Abstract
The natural polyester polyhydroxyalkanoate (PHA) is synthesized as an energy storage via thioester exchange reaction in microbial cells. The thermal and mechanical properties of PHA can be varied by modifying the monomeric composition, molecular weight, and chemical modification. To date, many efforts have been made to understand the polymerization mechanism and industrialization of PHA. PHA synthase (Acyltransferase; EC2.3) is the key player for making stereochemically regulated polyesters. PHA synthase should be one of the most important targets for the synthetic biology of PHA. In 2017, a major breakthrough occurred in the PHA research field, whereby the tertiary structures of two PHA synthases from the class I enzyme have been solved. Based on the crystal structures of the PHA synthases, the detailed reaction mechanism of PHA synthase is discussed in this chapter. Common and unique structural elements are extracted through structure- function relationships between both enzymes. Additionally, function-based studies of PHA synthases are introduced as another milestone. The discovery of a lactate-polymerizing enzyme (LPE) evolved from a PHA synthase is a typical case. The effectiveness of the evolutionary engineering of PHA synthases is demonstrated through case studies including the creation of new polyesters as well as tailor-made PHA production.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Beneficial mutation
- Domain structure
- Evolutionary engineering
- Lactate-polymerizing enzyme
- Open-closed form
- Polyhydroxyalkanoate
- Polyhydroxyalkanoate synthase
- Polymerization mechanism
- Substrate specificity
- Tertiary structure
7.1 Preface
Recently, bio-based materials have been synthesized through biorefinery technologies from naturally occurring polysaccharides or plant oils. Polyhydroxyalkanoates (PHAs) are bacterial storage polyesters that can be used as bio-based and biodegradable plastics and have attracted considerable research interests as an alternative to petroleum-derived plastics. Biodegradable and/or compostable plastic has been in increasing demand, particularly in the USA and Europe. In fact, KANEKA Company in Japan built a pilot-scale plant for PHA (Biodegradable Polymer PHBHTM) production in 2011. A representative polymer of PHA is a homopolymer of optically active (R)-3-hydroxybutyrate, poly(3-hydroxybutyrate) [P(3HB)], which was discovered by Dr. Maurice Lemoigne from the Institute of Pasteur, France, in the 1920s [1]. P(3HB) can be extracted from fermented bacterial cells, and the resulting product resembles some commodity plastics such as petroleum-derived polypropylene. Over 160 different monomeric constituents have been identified so far in the PHA family, giving rise to polymers with diverse properties [2, 3].
Generally, many biopolymers with ordered monomer sequences are synthesized via template-dependent polymerization. For instance, proteins are representative sequence-ordered polymers that are synthesized by incorporating amino acid monomers into assigned positions using mRNA as a template. On the other hand, PHAs are polymerized synthesized via polymerization in a non-template-dependent manner by catalysis of PHA synthase (acyltransferase), a key enzyme relevant for polymer synthesis [4]. In the process of PHA synthesis, the transfer of an acetyl group from one molecule to another is a fundamental biochemical process. PHA synthase belongs to the acetyltransferase superfamily, which catalyzes the transfer of the acetyl group from acetyl coenzyme A (as the “donor”) to a counterpart (as the “acceptor”). From the perspective of structural biology, the acetyltransferase superfamily is a case study on how a common acetyltransferase domain evolved to serve a wide variety of functions. The term acyl-XYZ is a general expression of acyl groups. Here we use the concrete terms fitting to individual cases like ‘acetyl’-CoA corresponding to the alkyl-chain-lengths.
In principle, monomers for PHA, 3-hydroxyalkanoic acids (3HAs), are randomly copolymerized with different monomeric constituents tightly depending on the substrate specificity of PHA synthase as well as monomer flux generated in bacterial cells. PHA synthase-catalyzed polymerization that can proceed in the water system should be the ultimate green chemistry over chemical polymerization, which frequently requires organic solvents and high-energy environments. Basically, the chemical polymerization proceeds via a release of water molecules upon ester-bond formation. Therefore, elimination of water molecules is necessary to obtain long-chain polymers. In this light, coenzyme A (CoA) of the PHA precursor is the preferable form for efficient operation of the thioester exchange reaction in a biological system to elongate the polymer chain. Thus, the polymer products tend to achieve ultrahigh molecular weights and have extremely high chirality.
In the green polymerization system, the creation of designer PHA synthases is an attractive project in order to create new polymers incorporating new monomers as well as tailor-made biosynthesis of the PHAs with designable properties. To this end, how can we engineer the PHA synthase? Since 1999, evolutionary engineering of the enzyme has been extensively applied to the alteration of functional properties without any tertiary structure information on PHA synthase [5, 6]. For the last few decades, beneficial mutations closely related to the activity and substrate specificity of three kinds of PHA synthases have been addressed through an evolutionary engineering approach [5, 6]. This evolutionary lineage of enzymes can be considered the most important key point for accomplishing the synthetic biology of polymeric material manufacturing. It should be of great interest to achieve the incorporation of new and unnatural monomers into the polymeric backbone. The word “unnatural” here means that the constituent itself is not natural among PHA members but is a naturally occurring substance. A typical example is lactic acid (LA). Given that a lactate-polymerizing enzyme (LPE) has been artificially evolved from one of the PHA synthases, LA-based polymers could be synthesized in vivo by using LPE [7,8,9]. This major breakthrough has opened the door to expand the diversity of monomers that can be incorporated as unnatural new building blocks into the polymeric backbones [10, 11].
Great efforts have long been made to perform a mechanistic study on PHA synthase- catalyzed polymerization via solving the crystal structure. Most currently, we have made another breakthrough, namely, the successful solution of tertiary structures of two enzymes that belong to class I PHA synthase [12]. One PHA synthase is derived from Cupriavidus necator (formally known as Ralstonia eutropha) [13,14,15], and another one is derived from Chromobacterium sp. USM2 [16]. The former is the most-studied PHA synthase, and the latter is a PHA synthase with the highest enzymatic activity reported to date. Such a monumental achievement would provide the PHA research community with the structure-function relationship of natural and evolved PHA synthases. In this chapter, first, the molecular basis and mechanistic study of PHA synthase will be described based on the solved crystal structures. As a second topic, achievements in the evolutionary engineering of PHA synthases will be discussed.
7.2 Mechanistic Studies of Polymerization for PHA Synthesis Based on Biochemical Findings
In the past, various biochemical studies were performed using the most-studied class I synthase from Cupriavidus necator (PhaCCn) and class III synthase from Chromatium vinosum (PhaCECv). Since all PHA synthases contain a PhaC box sequence ([GS]-X-C-X-[GA]-G) [17] and show a high sequence similarity to prokaryotic lipases, the catalytic mechanism of PhaC was proposed using lipase as a template [18]. Like PhaCs, lipases also belong to the superfamily of α-/β-hydrolase, which possesses a catalytic triad of Ser, Asp, and His.
Two types of catalytic mechanisms were proposed for PhaC: (i) the non-processive ping-pong model and (ii) the processive model [4]. In both proposals, the active site in PhaC is comprised of catalytic Cys, His, and Asp, forming a catalytic triad. The conserved Cys works as a nucleophile attacking the thiol group of the thioester of substrate acyl-CoA [19,20,21,22]. The catalytic His acts as a general base catalyst to the catalytic Cys, could assist in the deprotonation of the side-chain thiol group, and accelerates the nucleophilic reaction [18, 21]. Last, the negatively charged Asp was proposed to activate the 3-hydroxyl group of the substrate acyl-CoA, enhancing the nucleophilic reaction [18, 21]. In simpler terms, Cys is the nucleophile, His is the initiator, and Asp is the elongator.
The non-processive ping-pong model requires an active site that comprises two catalytic Cys residues from two PhaC monomers to come close together for chain transfer elongation of PHA, whereas in the processive model, only a single set of active sites from a single PhaC monomer is required. Both models share the same initiation step, where the Cys is activated through deprotonation, assisted by His, allowing the activated nucleophilic Cys to attack the thioester carbon, and subsequently releases CoA from the HB-CoA substrate as a by-product. This process forms a covalent intermediate of HB-Cys. In the ping-pong model, the second substrate HB-CoA enters the active site formed by the PhaC dimer and is covalently bound to the free Cys through the same initiation reaction. When both Cys residues are occupied, the Asp might act as a general base catalyst to attack the hydroxyl group of HB-Cys, which subsequently activates the HB bound to the first Cys to attack the thioester bond of the second HB-Cys, forming a (HB)2-Cys covalent intermediate. The free Cys will then attack another newly entered substrate similar to the initiation step. Again, the Asp from this active site will activate the substrate and initiate a chain transfer reaction, imitating a ping-pong game (Fig. 7.1a). In the processive model, the elongation step starts with the activation of HB-CoA by Asp, resulting in an attack on the thioester bond of HB-Cys. Then, a (HB)2-CoA non-covalent intermediate is formed. The unbound Cys is once again free to attack the thioester of (HB)2-CoA, forming the covalent intermediate of (HB)2-Cys, and releases CoA (Fig. 7.1b).
7.3 Tertiary Structures of PHA Synthases
The molecular structure of an enzyme always relates to its functions and provides valuable insights into understanding its working mechanism. The structural information is often useful for designing beneficial mutations and modifying substrate- binding pockets, with the goal of engineering a robust high-performance enzyme for industrial applications.
PHA synthase (PhaC) is the key enzyme involved in the polymerization reaction of PHA, a family of bacterial thermoplastic polyesters with properties like commodity plastics. PHA is being developed as a bio-based and biodegradable alternative for petrochemical plastics. Lack of structural information of PHA synthases has greatly hindered the progress in understanding its catalytic mechanism, which is important for the design and synthesis of superior bioplastics comparable to synthetic plastics. The most desired PhaC crystal structure is that of Cupriavidus necator H16 (formerly known as Alcaligenes eutrophus and Ralstonia eutropha) which is the model bacterium used in PHA research. Many studies on PHA biosynthesis have been done using this model bacterium. The PhaC of C. necator (PhaCCn) is grouped into the class I PHA synthase. PhaCs in this group are composed of an approximately 60 kDa single subunit polypeptide that forms a homodimer and polymerizes preferentially short-chain length (SCL) PHA monomers such as 3-hydroxybutyryl-CoA. In addition to class I, there are three other classes of PhaCs [12]. The crystal structure of the catalytic domain of C. necator PhaC (PhaCCn-CAT) at a resolution of 1.80 Å was recently published by two independent groups (PDB 5T6O [13] and PDB 5HZ2 [14]). At approximately the same time, the catalytic domain structure of another interesting class I PhaC from Chromobacterium sp. USM2 (PhaCCs) was also published. This latter structure of PhaCCs-CAT was determined at a much higher resolution of 1.48 Å (PDB 5XAV) [16]. Interestingly, all three crystals were grown after the partial degradation of the protein during crystallization. The full-length PhaCs could not be crystallized, probably because of the mobility of their N-terminal domain. In this chapter, we will discuss on the crystal structures of the catalytic domain of the PhaCs from Chromobacterium sp. USM2 and C. necator H16. This new structural information will then be compared with the proposed catalytic mechanisms of PHA synthases that are widely used to date [18, 21]. In addition, this chapter will also discuss the effects of mutations on the properties of PhaCs, especially with regard to the synthesis of new polyesters.
7.3.1 Crystal Structures of PhaC from Cupriavidus necator
The structures of the catalytic domain of the PhaC from C. necator (PhaCCn-CAT) were determined by two independent groups from the Massachusetts Institute of Technology (MIT) and Kyungpook National University (KNU). Both groups published almost the same crystal structures at 1.80 Å resolution [13, 14]. In the PhaCCn-CAT structure reported by Wittenborn et al. [13], the catalytic Cys319 had been mutated to Ala in order to improve protein stability. Their structure (PDB 5T6O) consisted of residues 201–368 and 378–589, whereas residues 369–377 were disordered (Fig. 7.2a). This disordered region was part of the protein structure that was flexible and, thus, not visible in the electron density map of the crystal. The other group was successful in determining the crystal structure of PhaCCn-CAT (PDB 5HZ2) using the wild-type PhaCCn [14]. The KNU group reported that the full- length PhaCCn protein succumbed to proteolysis, which occurred at Arg192. Therefore, by substituting the Arg192 with Ala, they claimed that the full-length PhaCCn crystal was successfully obtained, though it was diffracted poorly despite multiple trials [15]. The structure of PhaCCn-CAT from KNU consisted of residues 202–589 and displayed a D-loop, which appeared to be disordered in the previously reported structure by the group from MIT (Fig. 7.2b). The length between the catalytic Cys in a dimer of PhaCCn-CAT measures approximately 33 Å, while the total length of the dimeric PhaCCn-CAT is approximately 99 Å (~10 nm), with both the width and height at approximately 50 Å (~5 nm) each (Fig. 7.2a, b).
Crystal structures of dimeric PhaCCn-CAT
(a) PhaCCn-CAT was deposited in PDB as 5T6O; the catalytic Cys was mutated to Ala to improve protein stability. The disordered region is shown as a dotted line. (b) PhaCCn-CAT was deposited in PDB as 5HZ2, with a D-loop (green), which was derived as the disordered region in (a). (c) A disulfide bond was observed between a pair of non-conserved Cys residues in both structures of PhaCCn-CAT, which might stabilize a small opening for substrate entry. The distance of the catalytic Cys319 in the dimer is approximately 33 Å. PhaCCn-CAT (PDB 5T6O) is color-coded in pink and magenta; PhaCCn-CAT (PDB 5HZ2) is color-coded in orange and yellow
Overall, both the crystal structures of PhaCCn-CATs show the same conformation, adopting an α-/β-hydrolase fold core subdomain (residues 214–346, 472–589) and a CAP subdomain (residues 347–471) (Fig. 7.4c). PhaCCn-CAT forms dimers mediated by the CAP subdomains. The residues involved in the dimerization are Asp401, Val403, Val407, Val408, and Leu412 from one protomer as well as Ile357, Val360, and Glu364 from another protomer. Mutational analyses of these residues showed a lower dimer ratio and decreased activity compared to that of the wild-type PhaCCn [14]. The catalytic triad residues (Cys319, Asp480, and His508) of PhaCCn were positioned close to each other and formed hydrogen bonds in the active site [13, 14]. Unfortunately, the architecture of the catalytic triad was incomplete, as the Cys319 was mutated to Ala in 5T6O and the imidazole ring was flipped from the others in 5HZ2 (Fig. 7.3c). The flipped imidazole ring is not favorable, as it will affect the interaction between Asp with the Nδ atom of His [13, 16]. Additionally, another ring nitrogen atom, Nε of the His is positioned at a close distance (3.8 Å) to the Cys (Sγ) and, at the same time, bound to a water molecule (2.8 Å) in the structure of PhaCCs-CAT [16]. These observations would not be possible if the imidazole ring of the His was flipped around and changed the positions of the imidazole ring nitrogen atoms.
Crystal structures of dimeric PhaCCs-CAT
(a) The dimeric structure of PhaCCs-CAT comprises protomer A (core, pink; CAP, magenta) and protomer B (core, cyan; CAP, yellow). The interactions of the dimer are mediated mainly by the CAP subdomain. The N-terminal ends of both protomers are located at the same direction. (b) Water molecules (red spheres with surface dots) are observed around the nucleophilic Cys291. Part of the CAP subdomain has been removed to show the water cavity. Water molecules are divided into Site A (yellow residues) and Site B (cyan residues) by Cys291. Hydrophobic amino acids are mostly found in the cavity, except for His324, Tyr412 (Site A), and Asn220 (Site B). Hydrogen bonds involving catalytic triad residues are shown as dotted lines. (c) The overlay of the catalytic triad of all three PhaC-CATs. The Cys319 was mutated to Ala in PDB 5T6O (magenta sticks); the imidazole ring was flipped from others in PDB 5HZ2 (yellow sticks); the catalytic triad in PDB 5XAV shows a more accurate and complete architecture (orange sticks)
A detailed analysis of both crystal structures of PhaCCn-CAT revealed the presence of a non-conserved Cys-Cys disulfide bond between Cys382 and Cys438 (Fig. 7.2c) [16]. Physiologically, the occurrence of such a disulfide bond is not possible because the bacterial cytoplasm is maintained at a reduced state in the redox potential. It is highly possible that the observed disulfide bond formation in the crystal structures of PhaCCn-CAT is an artifact that occurred during crystallization. As it takes time for the crystals to grow, the proteins become oxidized and result in the formation of the disulfide bond. This disulfide bond is not observed in the structure of PhaCCs-CAT as PhaCCs does not possess such Cys residues. The artificial disulfide bond probably is the key factor in stabilizing the partially open form of PhaCCn-CAT through the unfolding of αA and ηA helices of PhaCCs-CAT into the D-loop of the PhaCCn-CAT. Through the opening, a possible substrate access channel filled with water was deduced from the structure [13]. Additionally, a putative product egress route was proposed; although the amino acid residues in the proposed route are conserved, a certain extent of conformational changes is required for the polymerization reaction to occur [13]. Another important feature of PhaC, its substrate specificity, was also explained using the crystal structure. A possible acyl moiety-binding pocket composed of mainly hydrophobic residues, i.e., Pro245, Ile252, Leu253, Phe318, Thr393, and Trp425, was proposed to stabilize the 3HB moiety of the substrate [14].
7.3.2 Crystal Structure of PhaC from Chromobacterium sp. USM2
In addition to the structure of PhaCCn-CAT from the most-studied class I PhaCCn, the structure of the most active PhaC from Chromobacterium sp. USM2 (PhaCCs-CAT) was determined, by a structural group from Nara Institute of Science and Technology (NAIST), at a resolution of 1.48 Å [16]. The catalytic domain of the PhaCCs-CAT is characterized by an amino acid sequence that belongs to the superfamily of α-/β-hydrolase. This domain exists as a globular structure. Structural analysis revealed that the PhaCCs-CAT (residues 175–567) is folded into α-/β-core subdomain (residues 186–318 and 439–562) and CAP subdomain (residues 319–438) (Fig. 7.3a, 7.4b). The α-/β-core subdomain adopts the regular α-/β-hydrolase fold with the central β-sheet displaying a left-handed superhelical twist and the first β-strand crossing the eighth β-strand (β9) at ~90° and the final strand (β11) at ~180°. The CAP subdomain projects topologically from the α-/β-core subdomain and completely covers the active site. As expected, the active site consists of the catalytic triad residues (Cys291, His477, and Asp447), which have been confirmed before by primary sequence analysis of many PHA synthases and mutation studies [23, 24].
Comparison of PhaCCn-CAT and PhaCCs-CAT
(a) The electrostatic surface potential representation of PhaCCn-CAT (PDB 5HZ2) and (b) PhaCCs-CAT (PDB 5XAV). Blue represents a basic/positively charged surface; red represents an acidic/negatively charged surface; white represents a neutral/hydrophobic surface. (c) The monomer structures of PhaCCn-CAT, the core subdomain (gray and blue), the CAP subdomain (yellow), and the LID region (green) are shown. (d) The monomer structures of PhaCCs-CAT, the core subdomain (cyan and blue), the CAP subdomain (magenta), and the LID region (green) are shown. A major difference is observed at the LID region (both green) and the non-conserved disulfide bond in (c)
A common characteristic of the members of α-/β-hydrolase superfamily is the possession of the important catalytic triad residues, which have been identified as Cys291, His477, and Asp447 in PhaCCs. Even though these residues are not close to each other in the primary amino acid sequence, they are brought closer together to form the catalytic pocket through hydrogen bonds (Fig. 7.3b, c). In the observed structure of PhaCCs-CAT, the catalytic pocket was completely covered by the CAP subdomain, which essentially blocks the substrates from entering. Therefore, this structure was called a closed conformation, unlike the partially open conformation that was observed for the C. necator PhaCCn-CAT. The 1.48 Å high-resolution structure of PhaCCs-CAT also allowed the visualization of water molecules both inside and outside of the catalytic pocket. The presence of the trapped free water molecules in the catalytic pocket is an indication that the LID region (residues 327–386), which is part of the CAP subdomain (residues 319–438), is a dynamic and flexible structure allowing water to get inside the catalytic pocket (Fig. 7.3b, 7.4d). Water molecules may play an important role in the polymerization process at the active site [25]. Water molecules may also play a role in the stabilization of the growing PHA chains [26], and the presence of water in PHA granules is well documented [27]. The water molecules were observed at both sides of the catalytic Cys and divided into two groups accordingly, namely, Site A and Site B. These sites are two possible cavities for the allocation of the acyl moiety of (R)-hydroxyalkanoyl-CoA substrate during the nucleophilic reaction. For instance, the acyl moiety, such as the 3HB moiety of the 3HB-CoA substrate, fits into these sites, and the thioester bond of R-hydroxyalkanoyl-CoA can be positioned close enough to the catalytic Cys, allowing the nucleophilic reaction to occur. This positioning of the acyl moiety also allows the formation of an acyl-S-intermediate and subsequent CoA release from the active site at the end of the nucleophilic reaction. Site A is formed by polar Tyr412, His324, and other hydrophobic residues, i.e., Val292, Leu321, Met407, and Ile449. On the other hand, Site B comprises nonpolar residues, Pro216, Pro217, Ile219, Leu224, and Met225, Phe292, Trp392, and only one polar residue, Asn220 [16]. The water molecules can also be used as a clue to search for the possible entry pathway for the water-soluble (R)-hydroxyalkanoyl-CoA substrates; however, this is not the case for PhaCCs-CAT, as the catalytic site is completely covered.
In general, PHA synthases are believed to be more active in the dimeric form and exist in a dynamic equilibrium between monomer and dimer in solution [28,29,30,31]. PhaCCs-CAT forms a face-to-face dimer with a pseudo-dyad axis with the N-terminal end of both protomers facing each other, suggesting that the N-terminal domain might enhance the oligomerization of PhaC through direct contact [16]. However, the exact function of the N-terminal domain remains unknown as it is absent in the current structure. The dimeric conformation of PhaCCs-CAT is most likely mediated by the CAP subdomain through intermolecular interactions such as hydrophobic, salt bridging, and aromatic stacking interactions. Two intimate contact areas can be identified from the structure, i.e., contact areas I and II. Contact area I forms a hydrophobic cluster with nonpolar residues, Leu369, Trp371, Pro386, Phe387, and Leu390, in both molecules A and B. Contact area II forms salt bridges (Arg365 and Glu329) inside the interface composed of Phe332, Phe333, His448, and Leu451. Notably, there is no connection between the two catalytic Cys in the current dimer structure because they are separated by quite a distance (the Sγ atom of Cys291 residues are at a distance of 28.1 Å) for the PHA chain transfer reaction to occur [16].
7.3.3 Structural Differences Between PhaCCn-CAT and PhaCCs-CAT
Since both PhaCCs and PhaCCn share an amino acid sequence identity of 46%, it would be interesting to compare the structure of PhaCCs-CAT with PhaCCn-CAT. In the structural comparison, the catalytic triad and α-/β-core subdomains of all three structures showed good alignment with each other [16]. However, some differences were noted. The first major difference is in the conformational changes of part of the CAP subdomain, known as the LID region (residues Pro327–Pro386 in PhaCCs-CAT; residues Thr335–Pro419 in PhaCCn-CAT) (Fig. 7.4c, d). The conformational changes are marked by two events, i.e., the unfolding of αA and ηA helices in PhaCCs-CAT into a flexible D-loop in PhaCCn-CAT and the folding of αB’ and ηB’ in PhaCCs-CAT into a long α4 helix in PhaCCn-CAT (Fig. 7.4c). These conformational changes result in a small opening for the substrate entry into the catalytic pocket, making PhaCCn-CAT a partially open structure [16].
The second difference between the crystal structures of PhaCCs-CAT and PhaCCn-CAT is their dimeric arrangement. In the structural comparison, by aligning one protomer (molecule A from both PhaC-CATs) from each dimer, another protomer (molecule B) of PhaCCn-CAT was swung ~40 Å and rotated ~120 Å from PhaCCs-CAT [16]. Additionally, the N-terminal ends of both protomers in PhaCCs-CAT are located at the same direction with a distance of 19.2 Å, whereas the N-termini of PhaCCn-CAT are distanced further at 55.1 Å, although still facing the same direction [16]. This phenomenon suggests that the N-terminal domain is involved in enhancing the dimerization of PhaCs through direct interaction, which was supported by the size exclusion analysis of full-length PhaCCs and PhaCCs-CAT [16]. In contrast, the small angle X-ray scattering data from full-length PhaCCn suggests that the N-terminal domains of PhaCCn (PhaCCn-ND) are localized at opposite sites from each other without direct contact [15].
Since the PHA granule is hydrophobic, it might be useful to identify the hydrophobic patches on the surface of PhaC-CATs to understand the localization of PhaCs on PHA granules. However, both PhaC-CATs showed evenly distributed charged surfaces, and no obvious hydrophobic patches were observed (Fig. 7.4a, b). The only difference between the two is that PhaCCn-CAT is more hydrophobic compared to that of PhaCCs-CAT, and this might be the reason for the better protein solubility and expression of PhaCCs. It is also worth mentioning that this information is based solely on the structure’s lack of an N-terminal domain. To obtain the full picture, the unique structures of the N-terminal domain of PhaCs are essential to fill the knowledge gaps.
7.3.4 Proposed Catalytic Mechanisms Based on Crystal Structures
The previously proposed non-processive ping-pong mechanism requires that the catalytic triad residues from both protomers in a dimeric PhaC come close to the interface for substrate transfer reactions (Fig. 7.1a). However, both the current crystal structures of PhaCCn and PhaCCs turn out to be different for such a reaction to occur. Therefore, the currently available structures of the catalytic domain of the 2 PhaCs favors the processive catalytic mechanism (Fig. 7.1b). However, if it is shown in the future that the full-length PhaC adopts a completely different conformation due to its flexible N-terminal domain, the reaction mechanism needs to be revisited.
All three crystal structures, however, do provide an accurate picture of the active site with the three conserved amino acid residues forming the catalytic triad (Cys-His-Asp). As expected, these three amino acids interact with each other through hydrogen bonds in the published crystal structures (PDB 5T6O, 5HZ2 & 5XAV) [13, 14, 16]. Among these three structures, 5XAV shows the most accurate picture of the catalytic triad because, in one structure (5T6O), the important catalytic Cys has been mutated to Ala to stabilize the protein. In the other structure (5HZ2), the imidazole ring of His is represented in a flipped form, which deviates from that shown in the other two structures (Fig. 7.3c). In the catalytic pocket, the position of the side-chain imidazole ring of the catalytic His is close enough to extract a hydrogen/proton from the side-chain thiol group of the catalytic center, Cys. In other words, the deprotonation of Cys allows nucleophilic attacks on the thioester carbon of the acyl-CoA substrate, which results in the formation of an acyl-Cys intermediate. The catalytic Asp was proposed to act as a general base, which is responsible for accelerating the deprotonation of the 3-hydroxyl group of the incoming acyl-CoA substrates in the PHA elongation step [18, 24].
The processive model involving a single set of catalytic triads. (a) In this model, the main difference is the role of His in the elongation, which was previously proposed to be the role of Asp. (b) In this model, the hydroxyl group of the subsequent 3HB-CoA substrate is involved in the elongation. (c) In this alternative model, the hydroxyl group of a second 3HB-CoA attacks the thioester bond of 3HB-Cys to produce (3HB)2-CoA. Then, this non-covalent intermediate leaves the active site
The new crystal structures from PhaCCn and PhaCCs have led to the proposition of three modified catalytic mechanisms [13, 14, 16]. While the initiation step involving acylation of Cys through His in the catalytic triad remains largely the same, the proposed elongation steps are different (Fig. 7.5). In the model based on the structure of the partially open form PhaCCn-CAT (PDB 5T6O), the major difference is the involvement of His in the activation of the hydroxyl group of incoming 3HB-CoA for product elongation (Fig. 5a) [13]. In the model based on the closed form of the PhaCCs-CAT structure (PDB 5XAV), the hydroxyl group from the second 3HB-CoA attacks the 3HB-Cys covalent intermediate forming the non-covalent intermediate (3HB)2-CoA. Then, the free thiol Cys attacks the thioester carbon of (3HB)2-CoA and forms a covalent intermediate of (3HB)2-Cys (Fig. 7.5b) [16]. The role of Asp in the activation of the incoming substrate is still elusive as the exact geometry of the Asp-substrate interaction is not revealed in the free-form structure. These two proposed mechanisms agree with previous reports of a covalent intermediate observed using saturated trimer CoA (sT-CoA), a substrate analogue of (3HB)3-CoA, in which the terminal hydroxyl group has been replaced by a hydrogen, making it a non-hydrolyzable analogue [19, 20, 32]. In the model based on the other structure of PhaCCn-CAT (PDB 5HZ2), the hydroxyl group of the subsequent substrates enters and attacks the 3HB-Cys covalent intermediate and forms a non- covalent intermediate (3HB)2-CoA, which then leaves the catalytic pocket. The cycle repeats with a new incoming 3HB-CoA, which forms a 3HB-Cys covalent intermediate in the catalytic pocket, followed by the reentry of previously released (3HB)2-CoA into the active site to produce (3HB)3-CoA and so on (Fig. 7.5c) [14]. However, this proposal contradicts the presence of sT-CoA, which has been shown to be stably bound to the catalytic Cys. In this case, the elongation process will stop once (3HB)3-CoA (like the non-hydrolyzable sT-CoA) interacts with the catalytic Cys; thus, the elongation process would require another active site through an intersubunit reaction. Although several modified catalytic mechanisms have been proposed based on the newly published structures of PhaCs, the validity of the proposals is still debatable. There are still many questions that remain unanswered, for instance, the exact role of Asp in the catalytic triad and the effect of the highly variable N-terminal domain, which is absent in all the structures.
It took almost three decades since the cloning of the first PhaC gene by three independent groups in the late 1980s until the successful crystallization of partial PhaC by another three independent groups. This achievement marks a significant milestone in the research and development of PHA even though the crystal structures of the two PhaCs are incomplete and different. The crystal structures of the catalytic domain of the class I PhaC from Chromobacterium sp. USM2 (PDB 5XAV) and C. necator (PDB 5T6O; PDB 5HZ2) share a high similarity in the α-/β- core subdomain, which contains the conserved catalytic triad (Cys-His-Asp). However, a big difference was observed in the CAP subdomain structures of the two PhaCs; in PhaCCs-CAT, the CAP subdomain was in a closed form, while in PhaCCn-CAT, it was in a partially open form, probably because of the artificial disulfide bridge in the latter. However, the open and closed forms do provide some insight into the possible conformational changes that may occur during the PhaC-substrate interactions. The crystal structure of a complete PhaC is much needed to get a better understanding of the polymerization mechanism and the subsequent PHA granule biogenesis.
7.4 Functional Alteration of PHA Synthase for Tailor-Made Polyester Synthesis
PHA synthase is a key enzyme for the polyester synthesis in terms of quality and quantity. So far, many researchers have focused on properties of the enzyme, such as activity and substrate specificity, to regulate the synthesized PHA characters (monomer composition and molecular weight) as well as its productivity (content in bacterial cells) (Fig. 7.6). In this chapter, the history of functional alteration of PHA synthases is summarized.
Polymer content depends on the enzyme activity. Monomer composition is governed by the enzyme substrate specificity. Molecular weight is affected by the protein expression.
As shown in Table 7.1, PHA synthases can be classified into three groups based on substrate specificity. PHA synthase derived from Ralstonia eutropha (= Cupriavidus necator) can polymerize short-chain-length-3-hydroxyacyl-coenzyme As (SCL-3HA-CoAs) such as 3-hydroxybutyryl-coenzyme A (3HB-CoA) and 3-hydroxyvaleryl-coenzyme A (3HV-CoA). On the other hand, PHA synthases derived from most Pseudomonads polymerize medium-chain-length-3-hydroxyacyl-coenzyme As (MCL-3HA-CoAs) with relatively long side chains. PHA synthases of Aeromonas caviae and Pseudomonas sp. 61–3 have wide substrate specificities and can polymerize both SCL- and MCL-3HA-CoAs [12].
Evolutionary engineering is a powerful method to change the properties of PHA synthase without any information on tertiary structure. So far, beneficial mutants of PHA synthases were gained by the combination of error-prone PCR-based mutagenesis and screening methods based on the Nile red stain [33]. At the initial stage starting from 1999, beneficial sites and amino acids related to the enzymatic performances (activity, substrate specificity, thermostability, and so on) were explored at the point mutation level by error-prone PCR-based mutagenesis. In the next stage, site-specific saturation mutagenesis at the hot positions was conducted in a parallel way to optimize the improvement of enzymatic performances.
This function-based enzyme engineering strategy was extensively applied to class I and class II PHA synthases. Table 7.2 and Fig. 7.7 show the summaries of beneficial mutations that provide the target enzymes with better functional performances. As stated in the following section, many beneficial mutations causing the improved enzyme performances were gained for the individual PHA synthases through the evolutional program. In Sections 7.3.1 and 7.3.2, typical functional alterations of PHA synthases are introduced.
Beneficial mutations of PHA synthases
PHA synthase consisted of α-/β-hydrolase fold region which plays a crucial catalytic role for enzyme activity exhibition. The N-terminal region is assumed to coordinate enzyme catalysis and substrate recognition in the polymerization [12]. C319 and C296 function as catalytic center sites of PHA synthases. Beneficial mutations related to activity, expression, thermostability, and substrate specificity are mapped on the whole regions of individual PHA synthases. A compatibility in activity improvement between F420S and F392S was confirmed at the corresponding positions (vertically dotted line). In a similar way, substitutions of amino acids at the Q481, A510, and A505 positions affect the enzyme substrate specificity
7.4.1 Improvement in Activity, Thermostability, and Expression
The first trial of in vitro evolution was conducted for the class I PHA synthase derived from R. eutropha (PhaCRe which is the same as PhaCCn described in 7.1–7.2) by error-prone PCR-based mutagenesis. PhaCRe is the most widely studied PHA synthase and was selected as the first target for the model study. Escherichia coli- carrying phaC Re, phaA Re, and phaB Re genes (phaA and phaB are monomer-supplying enzyme genes) can produce P(3-hydroxybutyrate) [P(3HB)], which is the most typical PHA (Fig. 7.9, Pathway I). Mutants of PhaCRes generated by error-prone PCR were expressed in E. coli cells together with PhaARe and PhaBRe, and then the recombinant strains produced P(3HB)s at various accumulation levels. There was a good correlation between P(3HB) accumulation level and PhaCRe activity for the selected seven mutants. This means that high-throughput screening (in vivo) is possible for exploring the beneficial mutants without a time-consuming enzymatic assay (in vitro). However, it was too difficult to gain the positive mutant enzyme starting from the highly active wild-type PHA synthase. Notably, the suppression mutagenesis approach was effective to obtain beneficial mutations causing enhanced activity of PhaCRe. In fact, two positive mutants were obtained by using one mutant harboring the Ser80Pro (S80P) mutation as a starter for the second mutation. The first mutant, Phe420Ser (F420S), exhibited a 2.4-fold increase in specific activity toward 3HB-CoA compared to the wild type [35]. This activity improvement contributed to the increased polymer productivity in vivo. This positive mutation presumably occurred due to the productive dimerization appropriate for enzyme activation of PhaCRe, considering the previous report on the mutation related to the lag-phase reduction in the PhaCRe reaction [29, 45]. To verify the adaptive possibility of a beneficial mutation across classes of PHA synthase, F420S of class I PhaCRe was applied to the class II PHA synthase of Pseudomonas sp. 61–3 (PhaCPs). Phe392Ser (F392S) of PhaCPs, which corresponds to F420S of PhaCRe (Fig. 7.7), resulted in an increased polymer content and an increased co-monomer fraction compared to wild-type PhaCPs [53], indicating that a beneficial mutation can work properly in different classes of PHA synthase. Another positive mutant, PhaCRe(G4D), was obtained with an increased expression level [44]. This improved expression was presumed to contribute to the increased polymer production. In addition, the molecular weight of P(3HB) polymerized by PhaCRe(G4D) tended to be higher, suggesting that G4D enhanced the polymer chain elongation. The higher expression of PhaCRe(G4D) was observed for not only various E. coli strains (JM109, DH5, and HB101) but also gram-positive Corynebacterium glutamicum, indicating that G4D is a host-independent-type mutation. Further substitution of the G4 residue with other amino acids by saturation mutagenesis yielded various other G4X mutants, which exhibited a higher P(3HB) content and higher molecular weights [44].
Thermostability of PHA synthase should be an important factor in order to realize sustainable production of the target polyesters during the cultivation. The variant of PhaCRe carrying the S80P mutation used for suppression mutagenesis exhibited higher thermostability, although the reason is unclear [35]. In the other study on the improvement of thermostability, Shue et al. attempted to carry out a chimeragenesis approach [59]. Thermophilic PhaC from Cupriavidus sp. stain S-6 (PhaCCsp) and mesophilic PhaCRe was combined, and one chimeric enzyme PhaCReβ, which was PhaCRe bearing 30 mutations derived from PhaCCsp, showed P(3HB) accumulation at 45 °C with high productivity. PhaCReβ exhibited increased thermostability, 127- fold compared to that of the wild type, indicating that the thermostability of PhaCCsp was successfully inherited by PhaCRe. In addition, Tajima et al. identified a natural thermostable PHA synthase from the thermotolerant bacterium Pseudomonas sp. SG4502 [61]. These studies indicate that improved thermostability can be achieved in both engineered and native PHA synthases.
7.4.2 Substrate Specificity Alteration (at the Side Chain)
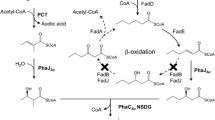
Substrate specificity alteration was successfully achieved mainly for two PHA synthases of A. caviae, class I PhaCAc and class II PhaCPs. These PHA synthases serve as key catalysts for synthesizing 3HB-based copolyesters with MCL-3HA monomers. By applying an in vitro evolutionary technique to a limited region of the phaC Ac gene, two beneficial mutations, N149S and D171G, were isolated [36]. These mutations exhibited an increase in the enzymatic activity toward 3HB-CoA compared to the wild-type enzyme in vitro assay and enhanced the accumulation of P(3HB-co-3-hydroxyhexanoate) [P(3HB-co-3HHx)] (6.5-fold and threefold) in vivo (Fig. 7.9, Pathways I and II). At the same time, the change in substrate specificity was demonstrated by data showing that the fraction of 3HHx (MCL-3HA monomer) was increased up to 18 mol% (N149S) and 16 mol% (D171G) from 10 mol% (wild type). Basically, class I PhaCAc preferentially polymerizes SCL- 3HA monomers over MCL-3HA monomers, indicating that the two mutants obtained a higher substrate specificity toward the MCL-3HA unit. The combination of Asn149Ser (N149S) and Asp171Gly (D171G) further increased the 3HHx fraction, probably due to the synergetic effect [50]. The PhaCAc(NSDG) mutant has been used as an industrial enzyme for P(3HB-co-3HHx) production at KANEKA Company. PhaCAc(NSDG) contributed the increase in the comonomer fraction of 3-hydroxy-4-methylvalerate harboring a branched side chain [53] and 3-hydroxy-2-methylbutyrate (3H2MB) harboring a beta-methylated side chain [68], indicating that the NSDG mutant has reactivity toward MCL-3HA monomers with various side-chain structures.
On the other hand, class II PhaCPs has a contrasted nature to PhaCAc, which means that PhaCPs preferentially polymerizes MCL-3HA monomers over SCL-3HA monomers. To reinforce the reactivity toward the 3HB monomer, in vitro evolutionary engineering based on error-prone PCR was employed. As a result, four beneficial mutations at position Glu130 (E130), Ser325 (S325), Ser477 (S477), and Gln481 (Q481), which are presumably located close to the active center Cys296, were identified [38, 40, 46, 47]. An in vitro activity assay revealed that E130 and S325 contribute to the enzyme activity, and the remaining Q477 and Q481 contribute to the substrate specificity. Via site-specific saturated mutagenesis and a combination of superior mutations, the activity toward 3HB-CoA was increased up to 720-fold. The 3HB fraction in P(3HA-co-3HB) was increased from 14 mol% (wild-type enzyme) to a maximum of 70 mol% (engineered enzyme) (Fig. 7.9, Pathways I and II), indicating that the molecular evolution of PHA synthase (PhaCPs) changed the enzyme substrate specificity.
7.5 Creation of New Polyesters by Engineered PHA Synthases
7.5.1 Alteration of Main-Chain Substrate Specificity (2-OH, LA Incorporation)
As described before, the substrate specificity of PHA synthase is closely related to the monomeric composition of the copolymers. In fact, many achievements were obtained for PHA synthases by enzyme evolutionary engineering (Table 7.1). Accordingly, tailor-made synthesis of copolymers with desired properties can be achieved by using engineered PHA synthases. These beneficial mutants mainly exhibit the alteration of side-chain-based substrate specificity represented by 3-hydroxyacyl (3-OH) monomer substrates. In contrast, the alteration of main-chain substrate specificity from 3-OH to 2-OH was one of the big challenges in the research field of PHA to expand biopolymer diversity. Lactate (LA) was selected as a first target of the 2-OH unit, and two pioneering works found a trace activity toward lactyl-CoA (LA-CoA) in PHA synthase from Allochromatium vinosum (class III) [72, 32]. However, at that time, unfortunately, the polymer-containing LA unit was not generated.
In 2008, the discovery of LPE paved the way for synthesis of the LA-based polymer [7]. First, Taguchi et al. attempted to explore natural and/or engineered PHA synthase(s) with a lactate-polymerizing activity based on the in vitro polymerization system using chemically synthesized LA-CoA as a monomer substrate. In the water-organic solvent two-phase in vitro system, LA-polymerizing activity was assayed based on the generation of a polymer-like precipitant. Among PHA synthases, only one engineered PHA synthase derived from Pseudomonas sp. 61–3, PhaCPs(STQK), termed LPE (lactate-polymerizing enzyme), clearly exhibited polymer- like precipitation (wild-type PhaCPs had a trace level) under the condition of the coexistence of LA-CoA and 3HB-CoA. Analysis of the precipitate revealed that LA was incorporated into the polymer chain together with the 3HB unit. The proof of concept for establishing that LPE was also functional in living cells was carried out. To transfer the LA-based polymer synthesis system into bacterial cells, an artificial metabolic pathway was constructed (Fig. 7.9). The key point was the utilization of propionyl-CoA transferase (PCT) for supplying the LA-CoA monomer, and PCT was expressed in E. coli together with LPE, PhaARe and PhaBRe. As a result, incorporation of the LA unit into the polymer chain was first found in E. coli. However, its molar fraction was too low, namely, 6 mol%. To enhance the LA fraction, a second-generation LPE was generated by adding the beneficial mutation (F392S) found in PhaCRe in the prototype LPE. This evolved LPE(STFSQK) exhibited further enhancements in the LA fraction in the copolymers as well as polymer production [53]. This LA enhancement was synergistically reinforced by combination of other strategies such as anaerobic cultivation for LA increase and metabolic pathway redirection (ΔpflAΔdld) [73]. These approaches allowed us to tune the LA fraction in the range of 0–99 mol%. Interestingly, an extremely high LA fraction (99.3 mol%) was observed in Corynebacterium glutamicum without 3HB-CoA- supplying enzyme genes, suggesting that the different monomer fluxes depended on the host cell [74]. These achievements imply the establishment of a microbial platform for the one-step production of polylactide (PLA) and its copolyesters by using LPE and evolved LPE.
It is of interest to promote the evolution of PhaCRe to LPE by introducing the mutation(s) that converted PhaCPs into LPE. Amino acid substitution at position Ala510 (A510) of PhaCRe, which corresponds to Gln481 (Q481) of LPE [PhaCPs(STQK)], was carried out (Fig. 7.7). Among 19 PhaCRe(A510X) mutants, 15 mutants synthesized P(LA-co-3HB), indicating that the 510 residue plays a critical role in LA polymerization. In addition, it was revealed that the LA-polymerizing ability could be transferred between different classes (from class II to class I) [63].
7.5.2 Diversification of Monomers for Creating New Polyesters
2HB and GL
After a major breakthrough in the enzyme reactivity, namely, the transition from 3-OH to 2-OH in the main chain of monomers, it is of interest to address the range of 2-OH monomers other than LA accessible by LPEs (STQK and STQKFS). First, 2-hydroxybutyrate (2HB) and glycolate (GL) should be targeted, as shown in Fig. 7.8. Homopolyesters and copolyesters containing 2HB and GL are bio-based polyesters like PLA. In particular, GL-based polyesters are often applied for biomedical materials. By blending the chiral homopolymers of the L-form and D-form, homopolymers of 2HB can form a stereocomplex in homogeneous combination and heterostereocomplex in heterogeneous combination with PLA.
Expansion of monomers by using native and engineered PHA synthases
Chemical structures of monomers are lined up based on the main chain and side chain. 3HB is the most common monomer in microbial polyesters (PHAs). Monomer structures are diversified from 3HB, as shown in the blue line. Transition from 3HB to LA could be achieved by using the engineered PHA synthase, PhaCPs(STQK)/lactate-polymerizing enzyme (LPE). Red arrows indicate the expansion of recognizable monomers by PHA synthases starting from LA
2HB-based polymers were successfully synthesized by PhaCRe (wild type), LPE(STQK), LPE(STSRQK), and the chimeric enzyme of PhaCAc and PhaCRe, termed AcRe12 [58, 62, 70, 71] (Fig. 7.8). In 2011, it was confirmed that PhaCRe polymerized P(2HB-co-3HB) in vitro from a mixture of (R)-2HB-CoA and (R)-3HB-CoA. However, at this time, the 2HB incorporation was very weak, and the polymerizing reaction was inhibited by the presence of (S)-2HB-CoA [58]. To explore a more efficient enzyme for the 2HB-based polymer production, the utilization of LPE was attempted. LPEs exhibit superior reactivity toward 2HB, and the P(2HB) homopolymer could be synthesized in vivo from endogenous racemic 2HB by expressing PCT and LPEs in E. coli [62, 70] (Fig. 7.9, Pathway IV). Recently, it was reported that chimeric enzyme AcRe12 can polymerize the block copolymer P(2HB-b-3HB) in vivo. This was the first report that chimeric PHA synthase possesses significant activity toward 2-OH monomers [71].
PHA biosynthesis pathways
The target PHAs are synthesized by the five metabolic pathways illustrated. Pathway I is an acetyl-CoA condensation pathway to generate (R)-3HB-CoA. Pathway II is composed of the β-oxidation pathway to produce (R)-3HA-CoAs. Pathway III is the new pathway for the synthesis of (R)-LA-CoA. Pathway IV supplies (R)-2HB-CoA and (R)-GL-CoA by the addition of the corresponding precursors. Pathway V is an amino acid derivative pathway to generate (R)-2HA-CoAs including an aromatic group
GL-based polymers were also successfully synthesized by LPE [57]. In addition to the 2HB-based polymer, the accumulation of P(GL-co-3HAs) with 17 mol% of GL fraction was confirmed in vivo using recombinant E. coli from exogenous glycolate and dodecanoate (Fig. 7.9, Pathway IV). Thus, LPE can recognize 2-OH monomers with no side chain and an ethyl side chain as well as methyl side chain (LA, 2-hydroxypropionate) (Fig. 7.8).
2H4MV, 2H3PhP, and 2H3MB
A side-chain variety of 2-OH monomers was further explored. Amino acid derivatives, which have a 2HA backbone with various side-chain structures corresponding with each amino acid, were the next candidates. Mizuno et al. first investigated the LPE acceptance of the 2-hydroxy-4-metylvalerate (2H4MV) unit derived from leucine supplied via 2H4MV dehydrogenase (LdhA) and 2H4MV-CoA transferase (HadA) reactions (Fig. 7.9, Pathway V). Further investigation determined that not only the 2H4MV unit but also 2-hydroxy-3-phenylpropionate (2H3PhP), 2-hydroxy-3-metylvalerate (2H3MB), and 2-hydroxy-3-metylvalerate (2H3MV) derived from phenylalanine, valine, and isoleucine were incorporated into the polymer as P(3HB-co-2HAs), suggesting that LPE(STQK) can recognize 2HA with bulky side chains [69].
7.6 Properties of Polyesters Produced by Native and Engineered PHA Synthases
The final goal of PHA research work is commercialization of the polymeric materials. The typical example is PHBH, P(3HB-co-3HHx), manufactured by KANEKA Company in Japan. In the fermentation process for production of the PHBH, NSDG mutant of PhaCAc has been actually utilized in the microbial platform. Additionally, PLA and its copolyesters can be biosynthesized by means of LPEs evolved from PHA synthases. In such a sense, engineering of the PHA synthase provides us the effective achievements related to the application of PHAs. Table 7.3 summarizes the up version and properties of PHA-related polyesters. Expansion of the variety of monomeric constituents has been extensively updated for creating new polyesters based on PHA synthase alteration.
aMelting temperature, bglass transition temperature, c3HA units (3-hydroxyoctanoate (< 1 mol%), 3-hydroxydecanoate (3 mol%), 3-hydroxydodecanoate (3 mol%), 3-hydroxy-cis-5-dodecanoate (< 1 mol%)), d2HA units (2HP (LA) (1 mol%), 2H3MB (1 mol%), 2H4MV (14 mol%), 2H3PhP (6 mol%))
7.7 Future Perspectives
The structure-function relation study of PHA synthase is crucial to better understand the reaction mechanism of polymerization for PHA synthesis and to improve PHA from the viewpoints of academic and industrial progress. In this sense, solution of the tertiary structures of two class I PHA synthases provides us with various insights into the aforementioned subject. In both crystal structures, one can find the common structural properties in the catalytic domain such as the conserved catalytic triad (Cys-His-Asp) in the α-/β-core subdomain. On the other hand, a big difference is observed in the CAP subdomain structures of the two PHA synthases. In particular, the presence of the open-closed mode found in PhaCCs is characteristic in creating the dynamic conformational change upon the PhaCCs-substrate interaction. Such structure-based interpretation is available for discussing the various enzyme functions. With the available structural information of PhaCs, another catalytic mechanism was proposed based on a modelled structure of class I PhaC from Aquitalea sp. USM4 [81]. Crystal structures of enzyme-substrate complexes together with the complete whole structures of PHA synthases should be required for further understanding the polymerization mechanism of PHA synthase and rational enzyme engineering.
Prior to the presentation of crystal structures of PHA synthases, evolutionary engineering has been applied to three kinds of PHA synthase focusing on the activity improvement, substrate specificity alteration, thermostability improvement, and expression improvement. Even with no information on enzyme tertiary structure, these achievements strongly evidence that evolutionary engineering is a powerful toolbox for improving the enzymatic properties related to PHA biosynthesis. For more than 20 years, a large library of class II-categorized PHA synthase mutants that result in alterations of monomeric composition, sequence regulation such as the development of a block copolymer [71] and the use of various molecular weights, and enhanced polymer production has been increasing. A typical case is the discovery of LPE, which can synthesize LA-based polymers. This case may be fortunate and now could be the time to take a great leap forward, as commented in the article [78]. Most recently, mechanistic model on polymerization of LA-based polymer has been demonstrated based on biochemical and kinetic studies [79]. The pioneering study has led to further expansion of the range of structural diversity of PHA members other than LA-based polymers. Along this line, one can expect to synthesize chiral copolymers with various monomer compositions, owing to the extremely high enantioselectivity and broad substrate specificity of the class II PHA synthase. In this chapter, it is a good starting point to have a discussion on the relationship between 3D-structure and mutation effects based on both milestones. For example, several key amino acid residues closely related to the enzymatic performances can be commonly addressed from four representative class I and class II PHA synthases [80].
For what is PHA synthase studied? The answer is simple. Needless to say, PHA synthase should be an essential machinery for PHA biosynthesis in practical applications. It seems to be clear that the evolutionary engineering of “biocatalysts” will be a key approach, analogous to the fact that the field of chemical polymeric materials has moved forward via the development of advanced “chemical catalysts” like Ziegler-Natta catalysts. With the current PHA synthase structure now unveiled, we are moving closer to tailor-made PHA to meet various needs.
References
Lemoigne M (1926) Products of dehydration and of polymerization of β-hydroxybutyric acid. Bull Soc Chim Biol (Paris) 8:770
Steinbüchel A, Doi Y (2002) Biopolymers. Vol. 3a (polyesters I) and 3b (polyesters II). WILEY-VCH Verlag GmbH, Weinheim
Anderson AJ, Dawes EA (1990) Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev 54:450–472
Stubbe J, Tian J, He A et al (2005) Nontemplate-dependent polymerization processes: polyhydroxyalkanoate synthases as a paradigm. Annu Rev Biochem 74:433–480
Taguchi S, Doi Y (2004) Evolution of polyhydroxyalkanoate (PHA) production system by “enzyme evolution”: successful case studies of directed evolution. Macromol Biosci 4:145–156
Nomura CT, Taguchi S (2007) PHA synthase engineering toward superbiocatalysts for custom-made biopolymers. Appl Microbiol Biotechnol 73:969–979
Taguchi S, Yamada M, Matsumoto K et al (2008) A microbial factory for lactate-based polyesters using a lactate-polymerizing enzyme. Proc Natl Acad Sci U S A 105(45):17323–17327
Taguchi S (2010) Current advances in microbial cell factories for lactate-based polyesters driven by lactate-polymerizing enzymes: towards the further creation of new LA-based polyesters. Polym Degrad Stab 95:1421–1428
Park SJ, Kim TW, Kim MK et al (2012) Advanced bacterial polyhydroxyalkanoates: towards a versatile and sustainable platform for unnatural tailor-made polyesters. Biotechnol Adv 30:1196–1206
Matsumoto K, Taguchi S (2013) Enzyme and metabolic engineering for the production of novel biopolymers: crossover of biotechnological and chemical processes. Curr Opin Biotechnol 24:1054–1060
Matsumoto K, Taguchi S (2013) Biosynthetic polyesters consisting of 2-hydroxyalkanoic acids: current challenges and unresolved questions. Appl Microbiol Biotechnol 97:8011–8021
Rehm BH (2003) Polyester synthases: natural catalysts for plastics. Biochem J 376:15–33
Wittenborn EC, Jost M, Wei Y et al (2016) Structure of the catalytic domain of the class I polyhydroxybutyrate synthase from Cupriavidus necator. J Biol Chem 291:25264–25277
Kim J, Kim Y-J, Choi SY et al (2017a) Crystal structure of Ralstonia eutropha polyhydroxyalkanoate synthase C-terminal domain and reaction mechanisms. Biotechnol J 12:1600648
Kim Y-J, Choi SY, Kim J et al (2017b) Structure and function of the N-terminal domain of Ralstonia eutropha polyhydroxyalkanoate synthase, and the proposed structure and mechanisms of the whole enzyme. Biotechnol J 12:1600649
Chek MF, Kim S-Y, Mori T et al (2017) Structure of polyhydroxyalkanoate (PHA) synthase PhaC from Chromobacterium sp. USM2, producing biodegradable plastics. Sci Rep 7(1):5312
Liebergesell M, Steinbüchel A (1992) Cloning and nucleotide sequences of genes relevant for biosynthesis of poly(3-hydroxybutyric acid) in Chromatium vinosum strain D. Eur J Biochem 209:135–150
Jia Y, Kappock TJ, Frick T et al (2000) Lipases provide a new mechanistic model for polyhydroxybutyrate (PHB) synthases: characterization of the functional residues in Chromatium vinosum PHB synthase. Biochemistry 39:3927–3936
Wodzinska J, Snell KD, Rhomberg A et al (1996) Polyhydroxybutyrate synthase: evidence for covalent catalysis. J Am Chem Soc 118:6319–6320
Muh U, Sinskey AJ, Kirby DP et al (1999) PHA synthase from Chromatium vinosum: cysteine 149 is involved in covalent catalysis. Biochemistry 38:826–837
Jia Y, Yuan W, Wodzinska J et al (2001) Mechanistic studies on class I polyhydroxybutyrate (PHB) synthase from Ralstonia eutropha: class I and III synthases share a similar catalytic mechanism. Biochemistry 40:1011–1019
Li P, Chakraborty S, Stubbe J (2009) Detection of covalent and non-covalent intermediates in the polymerization reaction catalyzed by a C149S–class III polyhydroxybutyrate synthase. Biochemistry 48:9202–9211
Amara AA, Steinbüchel A, Rehm BH (2002) In vivo evolution of the Aeromonas punctate polyhydroxyalkanoate (PHA) synthase: isolation and characterization of modified PHA synthases with enhanced activity. Appl Microbiol Biotechnol 59:477–482
Tian J, Sinskey AJ, Stubbe J (2005) Detection of intermediates from the polymerization reaction catalyzed by a D302A mutant of class III polyhydroxyalkanoate (PHA) synthase. Biochemistry 44:1495–1503
Kawaguchi Y, Doi Y (1992) Kinetics and mechanism of synthesis and degradation of poly(3- hydroxybutyrate) in Alcaligenes eutrophus. Macromolecules 25:2324–2329
Ellar D, Lundgren DG, Okamura K, Marchessault RH (1968) Morphology of poly-β- hydroxybutyrate granules. J Mol Biol 35:489–502
Lauzier C, Revol J-F, Marchessault RH (1992) Topotactic crystallization of isolated poly(β- hydroxybutyrate) granules from Alcaligenes eutrophus. FEMS Microbiol Lett 103:299–310
Gerngross TU, Snell KD, Peoples OP et al (1994) Overexpression and purification of the soluble polyhydroxyalkanoate synthase from Alcaligenes eutrophus: evidence for a required posttranslational modification for catalytic activity. Biochemistry 33:9311–9320
Wodzinska J, Snell KD, Rhomberg A et al (1996) Polyhydroxybutyrate synthase: evidence for covalent catalysis. J Am Chem Soc 118:6319–6320
Stubbe J, Tian J (2003) Polyhydroxyalkanoate (PHA) hemeostasis: the role of PHA synthase. Nat Prod Rep 20:445–457
Buckley RM, Stubbe J (2015) Chemistry with an artificial primer of polyhydroxybutyrate synthase suggests a mechanism for chain termination. Biochemistry 54:2117–2125
Yuan W, Jia Y, Tian J et al (2001) Class I and III polyhydroxyalkanoate synthases from Ralstonia eutropha and Allochromatium vinosum: characterization and substrate specificity studies. Arch Biochem Biophys 394:87–98
Spiekermann P, Rehm BH, Kalscheuer R et al (1999) A sensitive, viable-colony staining method using Nile red for direct screening of bacteria that accumulate polyhydroxyalkanoic acids and other lipid storage compounds. Arch Microbiol 171(2):73–80
Taguchi S, Maehara A, Takase K et al (2001) Analysis of mutational effects of a polyhydroxybutyrate (PHA) polymerase on bacterial PHB accumulation using an in vivo assay system. FEMS Micro Let 198:65–71
Taguchi S, Nakamura H, Hiraishi T et al (2002) In vitro evolution of a polyhydroxybutyrate synthase by intragenic suppression-type mutagenesis. J Biochem 131:801–806
Kichise T, Taguchi S, Doi Y (2002) Enhanced accumulation and changed monomer composition in polyhydroxyalkanoate (PHA) copolyester by in vitro evolution of Aeromonas caviae PHA synthase. Appl Environ Micobiol 68(5):2411–2419
Rehm BH, Antonio RV, Spiekermann P et al (2002) Molecular characterization of the poly(3-hydroxybutyrate) (PHB) synthase from Ralstonia eutropha: in vitro evolution, site- specific mutagenesis and development of a PHB synthase protein model. Biophys Acta 1594(1):178–190
Takase K, Taguchi S, Doi Y (2003) Enhanced synthesis of poly(3-hydroxybutyrate) in recombinant Escherichia coli by means of error-prone PCR mutagenesis, saturation mutagenesis, and in vitro recombination of the type II polyhydroxyalkanoate synthase gene. J Biochem 133:139–145
Solaiman DK (2003) Biosynthesis of medium-chain-length poly(hydroxyalkanoates) with altered composition by mutant hybrid PHA synthases. J Ind Microbiol Biotechnol 30(5):322–326
Takase K, Matsumoto K, Taguchi S et al (2004) Alteration of substrate chain-length specificity of type I synthase for polyhydroxyalkanoate biosynthesis by in vitro evolution: in vivo and in vitro enzyme assays. Biomacromolecules 5(2):480–485
Sheu DS, Lee CY (2004) Altering the substrate specificity of polyhydroxyalkanoate synthase 1 derived from pseudomonas putida GPo1 by localized semirandom mutagenesis. J bacterial 186(13):4177–4184
Niamsiri N, Delamarre SC, Kim YR et al (2004) Engineering of chimeric class II polyhydroxyalkanoate synthases. Appl Environ Microbiol 70(11):6789–6799
Tsuge T, Saito Y, Narike M et al (2004) Mutation effects of a conserved alanine (Ala510) in type I polyhydroxyalkanoate synthase from Ralstonia eutropha on polyester biosynthesis. Macromol Biosci 4(10):963–970
Normi YM, Hiraishi T, Taguchi S et al (2005) Characterization and properties of G4X mutants of Ralstonia eutropha PHA synthase for poly(3-hydroxybutyrate) biosynthesis in Escherichia coli. Macromol Biosci 5(3):197–206
Normi YM, Hiraishi T, Taguchi S et al (2005) Site-directed saturation mutagenesis at residue F420 and recombination with another beneficial mutation of Ralstonia eutropha polyhydroxyalaknoate synthase. Biotehcnol Lett 27(10):705–712
Matsumoto K, Takase K, Aoki E et al (2005) Synergistic effects of Glu130Asp substitution in the type II polyhydroxyalkanoate (PHA) synthase: enhancement of PHA production and alteration of polymer molecular weight. Biomactomolecules 6:99–104
Matsumoto K, Aoki E, Takase K et al (2006) In vivo and in vitro characterization of Ser477X mutations in polyhydroxyalkanoate (PHA) synthase 1 from Pseudomonas sp. 61-3: effects of beneficial mutations on enzymatic activity, substrate specificity, and molecular weight of PHA. Biomacromolecules 7(8):2436–2442
Tsuge T, Watanabe S, Sato S et al (2007) Variation in copolymer composition and molecular weight of polyhydroxyalkanoate generated by saturation mutagenesis of Aeromonas caviae PHA synthase. Macromol Biosci 7(6):846–854
Tsuge T, Watanabe S, Shimada D et al (2007) Combination of N149S and D171G mutations in Aeromonas caviae polyhydroxyalkanoate synthase and impact on polyhydroxyalkanoate biosynthesis. FEMS Microbiol Lett 277(2):217–222
Shozui F, Matsumoto K, Sakai T et al (2009) Engineering of polyhydroxyalkanoate synthase by Ser477X/Gln481X saturation mutagenesis for efficient production of 3-hydroxybutyrate- based copolyesters. Appl Microbiol Biotechnol 84(6):1117–1124
Matusmoto K, Takase K, Yamamoto Y et al (2009) Chimeric enzyme composed of polyhydroxyalkanoate (PHA) synthases from Ralstonia eutropha and Aeromonas caviae enhances production of PHAs in recombinant Eschericia coli. Biomacromolecules 10(4):682–685
Tanadachangseaeng N, Kitagawa A, Yamamoto T et al (2009) Identification, biosynthesis, and characterization of polyhydroxyalkanoate copolymer consisting of 3-hydroxybutyrate and 3-hydroxy-4-methylvalerate. Biomacromolecules 10(10):2866–2874
Yamada M, Matsumoto K, Shimizu K et al (2010) Adjustable mutations in lactate (LA)-polymerizing enzyme for the microbial production of LA-based polyesters with tailor-made monomer composition. Biomacromolecules 11(2):815–819
Shozui F, Sun J, Song Y et al (2010) A new beneficial mutation in Pseudomonas sp. 61-3 polyhydroxyalkanoate (PHA) synthase for enhanced cellular content 3-hydroxybutyrate-based PHA explored using its enzyme homolog as a mutation template. Biosci Biotechnol Biochem 74(8):1710–1712
Sun J, Shozui F, Yamada M et al (2010) Production of P(3-hydroxybutyrate-co-3-hydroxyhexanoate- co-3-hydroxyoctanoate) terpolymers using a chimeric PHA synthase in recombinant Ralstonia eutropha and Pseudomoans putida. Biosci Biotechnol Biochem 74(8):1716–1718
Shen XW, Shi ZY, Song G et al (2011) Engineering of polyhydroxyalkanoate (PHA) synthase phaC2Ps of Pseudomonas stutzeri via site-specific mutation for efficient production of PHA copolymers. Appl Microbiol Biotechnol 91(3):655–665
Matsumoto K, Ishiyama A, Sakai K et al (2011) Biosynthesis of glycolate-based polyesters containing medium-chain-length 3-hydroxyalkanoates in recombinant Escherichia coli expressing engineered polyhydroxyalkanoate synthase. J Biotechnol 156(3):214–217
Han X, Satoh Y, Satoh T et al (2011) Chemo-enzymatic synthesis of polyhydroxyalkanoate (PHA) incorporating 2-hydroxybutyrate by wild-type class I PHA synthase from Ralstonia eutropha. Appl Microbiol Biotechnol 92(3):509–517
Sheu DS, Chen WM, Lai YW et al (2012) Mutations derived from the thermophilic polyhydroxyalkanoate synthase PhaC enhance the thermostability and activity of PhaC from Cupriavidus necator H16. J Bacterial 194(10):2620–2629
Watanabe Y, Ichinomiya Y, Shimada D et al (2012) Development and validation of an HPLC- based screening method to acquire polyhydroxyalkanoate synthase mutants with altered substrate specificity. J sci Bioeng 113(3):286–292
Tajima K, Han X, Satoh Y et al (2012) In vitro synthesis of polyhydroxyalkanoate (PHA) incorporating lactate (LA) with a block sequence by using a newly engineered thermostable PHA synthase from Pseudomonas sp. SG4502 with acquired LA-polymerizing activity. Appl Microbiol Biotechnol 94(2):365–376
Matsumoto K, Terai S, Ishiyama A (2013) One-pot microbial production, mechanical properties, and enzymatic degradation of isotactic P[(R)-2-hydroxybutyrate] and its copolymer with (R)-lactate. Biomacromolecules 14(6):1913–1918
Ochi A, Matsumoto K, Ooba T et al (2013) Engineering of class I lactate-polymerizing polyhydroxyalkanoate synthases from Ralstonia eutropha that synthesize lactate-based polyester with a block nature. Appl Microbiol Biotechnol 97(8):3441–3447
Chuah JA, Tomizawa S, Yamada M et al (2013) Characterization of site-specific mutations in a short-chain-length/medium-chain-length polyhydroxyalkanoate synthase: in vivo and in vitro studies of enzymatic activity and substrate specificity. Appl Environ Microbiol 79(12):3813–3821
Chuah JA, Yamada M, Taguchi S et al (2013) Biosynthesis and characterization of polyhydroxyalkanoate containing 5-hydroxyalkanoate units: effects of 5HV units on biodegradability, cytotoxicity, mechanical and thermal properties. Polym Degrad Stabil 98(1):331–338
Chen YJ, Tsai PC, Hsu CH et al (2014) Critical residues of class II PHA synthase for expanding the substrate specificity and enhancing the biosynthesis of polyhydroxyalkanoate. Enzym Microb Technol 56:60–66
Watanabe Y, Ishizuka K, Furutate S et al (2015) Biosynthesis and characterization of novel poly(3-hydroxybutyrate-co-3-hydroxy-2-methylbutyrate): thermal behavior associated with α-carbon methylation. RSC Adv 5:58679–58685
Mizuno S, Hiroe A, Fukui T et al (2017) Fractionation and thermal characteristics of biosynthesized polyhydroxyalkanoates bearing aromatic groups as side chains. Polym J 49:557–565
Mizuno S, Enda Y, Saika A et al (2017) Biosynthesis of polyhydroxyalkanoates containing 2-hydroxy-4-methylvalerate and 2-hydroxy-3-phenylpropionate units from a related or unrelated carbon source. J Biosci Bioeng 125(3):295–300
Hori C, Oishi K, Matsumoto K et al (2018) Site-directed saturation mutagenesis of polyhydroxyalkanote synthase for efficient microbial production of poly[(R)-2-hydroxybutyrate]. J Biosci Bioeng 125(6):632–636
Matsumoto K, Hori C, Takaya M et al (2018) Dynamic changes of intracellular monomer levels regulate block sequence of polyhydroxyalkanoates in engineered Escherichia coli. Biomacromolecules 19(2):662–671
Steinbüchel A, Valentin HE (1995) Diversity of bacterial polyhydroxyalkanoic acids. FEMS Microbiol Lett 128(3):219–228
Nduko JM, Matsumoto K, Ooi T et al (2014) Enhanced production of poly(lactate-co-3- hydroxybutyrate) from xylose in engineered Escherichia coli overexpressing a galactitol transporter. Appl Microbiol Biotechnol 98(6):2453–2460
Song Y, Matsumoto K, Yamada M et al (2012) Engineered Corynebacterium glutamicum as an endotoxin-free platform strain for lactate-based polyester production. Appl Microbiol Biotechnol 93(5):1917–1925
Tsuge T (2002) Metabolic improvements and use of inexpensive carbon sources in microbial production of polyhydroxyalkanoates. J Biosci Bioeng 94(6):579–584
Mizuno S, Katsumata S, Hiroe A et al (2014) Biosynthesis and thermal characterization of polyhydroxyalkanoates bearing phenyl and phenylalkyl side groups. Polym Degrad Stabil 109:379–384
Hiroe A, Ishii N, Ishii D et al (2016) Uniformity of monomer composition and material properties of medium-chain-length polyhydroxyalkanoates biosynthesized from pure and crude fatty acids. ACS Sustain Chem Eng 4(12):6905–6911
Taguchi S (2017) Designer enzyme for green materials innovation: lactate-polymerizing enzyme as a key catalyst. Fron Chem Sci Eng 11(1):139–142
Matsumoto K, Iijima M, Hori C et al (2018) In vitro analysis of d-lactyl-CoA-polymerizing polyhydroxyalkanoate synthase in polylactate and poly(lactate-co-3-hydroxybutyrate). Biomacromolecules 19(7):2889–2895
Chek MF, Hiroe A, Hakoshima T et al (2019) PHA synthase (PhaC): interpreting the function of bioplastic-producing enzyme from a structural perspective. Appl Microbiol Biotechnol 103(3):1131–1141
Teh A-H, Chiam N-C, Furusawa G, Sudesh K (2018) Modelling of polyhydroxyalkanoate synthase from Aquitalea sp. USM4 suggests a novel mechanism for polymer elongation. Int J Biol Macromol 119:438–445
Acknowledgments
This work was supported by CREST, JST (JPMJCR12B4 to ST), MIRAI, JST (JPMJMI17EC to AH), A-STEP, JST (29A1027 to TH), as well as APEX Delivering Excellence Grant 2012 (Grant No.: 1002/PBIOLOGI/910322) from Universiti Sains Malaysia.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Hiroe, A., Chek, M.F., Hakoshima, T., Sudesh, K., Taguchi, S. (2019). Synthesis of Polyesters III: Acyltransferase as Catalyst. In: Kobayashi, S., Uyama, H., Kadokawa, Ji. (eds) Enzymatic Polymerization towards Green Polymer Chemistry. Green Chemistry and Sustainable Technology. Springer, Singapore. https://doi.org/10.1007/978-981-13-3813-7_7
Download citation
DOI: https://doi.org/10.1007/978-981-13-3813-7_7
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-3812-0
Online ISBN: 978-981-13-3813-7
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)