s, Blackburn and Greider showed that cellular extract possesses enzymatic activities capable of adding tandem hexanucleotides repeats to natural chromosome ends that eventually leads to the discovery of telomerase for which Blackburn in 2009 shared the Nobel Prize in Physiology along with Greider and Szostak (Szostak and Blackburn 1982).
2 Telomere Structure and Function
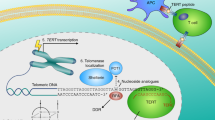
In vertebrates, telomeres are very specialized nucleoprotein structures spanning about 10–50 kb in length and are composed of tandem repeats of the hexanucleotides, 5′-(TTAGGG)n-3′ (Fig. 14.1). According to species, types, and nature of cells, telomere length ranges from 10 kb to 50 kb in both human and mus musculus (de Lange et al. 1990; Kipling and Cooke 1990). It is a single–double-stranded hybrid structure in which approximately 10–15 kb (in human at birth) long is double-stranded nucleotide sequences and 75–200 single-stranded DNA sequences rich in guanine nucleotides are present at 3′ chromosomal ends (Doksani et al. 2013). This G-rich single-stranded overhang plays crucial role in telomere capping and conformation stabilization by forming higher-order structure. Telomere protects chromosomal ends from genome surveillance machinery, nucleolytic attacks, and recombination proteins by burying, otherwise double-stranded breaks through generation of stabilized secondary structures. Among these structures, one is the formation of circular structure called a T loop in which the single-stranded part of telomere folds back on the double-stranded repeats and invades them by displacing Watson–Crick base pairing. Further, G-strand overhang by displacing normal Watson–Crick base pairing results in the generation of a stabilized structure called as displacement loop (D loop). Additionally, others in the category of secondary structures are G-quadruplexes. In vitro experiments revealed that G-rich region of telomere DNA forms a four stranded stabilized structure known as G-quadruplex (or G-quartet). This motif forms a stable planar four-stranded structure by stacking upon each other and further trapped a cation by coordinating with carbonyl group of each guanine residue. This coordination and trapping of cation further stabilizes the telomere structure. Few experimental studies have indicated that interaction of G-quadruplex and telomerase RNA template might influence telomerase activity at chromosomal ends and hence provided evidences of being used as a suitable therapeutic target (Kim et al. 2003; Shin-ya et al. 2001).
3 Telomeric Proteins
Secondary structures of telomere maintain the chromosomal capping with the help of a number of proteins that directly or indirectly localized to telomere (Smith et al. 2020). These nucleoprotein structures together play crucial roles in evading DNA repair and recombination (DDR) mechanisms. In mammals, these proteins on the basis of their localization on telomere were divided into three different categories. The initial two categories were structural proteins, and together were called as shelterin proteins (Doksani et al. 2013). First category of proteins directly interact with telomeric DNA using specialized DNA-binding motifs present in their primary sequence. In this category, two proteins including TTAGGG telomere repeat binding factor 1 (TRF1) and telomere repeat binding factor 2 (TRF 2) attach directly to double-stranded telomeric DNA, whereas single-stranded structure is guarded by protection of telomere 1 (POT1) protein (Doksani et al. 2013). The TRF1 protein negatively regulates telomere length, whereas TRF2 after binding at chromosomal termini suppresses DDR pathways and prevents their recognition as double-stranded breaks. These three proteins perform special functions to ensure telomere stability. Such as, TRF2 promotes T-loop formation and prevents the activation of ataxia telangiectasia mutated (ATM)-dependent DDR pathways and nonhomologous end joining (NHEJ) (Arnoult and Karlseder 2015). TRF1 plays significant role in the replication of telomere DNA (Zimmermann et al. 2014). Further, POT1 suppresses ataxia telangiectasia and Rad3-related protein (ATR)-dependent DDR pathways by inhibiting access of replication protein A (RPA) at single-stranded telomeric DNA sequences (Denchi and de Lange 2007). Second category of proteins interact with above mentioned telomere binding proteins and form a multiprotein complex involved in telomere length regulation. Three unique proteins, namely POT1 and TIN2 organizing protein 1 (TPP1), TRF2 interacting protein 1 (RAP1) and TRF1 and TRF2 interacting nuclear protein 2 (TIN2) along with above proteins, associate with each other to form highly specialized structure known as Shelterin complex (de Lange 2005; Doksani et al. 2013). TIN2 promotes the integrity and stability of this complex by acting as a scaffold protein on which TRF1, TRF2, and TPP1/POT1 heterodimers are attached (Ye 2004). This attachment further stabilizes TRF1 and TRF2 interaction with telomeric DNA (Frescas and de Lange 2014a, b). RAP1 by attaching with TRF2 increases the efficiency of interaction between TRF2 and telomeric DNA (Janoušková et al. 2015). Additionally, few shelterin complex associated proteins that regulate the telomere structure and its length include Pin2/TRF1-interacting factor (PINXI) and tankyrases 1 and 2 (Smith 1998; Zhou and Lu 2001). PINXI is a negative regulator of telomere length, while tankyrases 1 and 2 both positively regulate the telomere length by recruiting telomerase on the telomere. Final class of proteins includes those which regulate the biological processes including DNA damage regulators. Similar to above-discussed second class, these proteins do not bind directly to the telomeric DNA but otherwise require telomere-associated factors to localize on the telomeres. It includes MRE11/RAD50/NBS1 (MRN) complex, Ku70/86 heterodimer, RecQ helicases, Werner (WRN), and Bloom (BLM); these proteins play crucial roles in maintaining telomere homeostasis. In addition, some proteins implicitly recognize telomere dysfunction such as ATM, ATR kinases, 53BP1 (p53-binding protein 1), γH2AX, and RAD17 (radiation sensitive 17) (Lillard-Wetherell et al. 2004; Opresko et al. 2002). These proteins, in response to critically shortened telomeres, are recruited to special location at telomeres known as telomere dysfunction induced foci (TIF) and induce senescence and/or apoptosis.
It has been observed that telomere length quantitatively associates with expression levels of telomere-associated proteins. Dysregulation of telomere is a very common observation seen in many cancers; it has been observed that with alteration in telomere length there is simultaneous alteration in expression levels of these proteins. Butler et al. reported that there were increased mRNA levels of TRF1, TRF2, POT1, and TIN2 mRNA, but not TERT mRNA with decrease telomere contents in the breast carcinoma patients (Butler et al. 2012). Similarly, in 2010, Hu et al. reported in gastric cancer patients that increased protein levels of TRF1, TRF2, TIN2, and TERT molecules correlate negatively with the telomere length (Hu et al. 2010).
In our research laboratory, we studied the association between expression levels of shelterin complex and its associated molecules and telomere length in multiple myeloma patients. We observed significantly decreased telomere length and increased telomerase activity in bone marrow samples of myeloma patients in comparison with age-matched controls (Fig. 14.2). Further, we demonstrated significantly increased mRNA and protein expression of TRF1, TRF2, POT1, RAP1, TPP1, TIN2, and TANK1 in myeloma patients while drastically decreased molecular expression of PINX1 in these patients was observed. At mRNA levels, expression of these molecules correlated significantly with each other and with patients’ stages. Further, mRNA expression also correlated negatively with telomere length in myeloma patients (Kumar et al. 2018a, b). Thus, these results suggest that there is a strong association between telomere length and expression of shelterin complex molecules in most of cancers and projected these molecules as suitable theranostic markers.
(a) Representative image showing southern blot analyses in MM patients (all stages) and controls. Lane 1—ladder showing molecular weights on left side, lane 2–4—stage III MM patients, lane 5–7—stage II MM patients, lane 8–10—stage I MM patients, and lane 11–15—control subjects; (b) Box and Whisker plot showing mTL determined using the telotool software in southern blot experiment in controls and MM patients (total patients and all stages); (c) Box and Whisker plot showing RTA in total cell lysate of controls and MM patients (total patients and all stages): MM—multiple myeloma; mTL—mean telomere length; RTA—relative telomerase activity. “*” is denoted as p < 0.05 to determine the level of significance. (Adapted with permission from (Kumar et al. 2018a, b)
4 Telomere and Mechanism of Cellular Mortality
Telomere length maintenance is of utmost importance for normal cells to grow, sustain, and divide. Telomere length shortening along with other potential oncogenic changes contributes to genomic instability which ultimately leads to initiation of early stage cancer. Literature reported that normal somatic cells divide for a limited number of cell divisions. This is because our classical replication machinery is incapable to replicate the end part of chromosome which is also called as “end-replication problem”. Due to end replication problem after each round of cell division, a part of telomere (in humans 50–150 bp) breaks off, thereby leading to telomere shortening. Once the telomere becomes critically shortened, they are more prone to induce damage to the genome or thus will undergo senescence by activating DNA repair and recombination pathways. Thus, this capacity of limited cell division correlates well with the telomere length. Further, the average age of an individual depends weakly on the mean telomere length of their somatic cells. Nevertheless, these observations implied the role of telomere as a molecular clock which limits the cellular lifespan on the basis of number of cellular divisions.
Inside cell at molecular level, two crucial barriers which prevent immortalization and ultimately malignant transformation are replicative senescence (M1) and crisis (M2) (Wright et al. 1989). It has been seen that human somatic cells cultured in vitro for extended period of time ultimately ceases to divide by entering nonproliferative though metabolically active state known as mortality stage 1 (M1) or also known as replicative senescence (Fig. 14.3). This M1 stage involves critically shortened length of telomeres which ultimately leads to abrogation of cellular proliferation. Some studies suggested that this inhibition can be made by uncapping of single telomere, while majority suggested that telomere state rather than telomere length determines the fate of those cells. Majority of normal somatic cells stay either in the M1 state for their entire life or undergo apoptosis by activating apoptosis-inducing pathways. While in the existence of cancer-inducing changes, M1 stage can be by-passed by deregulating tumor suppressor pathways which predominantly involve inactivation of p53 and Rb genes, thereby underlining their potential in maintaining these cells at senescence or M1 stage. Once the ablation of these genes or pathways occurs, cells continue to proliferate until they reach a new dysfunctional state termed as mortality stage 2 (M2 stage) or crisis stage. This stage is accompanied by critically shortened or naked telomeres leading to formation of multiple TIF characterized by breakage-fusion-bridge (BFB) formation. This stage also involves massive cellular apoptosis along with continued DNA replication, thereby indicating that cells entering M2 stage are no longer able to stop the cell cycle progression (Hayashi et al. 2015). However, a rare clone (1 in 107 cells) progresses toward immortalization (Castro-Vega et al. 2015). These surviving cells will maintain their telomere lengths by expressing detectable levels of telomerase or through a totally different lesser-known mechanism called as alternative lengthening of telomeres (ALT) (Shay and Wright 2011). Though ALT method has been seen in only about 15% of malignancies, the rest 85% of human cancers involve reactivation of telomerase.
Schematic representation of telomere length dynamics in different cell types. Embryonic cells and germ cells maintain their telomere length throughout their life, while pluripotent stem cells have reduced rate of telomere loss and therefore survive for longer duration. In contrast, most somatic cells lack telomerase activity and thereby divide for a limited number of cycles and then undergo senescence. Some cells are able to bypass senescence and reach crisis stage which involves wide-scale genomic instability and apoptosis in cells. One of ten million cells escapes the crisis stage by reactivating telomerase and become a cancerous cell with limitless replicative potential
5 Structure and Function of Telomerase
Telomerase is a highly specialized ribonucleoprotein structure that helps in maintenance of telomere length and ultimately genome integrity by enzymatically repairing ends of the chromosome (Nguyen et al. 2019; Roake and Artandi 2020). In the eukaryotic evolution, two critical drawbacks arose because of linear chromosomes in the genomic material which could deprotect eukaryotic cells genomic integrity (de Lange 2015). First, cellular DNA polymerase is incapable to completely replicate their lagging strand of linear DNA known as “end-replication problem” (Watson 1972). These unreplicated ends can be recognized by DNA repair and recombination machinery as damaged DNA whose repair results in the loss of genetic information (Olovnikov 1973). Second, these unreplicated ends are always prone to be targeted by endonuclease activity which is known as “end-protection problem.” Some special mechanisms were needed to protect these chromosomes termini from nuclease activity and to prevent their shortening. In the late 1980s, telomerase enzyme discovery unfurled the end-replication mystery. The ribonucleoprotein complex, telomerase, using RNA template adds repetitive DNA sequences and maintains the telomere length (Greider and Blackburn 1989). Then, telomeric proteins in a sequence-specific manner attach to these telomeric repeats and form a protective cap around the chromosome, thus solving the “end-protection problem” (de Lange 2009).
In 1985, the enzyme which can de novo synthesize and elongate telomeric DNA was discovered as telomerase (Greider and Blackburn 1985, 1989). Later on, in 1997, it was identified and characterized (Harrington et al. 1997). This enzyme is a large ribonucleoprotein complex which though error-prone progressively adds DNA repeats (TTAGGG) to the telomere. It is a RNA-dependent DNA polymerase which comprises two minimally functional subunits: an RNA component called as telomerase RNA (TR) and a functional protein known as telomerase reverse transcriptase (hTERT). In humans, hTR (human telomerase RNA) is synthesized from TERC gene localized on 3q26 region of the chromosome, whereas hTERT protein is encoded from TERT gene present on 5p15.33 region of the chromosome 5 (Cong 1999; Feng et al. 1995). Other proteins implicated in the generation of holoenzyme are Dyskerin, Gar1, Nhp2, Pontin, Reptin, Tcab1, and telomerase protein component (TEP1). These proteins play crucial roles in proper assembly and its recruitment to the telomere (Cohen et al. 2007; Saito et al. 1997; Venteicher et al. 2008). Additionally, Es1p and Es3p (Ku heterodimer) are required for proper assembly and maturation of telomerase protein complex (Liu et al. 2004).
Evolutionary, hTERT is highly conserved and transcriptionally regulated protein whose expression levels correlate significantly with telomerase activity, thereby presenting as rate limiting and important determinant for enzymatic activity (Cong et al. 2002). In contrast, hTR is ubiquitously and constitutively expressed mRNA which is required for de novo synthesis of telomeric DNA (Cong et al. 2002). Further, hTERT comprises four domains: telomerase “essential” N-terminal (TEN) domain, telomerase RNA-binding domain (TRBD), reverse transcriptase (RT) domain, and C-terminal extension (CTE) or thumb domain. The N-terminal TEN domain binds with single-stranded telomeric DNA and also interacts with hTR (O’Connor et al. 2005; Robart and Collins 2011). TRBD domain is made of all helical domains where α-helices arranged to form asymmetric halves which collectively forms TRBD RNA-binding pocket which can binds with both single-stranded and paired RNA (Rouda and Skordalakes 2007). RT domain is the most evolutionary conserved and characterized domain that forms the catalytic/enzymatic domain of TERT (Lue et al. 2003). The hTERT CTE domain is made up of three highly conserved regions required for formation of stable RNA–DNA duplex. Mutation in these regions can lead to multiple human diseases including aplastic anemia, dyskeratosis congenital and idiopathic pulmonary fibrosis as they lead to decreased telomerase activity and processivity by disrupting interaction between RT and DNA (Hoffman et al. 2017). hTR is a very versatile component that shows sequence and size divergence in wide variety of eukaryotic species. In vertebrates, the size of hTR ranges from ~310 to 560 nt (Xie et al. 2008), while in yeast the hTR’s size ranges between ~780 and 1820 nt (Gunisova et al. 2009). At one time, ~1.5 repeats of the RNA template domain of hTR are copied and incorporated into the growing telomere DNA as a complementary sequence. The RNA template domain comprises two distinct segments: 3′ region helps in pairing with the DNA sequence, while 5′ region provides the template which is used by the hTERT for telomeric DNA synthesis (Greider 1991). The template region helps in telomerase processivity, regulating telomerase enzymatic activity and the template utilization. Initially, both hTERT and hTR are located in different nucleolar compartment, but with beginning of S-phase, hTERT is colocalized with hTR in the cajal bodies. Further, loading of human telomerase onto the telomere sequence requires TIN2 and TPP1 shelterin complex proteins already present on the double-stranded region through TRF1 and TRF2 (Abreu et al. 2010; de Lange 2010). Additionally, TPP1 interacts with single-stranded DNA-binding protein POT1, thereby forming a TPP1-POT1 complex essential for the telomerase activity (Broccoli et al. 1995).
6 Implications of Telomerase in Cancer
Due to stringent regulation of hTERT, telomerase activity ranges from very low to absent in normal somatic cells, while it is higher in stem cells, germline, and other rapidly renewing cells (Broccoli et al. 1995). However, some mitotically active cells such as endometrial tissue, hair follicles, and proliferative tissue of intestinal crypts demonstrate significantly increased telomerase activity (Brien et al. 1997; Broccoli et al. 1995; Ramirez et al. 1997). Telomere length and telomerase activity show significantly great variation between embryonic stem cells and normal somatic cells. Embryonic stem cells consistently maintained their telomere length by displaying considerably higher telomerase activity, whereas normal somatic cells show progressive decrease in telomere length due to lack of telomerase activity. This property of selective expression of hTERT has recognized its potential as suitable diagnostic and prognostic biomarker in various cancers including bladder, prostate, thyroid, breast, colon, cervical, gastric, lung, and myeloma cancer (Fernández-Marcelo et al. 2015; Glybochko et al. 2014; Kulić et al. 2016; Kumar et al. 2018a, b; Sharma et al. 2007; Tahara et al. 1995; Umbricht et al. 1997; Wu et al. 2000).
Cancer occurs when normal cells undergo genomic instability due to multiple genetic mutations and acquires the ability to proliferate indefinitely by bypassing the senescence via reactivation of telomerase or other less explored mechanisms such as ALT pathway (Shay and Wright 2011). Telomerase levels are upregulated in >85% of malignancies, while only 10–15% neoplasias follow mechanism like ALT for continued proliferation. In the last 20 years, exemplary research has been taking place in identifying different mechanisms regulating telomerase upregulation in cancer. These mechanisms include majorly hTERT promoter mutation (31%) and hTERT promoter methylation (53%), whereas minor pathways for upregulation were hTERT gene amplification (3%) and gene rearrangements (3%) (Elisabeth Naderlinger and Klaus Holzmann 2017) (Fig. 14.4).
Regulation of hTERT gene expression. Most common modes of hTERT gene expression regulators are promoter mutation, promoter methylation, and miRNA expression. Promoter methylation prevents the binding of transcriptional repressor, whereas promoter mutation generates novel binding sites for transcriptional activators. miRNA, however, regulates hTERT translation by binding to the 3′-UTR regions of the hTERT mRNA
6.1 Role of Gene Amplification and Rearrangements
During oncogenesis, gene amplification has been observed as one of the most important mechanisms leading to gain or loss of genetic material. Gene amplification involves increased gene copy number which leads to enhanced gene expression. Multiple models including replication error, telomere dysfunction, and occurrence of fragile sites in chromosome lead to increased expression (Albertson 2006). Clinically, poor disease outcome in multiple malignancies including breast and thyroid cancers significantly correlated with upregulated hTERT expression due to increased copy number (Piscuoglio et al. 2016; Wang et al. 2016). A study involving a cohort of multiple cancers demonstrated that only 3% of hTERT expressing tumors has gene amplification, thereby suggesting other important mechanisms in deregulating hTERT expression (Zhang et al. 2000). Another potential mechanism for hTERT upregulation is genomic rearrangements by placing activators and enhancers at the promoter region of hTERT gene locus (Peifer et al. 2015). In 2016, Kawashima et al. reported that hTERT upregulation through genomic rearrangements correlated with poor clinical outcomes in neuroblastoma patients (Kawashima et al. 2016).
6.2 Role of Promoter Mutations in hTERT Regulation
Genetic alterations such as mutation in TERT promoter (TERTpMut) are unique but frequent, leading to increased hTERT expression and telomerase activation. The 260 bp long core hTERT promoter comprises several transcription factors binding motifs involved in regulation of gene activation and transcription (Kyo et al. 2008). Two landmarked studies in 2013 uncovered the potential of hTERT promoter mutation at two noncoding regions in both familial and sporadic melanomas (Horn et al. 2013; Huang et al. 2013). These mutations which were observed at −124 bp and −146 bp positions upstream to the transcription start site involved C > T transitions (at positions 1,295,228 (C228T) and 1,295,250 (C250T) on chromosome 5) and thereby generate a novel nucleotide stretch suitable for binding with E26 transformation-specific (ETS) transcriptional factor involved in both activation or repression of hTERT. Since then, studies have shown significant association of TERTpMut with distinct tumors including glioblastoma, thyroid cancer, bladder cancer, etc. (Vinagre et al. 2013). The multifarious transcriptional controlling of hTERT gene is evidenced by the fact that its promoter region contains binding motifs for both enhancers and suppressors transcription factors. Widespread occurrence of TERTpMut at difference stages and in grades of various carcinomas supported this as crucial early event during carcinogenesis (Kinde et al. 2013; Wang et al. 2014).
Clinically, biomarker utility of hTERT was observed when tumors carrying TERTpMut expressed high hTERT mRNA and proteins levels in comparison with tumors having wild-type promoter (Jin et al. 2018; Leão et al. 2018; Vinagre et al. 2013). In 2018, Spiegl-Kreinecker et al. reported the involvement of hTERT promoter mutation in poor prognosis and cellular immortalization in meningioma (Spiegl-Kreinecker et al. 2018). A study reported by Wu et al. reported that simultaneous occurrence of mutation in TP/RB1 and TERTpMut might play significant role in the disease progression (Wu et al. 2014). Barczak et al. (2017) have emphasized that 36% of head and neck cancer patients had hTERT C250T promoter mutation in early stage tumors (Barczak et al. 2017). Further, in urothelial bladder carcinoma, the identification of TERTpMut in urine and tissue samples projected its role as non-invasive diagnostic and prognostic biomarker. Additionally, a report has shown that apart from disease prediction or outcome, TERTpMut was a better predictor of response to other adjuvant therapies including radiotherapy resistance (Gao et al. 2016). Yuan et al. (2016) in a meta-analysis reported the prognostic significance of TERTpMut along with clinical characteristics such as patient’s age, gender, and distant metastasis in nonsmall cell lung carcinoma (NSCLC) (Yuan et al. 2016). Literature survey in adult gliomas reported that frequency of TERTpMut is highest in glioblastomas (70%), followed by oligodendrogliomas (60%) and then oligoastrocytomas (35%) (Killela et al. 2013). In urological malignancies, TERTpMut varies widely ranging from 85% in urothelial carcinoma of bladder to 9% in renal cell carcinoma to a complete absence in testicular and prostate cancers. Some cancers such as colorectal carcinoma, prostate cancer, and testicular carcinoma do not harbor TERTpMut but display telomerase activation and self-renewal properties, thereby suggesting other mechanisms of its activation.
6.3 Role of Promoter Methylation in hTERT Regulation
In contrast to above mechanisms, epigenetic process such as DNA methylation is a very stable and frequent mode of gene expression regulation. In whole genome, this occurs at CpG sites generally located in the noncoding regions. Methylation is performed by DNA methyltransferases which adds a methyl group on the 5-carbon of cytosine which is always preceded to a guanine base. Generally, CpG dinucleotide repeats are widely distributed throughout genome with some specific clustered regions known as CpG islands. Most of these methylated CpG islands are located in the intergenic region specifically in the promoter part upstream to the transcription initiation site. Thus, DNA methylation at the promoter region plays crucial role in gene expression regulation. In general, genes which are having hypermethylated promoters are transcriptionally silenced, while those genes having less methylations are transcriptionally active genes. It is because hypermethylated DNA in the promoter region interferes with the proper positioning of transcription activators or chromatin conformation (Baylin and Jones 2011). Research established that during cancer progression, there is an elevated hypermethylation of CpG islands in the promoter region of oncogenes, thereby proposing them as a suitable hallmark of oncogenesis (Bartlett et al. 2013). Some studies have demonstrated contradictory results where hypomethylation of CpG islands in the hTERT promoter suppresses hTERT expression, while others have reported that hTERT overexpressing cancer cells displays promoter hypermethylation (Guilleret et al. 2002; Shin et al. 2003). Later on, it was observed that the region involving hTERT core promoter was hypomethylated and promotes hTERT overexpression, whereas region upstream to the core promoter was hypermethylated which did not allow repressors to bind and decrease their expression (Zinn et al. 2007). A recent report by Tsujioka et al. evidenced that hTERT promoter methylation occurs mainly in the core promoter which generates binding sites for ETS family of transcription activating factors (Tsujioka et al. 2015). Further CTCF, a transcription repressive factor, interacts with core promoter and decreases hTERT transcription by compacting chromatin organization. Therefore, promoter DNA methylation prevents binding of CTCF, thereby enhancing the telomerase expression (Renaud 2005). In addition to that, c-myc (transcription activator) binds to the hTERT core promoter in a hypomethylation state and activates telomerase activity and ultimately promotes cellular proliferation and differentiation (Wu et al. 1999a, b).
Clinically, hypermethylation of region upstream to the hTERT core promoter has been observed in the multiple cancer types including brain, prostate, urothelium, colon, and blood. Svahn et al. in 2018 reported that methylation of hTERT promoter is significantly correlated with poor outcomes in adrenocortical carcinoma (Svahn et al. 2018). In a study done in pediatric gliomas, methylation of hTERT promoter is considered as a promising biomarker of tumor progression (Castelo-Branco et al. 2013). THOR stands for TERT hypermethylated oncological region, which is 100% specific and 96% sensitive in detecting malignant neoplasm positive for hTERT expression. The identification of THOR methylation has demonstrated an unmatched potential to be used as promising diagnostic and prognostic biomarker in multiple cancers including thyroid cancer, acute myeloid leukemia/myelodysplastic syndrome, esophageal carcinoma, meningioma. and hepatocellular carcinoma (Castelo-Branco et al. 2013; Deng et al. 2015; Fürtjes et al. 2016; Wang et al. 2016; Zhang et al. 2015; Zhao et al. 2016). In these cancers, hTERT methylation pattern was positively correlating with hTERT reactivation and its expression and in many cases associated significantly with worst clinical outcomes.
6.4 Role of MicroRNAs in hTERT Regulation
MicroRNAs (miRNAs) are small, endogenously synthesized 20–25 nucleotides long noncoding RNA molecules known to regulate gene expression. They also play critical roles in regulation of various pathophysiological processes including cellular proliferation, apoptosis, and differentiation in several diseases and are implicated in genome instability by acting as oncogenic and suppressor drivers (Li et al. 2015; Vincent et al. 2014). Functionally, they regulate the posttranscriptional gene silencing by mRNA degradation and repressing translation. During cancer occurrence, decreased expression of miRNAs in tumor tissue indicates their suppressive nature, as decreased levels contribute to tumorigenesis. Further, overexpression of miRNAs (oncomiRNAs) regulated tumor suppressor genes also leads to occurrence of tumors (Cho 2007; Eckburg et al. 2020). Therefore, their functions as tumor suppressors or oncogenes depend on targeted genes. Multiple miRNAs discovered so far demonstrate enormous potential to regulate hTERT translation in many cancers. miRNAs and hTERT expressions are inversely correlated, thus during tumorigenesis high hTERT expression will have downregulated levels of respective miRNAs (Cho 2007; Hrdličková et al. 2014). miRNAs regulate the hTERT expression in two ways: In the direct mode, they attach to 3′ untranslated region (3′-UTR) of hTERT and inhibit its translation (Bai et al. 2017). Study reported in thyroid carcinoma cells that decreased expression of miR-138 was inversely correlated with hTERT overexpression; further induced expression of miR-138 significantly decreased hTERT protein levels (Mitomo et al. 2008). Additionally, another report demonstrated that interaction of let-7 g*, miR-133a, miR-342-5p, and miR-491-5p with the 3′-UTR region of hTERT mRNA significantly downregulated telomerase activity and inhibited cellular proliferation (Hrdličková et al. 2014). Furthermore, miR-1182 was also found to be downregulated in tumor tissues and cell lines of bladder cancer and restoration of miR-1182 level inhibited cellular proliferation and its invasion (Zhou et al. 2016). Indirectly, miRNAs regulate hTERT transcription by regulating transcription factors required for hTERT gene expression such as c-myc. For example, in esophageal squamous cell carcinoma, miR-1294 downregulates c-myc levels (Liu et al. 2015a, b). Further, miR-34a, a well-known tumor suppressor, decreases telomerase activity and induces cellular senescence by targeting c-myc/FoxM1 pathway in hepatocellular cancer cells (Xu et al. 2015). Due to high stability in tissue and body fluids, biomarker and therapeutic potential of miRNAs have been highlighted in multiple studies (Weber et al. 2010). In bladder, gastric and ovarian cancers downregulated levels of miR-1182, miR-1207-5p, miR-1266, miR-532, and miR-3064 associated poorly with clinical outcomes. In bladder cancer, increased expression of miR-1182 sensitizes cancer cells to chemotherapeutic drug cisplatin, thereby leading to induce better patients’ response during or after treatment (Zhou et al. 2016). Potential of miRNA targeting hTERT and other factors crucial for telomere pathway holds great prospects to be used as promising therapeutic approach to suppress telomerase activity and inhibits other cancer pathways in future (Rupaimoole and Slack 2017).
7 Therapeutic Implications of Telomerase in Cancer
Majority of human tumors (>85% of cancers) and tumor cell lines showed increased telomerase expression. Telomerase is critically involved in telomere length maintenance and limitless cancer cells proliferation. Further, in contrast to its expression in tumor cells, normal cells display minimal to complete absence of telomerase expression. Thus, this property of selective telomerase expression could be exploited as important biomarker and further generates the possibility of development of telomerase inhibitors as anticancer agents (Guterres and Villanueva 2020). Due to its structural and functional complexity, it can be targeted in many ways. Some telomerase inhibitors target its catalytic components, whereas other targets its RNA template part. These telomerase inhibitors range from natural compounds to synthetic products along with artificially created oligonucleotides. These compounds include terpenes, alkaloids, polyphenols, xanthones, and artificial products (Imetelstat, G-quadruplex stabilizers) which inhibit the telomerase expression and/or activity and thus restrained cellular proliferation (Fig. 14.5). Therapeutic implications of telomerase in various cancers by these products are discussed below.
7.1 Polyphenols
Curcumin, a major component of rhizome in turmeric (Curcuma longa L.), is a phenolic compound of medicinal importance. Literature reported that curcumin significantly induced apoptosis and showed anti-inflammatory, antioxidant, and neuroprotective activities (Griffiths et al. 2016). In 2006, Cui et al. reported that curcumin inhibited cells proliferation and decreased telomerase activity in a dose-dependent manner in multiple cancer cell types (Bel7402, HL60, and SGC7901) (Cui et al. 2006). Similarly, Ramachandran et al. reported that curcumin at 50–100 μM doses decreased telomerase expression and its activity in MCF-7 cancer cells in a c-myc-independent manner (Ramachandran et al. 2002). Further, Lee and Chung demonstrated that curcumin also induced cellular apoptosis by inhibiting the nuclear translocation of hTERT, thereby suppressing telomerase activity (Lee and Chung 2010).
Similarly, quercetin is a naturally occurring polyphenol found in most fruits, vegetables, green tea, and food grains. Multiple studies unearthed antiproliferative and pro-apoptotic properties of quercetin in various cancer cells (Lou et al. 2016; Ren et al. 2017). Further, multiple leukemic cells lines showed significantly decreased cell viability and telomerase activity after treatment with quercetin, thereby supporting its potential as suitable therapeutic target (Avci et al. 2011). Furthermore, several studies showed that quercetin dose dependently decreased the hTERT expression, lowered telomerase activity, and induced apoptosis in multiple cancer cells including lung (Wang et al. 2003), stomach (Wei et al. 2007), brain (Zamin et al. 2009), colon (Behjati et al. 2017), gastric cancer (Wei et al. 2007), and nasopharyngeal (Zheng and Chen 2017).
Resveratrol (3,5,40-trihydroxy-trans-stilbene) is a naturally occurring phenolic phytoalexin compound obtained from many plants and fruits. Chen, RJ et al. reported that resveratrol decreased cell viability in lung cancer cells by downregulating expression and activity of telomerase via modulating p53 gene expression (Chen et al. 2017). In another study, the effects of resveratrol on declining hTERT mRNA expression and telomerase activity in colorectal cancer cells were reported (Fuggetta et al. 2006). Similarly, Wang XY et al. reported that in colorectal cancer cells, resveratrol decreased cellular proliferation by inhibiting the hTERT promoter activity (Wang et al. 2010). Pterostilbene, a natural analog of resveratrol, induces apoptosis in cancer cells by binding and blocking the active site of telomerase (Tippani et al. 2014).
Tannic acid is naturally occurring polyphenol compound present in red wine, grapes, beans, tea, coffee, nuts, vegetables, and fruits. In a study published by Cosan et al. significant apoptosis and reduction in telomerase activity by tannic acid have been reported in breast and colon cancer cells (Turgut Cosan et al. 2011). Epigallocatechin-3-gallate (EGCG), one of the well-known tannic acids, demonstrates significant anticancer potential by inducing apoptosis through mitochondrial membrane potential, activating caspase-3 expression, and inhibiting telomerase activity in cancer cells (Gurung et al. 2015; Liu et al. 2017). Further, studies reported that it has also decreased hTERT and c-myc gene molecular expression (mRNA and protein) in various cancer cells (Liu et al. 2017; Zhang et al. 2014).
7.2 Alkaloids
Boldine (1,10-dimethoxy-2,9-dihydroxy aporphine) is a naturally occurring aporphine alkaloid synthesized in boldo tree (Peumus boldus) and also in lindera (Lindera aggregata). Boldine, dose, and time dependently induced cell death and exhibited antitumor effects against multiple cancer cell lines including bladder (T24), brain (U138-MG, U87-MG, and C6), and hepatocarcinoma (HepG-2) cells. Studies reported that induction of cell death is due to decreased hTERT gene expression along with lowered telomerase activity (Gerhardt et al. 2014; Kazemi Noureini and Wink 2015).
Similarly, berberine is an alkaloid extracted from the rhizome, stem barks, and roots of Berberis vulgaris (barberry) and many other plants. Wu et al. and Naasani et al. showed that berberine displayed time-dependent increase in apoptosis and decreased telomerase activity in human leukemic cancer cells (Naasani et al. 1999; Wu et al. 1999a, b). Further, berberine also increased the formation of G-quadruplexes at telomeres leading to cellular growth arrests and thus demonstrated anticancer potential (Franceschin et al. 2006; Ji et al. 2012).
7.3 Terpenes
Pristimerin is a triterpene extracted from Celastraceae and Hippocrateaceae families of plants and is known to display chemopreventive potential. This drug demonstrated significant antiproliferative potential against multiple human cancer cells (Cevatemre et al. 2018; Tiedemann et al. 2009). Liu et al. reported that in prostate cancer cells, pristimerin decreased telomerase activity by inhibiting expression of transcription factors involved in regulation of hTERT gene transcription (Liu et al. 2015a, b). Further, in another study performed on pancreatic duct adenocarcinoma cells, pristimerin substantially decreased cell proliferation by both arresting and inducing apoptosis along with decreased telomerase activity (Deeb et al. 2015). Similarly, oleanane (methyl-2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oate) is a triterpene which is known to have substantial anti-inflammatory, pro-apoptotic activities and thus demonstrated anticancer potential against pancreatic ductal adenocarcinoma cancer cells (Deeb et al. 2013).
In our laboratory, we have studied anticancer potential of Tanshinone I (TanI), a diterpene, on myeloma cancer cell lines (RPMI 8226 and U266). We observed that Tanshinone I alone or in combination lenalidomide time and dose dependently induced apoptosis in both myeloma cells lines. Similarly, very low doses (2.0 μM) of TanI significantly decreased telomerase activity in both myeloma cancer cells at 24-h time point (Fig. 14.6). Further, TanI alone and in association with lenalidomide significantly decreased the molecular expression (mRNA and protein) of shelterin complex and its associated factors in myeloma cancer cells (Kumar et al. 2018a).
Determination of relative telomerase activity after treatment of myeloma cells (RPMI 8226 and U266) with Tanshinone I for 24 hr. (Adapted with permission from Ref. (Kumar et al. 2018a))
7.4 Xanthones
In this category, two important secondary metabolites are gambogic acid and gambogenic acid extracted from the resin of Garcinia hanburyi tree. Due to their unique color, they are generally used as coloring substances. Studies have reported that these xanthones induced cytotoxicity in cancer cells in both dose- and time-dependent manner (Pan et al. 2017; Zhao et al. 2017). Additionally, report suggested that these xanthones induced apoptosis and decreased telomerase expression and activity by inhibiting binding of transcriptional activators at the hTERT promoter region (Yu et al. 2006).
Though these natural compounds demonstrated significant anticancer potentials and drastically reduced telomerase levels in cancer cells, till today very few of them have been used as chemotherapeutic agents. Thus, synthetic drugs have provided significant hopes as anti-telomerase inhibitors against cancer cells.
7.5 Currently Used Inhibitors of Telomerase
Recently, a large number of chemically generated drugs targeting different sites of telomerase have been developed to fight against cancer cells. Among all, GRN163L, also called as Imetelstat, is by far the most widely used and successful chemotherapeutic drug that targets telomerase. It is 13-mer antisense oligonucleotides, which form a complementary pairing with the RNA template of the telomerase and inhibits its action (Chiappori et al. 2015). Hochreiter et al. observed that GRN163L dose dependently decreased telomerase activity and also reduced tumorigenicity by causing reduction in cell growth, metastasis, and invasiveness of breast cancer cells (Hochreiter et al. 2006). Similarly, in the breast and pancreatic cancer cells, GRN163L treatment resulted in the significant induction of apoptosis and impaired cellular growth. Simultaneously, the inhibitory effect was much more pronounced with the cancer cells with critically shortened telomeres (Burchett et al. 2014). Due to nonspecific toxicity and longer duration of treatment, these drugs showed limited clinical efficacy despite excellent inhibitory potential.
Another strategy aimed to inhibit the telomerase involved increased stabilization and generation of endogenous higher-order telomeric structures such as G-quadruplex. These naturally occurring secondary structures are rich in guanine, and they form planar structures which impede the movement of replication machinery through replication fork (Maestroni et al. 2017). Normally, these structures are resolved by telomerase, but utilization of G-quadruplex stabilizing ligands will prevent the access of telomerase and thus holds great promises for various malignant and progressive cancers. These ligands including telomestatin, BRACO-19, and RHPS4 significantly increased the G-quadruplex stability and ultimately enhanced the DDR pathways in cancer cells (Burger et al. 2005; Cookson et al. 2005). There are few indirect treatments available for targeting telomere or telomerase which includes tankyrase inhibitors and shelterin components inhibitors which have also shown promising effects in limiting the length of telomere and activating various DNA repair pathways.
8 Conclusion
At chromosomal ends, telomeres in association with telomeric proteins are organized into highly specialized nucleoprotein structures. These structures play significant role in maintaining telomere homeostasis. Normal somatic cells display limited replicative capacity due to end replication problem. However, for cancer cells to undergo continued proliferation, the maintenance of telomere length is critical. This cellular self-renewal capacity of cancer cells is regulated by reactivation of telomerase. Studies showed in this chapter supported that different mechanisms are involved in regulation of telomerase expression and reactivation. Literature supported that different genetic (promoter mutation) and epigenetic mechanisms (promoter methylation and miRNAs) are involved in reactivation of telomerase and further projected them as suitable diagnostic, prognostic, and therapeutic markers. Future research is focusing on cues and signals targeting these epigenetic changes and how these methylation patterns can be used as therapeutic targets. Further, due to selective expression of telomerase in cancer cells plethora of compounds targeting telomerase have been explored. Current literature provided ample evidences that there is a plethora of natural compounds that regulate telomerase levels at multiple genetic and epigenetic levels and also inhibits cellular proliferation and induces apoptosis. Few natural compounds such as resveratrol and curcumin and synthetic compounds such as imetelstat and G-quadruplex stabilizers have shown promising anticancer potential against multitude of cancers. This chapter concludes the telomerase biomarker and therapeutic implications in cancers and projected it as a suitable theranostic marker in future.
References
Abreu E, Aritonovska E, Reichenbach P, Cristofari G, Culp B, Terns RM, Lingner J, Terns MP (2010) TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol Cell Biol 30(12):2971–2982. https://doi.org/10.1128/MCB.00240-10
Albertson DG (2006) Gene amplification in cancer. Trends Genet 22(8):447–455. https://doi.org/10.1016/j.tig.2006.06.007
Arnoult N, Karlseder J (2015) Complex interactions between the DNA-damage response and mammalian telomeres. Nat Struct Mol Biol 22(11):859–866. https://doi.org/10.1038/nsmb.3092
Avci CB, Yilmaz S, Dogan ZO, Saydam G, Dodurga Y, Ekiz HA, Kartal M, Sahin F, Baran Y, Gunduz C (2011) Quercetin-induced apoptosis involves increased hTERT enzyme activity of leukemic cells. Hematology 16(5):303–307. https://doi.org/10.1179/102453311X13085644680104
Bai L, Wang H, Wang A-H, Zhang L-Y, Bai J (2017) MicroRNA-532 and microRNA-3064 inhibit cell proliferation and invasion by acting as direct regulators of human telomerase reverse transcriptase in ovarian cancer. PLoS One 12(3):e0173912. https://doi.org/10.1371/journal.pone.0173912
Barczak W, Suchorska WM, Sobecka A, Bednarowicz K, Machczynski P, Golusinski P, Rubis B, Masternak MM, Golusinski W (2017) HTERT C250T promoter mutation and telomere length as a molecular markers of cancer progression in patients with head and neck cancer. Mol Med Rep 16(1):441–446. https://doi.org/10.3892/mmr.2017.6590
Bartlett TE, Zaikin A, Olhede SC, West J, Teschendorff AE, Widschwendter M (2013) Corruption of the intra-gene DNA methylation architecture is a hallmark of cancer. PLoS One 8(7):e68285. https://doi.org/10.1371/journal.pone.0068285
Baylin SB, Jones PA (2011) A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer 11(10):726–734. https://doi.org/10.1038/nrc3130
Behjati M, Hashemi M, Kazemi M, Salehi M, Javanmard S (2017) Evaluation of energy balance on Human Telomerase Reverse Transcriptase (hTERT) alternative splicing by semi-quantitative RT-PCR in human umbilical vein endothelial cells. Adv Biomed Res 6(1):43. https://doi.org/10.4103/2277-9175.204591
Blackburn EH, Gall JG (1978) A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol 120(1):33–53. https://doi.org/10.1016/0022-2836(78)90294-2
Brien TP, Kallakury BV, Lowry CV, Ambros RA, Muraca PJ, Malfetano JH, Ross JS (1997) Telomerase activity in benign endometrium and endometrial carcinoma. Cancer Res 57(13):2760–2764
Broccoli D, Young JW, de Lange T (1995) Telomerase activity in normal and malignant hematopoietic cells. Proc Natl Acad Sci 92(20):9082–9086. https://doi.org/10.1073/pnas.92.20.9082
Burchett KM, Yan Y, Ouellette MM (2014) Telomerase Inhibitor Imetelstat (GRN163L) limits the lifespan of human pancreatic cancer cells. PLoS One 9(1):e85155. https://doi.org/10.1371/journal.pone.0085155
Burger AM, Dai F, Schultes CM, Reszka AP, Moore MJ, Double JA, Neidle S (2005) The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res 65(4):1489–1496. https://doi.org/10.1158/0008-5472.CAN-04-2910
Butler KS, Hines WC, Heaphy CM, Griffith JK (2012) Coordinate regulation between expression levels of telomere-binding proteins and telomere length in breast carcinomas. Cancer Med 1(2):165–175. https://doi.org/10.1002/cam4.14
Castelo-Branco P, Choufani S, Mack S, Gallagher D, Zhang C, Lipman T, Zhukova N, Walker EJ, Martin D, Merino D, Wasserman JD, Elizabeth C, Alon N, Zhang L, Hovestadt V, Kool M, Jones DT, Zadeh G, Croul S et al (2013) Methylation of the TERT promoter and risk stratification of childhood brain tumours: an integrative genomic and molecular study. Lancet Oncol 14(6):534–542. https://doi.org/10.1016/S1470-2045(13)70110-4
Castro-Vega LJ, Jouravleva K, Ortiz-Montero P, Liu W-Y, Galeano JL, Romero M, Popova T, Bacchetti S, Vernot JP, Londoño-Vallejo A (2015) The senescent microenvironment promotes the emergence of heterogeneous cancer stem-like cells. Carcinogenesis 36(10):1180–1192. https://doi.org/10.1093/carcin/bgv101
Cevatemre B, Erkısa M, Aztopal N, Karakas D, Alper P, Tsimplouli C, Sereti E, Dimas K, Armutak EII, Gurevin EG, Uvez A, Mori M, Berardozzi S, Ingallina C, D’Acquarica I, Botta B, Ozpolat B, Ulukaya E (2018) A promising natural product, pristimerin, results in cytotoxicity against breast cancer stem cells in vitro and xenografts in vivo through apoptosis and an incomplete autophagy in breast cancer. Pharmacol Res 129:500–514. https://doi.org/10.1016/j.phrs.2017.11.027
Chen R-J, Wu P-H, Ho C-T, Way T-D, Pan M-H, Chen H-M, Ho Y-S, Wang Y-J (2017) P53-dependent downregulation of hTERT protein expression and telomerase activity induces senescence in lung cancer cells as a result of pterostilbene treatment. Cell Death Dis 8(8):e2985–e2985. https://doi.org/10.1038/cddis.2017.333
Chiappori AA, Kolevska T, Spigel DR, Hager S, Rarick M, Gadgeel S, Blais N, Von Pawel J, Hart L, Reck M, Bassett E, Burington B, Schiller JH (2015) A randomized phase II study of the telomerase inhibitor imetelstat as maintenance therapy for advanced non-small-cell lung cancer. Ann Oncol 26(2):354–362. https://doi.org/10.1093/annonc/mdu550
Cho WC (2007) OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer 6(1):60. https://doi.org/10.1186/1476-4598-6-60
Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR (2007) Protein composition of catalytically active human telomerase from immortal cells. Science 315(5820):1850–1853. https://doi.org/10.1126/science.1138596
Cong Y (1999) The human telomerase catalytic subunit hTERT: organization of the gene and characterization of the promoter. Hum Mol Genet 8(1):137–142. https://doi.org/10.1093/hmg/8.1.137
Cong Y-S, Wright WE, Shay JW (2002) Human telomerase and its regulation. Microbiol Mol Biol Rev 66(3):407–425. https://doi.org/10.1128/MMBR.66.3.407-425.2002
Cookson JC, Dai F, Smith V, Heald RA, Laughton CA, Stevens MFG, Burger AM (2005) Pharmacodynamics of the G-quadruplex-stabilizing telomerase inhibitor 3,11-Difluoro-6,8,13-trimethyl-8 H -quino[4,3,2- kl ]acridinium methosulfate (RHPS4) in vitro: activity in human tumor cells correlates with telomere length and can be enhanced, or antagonized, with cytotoxic agents. Mol Pharmacol 68(6):1551–1558. https://doi.org/10.1124/mol.105.013300
Cui S-X, Qu X-J, Xie Y-Y, Zhou L, Nakata M, Makuuchi M, Tang W (2006) Curcumin inhibits telomerase activity in human cancer cell lines. Int J Mol Med 18(2):227–231
de Lange T (2005) Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev 19(18):2100–2110. https://doi.org/10.1101/gad.1346005
de Lange T (2009) How telomeres solve the end-protection problem. Science 326(5955):948–952. https://doi.org/10.1126/science.1170633
de Lange, T., 2010. How Shelterin solves the telomere end-protection problem. Cold Spring Harb Symp Quant Biol, 75(0), 167–177. doi: https://doi.org/10.1101/sqb.2010.75.017
de Lange T (2015) A loopy view of telomere evolution. Front Genet 6. https://doi.org/10.3389/fgene.2015.00321
de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, Varmus HE (1990) Structure and variability of human chromosome ends. Mol Cell Biol 10(2):518–527. https://doi.org/10.1128/mcb.10.2.518
Deeb D, Gao X, Liu Y, Varma N, Arbab A, Gautam S (2013) Inhibition of telomerase activity by oleanane triterpenoid CDDO-Me in pancreatic cancer cells is ROS-dependent. Molecules 18(3):3250–3265. https://doi.org/10.3390/molecules18033250
Deeb D, Gao X, Liu Y, Pindolia K, Gautam SC (2015) Inhibition of hTERT/telomerase contributes to the antitumor activity of pristimerin in pancreatic ductal adenocarcinoma cells. Oncol Rep 34(1):518–524. https://doi.org/10.3892/or.2015.3989
Denchi EL, de Lange T (2007) Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature 448(7157):1068–1071. https://doi.org/10.1038/nature06065
Deng J, Zhou D, Zhang J, Chen Y, Wang C, Liu Y, Zhao K (2015) Aberrant methylation of the TERT promoter in esophageal squamous cell carcinoma. Cancer Genet 208(12):602–609. https://doi.org/10.1016/j.cancergen.2015.10.004
Doksani Y, Wu JY, de Lange T, Zhuang X (2013) Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell 155(2):345–356. https://doi.org/10.1016/j.cell.2013.09.048
Eckburg A, Dein J, Berei J, Schrank Z, Puri N (2020) Oligonucleotides and microRNAs targeting telomerase subunits in cancer therapy. Cancers 12(9):2337. https://doi.org/10.3390/cancers12092337
Feng J, Funk W, Wang S, Weinrich S, Avilion A, Chiu C, Adams R, Chang E, Allsopp R, Yu J et al (1995) The RNA component of human telomerase. Science 269(5228):1236–1241. https://doi.org/10.1126/science.7544491
Fernández-Marcelo T, Gómez A, Pascua I, de Juan C, Head J, Hernando F, Jarabo J-R, Calatayud J, Torres-García A-J, Iniesta P (2015) Telomere length and telomerase activity in non-small cell lung cancer prognosis: clinical usefulness of a specific telomere status. J Exp Clin Cancer Res 34(1):78. https://doi.org/10.1186/s13046-015-0195-9
Franceschin M, Rossetti L, D’Ambrosio A, Schirripa S, Bianco A, Ortaggi G, Savino M, Schultes C, Neidle S (2006) Natural and synthetic G-quadruplex interactive berberine derivatives. Bioorg Med Chem Lett 16(6):1707–1711. https://doi.org/10.1016/j.bmcl.2005.12.001
Frescas, D., and de Lange, T., 2014a. Binding of TPP1 Protein to TIN2 protein is required for POT1a,b protein-mediated telomere protection. J Biol Chem, 289(35), 24180–24187. doi: https://doi.org/10.1074/jbc.M114.592592
Frescas D, de Lange T (2014b) TRF2-Tethered TIN2 can mediate telomere protection by TPP1/POT1. Mol Cell Biol 34(7):1349–1362. https://doi.org/10.1128/MCB.01052-13
Fuggetta MP, Lanzilli G, Tricarico M, Cottarelli A, Falchetti R, Ravagnan G, Bonmassar E (2006) Effect of resveratrol on proliferation and telomerase activity of human colon cancer cells in vitro. J Exp Clin Cancer Res: CR 25(2):189–193
Fürtjes G, Köchling M, Peetz-Dienhart S, Wagner A, Heß K, Hasselblatt M, Senner V, Stummer W, Paulus W, Brokinkel B (2016) HTERT promoter methylation in meningiomas and central nervous hemangiopericytomas. J Neuro-Oncol 130(1):79–87. https://doi.org/10.1007/s11060-016-2226-6
Gao, K., Li, G., Qu, Y., Wang, M., Cui, B., Ji, M., Shi, B., and Hou, P., 2016. TERT promoter mutations and long telomere length predict poor survival and radiotherapy resistance in gliomas. Oncotarget 7(8), 8712–8725. doi: https://doi.org/10.18632/oncotarget.6007
Gerhardt D, Bertola G, Dietrich F, Figueiró F, Zanotto-Filho A, Moreira Fonseca JC, Morrone FB, Barrios CH, Battastini AMO, Salbego CG (2014) Boldine induces cell cycle arrest and apoptosis in T24 human bladder cancer cell line via regulation of ERK, AKT, and GSK-3β. Urol Oncol: Semin Original Investig 32(1):36.e1–36.e9. https://doi.org/10.1016/j.urolonc.2013.02.012
Glybochko PV, Zezerov EG, Glukhov AI, Alyaev YG, Severin SE, Polyakovsky KA, Varshavsky VA, Severin ES, Vinarov AZ (2014) Telomerase as a tumor marker in diagnosis of prostatic intraepithelial neoplasia and prostate cancer: telomerase-A tumor marker. Prostate 74(10):1043–1051. https://doi.org/10.1002/pros.22823
Greider CW (1991) Telomerase is processive. Mol Cell Biol 11(9):4572–4580. https://doi.org/10.1128/MCB.11.9.4572
Greider CW, Blackburn EH (1985) Identification of a specific telomere terminal transferase activity in tetrahymena extracts. Cell 43(2):405–413. https://doi.org/10.1016/0092-8674(85)90170-9
Greider CW, Blackburn EH (1989) A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature 337(6205):331–337. https://doi.org/10.1038/337331a0
Griffiths K, Aggarwal B, Singh R, Buttar H, Wilson D, De Meester F (2016) Food antioxidants and their anti-inflammatory properties: a potential role in cardiovascular diseases and cancer prevention. Diseases 4(4):28. https://doi.org/10.3390/diseases4030028
Guilleret I, Yan P, Grange F, Braunschweig R, Bosman FT, Benhattar J (2002) Hypermethylation of the human telomerase catalytic subunit (hTERT) gene correlates with telomerase activity. Int J Cancer 101(4):335–341. https://doi.org/10.1002/ijc.10593
Gunisova S, Elboher E, Nosek J, Gorkovoy V, Brown Y, Lucier J-F, Laterreur N, Wellinger RJ, Tzfati Y, Tomaska L (2009) Identification and comparative analysis of telomerase RNAs from Candida species reveal conservation of functional elements. RNA 15(4):546–559. https://doi.org/10.1261/rna.1194009
Gurung RL, Lim SN, Low GKM, Hande MP (2015) MST-312 Alters telomere dynamics, gene expression profiles and growth in human breast cancer cells. J Nutrigenet Nutrigenomics 7(4–6):283–298. https://doi.org/10.1159/000381346
Guterres AN, Villanueva J (2020) Targeting telomerase for cancer therapy. Oncogene 39(36):5811–5824. https://doi.org/10.1038/s41388-020-01405-w
Harrington L, Zhou W, McPhail T, Oulton R, Yeung DSK, Mar V, Bass MB, Robinson MO (1997) Human telomerase contains evolutionarily conserved catalytic and structural subunits. Genes Dev 11(23):3109–3115. https://doi.org/10.1101/gad.11.23.3109
Hayashi MT, Cesare AJ, Rivera T, Karlseder J (2015) Cell death during crisis is mediated by mitotic telomere deprotection. Nature 522(7557):492–496. https://doi.org/10.1038/nature14513
Hochreiter AE, Xiao H, Goldblatt EM, Gryaznov SM, Miller KD, Badve S, Sledge GW, Herbert B-S (2006) Telomerase template antagonist GRN163L disrupts telomere maintenance, tumor growth, and metastasis of breast cancer. Clin Cancer Res 12(10):3184–3192. https://doi.org/10.1158/1078-0432.CCR-05-2760
Hoffman H, Rice C, Skordalakes E (2017) Structural analysis reveals the deleterious effects of telomerase mutations in bone marrow failure syndromes. J Biol Chem 292(11):4593–4601. https://doi.org/10.1074/jbc.M116.771204
Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, Schadendorf D, Kumar R (2013) TERT Promoter mutations in familial and sporadic melanoma. Science 339(6122):959–961. https://doi.org/10.1126/science.1230062
Hrdličková R, Nehyba J, Bargmann W, Bose HR (2014) Multiple tumor suppressor microRNAs regulate telomerase and TCF7, an important transcriptional regulator of the Wnt pathway. PLoS One 9(2):e86990. https://doi.org/10.1371/journal.pone.0086990
Hu H, Zhang Y, Zou M, Yang S, Liang X-Q (2010) Expression of TRF1, TRF2, TIN2, TERT, KU70, and BRCA1 proteins is associated with telomere shortening and may contribute to multistage carcinogenesis of gastric cancer. J Cancer Res Clin Oncol 136(9):1407–1414. https://doi.org/10.1007/s00432-010-0795-x
Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA (2013) Highly recurrent TERT promoter mutations in human melanoma. Science 339(6122):957–959. https://doi.org/10.1126/science.1229259
Janoušková E, Nečasová I, Pavloušková J, Zimmermann M, Hluchý M, Marini V, Nováková M, Hofr C (2015) Human Rap1 modulates TRF2 attraction to telomeric DNA. Nucleic Acids Res 43(5):2691–2700. https://doi.org/10.1093/nar/gkv097
Ji X, Sun H, Zhou H, Xiang J, Tang Y, Zhao C (2012) The interaction of telomeric DNA and C-myc22 G-Quadruplex with 11 natural alkaloids. Nucleic Acid Ther 22(2):127–136. https://doi.org/10.1089/nat.2012.0342
Jin A, Xu J, Wang Y (2018) The role of TERT promoter mutations in postoperative and preoperative diagnosis and prognosis in thyroid cancer. Medicine 97(29):e11548. https://doi.org/10.1097/MD.0000000000011548
Kawashima M, Kojima M, Ueda Y, Kurihara S, Hiyama E (2016) Telomere biology including TERT rearrangements in neuroblastoma: a useful indicator for surgical treatments. J Pediatr Surg 51(12):2080–2085. https://doi.org/10.1016/j.jpedsurg.2016.09.042
Kazemi Noureini S, Wink M (2015) Dose-dependent cytotoxic effects of boldine in HepG-2 cells—telomerase inhibition and apoptosis induction. Molecules 20(3):3730–3743. https://doi.org/10.3390/molecules20033730
Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Friedman AH, Friedman H, Gallia GL, Giovanella BC, Grollman AP, He T-C, He Y, Hruban RH, Jallo GI, Mandahl N, Meeker AK, Mertens F, Netto GJ et al (2013) TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci 110(15):6021–6026. https://doi.org/10.1073/pnas.1303607110
Kim M-Y, Gleason-Guzman M, Izbicka E, Nishioka D, Hurley LH (2003) The different biological effects of telomestatin and TMPyP4 can be attributed to their selectivity for interaction with intramolecular or intermolecular G-quadruplex structures. Cancer Res 63(12):3247–3256
Kinde, I., Munari, E., Faraj, S. F., Hruban, R. H., Schoenberg, M., Bivalacqua, T., Allaf, M., Springer, S., Wang, Y., Diaz, L. A., Kinzler, K. W., Vogelstein, B., Papadopoulos, N., Netto, G. J., 2013. TERT Promoter mutations occur early in urothelial neoplasia and are biomarkers of early disease and disease recurrence in Urine. Cancer Res, 73(24), 7162–7167. doi: https://doi.org/10.1158/0008-5472.CAN-13-2498
Kipling D, Cooke HJ (1990) Hypervariable ultra-long telomeres in mice. Nature 347(6291):400–402. https://doi.org/10.1038/347400a0
Klobutcher LA, Swanton MT, Donini P, Prescott DM (1981) All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc Natl Acad Sci 78(5):3015–3019. https://doi.org/10.1073/pnas.78.5.3015
Kulić A, Plavetić ND, Gamulin S, Jakić-Razumović J, Vrbanec D, Sirotković-Skerlev M (2016) Telomerase activity in breast cancer patients: association with poor prognosis and more aggressive phenotype. Med Oncol 33(3):23. https://doi.org/10.1007/s12032-016-0736-x
Kumar R, Gupta N, Himani, and Sharma, A. (2018a) Novel combination of tanshinone I and lenalidomide induces chemo-sensitivity in myeloma cells by modulating telomerase activity and expression of shelterin complex and its associated molecules. Mol Biol Rep 45(6):2429–2439. https://doi.org/10.1007/s11033-018-4409-z
Kumar R, Khan R, Gupta N, Seth T, Sharma A, Kalaivani M, Sharma A (2018b) Identifying the biomarker potential of telomerase activity and shelterin complex molecule, telomeric repeat binding factor 2 (TERF2), in multiple myeloma. Leuk Lymphoma 59(7):1677–1689. https://doi.org/10.1080/10428194.2017.1387915
Kyo S, Takakura M, Fujiwara T, Inoue M (2008) Understanding and exploiting hTERT promoter regulation for diagnosis and treatment of human cancers. Cancer Sci 99(8):1528–1538. https://doi.org/10.1111/j.1349-7006.2008.00878.x
Leão R, Apolónio JD, Lee D, Figueiredo A, Tabori U, Castelo-Branco P (2018) Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: clinical impacts in cancer. J Biomed Sci 25(1):22. https://doi.org/10.1186/s12929-018-0422-8
Lee JH, Chung IK (2010) Curcumin inhibits nuclear localization of telomerase by dissociating the Hsp90 co-chaperone p23 from hTERT. Cancer Lett 290(1):76–86. https://doi.org/10.1016/j.canlet.2009.08.026
Li J, Lei H, Xu Y, Tao Z (2015) MiR-512-5p Suppresses tumor growth by targeting hTERT in telomerase positive head and neck squamous cell carcinoma in vitro and in vivo. PLoS One 10(8):e0135265. https://doi.org/10.1371/journal.pone.0135265
Lillard-Wetherell K, Machwe A, Langland GT, Combs KA, Behbehani GK, Schonberg SA, German J, Turchi JJ, Orren DK, Groden J (2004) Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum Mol Genet 13(17):1919–1932. https://doi.org/10.1093/hmg/ddh193
Liu L, Lai S, Andrews LG, Tollefsbol TO (2004) Genetic and epigenetic modulation of telomerase activity in development and disease. Gene 340(1):1–10. https://doi.org/10.1016/j.gene.2004.06.011
Liu YB, Gao X, Deeb D, Pindolia K, Gautam SC (2015a) Role of telomerase in anticancer activity of pristimerin in prostate cancer cells. J Exp Ther Oncol 11(1):41–49
Liu K, Li L, Rusidanmu A, Wang Y, Lv X (2015b) Down-regulation of MiR-1294 is related to dismal prognosis of patients with esophageal squamous cell carcinoma through elevating C-MYC expression. Cell Physiol Biochem 36(1):100–110. https://doi.org/10.1159/000374056
Liu L, Zuo J, Wang G (2017) Epigallocatechin-3-gallate suppresses cell proliferation and promotes apoptosis in Ec9706 and Eca109 esophageal carcinoma cells. Oncol Lett 14(4):4391–4395. https://doi.org/10.3892/ol.2017.6712
Lou M, Zhang L, Ji P, Feng F, Liu J, Yang C, Li B, Wang L (2016) Quercetin nanoparticles induced autophagy and apoptosis through AKT/ERK/Caspase-3 signaling pathway in human neuroglioma cells: in vitro and in vivo. Biomed Pharmacother 84:1–9. https://doi.org/10.1016/j.biopha.2016.08.055
Lue NF, Lin Y-C, Mian IS (2003) A conserved telomerase motif within the catalytic domain of telomerase reverse transcriptase is specifically required for repeat addition processivity. Mol Cell Biol 23(23):8440–8449. https://doi.org/10.1128/MCB.23.23.8440-8449.2003
Maestroni L, Matmati S, Coulon S (2017) Solving the telomere replication problem. Genes 8(2):55. https://doi.org/10.3390/genes8020055
McClintock B (1941) The stability of broken ends of chromosomes in Zea Mays. Genetics 26(2):234–282
Mitomo S, Maesawa C, Ogasawara S, Iwaya T, Shibazaki M, Yashima-Abo A, Kotani K, Oikawa H, Sakurai E, Izutsu N, Kato K, Komatsu H, Ikeda K, Wakabayashi G, Masuda T (2008) Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci 99(2):280–286. https://doi.org/10.1111/j.1349-7006.2007.00666.x
Muller HJ (1938) The remaking of chromosomes. Collect Nat 13
Naasani I, Seimiya H, Yamori T, Tsuruo T (1999) FJ5002: a potent telomerase inhibitor identified by exploiting the disease-oriented screening program with COMPARE analysis. Cancer Res 59(16):4004–4011
Naderlinger E, Holzmann K (2017) Epigenetic regulation of telomere maintenance for therapeutic interventions in gliomas. Genes 8(5):145. https://doi.org/10.3390/genes8050145
Nguyen THD, Collins K, Nogales E (2019) Telomerase structures and regulation: shedding light on the chromosome end. Curr Opin Struct Biol 55:185–193. https://doi.org/10.1016/j.sbi.2019.04.009
O’Connor CM, Lai CK, Collins K (2005) Two purified domains of telomerase reverse transcriptase reconstitute sequence-specific interactions with RNA. J Biol Chem 280(17):17533–17539. https://doi.org/10.1074/jbc.M501211200
Olovnikov AM (1973) A theory of marginotomy. J Theor Biol 41(1):181–190. https://doi.org/10.1016/0022-5193(73)90198-7
Opresko PL, von Kobbe C, Laine J-P, Harrigan J, Hickson ID, Bohr VA (2002) Telomere-binding protein TRF2 binds to and stimulates the Werner and bloom syndrome helicases. J Biol Chem 277(43):41110–41119. https://doi.org/10.1074/jbc.M205396200
Pan H, Jansson KH, Beshiri ML, Yin J, Fang L, Agarwal S, Nguyen H, Corey E, Zhang Y, Liu J, Fan H, Lin H, Kelly K (2017) Gambogic acid inhibits thioredoxin activity and induces ROS-mediated cell death in castration-resistant prostate cancer. Oncotarget 8(44):77181–77194. https://doi.org/10.18632/oncotarget.20424
Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R, Krämer A, Roncaioli JL, Sand F, Heuckmann JM, Ikram F, Schmidt R, Ackermann S, Engesser A, Kahlert Y, Vogel W, Altmüller J, Nürnberg P, Thierry-Mieg J et al (2015) Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature 526(7575):700–704. https://doi.org/10.1038/nature14980
Piscuoglio S, Ng CK, Murray M, Burke KA, Edelweiss M, Geyer FC, Macedo GS, Inagaki A, Papanastasiou AD, Martelotto LG, Marchio C, Lim RS, Ioris RA, Nahar PK, Bruijn ID, Smyth L, Akram M, Ross D, Petrini JH et al (2016) Massively parallel sequencing of phyllodes tumours of the breast reveals actionable mutations, and TERT promoter hotspot mutations and TERT gene amplification as likely drivers of progression: TERT alterations in Phyllodes tumors. J Pathol 238(4):508–518. https://doi.org/10.1002/path.4672
Ramachandran C, Fonseca HB, Jhabvala P, Escalon EA, Melnick SJ (2002) Curcumin inhibits telomerase activity through human telomerase reverse transcriptase in MCF-7 breast cancer cell line. Cancer Lett 184(1):1–6. https://doi.org/10.1016/S0304-3835(02)00192-1
Ramirez RD, Wright WE, Shay JW, Taylor RS (1997) Telomerase activity concentrates in the mitotically active segments of human hair follicles. J Investig Dermatol 108(1):113–117. https://doi.org/10.1111/1523-1747.ep12285654
Ren K-W, Li Y-H, Wu G, Ren J-Z, Lu H-B, Li Z-M, Han X-W (2017) Quercetin nanoparticles display antitumor activity via proliferation inhibition and apoptosis induction in liver cancer cells. Int J Oncol 50(4):1299–1311. https://doi.org/10.3892/ijo.2017.3886
Renaud S (2005) CTCF binds the proximal exonic region of hTERT and inhibits its transcription. Nucleic Acids Res 33(21):6850–6860. https://doi.org/10.1093/nar/gki989
Roake CM, Artandi SE (2020) Regulation of human telomerase in homeostasis and disease. Nat Rev Mol Cell Biol 21(7):384–397. https://doi.org/10.1038/s41580-020-0234-z
Robart AR, Collins K (2011) Human telomerase domain interactions capture DNA for TEN domain-dependent processive elongation. Mol Cell 42(3):308–318. https://doi.org/10.1016/j.molcel.2011.03.012
Rouda S, Skordalakes E (2007) Structure of the RNA-binding domain of telomerase: implications for RNA recognition and binding. Structure 15(11):1403–1412. https://doi.org/10.1016/j.str.2007.09.007
Rupaimoole R, Slack FJ (2017) MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16(3):203–222. https://doi.org/10.1038/nrd.2016.246
Saito T, Matsuda Y, Suzuki T, Hayashi A, Yuan X, Saito M, Nakayama J, Hori T, Ishikawa F (1997) comparative gene mapping of the human and mouse TEP1 genes, which encode one protein component of telomerases. Genomics 46(1):46–50. https://doi.org/10.1006/geno.1997.5005
Sharma A, Rajappa M, Saxena A, Sharma M (2007) Telomerase activity as a tumor marker in Indian women with cervical intraepithelial neoplasia and cervical cancer. Mol Diag Ther 11(3):193–201. https://doi.org/10.1007/BF03256241
Shay JW, Wright WE (2011) Role of telomeres and telomerase in cancer. Semin Cancer Biol 21(6):349–353. https://doi.org/10.1016/j.semcancer.2011.10.001
Shin K-H, Kang MK, Dicterow E, Park N-H (2003) Hypermethylation of the hTERT promoter inhibits the expression of telomerase activity in normal oral fibroblasts and senescent normal oral keratinocytes. Br J Cancer 89(8):1473–1478. https://doi.org/10.1038/sj.bjc.6601291
Shin-ya K, Wierzba K, Matsuo K, Ohtani T, Yamada Y, Furihata K, Hayakawa Y, Seto H (2001) Telomestatin, a Novel telomerase inhibitor from Streptomyces anulatus. J Am Chem Soc 123(6):1262–1263. https://doi.org/10.1021/ja005780q
Smith S (1998) Tankyrase, a Poly(ADP-Ribose) Polymerase at human telomeres. Science 282(5393):1484–1487. https://doi.org/10.1126/science.282.5393.1484
Smith EM, Pendlebury DF, Nandakumar J (2020) Structural biology of telomeres and telomerase. Cell Mol Life Sci 77(1):61–79. https://doi.org/10.1007/s00018-019-03369-x
Spiegl-Kreinecker, S., Lötsch, D., Neumayer, K., Kastler, L., Gojo, J., Pirker, C., Pichler, J., Weis, S., Kumar, R., Webersinke, G., Gruber, A., and Berger, W., 2018. TERT promoter mutations are associated with poor prognosis and cell immortalization in meningioma. Neuro-Oncology, 20(12), 1584–1593. doi: https://doi.org/10.1093/neuonc/noy104
Svahn F, Paulsson J, Stenman A, Fotouhi O, Mu N, Murtha T, Korah R, Carling T, Bäckdahl M, Wang N, Juhlin C, Larsson C (2018) TERT promoter hypermethylation is associated with poor prognosis in adrenocortical carcinoma. Int J Mol Med. https://doi.org/10.3892/ijmm.2018.3735
Szostak JW, Blackburn EH (1982) Cloning yeast telomeres on linear plasmid vectors. Cell 29(1):245–255. https://doi.org/10.1016/0092-8674(82)90109-X
Tahara H, Kuniyasu H, Yokozaki H, Yasui W, Shay JW, Ide T, Tahara E (1995) Telomerase activity in preneoplastic and neoplastic gastric and colorectal lesions. Clin Cancer Res 1(11):1245–1251
Tiedemann RE, Schmidt J, Keats JJ, Shi C-X, Zhu YX, Palmer SE, Mao X, Schimmer AD, Stewart AK (2009) Identification of a potent natural triterpenoid inhibitor of proteosome chymotrypsin-like activity and NF-κB with antimyeloma activity in vitro and in vivo. Blood 113(17):4027–4037. https://doi.org/10.1182/blood-2008-09-179796
Tippani R, Prakhya L, Porika M, Sirisha K, Abbagani S, Thammidala C (2014) Pterostilbene as a potential novel telomerase inhibitor: molecular docking studies and its in vitro evaluation. Curr Pharm Biotechnol 14(12):1027–1035. https://doi.org/10.2174/1389201015666140113112820
Tsujioka T, Yokoi A, Itano Y, Takahashi K, Ouchida M, Okamoto S, Kondo T, Suemori S, Tohyama Y, Tohyama K (2015) Five-aza-2′-deoxycytidine-induced hypomethylation of cholesterol 25-hydroxylase gene is responsible for cell death of myelodysplasia/leukemia cells. Sci Rep 5(1):16709. https://doi.org/10.1038/srep16709
Turgut Cosan D, Soyocak A, Basaran A, Degirmenci İ, Gunes HV, Mutlu Sahin F (2011) Effects of various agents on DNA fragmentation and telomerase enzyme activities in adenocarcinoma cell lines. Mol Biol Rep 38(4):2463–2469. https://doi.org/10.1007/s11033-010-0382-x
Umbricht CB, Saji M, Westra WH, Udelsman R, Zeiger MA, Sukumar S (1997) Telomerase activity: a marker to distinguish follicular thyroid adenoma from carcinoma. Cancer Res 57(11):2144–2147
Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE (2008) Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 132(6):945. https://doi.org/10.1016/j.cell.2008.01.019
Vinagre J, Almeida A, Pópulo H, Batista R, Lyra J, Pinto V, Coelho R, Celestino R, Prazeres H, Lima L, Melo M, da Rocha AG, Preto A, Castro P, Castro L, Pardal F, Lopes JM, Santos LL, Reis RM (2013) Frequency of TERT promoter mutations in human cancers. Nat Commun 4(1):2185. https://doi.org/10.1038/ncomms3185
Vincent K, Pichler M, Lee G-W, Ling H (2014) MicroRNAs, genomic instability and cancer. Int J Mol Sci 15(8):14475–14491. https://doi.org/10.3390/ijms150814475
Wang J, Zhang P, Tu Z (2003) Effects of quercetin on proliferation of lung cancer cell line A549 by down-regulating hTERT gene expression. J Third Mil Med Univ 24
Wang, X. Y., Fan, Y., Zhang, Y. L., Zhong, X. M. (2010) Effect of resveratrol on promoter and human telomerase reverse transcriptase(hTERT) expression of human colorectal carcinoma cell—《Journal of Jiangsu University(Medicine Edition)》2010年01期. https://en.cnki.com.cn/Article_en/CJFDTotal-ZJYZ201001015.htm
Wang, N., Liu, T., Sofiadis, A., Juhlin, C. C., Zedenius, J., Höög, A., Larsson, C., and Xu, D., 2014. TERT promoter mutation as an early genetic event activating telomerase in follicular thyroid adenoma (FTA) and atypical FTA: TERT Promoter Mutation in FTA/AFTA. Cancer, 120(19), 2965–2979. doi: https://doi.org/10.1002/cncr.28800
Wang N, Kjellin H, Sofiadis A, Fotouhi O, Juhlin CC, Bäckdahl M, Zedenius J, Xu D, Lehtiö J, Larsson C (2016) Genetic and epigenetic background and protein expression profiles in relation to telomerase activation in medullary thyroid carcinoma. Oncotarget 7(16):21332–21346. https://doi.org/10.18632/oncotarget.7237
Watson JD (1972) Origin of concatemeric T7DNA. Nat New Biol 239(94):197–201. https://doi.org/10.1038/newbio239197a0
Weber JA, Baxter DH, Zhang S, Huang DY, How Huang K, Jen Lee M, Galas DJ, Wang K (2010) The MicroRNA spectrum in 12 body fluids. Clin Chem 56(11):1733–1741. https://doi.org/10.1373/clinchem.2010.147405
Wei, J. W., Fan, Y., Y. L. Zhang, Wu, Y., Wang, X. Y., & Chen, P. (2007) Effects of Quercetin on telomerase activity and apoptosis in gastric cancer cells.《Shandong Medical Journal》2007年35期. https://en.cnki.com.cn/Article_en/CJFDTotal-SDYY200735008.htm
Wright WE, Pereira-Smith OM, Shay JW (1989) Reversible cellular senescence: implications for immortalization of normal human diploid fibroblasts. Mol Cell Biol 9(7):3088–3092. https://doi.org/10.1128/MCB.9.7.3088
Wu K-J, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, Dalla-Favera R (1999a) Direct activation of TERT transcription by c-MYC. Nat Genet 21(2):220–224. https://doi.org/10.1038/6010
Wu HL, Hsu CY, Liu WH, Yung BY (1999b) Berberine-induced apoptosis of human leukemia HL-60 cells is associated with down-regulation of nucleophosmin/B23 and telomerase activity. Int J Cancer 81(6):923–929. https://doi.org/10.1002/(sici)1097-0215(19990611)81:6<923::aid-ijc14>3.0.co;2-d
Wu Y, Tang Y, Jiang ZQ (2000) Diagnosis of human bladder cancer by detecting the telomerase activity in exfoliated urothelial cells. Hunan Yi Ke Da Xue Xue Bao = Hunan Yike Daxue Xuebao = Bulletin of Hunan Medical University 25(6):599–600
Wu S, Huang P, Li C, Huang Y, Li X, Wang Y, Chen C, Lv Z, Tang A, Sun X, Lu J, Li W, Zhou J, Gui Y, Zhou F, Wang D, Cai Z (2014) Telomerase reverse transcriptase gene promoter mutations help discern the origin of urogenital tumors: a genomic and molecular study. Eur Urol 65(2):274–277. https://doi.org/10.1016/j.eururo.2013.10.038
Xie M, Mosig A, Qi X, Li Y, Stadler PF, Chen JJ-L (2008) Structure and function of the smallest vertebrate telomerase RNA from teleost fish. J Biol Chem 283(4):2049–2059. https://doi.org/10.1074/jbc.M708032200
Xu X, Chen W, Miao R, Zhou Y, Wang Z, Zhang L, Wan Y, Dong Y, Qu K, Liu C (2015) MiR-34a induces cellular senescence via modulation of telomerase activity in human hepatocellular carcinoma by targeting FoxM1/c-Myc pathway. Oncotarget 6(6):3988–4004. https://doi.org/10.18632/oncotarget.2905
Ye JZ-S (2004) POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev 18(14):1649–1654. https://doi.org/10.1101/gad.1215404
Yu J, Guo Q-L, You Q-D, Zhao L, Gu H-Y, Yang Y, Zhang H-W, Tan Z, Wang X (2006) Gambogic acid-induced G2/M phase cell-cycle arrest via disturbing CDK7-mediated phosphorylation of CDC2/p34 in human gastric carcinoma BGC-823 cells. Carcinogenesis 28(3):632–638. https://doi.org/10.1093/carcin/bgl168
Yuan P, Cao J, Abuduwufuer A, Wang L-M, Yuan X-S, Lv W, Hu J (2016) Clinical characteristics and prognostic significance of TERT promoter mutations in cancer: a cohort study and a meta-analysis. PLoS One 11(1):e0146803. https://doi.org/10.1371/journal.pone.0146803
Zamin LL, Filippi-Chiela EC, Dillenburg-Pilla P, Horn F, Salbego C, Lenz G (2009) Resveratrol and quercetin cooperate to induce senescence-like growth arrest in C6 rat glioma cells. Cancer Sci 100(9):1655–1662. https://doi.org/10.1111/j.1349-7006.2009.01215.x
Zhang A, Zheng C, Lindvall C, Hou M, Ekedahl J, Lewensohn R, Yan Z, Yang X, Henriksson M, Blennow E, Nordenskjöld M, Zetterberg A, Björkholm M, Gruber A, Xu D (2000) Frequent amplification of the telomerase reverse transcriptase gene in human tumors. Cancer Res 60(22):6230–6235
Zhang W, Yang P, Gao F, Yang J, Yao K (2014) Effects of epigallocatechin gallate on the proliferation and apoptosis of the nasopharyngeal carcinoma cell line CNE2. Exp Ther Med 8(6):1783–1788. https://doi.org/10.3892/etm.2014.2020
Zhang H, Weng X, Ye J, He L, Zhou D, Liu Y (2015) Promoter hypermethylation of TERT is associated with hepatocellular carcinoma in the Han Chinese population. Clin Res Hepatol Gastroenterol 39(5):600–609. https://doi.org/10.1016/j.clinre.2015.01.002
Zhao X, Tian X, Kajigaya S, Cantilena CR, Strickland S, Savani BN, Mohan S, Feng X, Keyvanfar K, Dunavin N, Townsley DM, Dumitriu B, Battiwalla M, Rezvani K, Young NS, Barrett AJ, Ito S (2016) Epigenetic landscape of the TERT promoter: a potential biomarker for high risk AML/MDS. Br J Haematol 175(3):427–439. https://doi.org/10.1111/bjh.14244
Zhao T, Wang H-J, Zhao W-W, Sun Y-L, Hu L-K (2017) Gambogic acid improves non-small cell lung cancer progression by inhibition of mTOR signaling pathway. Kaohsiung J Med Sci 33(11):543–549. https://doi.org/10.1016/j.kjms.2017.06.013
Zheng D-S, Chen L-S (2017) Triterpenoids from Ganoderma lucidum inhibit the activation of EBV antigens as telomerase inhibitors. Exp Ther Med 14(4):3273–3278. https://doi.org/10.3892/etm.2017.4883
Zhou XZ, Lu KP (2001) The Pin2/TRF1-interacting protein PinX1 Is a potent telomerase inhibitor. Cell 107(3):347–359. https://doi.org/10.1016/S0092-8674(01)00538-4
Zhou J, Dai W, Song J (2016) MiR-1182 inhibits growth and mediates the chemosensitivity of bladder cancer by targeting hTERT. Biochem Biophys Res Commun 470(2):445–452. https://doi.org/10.1016/j.bbrc.2016.01.014
Zimmermann M, Kibe T, Kabir S, de Lange T (2014) TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev 28(22):2477–2491. https://doi.org/10.1101/gad.251611.114
Zinn RL, Pruitt K, Eguchi S, Baylin SB, Herman JG (2007) HTERT Is Expressed in Cancer Cell Lines Despite Promoter DNA methylation by preservation of unmethylated DNA and active chromatin around the transcription start site. Cancer Res 67(1):194–201. https://doi.org/10.1158/0008-5472.CAN-06-3396
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Kumar, R., Gupta, N., Sharma, A. (2022). Telomerase and Its Therapeutic Implications in Cancer. In: Basu, S.K., Panda, C.K., Goswami, S. (eds) Cancer Diagnostics and Therapeutics . Springer, Singapore. https://doi.org/10.1007/978-981-16-4752-9_14
Download citation
DOI: https://doi.org/10.1007/978-981-16-4752-9_14
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-4751-2
Online ISBN: 978-981-16-4752-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)