Abstract
NMR spectroscopy has become substantial in the elucidation of RNA structures and their complexes with other nucleic acids, proteins or small molecules. Almost half of the RNA structures deposited in the Protein Data Bank were determined by NMR spectroscopy, whereas NMR accounts for only 11% for proteins. Recent improvements in isotope labeling of RNA have strongly contributed to the high impact of NMR in RNA structure determination. In this book chapter, we review the advances in isotope labeling of RNA focusing on larger RNAs. We start by discussing several ways for the production and purification of large quantities of pure isotope labeled RNA. We continue by reviewing different strategies for selective deuteration of nucleotides. Finally, we present a comparison of several approaches for segmental isotope labeling of RNA. Selective deuteration of nucleotides in combination with segmental isotope labeling is paving the path for studying RNAs of ever increasing size.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- High Performance Liquid Chromatography
- Hepatitis Delta Virus
- Hammerhead Ribozyme
- Large RNAs
- Cyclic Phosphate
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
RNA has become widely recognized not only as protein coding information carrier but also as an important regulator of gene expression such as riboswitches, miRNAs or large non-coding RNAs [1]. It has been shown that with increasing complexity of the organism the extent of non protein coding DNA is increasing drastically [2]. In humans, 98.8% of the DNA is not coding for proteins but is mainly transcribed into RNA. The ratio between the number of different proteins and RNAs in the cell is in strong contrast to the ratio of structures deposited in the Protein Data Bank. As of April 2012 only 1.1% of the structures are RNA molecules, whereas proteins account for 92.6%. This discrepancy is due to difficulties in crystallizing and phasing RNA for X-ray crystallography. The strong negative charges on the RNA backbone as well as the dynamic nature of the RNA molecules often impede crystallization. NMR spectroscopy, on the other hand, has proven to have high potential to determine RNA structures. The percentage of RNA structures elucidated by NMR (45%) is almost equal to the ones elucidated by X-ray crystallography (52%). Small RNAs (<40 nucleotides) can be studied using uniformly 1H, 13C, 15N-labeled nucleotides with different nucleotide specific labeling schemes [3–5]. However, larger biologically relevant RNA structures are difficult to tackle due to the severe spectral overlap observed in NMR spectra of RNA [6]. Furthermore, strong relaxation of important RNA resonances due to dipolar interactions (mostly 1H-13C in 13C-labeled nucleotides or 1H-1H in the sugar or the pyrimidine H5-H6 proton pair) limited the studies of larger RNAs. Improvements in isotope labeling of RNA, especially site-specific deuteration and segmental labeling of RNA, have opened the avenue for studying RNA molecules of ever increasing size by NMR spectroscopy. NMR spectroscopy is therefore expected to become more and more important in the elucidation of RNA structures and their complexes with other nucleic acids, small molecules or proteins.
In this book chapter, we discuss several aspects of isotope labeling of RNA focusing on larger RNAs. We start by comparing the different methods for the production of RNA that are chemical synthesis, in vitro enzymatic transcription and in vivo RNA production. We also point out different possibilities that allow obtaining RNAs with homogenous termini. In a second part, we present different approaches to purify RNA in both a native and denaturing manner. The third part focuses on several strategies for selective deuteration/protonation of nucleotides leading to a strong reduction of the spectral overlap and also to a reduction of transverse relaxation. The review ends with an extensive description of different strategies for segmental isotopic labeling of RNA that are becoming essential for studying RNAs of moderate to large size by NMR spectroscopy, especially in combination with measurements of residual dipolar couplings, paramagnetic relaxation enhancement [5, 7], electron paramagnetic resonance (our unpublished results) and small angle X-ray scattering [8].
2 RNA Synthesis
RNA can be synthesized in three different ways depending on the length and the requirements for isotope labeling: chemical synthesis, in vitroenzymatic transcription and in vivoproduction of RNA (see Fig. 7.1a).
RNA production.(a) RNA synthesis can be performed either by chemical synthesis, in vitrotranscription or in vivoRNA production. (b) Methods for generating homogenous 5′- and 3′-termini. (c) Methods for preparative RNA purification (Adapted with permission from Dominguez et al. [5])
2.1 Chemical Synthesis
Chemical synthesis is the method of choice for preparing small RNAs, as in vitroenzymatic synthesis of RNAs smaller than ten nucleotides has been reported not to be successful [9], except in one case [10]. Chemical synthesis of RNA is reported for RNAs up to 80 nucleotides [11–13]. However, low yields and high costs for larger RNAs make the chemical synthesis suitable only for short RNAs (<20 nts). A unique advantage of chemical synthesis is the possibility of introducing modified nucleotides at desired positions. For example, introduction of thiouridines at specific positions in an RNA allows the attachment of nitroxide spin-labels for measuring paramagnetic relaxation enhancement [14]. Moreover, a protocol for synthesizing short RNAs, that are selectively 13C-labeled on sugar carbons has been developed [15] and used to solve the structure of several protein-RNA complexes [16–20], but unfortunately isotope labeled phosphoamidites are not yet commercially available. Introduction of modified nucleotides into larger RNAs can be achieved by enzymatic ligation of small synthetic RNAs (see last section in this chapter) [21, 22].
2.2 In VitroEnzymatic Transcription
In vitroenzymatic transcription using SP6, T3 or T7 RNA polymerases is the most widely used method for the production of RNAs larger than 12 nucleotides [23–26]. The possibility of incorporating commercially available 13C- and/or 15N- labeled, perdeuterated or even partially deuterated nucleoside triphosphates (NTPs) allows production of RNAs suitable for heteronuclear multidimensional NMR [27–31]. T7 RNA polymerase can be obtained commercially or produced in-house by overexpressing a His-tagged T7 RNA polymerase in E. coli[32]. Transcription reactions should be first optimized on small scale reactions by changing concentrations of MgCl2, DNA, NTPs and T7 RNA polymerase and testing the influence of the addition of pyrophosphatase and/or guanine monophosphate (GMP). The best condition can be scaled-up to a large scale reaction of for example 10 ml, which typically yields around 500 nmol of RNA. Unlabeled and uniformly labeled NTPs are commercially available but can also be produced in-house [28]. Transcription using T7 RNA polymerase can be performed from chemically synthesized double-stranded DNA templates or from linearized plasmids. Since only the 18 nts T7 promoter on the top-strand is sufficient for transcription, the same top-strand can be used for any transcription. However, it has been observed specially for structured RNAs that higher yields are obtained when fully double-stranded DNA is used [33]. The first nucleotide, which is incorporated, must be a guanine. Transcription efficiency is highly dependent on the starting six nucleotides. Excellent starting sequences are native starting sequences of the T7 RNA polymerase, such as GGGAGA, GGGAUC, GGCAAC or GGCGCU [23]. Besides the 5′ sequence requirements, another drawback of T7 in vitrotranscription is the 3′ and 5′ inhomogeneity. More than 30% of untemplated 5′ nucleotides have been observed for sequences starting with 4–5 consecutive guanines, whereas a sequence starting with GCG showed no detectable 5′ inhomogeneity [34]. More severe is the 3′ inhomogeneity, where up to six additional nucleotides can be added. An overview of several methods to overcome 5′ and 3′ inhomogeneity is presented in Fig. 7.1b.
The problem of 5′ and 3′ inhomogeneity can be circumvented by incorporation of a ribozyme sequence in cis, which cleaves co-transcriptionally leading to an homogenous 5′-hydroxyl or a 2′,3′-cyclic phosphate end, respectively (see Fig. 7.6dstep 1) [31, 35]. Concerning 5′-inhomogeneity, hammerhead ribozymes are interesting because they have no sequence requirements [26]. When placed 5′ to the RNA of interest, they allow cleavage of the RNA with MgCl2as cofactor almost to completion [26, 36, 37]. Concerning 3′-inhomogeneity, the hepatitis delta virus (HDV) RNA ribozyme, that has no sequence requirements [38, 39], or the NeurosporaVarkud satellite (VS) ribozyme that has minimal sequence requirements (VS will cut efficiently after any nucleotide other than cytosine) can be efficiently used [36, 40]. It has been shown that hammerhead ribozymes [41] and VS ribozymes [36] can be added in transthereby saving isotope labeled NTPs that otherwise would be used to produce the ribozyme incorporated in cis(see Fig. 7.6dstep 1).
In addition to ribozymes, DNAzymes have been developed by in vitroevolution as engineering tools [42–44]. The 10–23 family of DNAzymes cleaves between a purine and a pyrimidine, which is the only sequence requirement. Cleavage results in a 5′-hydroxyl group and a 2′,3′-cyclic phosphate similarly to small ribozymes. Moreover, it has been shown that RNAs can be cleaved sequence-specifically by RNase H, when the RNA of interest is hybridized with a 2′-O-methyl-RNA/DNA chimera [45, 46]. In contrast to ribozyme and DNAzyme mediated RNA cleavage, RNase H produces 5′-monophosphates and 3′-hydroxyl groups (see Fig. 7.6dstep 2). Another approach is the use of a DNA template strand for transcription, in which the two 5′ nucleotides are modified with C2′-methoxyls. This dramatically reduced 3′-end inhomogeneities [47].
2.3 In VivoProduction of RNA
Dardel and co-workers developed an in vivomethod using a tRNA scaffold to protect the RNA from cellular RNases for the production of milligram quantities of RNA for NMR studies [48, 49]. The tRNA scaffold can be removed either by DNAzymes or by sequence-specific RNase H cleavage [42, 44–46]. Using this method, a reasonable amount of RNA for NMR studies (0.8 μmol) was obtained from 2 l of E. coliculture grown on 15N/13C-labeled medium.
3 RNA Purification
RNA obtained by in vitroenzymatic or in vivotranscription must be purified from proteins (T7 RNA polymerase, pyrophosphatase) and abortive transcription products (a large number of smaller oligoribonucleotides of two to six nucleotides in length are generated during transcription due to abortive initiation events) as well as from unused NTPs. In addition, RNA with one or two additional nucleotides arising from untemplated nucleotide addition at the 3′ end must be removed, when a homogenous RNA is required. An overview of different purification methods is presented in Fig. 7.1c.
Denaturing polyacrylamide gel electrophoresis (PAGE) is the most commonly used purification method for large quantities of RNA needed for NMR spectroscopy. Single nucleotide resolution for preparative scales is typically achieved for RNAs up to 30 nucleotides. However, this procedure is laborious and suffers from low recovery yields, especially with larger RNAs. Additionally, PAGE requires the RNA to be denatured and refolded after purification, which might lead to aggregation and dimerization of the RNA [50]. Furthermore, the RNA is not free of low-molecular-weight acrylamide contaminants, which might interact with RNA and also compromise NMR spectral analysis [51]. Therefore, different chromatographic methods have been developed to purify RNA. Frederick and coworkers proposed purifying RNA by non-denaturing anion-exchange chromatography [52]. Depending on the salt type of the elution buffer (NaCl, CsCl or MgCl2) they could separate RNAs with different conformations. Recently, Lukavsky and co-workers showed that weak anion-exchange fast protein liquid chromatography (FPLC) under non-denaturing conditions can be used to separate the desired RNA product from the T7 RNA polymerase, unincorporated rNTPs, small abortive transcripts and the plasmid DNA template [53]. Rapid purification of homogeneous RNAs can be achieved by using trans-acting hammerhead ribozymes in combination with anion-exchange high performance liquid chromatography (HPLC) chromatography at high temperature (90°C) [41]. In our laboratory, we are using an anion exchange HPLC under denaturing conditions (6 M Urea) at high temperature (85°C) allowing separation of RNAs to almost single nucleotide resolution up to 40 nucleotides. These harsh denaturing conditions also allow the separation of an RNA from a long DNA splint used in splinted ligation [37]. The eluted RNA is subsequently liberated from urea using butanol extraction [54]. Certain biologically relevant RNAs might fold into different conformations or might form multimers, which can be separated by purifying them under non-denaturing conditions using size-exclusion FPLC [50, 51, 55]. In addition to reverse-phase HPLC [56], the use of affinity chromatography has been described [57–60]. Batey and Kieft developed a sophisticated approach, where an affinity tag is attached to the 3′-end of the RNA by a glucosamine-6-phosphate activated (glmS)ribozyme [59]. The affinity tag is based on two RNA stem-loops having high affinity for the MS2 coat protein fused to a 6×His-tagged MBP, which binds to a Ni2+-affinity column. Elution of the RNA can be achieved by activating the ribozyme with addition of GlcN6P. Affinity purification based on aptamer tags binding Sephadex or Streptavidin have also been proposed [48, 60].
Depending on the purification method, the RNA can either be eluted directly into NMR buffer or needs to be exchanged into a suitable buffer. Buffer exchange can be performed by dialysis or by washing and concentrating the RNA with ultracentrifugation with an appropriate molecular weight cut-off (MWCO). Dialysis bags and ultracentrifugation filter devices with 1,000 Da MWCO are commercially available and are appropriate for RNAs produced by in vitrotranscription. The RNAs can be lyophilized and subsequently resuspended into NMR buffer. Typical NMR buffers for RNA are 10–50 mM sodium phosphate at pH = 5.5–6.
4 Deuteration Strategies for Larger RNAs
Small RNAs can be studied using uniformly 1H, 13C, 15N-labeled nucleotides with different nucleotide specific labeling schemes [3–5]. However, once the size of RNA oligonucleotides exceeds 40 nucleotides, NMR spectroscopic studies suffer from two major problems, which make unambiguous resonance assignments increasingly difficult [61]. First, this is caused by extensive overlap of the six ribose protons, of which five resonate within a narrow window of 1 ppm. Often, it is also impossible to gain additional resolution from the attached ribose carbon atoms, which also resonate in defined, but again narrow spectral regions. In turn, this leads to unresolvable resonance overlap in through-bond and through-space 3D-type NMR experiments and thus makes resonance assignments impossible. Second, larger RNA molecules are often elongated and tumble slowly in solution and thus 1H-13C dipolar relaxation is more pronounced. This results in increased proton and carbon linewidths, which additionally aggravate the existing resonance overlap and often broaden resonances beyond detection.
For larger proteins, partial or full deuteration helps to overcome short transverse relaxation times caused by dipolar 1H-1H and 1H-13C interactions and thus allows for efficient magnetization transfer along the protein backbone and into the side chains. This approach makes resonance assignments even for very large proteins feasible [62]. In larger RNA molecules, on the other hand, short transverse relaxation times are mainly caused by dipolar 1H-13C interactions and not so much by dipolar 1H-1H interactions due to the much lower proton density in RNA as compared to proteins [63]. Moreover, resonance assignments of RNA heavily rely on 3D 1H-13C correlation NMR spectra and therefore full deuteration is not an option for resonance assignments of large RNAs. For these reasons, different approaches have been developed to overcome the problem of resonance overlap in large RNAs using site-specific ribose or base labeling.
4.1 Site-Specific Ribose Deuteration
RNA consists of four types of residues, namely adenosine, guanosine, cytidine, and uridine, which differ by the nature of their base, but all contain the same ribose moiety. Hence, most extensive overlap in large RNAs is observed for the ribose protons H2′-H5′/H5″, which typically resonate between 4 and 5 ppm as well as their attached carbon atoms C2′-C5′, which resonate in specific, but narrow spectral regions from 60 to 85 ppm [64]. To resolve this overlap, selective labeling methods are required rather than uniform or residue-type specific labeling schemes as used for proteins.
Several site-specific labeling schemes have been developed for the ribose moiety of RNA: Site-specific deuteration in the ribose ring is used for spectral simplification of the crowded ribose region. Moreover, a combination of uniform or selective 13C-labeling with site-specific deuteration allows for spectral editing and helps to reduce the dipole-dipole interaction induced relaxation for improved efficiency of 1H-13C correlation experiments. While chemical synthesis would allow to precisely control the placement of specific isotopic labels in the ribose ring, this method requires expensive labeled precursors, multistep synthesis and often results in poor overall yields. Enzymatic synthesis with enzymes from the pentose phosphate pathway, on the other hand, can use glucose with a variety of labeling schemes and thus is more cost effective and results in higher overall yields. This method has been pioneered by the Williamson lab and some of these different possible labeling schemes have been employed to determine structures of large RNAs [29, 65].
Starting from fully deuterated glucose with or without 13C-labeling, enzymes from the pentose pathway are used to prepare 5-phospho-D-ribosyl α-1-pyrophosphate, which can be linked to the corresponding bases using enzymes from the nucleotide biosynthesis and salvage pathways. In addition, specific enzymes can be used in the first reaction steps to back-exchange the H1′ and/or H2′ position in the resulting ribose moiety, while H3′, H4′, H5′ and H5″ positions remain deuterated (Fig. 7.2a). This labeling scheme leads to great spectral simplification in the crowded ribose–ribose and ribose–base regions in 2D NOESY spectra, but retains important structural information. The sugar pucker can still be determined from H1′-H2′ cross peaks in 2D TOCSY and DQF COSY spectra and the cross peaks intensities of sequential H1′-base and H2′-base connectivities are indicative of canonical A-form or non-canonical RNA conformation. If combined with 13C-labeling, additional resolution of the H1′ and H2′ protons is gained from the attached carbon atoms in 3D 1H-13C correlation NMR experiments. In addition, this labeling scheme allows editing of C2′-H2′ correlations, which can overlap with C3′-H3′ correlations and thus makes unambiguous assignments possible, when through-bond HCCH-type experiments fail due to short transverse relaxation times [65].
Site-specific ribose and base labeling schemes.(a) Ribose moiety protonated in the H1′ and H2′ positions and deuterated in the H3′, H4′, H5′/H5″ positions from the Williamson group [65]. Deuterated positions are highlighted in red. This labeling scheme is also commercially available for all four nucleotides. (b) Ribose moiety protonated in the H2′ and H5′ positions and deuterated in the H1′, H3′, H4′, H5″ positions from the Wijmenga group [30]. Deuterated positions are highlighted in red. (c): Adenosine and guanosine nucleotides with the purine base moiety selectively protonated in the H8 position and a perdeuterated ribose moiety prepared from perdeuterated rNTPs following procedures described in [31]. Deuterated positions are highlighted in red. (d) Cytidine and uridine nucleotides with the pyrimidine base moiety selectively deuterated in the C5 position and a fully protonated ribose moiety prepared from fully protonated rNTPs following procedures described in [68]. Deuterated positions are highlighted in red
The Butcher group has successfully applied this labeling scheme to determine the structure of a 30 kDa GAAA tetraloop receptor complex [66]. Using uniform as well as nucleotide-specific 13C, 15N-labeling together with 3D NOESY and 2D filter/edited NOESY experiments, it was not possible to assign many NOEs in the ribose region which crucially defined the interaction interface. Selective deuteration of the ribose H3′-H5′/H5″ protons together with pyrimidine base H5 deuteration, on the other hand, allowed to assign many NOEs in otherwise crowded spectral regions (Fig. 7.3) and also helped to differentiate H2′ and H3′ chemical shifts, which were overlapped even in 13C-edited NOESY spectra. The authors also point out a minor disadvantage of this labeling scheme which results from the lack of NOEs from H4′ and H5′/H5″ protons, which often yield important structural information in non-canonical regions of the RNA molecule, such as bulges and loops [66]. But in many cases, non-A-form torsion angles or unusual stacking interactions in these regions result in unique 13C and 1H chemical shifts for these resonances and thus allow assignments even without selective labeling.
Relief of spectral crowding by site-specific ribose deuteration.(a) Secondary structure of a 30 kDa GAAA tetraloop-receptor RNA dimer. The helical regions are shown in black, the tetraloop is shown in red, and the receptor in green. Numbering according to [66]. (b, c) Region of the 2D NOESY spectrum obtained for the fully protonated in (b) or selectively ribose deuterated in (c) tetraloop-receptor RNA dimer. Deuteration of the ribose H3′, H4′, H5′ and H5″ positions (see Fig. 7.2a) reliefs the severe overlap in the ribose – base region of the 2D NOESY spectrum and allows sequential assignments through H2′ – base connectivities (Reprinted with permission from Davis et al. [66])
A partial deuteration scheme for the ribose moiety has also been described by the Wijmenga group. Again, starting from fully deuterated and 13C -labeled glucose, stereochemical H-exchange is used to selectively back-exchange the H2′ and H5′ position in the resulting ribose moiety, while the H1′, H3′, H4′ and H5″ positions remain deuterated (Fig. 7.2b) [30]. This labeling scheme preserves important structural information from H2′-base connectivities, the H2′-H5′ intra-sugar NOE contacts can provide sugar pucker information and sequential H2′-H5′ NOE contacts constrain the RNA backbone. The main advantage is that in addition, important torsion angle information is accessible through J H5′-C3′coupling (γ torsion angle) and J C4′Pand J H5′Pcouplings (β torsion angle). This scheme has been successfully demonstrated on a 31 nucleotide RNA, but still awaits an application to larger RNA systems [30].
4.2 Site-Specific Base Deuteration
Spectral simplification of the ribose–base region in NOESY spectra can also be achieved by site-specific deuteration of the purine and pyrimidine base moieties. The purine C8 and the pyrimidine C5 position of 5′ rNMPs can be deuterated using bisulfite modification under basic conditions before conversion to 5′ rNTPs [67, 68]. Likewise, protons can be selectively introduced at these positions in deuterated NMPs (Fig. 7.2c, d). The purine C8 position can also be efficiently exchanged in rNTPs rather than rNMPs under basic conditions without significant hydrolysis and degradation. Incubation of rATP or rGTP with triethylamine (TEA) at 60°C for 5 days or 24 h, respectively, leads to essentially complete exchange in the C8 position and the resulting rNTPs can be used directly for in vitrotranscription after removal of volatile TEA by lyophilization [31]. Exchange at the pyrimidine C6 position of rCMP or rUMP is less straightforward, but can be achieved with up to 70% efficiency in alkaline solution with DMSO before the rNMPs start to decompose [68].
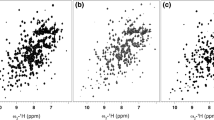
Purine C8 deuteration has been used prior to 13C,15N-labeling of RNA to distinguish adenine from guanine H8 protons and both of them from adenine H2 protons [67]. Deuteration of the C5 position of pyrimidines eliminates the strong H5-H6 crosspeak in NOESY and TOCSY spectra and by labeling RNA with either C5-deuterated uracil or cytosine their H6 protons can be discriminated through the absence of H5-H6 correlations for the C5-deuterated pyrimidine (Fig. 7.2d). In addition, this is advantageous for spectral simplification in large RNAs, since strong crosspeaks arising from the H5-H6 protons can obscure important sequential connectivities in the H1′ – base region of 2D and 3D NOESY spectra. The Lukavsky group has recently employed this labeling scheme to unambiguously assign the 48 nts K10transport signal RNA (Fig. 7.4a) [69]. This RNA displayed a high sequence redundancy containing 14 Watson-Crick A:U base pairs and a total of 18 adenosine and 19 uridine residues. Nevertheless, almost all adenine C2-H2 and C8-H8 crosspeaks were resolved in 1H-13C correlation NMR experiments, while uracil C6-H6 correlations showed a high degree of overlap (Fig. 7.4b), which could not be resolved in 3D NOESY-HSQC spectra. Likewise, unambiguous resonance assignments using homonuclear 2D NOESY spectra were also compromised by the prominent H5-H6 crosspeaks from the 19 uridine and 6 cytidine residues (Fig. 7.4c). Site-specific deuteration of the pyrimidine H5 positions helped to edit for the H1′–base connectivities, which allowed unambiguous, sequential assignments of a run of five consecutive uridine residues (Fig. 7.4d) [69]. In addition, this labeling scheme eliminates the nearby 1H,1H dipolar coupling partner of the pyrimidine H6 proton and subsequently leads to line narrowing of NOESY crosspeaks involving the pyrimidine H6 protons.
RNA labelling with C5 deuterated pyrimidines simplifies 2D NOESY spectra.(a) Secondary structure of wild-type K10TLS RNA. Numbering according to [69]. Outlined nucleotides were added to improve transcription efficiency. The three helical segments are separated by single nucleotide bulges (C33 and A37). (b) 1H,13C-ct-HSQC (constant-time heteronuclear single quantum coherence) spectrum of aromatic pyrimidine C6-H6 and purine C8-H8 correlations in 13C, 15N-labeled K10-wtRNA. Most aromatic resonances (labeled according to a) display excellent chemical shift dispersion, except for several uracil C6-H6 resonances, which show severe chemical shift overlap in both the carbon and proton dimensions (indicated by an oval). (c) Overlay of a homonuclear TOCSY (green) and NOESY (purple) spectrum of unlabeled K10-wtRNA presenting a crowded region of the aromatic-anomeric walk. Strong NOESY cross-peaks from pyrimidine H5-H6 protons (also indicated as green TOCSY peaks) obscure part of the aromatic-anomeric walk indicated with orange lines(e.g. U13 or U14). Intraresidue correlations are labeled with residue numbers according to (a). (d) Overlay of a homonuclear TOCSY (green) and NOESY (purple) spectrum of site-specifically pyrimidine C5 deuterated K10-wtRNA showing the same part of the aromatic-anomeric walk as in (c). Strong NOESY cross peaks (purple) from pyrimidine H5-H6 protons are now absent due to the site-specific pyrimidine C5 deuteration and only two residual TOCSY peaks (green) are visible as indicated by an asterix. The “pyrimidine H5-H6 crosspeak-free” aromatic-anomeric walk is indicated with orange lines(Reprinted with permission from Bullock et al. [69])
The Summers group makes extensive use of different combinations of perdeuterated, fully protonated and partially deuterated rNTPs to achieve assignments of very large RNA systems. These labeling schemes are often combined with segmental labeling using techniques outlined in the next section [31]. Unambiguous assignments for the 101 nts core encapsidation signal of the moloneymurine leukemia virus (MoMuLV) RNA were accomplished by structure determination of individual stem-loops complemented with conventional 3D and 4D 13C-edited NOESY experiments using nucleotide-specific 13C,15N-labeling on the entire 101 nts RNA [70, 71]. The assignments were completed with 2D NOESY spectra collected on four different nucleotide-specifically protonated samples with the remaining nucleotides being perdeuterated. This labeling scheme allows editing of intra-nucleotide NOEs for the protonated residue type and identifies inter-nucleotide NOEs between protonated residues if they are adjacent in sequence or close in space as a result of tertiary interactions [70].
For the assignments of the 132 nts double kissing hairpin of the MoMuLV 5′ untranslated region (Fig. 7.5a), this approach was extended using RNA samples with specifically C6/C8-protonated and perdeuterated or C8-deuterated and fully protonated nucleotides in combination with perdeuterated or fully protonated nucleotides (Fig. 7.2c) [72]. For instance, a 2D NOESY spectrum collected on an RNA sample containing perdeuterated uridine and cytosine, fully protonated guanosine and C8-protonated, perdeuterated adenosine edits the following structural information: intra ribose – base crosspeaks for all guanosines, inter ribose – base crosspeaks for sequential guanosines (Gi-riboseto Gi+1-H8), inter ribose – base crosspeaks for sequential guanosine-adenosine pairs (Gi-riboseto Ai+1-H8) and sequential H8 – H8 NOEs for all guanosines and adenosines (for example see Fig. 7.5b, c). Depending on the chemical shift dispersion of the aromatic protons and the number of different sequential nucleoside pairs, several combinations are required to obtain full assignments of a large RNA. In the case of the 132 nts double kissing hairpin, six different labeling combinations were required to obtain 1,248 unique NOE-derived 1H-1H distance restraints. Together with additional restraints which aid to maintain A-form conformation in helices, this yielded a well-defined structure of this large RNA with an r.m.s.d of 0.4 Å for the final ensemble of 20 structures [72]. This represents the largest RNA determined by solution NMR spectroscopy to date.
H8 enrichment of perdeuterated purines simplifies 2D NOESY spectra.(a) Secondary structure of the stem loops C and D of the MoMuLV 5′UTR. Numbering according to [72]. (b) 2D NOESY spectrum of the fully protonated stem loop CD dimer displays severe resonance overlap in the H1′ – base region. (c) 2D NOESY spectrum of the stem loop CD dimer transcribed using fully protonated GTP, H8-protonated, perdeuterated ATP, and perdeuterated CTP and UTP. Black linesdenote inter-guanosine H1′ – base connectivities and red linesdenote adenosine H8 to guanosine H1′ connectivities (For detailed explanation see text. Reprinted with permission from Miyazaki et al. [72])
Many of the presented labeling schemes can be implemented using commercially available rNTPs. Fully protonated, fully deuterated, 15N-, 13C- or 13C,15N-labeled rNTPs and rNMPs can be purchased and converted into rNTPs selectively protonated or deuterated at various base positions. The attractive labeling scheme from the Williamson group with deuterated H3′, H4′ and H5′/H5″ positions in the ribose moiety and the H5 position in pyrimidine bases is also commercially available and other labeling schemes might be available upon request [29, 65].
5 Segmental Isotope Labeling for Large RNAs
Although signal overlap can be substantially reduced using selectively labeled nucleotides or RNAs containing nucleotide-specific labeling schemes, larger RNA structures are difficult or impossible to tackle due to the tremendous spectral overlap [6]. Segmental isotopic labeling of RNA is therefore essential to study RNAs of moderate to large size by NMR spectroscopy. Firstly, it allows verifying whether a small structural RNA element such as a hairpin that can be studied by standard NMR methods retains the same structure as in the larger, biological active RNA. One can, for example, ligate a small isotopically labeled fragment produced by chemical synthesis or in vitro transcription to a larger unlabeled fragment. Using this approach, Puglisi and co-workers could show that a 25 kDa RNA domain adopts the same structure in isolation as found in the context of the entire 100 kDa natural RNA [73, 74]. Very recently, the group of Summers segmentally isotope labeled 29 nts at the 3′ end of the intact 230 kDa HIV-1 5′-leader [75] and they could detect structures that regulate HIV-1 genome packaging. Secondly, segmental isotope labeling is reducing the number of resonances in 13C- or 15N-edited correlation spectra and therefore the spectral overlap. Properly choosing the segmental isotope labeling strategy allows the measurements of residual dipolar couplings and paramagnetic relaxation enhancement of enough RNA resonances to extract structural information (our unpublished results and references [7, 73]).
RNA segmental labeling can be also very useful for other biophysical techniques. Site-specific incorporation of heavy atoms into internal positions within longer RNAs showed to have high potential for solving phases in X-ray crystallography [21, 76]. Similarly, single molecule experiments with RNA often require the incorporation of modified nucleotides at specific positions within a long RNA to study its structure or folding [22]. Finally, we could show using segmental labeling that introduction of two nitroxyl radical tags into specific positions of a longer RNA allows the measurement of long-range distances by pulse electron paramagnetic resonance (our unpublished results). Therefore, enzymatic ligation between a short synthetic RNA containing modified nucleotides and longer fragments produced by in vitrotranscription is expected to become a method of choice for studying biologically important RNAs (unpublished results and reference [77]). However, methods for incorporating modified nucleotides into internal positions of longer RNAs (>100 nts) for structural studies or segmental isotope labeling for NMR structural studies remained very time-consuming, costly as the yield they provide is low and not always applicable because of the sequence-dependence of most protocols [31, 35]. In the following sections, we will first describe the principle and shortly present several methods for segmental isotope labeling by emphasizing their advantages and disadvantages. Then we will describe our recently developed alternative approach for segmental labeling of RNA by which we can obtain very rapidly (5–7 days) high amounts (up to ten-fold higher than previously reported) of segmentally labeled RNAs without sequence requirements [37]. We will finish this chapter by providing a detailed protocol of our recently published method.
5.1 General Principle
Basically, two or several RNA fragments that are unlabeled, uniformly isotope labeled or are containing modified or specifically labeled nucleotides at specific positions in the RNA sequence are combined and ligated to obtain the full length RNA of interest. The RNA fragments can be produced by chemical synthesis, in vitroenzymatic transcription or in vivodepending on their required property (see first section). The RNA fragments have to fulfill certain requirements such that they can be ligated without self-ligation or ligation in the wrong sequential order. RNA ligation can be performed by T4 RNA or T4 DNA ligase [78–80] or by using a deoxyribozyme that catalyzes RNA ligation [81]. Both T4 RNA and T4 DNA ligases require a 5′-monophosphate on the donor fragment and 3′-hydroxyl on the acceptor fragment at the site of ligation (see Fig. 7.6dstep3), whereas the deoxyribozyme catalyzes a ligation reaction with a 5′-triphosphate on the donor fragment and with a 3′-hydroxyl on the acceptor fragment. Ligation with DNA ligase, which recognizes a nicked double-stranded substrate, is performed by annealing a DNA oligonucleotide or a 2′-O-methyl-RNA/DNA chimera to the site of ligation [78]. Unlike DNA ligase, RNA ligase requires a single-stranded site of ligation. Preferentially, the acceptor and the donor are brought together by base-pairing such that the site of ligation is in a hairpin loop [37, 80, 82]. However, it has been shown that RNA ligase can also be used in combination with a DNA oligonucleotide annealing with the site of ligation designed to mimic the natural substrate of RNA ligase [83]. To prevent self-ligation or ligation of the fragments in the improper sequential order (especially using T4 RNA ligase), the acceptor fragment should contain a hydroxyl group both at its 5′ and 3′ ends, whereas the donor fragment should have a monophosphate at the ligation site and a monophosphate or a 2′,3′-cyclic phosphate at the 3′-end (see Fig. 7.6dstep3). As discussed in the first section, RNAs obtained by in vitro transcription contain a 5′-terminus with a tri-phosphate and an inhomogenous 3′-hydroxyl terminus. Ribozymes engineered at the 3′-end producing homogenous 2′,3′-cyclic phosphates can thus be used to generate 3′-ends of both acceptor and donor fragment, whereupon the acceptor 3′-end has to be further dephosphorylated using T4 polynucleotide kinase (PNK), which has 3′-phosphatase activity [31, 75, 84, 85]. Hammerhead ribozymes located 5′ to the acceptor fragment generate the correct 5′-hydroxyl end, whereas the 5′ donor end generated by a hammerhead ribozyme has to be phosphorylated by T4 PNK. To generate two samples, in which one segment is labeled and the second one is unlabeled, two unlabeled and two labeled transcription reactions must be performed. In addition, the use of T4 PNK is an additional costly step, which also requires an additional purification.
Different approaches for segmental isotope labeling of RNA.The approaches by Xu et al. (a), Tzakos et al. (b), Nelissen et al. (c) and Duss et al. (d) are explained in the main text. An overview of the four methods is presented in (e). (a–d) The isotope labeled material is highlighted in red, the unlabeled material in black. The G or the UX at the 5′- or the 3′-end of certain fragments indicate sequence requirements. The G at the 5′-end of certain RNA fragments represents a good transcription start site. N+++ stands for an inhomogenous 3′-end. P-2′/3′ indicates a 2′/3′-cyclic phosphate. In the 2′-O-methyl RNA/DNA chimeras (a, d) the DNA is in dark blueand the 2′-O-methyl RNA in light blue. Scissorsindicate RNase H cleavage sites. The orange starsin (c) represent selectively deuterated uridines. (d) The correctly protected termini of both acceptor and donor fragments are encircled in green. E: To compare the different approaches the yield was calculated to be the amount obtained for two segmental isotopically labeled samples, in which only one fragment is isotopically labeled starting from one 20 ml labeled transcription reaction (4.5 mM in each NTP): Fig. 7.6 (continued) For the approach of Duss et al., the yields for the two-piece ligation (d, one RNase H cleavage and one ligation reaction) or for the four-piece ligation (Fig. 7.7; two RNase H cleavages and one ligation reaction) were determined by starting with each 400 nmol of labeled and 400 or 1,200 nmol of unlabeled RNA (obtained from one 20 ml labeled and one 20 or 60 ml unlabeled transcription reaction for the two-fragments or the four-fragments ligation, respectively) and determining the final yield assuming typical yields (after purification) for RNase H cleavage of 50–90% and T4 DNA/RNA ligation of 50–85%. In the approach of Nellisen et al., they start from 665 nmol of isotopically labeled fragment and obtain 173 nmol of onesegmentally labeled RNA (using the more efficient two-pot ligation), in which only the middle fragment is isotopically labeled. It is not mentioned from which volume of transcription reaction this was obtained. Considering the fact that PAGE purification was used to purify the fragments and that the yield of the fragments is RNA sequence dependent, it can be assumed that the 665 nmols of labeled RNA fragment were obtained from >60 ml transcription reaction (at a concentration of 4.5 mM for each NTP). To obtain twosegmentally labeled samples, in which a different segment is labeled, 120 ml transcription reaction would be required to obtain 173 nmol of each sample. Therefore, they would obtain <30 nmol of two segmentally isotope labeled RNAs from one 20 ml labeled transcription reaction. The time requirement is without cloning and template plasmid production, however it includes the time needed for small scale optimization reactions required for a new system under study. If not mentioned by the authors, 1 day for each small scale optimization (transcription, RNase H cleavage or ligation) and 3–4 days for PAGE purification is assumed (dand eare adapted with permission from Duss et al. [37])
In the last 15 years, several groups came up with elegant approaches to improve the efficiency and practicability of segmental isotope labeling of RNA. In the following sections, we will present their principles and discuss their advantages and disadvantages.
5.2 Previous Methods
Crothers and co-workers showed that sequence-specific RNase H cleavage of an unlabeled and a labeled RNA can be followed by direct cross re-ligation of a labeled with an unlabeled fragment using T4 DNA ligase (see Fig. 7.6a) [86]. This method allows for multiple segmental isotope labeling and requires only one labeled and one unlabeled transcription reaction. However, this method has 5′ sequence requirements in that the 5′-end has to be a good transcription start site. Therefore, the final yield depends on the 5′-end. Furthermore, the method suffers from 3′-inhomogeneities. Because the four termini of the two fragments to be ligated are not correctly protected ligation can only be performed using splinted T4 DNA ligation as non-splinted ligation would lead to undesired side-products (self-ligation, ligation in the wrong sequential order, multimerization, circularization).
Lukavsky and co-workers used a plasmid encoding the 3′ donor fragment followed by a hammerhead ribozyme, which is connected by a flexible linker to a second hammerhead ribozyme preceding the 5′ acceptor fragment yielding a terminal 3′-hydroxyl after transcription (see Fig. 7.6b) [73, 87, 88]. If the transcription reaction is primed with GMP, both fragments are correctly protected for ligation with T4 RNA ligase (or T4 DNA ligase), the 5′ acceptor fragment being protected in both 5′- and 3′-ends with hydroxyl groups and the 3′ donor fragment being protected by a 5′-phosphate and a 2′,3′-cyclic phosphate. An additional advantage is that this approach requires only one labeled and one unlabeled transcription reaction. The drawbacks of this technique are that a G is present at the 3′ of the ligation site and a U at the penultimate position of the RNA. Furthermore, transcription can potentially generate an inhomogenous 3′-end of the acceptor fragment leading to possible incorporation of additional nucleotides at the site of ligation, especially when using RNA ligase. Finally, this technique does not allow for multiple segmental isotope labeling. It is therefore not possible to label a fragment within an RNA.
Wijmenga and co-workers introduced selectively deuterated uridine residues into the central position of a 20 kDa RNA (see Fig. 7.6c) [89]. The three fragments (two unlabeled and one transcribed with selectively deuterated, uniformly 13C/15N-labeled UTP and otherwise unlabeled NTPs) were transcribed separately. Subsequently, the three fragments were either ligated in a two-step protocol first with T4 DNA ligase followed by T4 RNA ligation or by a one-step protocol using only T4 RNA ligase. The ligation sites have to be designed in such a way that the three fragments fold into the target or target-like structure for preventing undesired side-product formation (self-ligation, ligation in the wrong sequential order, multimerization, circularization) as the termini are not correctly protected. Furthermore, the different fragments have to be purified by PAGE to prevent or minimize the introduction of non-native nucleotides at the sites of ligation. As the three fragments are obtained from three separate transcription reactions every fragment has to start with a guanosine and the final yield strongly depends on the specific starting sequence of the several fragments. As demonstrated for this specific example, multiple segmental isotope labeling is possible. However, its practicability is limited to cases for which the fragments can fold into target-like structures and start with a guanosine. A similar approach, using both T4 DNA and RNA ligase for ligating multiple chemically synthesized RNA fragments, has allowed introducing site-specific 2′-methylseleno labels into long RNAs [21, 82].
Very recently, the site-specific introduction of a single isotopically labeled guanosine residue into a long RNA has been presented. This method consists of two simple enzymatic reactions. Firstly, a group I self-splicing intron transfers an isotopically labeled GMP into an RNA of interest generating a 5′-residue labeled fragment. This 3′-fragment is then ligated to an unlabeled 5′ fragment using T4 DNA splinted ligation [90].
A comparison of the different methods for segmental isotope labeling of RNA summarizing their strong and weak points is shown in Fig. 7.6e.
The techniques presented in this section resulted in a so far very limited use of segmental isotope labeling in NMR studies of RNA [74, 75]. Reasons are:
-
the reported low yields (less than 30 nmol of segmentally labeled RNA were obtained starting from 20 ml labeled transcription reactions (see Fig. 7.6e))
-
the sequence requirements at the sites of ligation (such as the need of a guanosine 3′ to the ligation site [88]) or that all [89] or some [86] fragments must start with a guanosine
-
the requirement that the different fragments to be ligated need to fold into a target-like structure for efficient ligation [89]
-
the tedious purification steps by PAGE
-
the need to use additional enzymes such as T4 polynucleotide kinase for engineering the 3′- and 5′-termini [31, 75]
-
the inability or difficultly to introduce labeled fragments within and not only at the end of an RNA
In the next section, we will discuss an approach for segmental isotope labeling of RNA that we recently published [37]. This approach allows for multiple segmental isotope labeling and does not suffer from sequence requirements. High yields are obtained within minimal preparation times paving the path for studying RNAs of biologically relevant size.
5.3 Fast, Efficient and Sequence-Independent Method for Flexible Multiple Isotope Labeling of RNA
5.3.1 Principle
This approach is based on the transcription of two full-length RNAs with identical sequence, one isotopically labeled and one unlabeled (Fig. 7.6d) [37]. The transcribed RNAs are flanked at the 5′-end by a hammerhead (HH) ribozyme in cisand at the 3′-end by the minimal sequence required by the NeurosporaVarkud satellite ribozyme for cleavage in trans[36] (step 1 in Fig. 7.6d) or an HDV ribozyme having no sequence requirements when placed at the 3′-end [38, 39]. Both ribozymes (in cisor in trans) cleave co-transcriptionally leading to two homogenous termini, a 5′-hydroxyl and 2′/3′-cyclic phosphate for the full-length RNA. After purification, the two transcribed RNAs are cleaved site-specifically by RNase H using a guide 2′-O-methyl-RNA/ DNA splint yielding an acceptor fragment (5′-fragment) with two hydroxyl termini and a donor fragment (3′-fragment) with a phosphate at its 5′-end and a cyclic 2′/3′-phosphate at its 3′-end [45] (step 2 in Fig. 7.6d). After separation of the two fragments of each cleavage reaction, the subsequent two cross-religations between the labeled fragment and the unlabeled fragment using T4 RNA or T4 DNA ligase results in two segmentally labeled RNAs, in which either the 5′- or the 3′-fragment is isotopically labeled [86] (step 3 in Fig. 7.6d). Each reaction step is followed by a fast and efficient denaturing anion-exchange HPLC purification followed by an n-butanol extraction or a dialysis to remove urea and salts. For a two-piece ligation, 5–7 days are required in total, whereas 2–3 days are needed for step 1 (1 day transcription optimization, 1–2 days large scale transcription and purification), 1.5–2 days for step 2 (0.5–1 day RNase H cleavage optimization, 1 day large scale cleavage and purification) and 1.5–2 days for step 3 (0.5–1 day ligation optimization, 1 day large scale ligation and purification).
One main advantage of this method compared to others is that one obtains from only one labeled transcription reaction two homogenous fragments, which are correctly engineered at all four termini (Fig. 7.6dafter step 2). This is essential to obtain the highest possible yields during the ligation step (Fig. 7.6dstep 3) since no self-ligation or ligation in the wrong sequential order is possible and only the correct product can be obtained. Furthermore, there are no sequence restrictions on the identity of the fragments. This method is flexible in that it allows not only ligating two differentially labeled fragments but also allows to introduce any labeled fragment withinan unlabeled RNA via three- or more-piece ligations. The method was applied on a 72 nts non-coding RNA containing four stem-loops, for which four different NMR samples could be obtained with one of the four stem-loops isotopically labeled at the time (see Fig. 7.7) [37, 91]. For generating these four samples, only one labeled transcription reaction was required.
Principle, reaction efficiencies and NMR evidence for isotope labeling of each stem-loop of the RsmZ RNA separately.(a) Sequence-specific RNase H cleavages to obtain all four isotopically labeled stem-loop fragments. The yields of the cleavage reactions before HPLC purification are indicated, the values in brackets are expressing the yield after purification. The different stem-loops are colored (SL1: magenta, SL2: green, SL3: orange, SL4: cyan). (b) Splinted T4 DNA ligase mediated ligations of isotope labeled (in color) and unlabeled (in black) fragments. The unlabeled fragments were obtained in a similar way as the labeled fragments. (c) NMR evidence for the successful segmental isotope labeling of each stem-loop separately. 1H-15N-HSQC NMR spectrum of the uniformly 15N-labeled RsmZ RNA (left) and overlay of the 1H-15N-HSQC NMR spectra of the four segmentally labeled RsmZ RNAs with each stem-loop labeled separately (right). The spectra were recorded on a Bruker 600 MHz spectrometer at 10°C (Reprinted with permission from Duss et al. [37])
5.3.2 Protocol
5.3.2.1 Vector Construction and Plasmid Amplification
-
1.
DNA construct design: The construct used for co-transcriptional cleavage of the target RNA (step 1 Fig. 7.6d) is composed of a T7 promoter, an optimal transcription starting sequence (for example GGGAUC, see Milligan et al. 1987 [23]), a HH ribozyme [36], the coding sequence of the target RNA, a minimal sequence for recognizing the VS RNA in trans[40] and finally a BamHI restriction site for linearization. If the target RNA has a similar length like the HH ribozyme additional nucleotides (such as an MS2 stem-loop structure) can be included preceding the 5′ HH ribozyme to extend the length of the fragment containing the HH ribozyme. This allows for an optimal separation of the target RNA from the 5′HH during anion-exchange HPLC. Instead of a VS ribozyme (which has minimal sequence requirements by cutting after any nucleotide other than a cytosine) a hepatitis delta virus (HDV) ribozyme can be used at the 3′ end. The HDV ribozyme has no sequence requirements when placed at the 3′ end. To check that both HH and VS ribozyme are predicted to fold properly M-fold [92] should be used to check the secondary structure of the entire transcribed RNA.
-
2.
Synthesis of insert by PCR: The full insert (for example 300 nts) is obtained by three consecutive PCR reactions. A first PCR reaction is performed using only one overlapping primer pair and no template. After purification, this first PCR product is used as template for a second PCR reaction using primers extending on both ends. After a second purification, this extended primer PCR is repeated once by again using the product of the previous reaction as template for the next PCR to finally obtain the insert sequence.
-
3.
The insert is double digested and purified.
-
4.
The purified double digested insert is ligated with a doubly digested pUC19 plasmid and subsequently transformed into appropriate competent cells.
-
5.
After sequencing, the plasmid is amplified by performing a large plasmid purification to obtain typically 5–8 mg of DNA (QIAGEN plasmid Giga kit).
-
6.
The plasmid is linearized using 0.2 U BamHI/μg of DNA doing an overnight digestion. No further purification is required for transcription.
-
7.
The DNA sequence coding for the VS ribozyme RNA [40] is obtained with the same extended primer PCR as described above, is also cloned into a pUC19 vector, amplified using a large plasmid purification and finally linearized.
5.3.2.2 RNA Purification
After every reaction step (transcription, RNase H cleavage and T4 RNA or DNA ligation; see below) an RNA purification is required (see Fig. 7.8).
Protocol for multiple segmental isotope labeling of RNA.(a, b) Co-transcriptional ribozyme cleavage and its purification. (a) Denaturing anion-exchange HPLC profile of a 10 ml transcription mix (which corresponds to 200 nmol of a 72 nts RNA product after purification) and (b) analytical 16% denaturing PAGE gel of the corresponding elution fractions. The different fragments obtained by co-transcriptional ribozyme cleavage are shown on the topof their corresponding peak (blue: target 72nts RNA, red: hammerhead ribozyme, green: 24 nts VS stem-loop sequence required for VS ribozyme cleavage in trans, orange: VS ribozyme). (c) Preparative scale (120 nmol) denaturing anion-exchange HPLC profile of fragments obtained by site-specific RNase H cleavage of a 40 nts RNA (SL12) to obtain SL1 (16 nts) and SL2 (24 nts) using only 5% chimera (14 nts). The different fragments obtained by RNase H cleavage are shown on the topof their corresponding peak. The chimera used for this cleavage is shown on the bottom. Its nucleotides are coloredaccording to the stem-loops they are hybridizing to. The nucleotides in the chimeras are either DNA (underlined) or 2′-O-methyl-RNA (Am, Cm, Gm, Um). (d) Analytical 16% denaturing PAGE gel of a non-splinted ligation reaction of a 29 nts RNA (5′-RNA) and a 43 nts RNA (3′-RNA) using T4 RNA ligase. In the ligation scheme the unlabeled RNA is in blackand the labeled RNA in red(Adapted with permission from Duss et al. [37])
-
1.
The purification is performed by anion-exchange chromatography on a preparative Dionex DNAPac PA-100 column (22 × 250 mm) at 85°C. Flow rate: 20 ml/min; eluent A: 12.5 mM Tris–HCl (pH = 8.0), 6 M urea; eluent B: 12.5 mM Tris–HCl (pH = 8.0), 0.5 M NaClO4, 6 M urea; detection at 260 nm; 30–75% B gradient within 18 min.
-
2.
Fractions containing the purified RNA are determined by 16% urea acrylamide gels
-
3.
Fractions are liberated from urea and desalted by dialysis against water or by n-butanol extraction of the aqueous phase until RNA precipitation [54]. The RNA precipitate is redissolved into 1 ml of water and precipitated with a 30–50 ml of n-butanol followed by centrifugation. The last step is repeated twice more. The final precipitate is resuspended in a few hundred microliters of water and freeze-dried overnight. The lyophilized RNA is dissolved into an appropriate buffer.
5.3.2.3 RNA Transcription and Co-transcriptional Ribozyme Cleavage (Step 1 Fig. 7.6d)
-
1.
Transcription yields and ribozyme cleavage efficiencies are optimized on 40 μl small scale reactions with changing concentrations of MgCl2, plasmid DNA, NTPs and T7 polymerase and testing the influence of the addition of pyrophosphatase and/or GMP.
-
2.
The best condition is scaled-up to a large scale reaction of 5–20 ml. A typical reaction contains 42.5 mM MgCl2, 4.5 mM of each NTP, 33 ng/μl linearized plasmid, 10 μM separately transcribed VS RNA and 1.7 μM in-houseproduced T7 Polymerase [32] in a transcription buffer containing 40 mM Tris–HCl pH = 8.0, 1 mM spermidine, 0.01% Triton X-100 and 5 mM DTT. After 4–6 h of transcription it is usually sufficient to heat the reaction mix to 65°C for 15 min to complete the ribozyme cleavage. However, it might be necessary to perform several cycles of thermal cycling if the ribozyme cleavage efficiencies are unsatisfactory. Finally, the reaction is stopped with the addition of 100 mM EDTA pH = 8.0.
-
3.
After filtration of the RNA using a 0.22 μm filter the RNA is purified by anion-exchange HPLC followed by n-butanol extraction (see Fig. 7.8a, b).
Note: The labeled NTPs are commercially available but can also be prepared in-houseaccording to the protocol from Batey et al. [28].
5.3.2.4 Sequence-Specific RNase H Cleavage (Step 2 Fig. 7.6d)
-
1.
Sequence-specific RNase H cleavage is performed by annealing a 2′-O-methyl-RNA/DNA chimera to the site of ligation [93]. An example of a chimera is shown in Fig. 7.8c.
-
2.
The best conditions for cleavage are determined by small scale reactions (typically 500 pmol RNA in 15 ul reaction volume) mainly optimizing the RNase H enzyme concentration (NEB, or in-house produced [49]) and the ratio between the RNA and the 2′-O-methyl-RNA/DNA chimera. The reaction temperature and the reaction time do usually not have a big influence on the cleavage specificities. Most reactions are conducted for 1 h at 37°C. However, some reactions require cleavage at 4°C to minimize unspecific cleavage. Most RNase H reactions are performed using only 5% stoichiometric amount of 2′-O-methyl-RNA/ DNA chimera. However, some reactions are less sensitive to unspecific cleavage when using stoichiometric amounts of chimera.
-
3.
The best conditions are scaled up to a large scale reaction (20–200 nmol). The only parameter that cannot be scaled up is the RNase H enzyme concentration that has to be down-scaled ten times compared to the small scale reaction to prevent potential unspecific cleavage. A typical reaction to cleave 200 nmol of RNA is performed in 6 ml volume containing 33 μM RNA, 1.65 μM chimera, 80 nM in-houseproduced RNase H in 50 mM Tris–HCl pH = 7.5, 100 mM NaCl and 10 mM MgCl2.
-
4.
The reactions are directly loaded onto the anion-exchange HPLC followed by n-butanol extraction and lyophilization (see Fig. 7.8c).
5.3.2.5 RNA Ligation Using T4 RNA and DNA Ligase (Step 3 Fig. 7.6d)
-
1.
Non-splinted T4 RNA based ligations are first performed on small scale reactions (typically 400 pmol RNA fragments in 10 ul reaction volume) mainly optimizing the T4 RNA enzyme concentration (NEB) and testing the addition of BSA (see Fig. 7.8d).
-
2.
Splinted T4 DNA based small scale ligation reactions (typically 200 pmol RNA fragments in 20 ul reaction volume) are performed by optimizing the T4 DNA enzyme concentration (NEB, fermentas or in-house), the reaction time, the reaction temperature and testing the influence of PEG-4000. The DNA splints, which are added in a 1–1.2-fold excess in respect to the RNA fragments, are usually annealed to the RNA fragments prior to ligation. However, we found that depending on the secondary structure of the RNA and the DNA splints annealing is not required.
-
3.
The best reaction conditions are scaled up for the large scale reactions. A typical large scale ligation reaction (e.g. 100 nmol of RNA fragments) using T4 RNA ligase is 40 μM in both RNA fragments, 1× in NEB ligation buffer (50 mM Tris–HCl pH = 7.8, 1 mM ATP, 10 mM MgCl2, 10 mM DTT), 1× in BSA using 5U T4 RNA ligase per nmol of RNA to be ligated. The reaction is performed for 2 h at 37°C.
-
4.
A typical large scale ligation reaction (e.g. 100 nmol of RNA fragments) using T4 DNA ligase is 10 μM in RNA fragments, 15 μM in DNA splint oligo, 10% in PEG-4000, 40 mM Tris–HCl pH = 7.8, 0.5 mM ATP, 10 mM MgCl2, 10 mM DTT, 50U T4 DNA ligase (fermentas) per nmol of RNA to be ligated or 2 μM final concentration of in-houseproduced T4 DNA ligase. The reaction is performed for 2–6 h at 37°C.
-
5.
The reactions are subjected to HPLC purification followed by n-butanol extraction and lyophilization.
5.3.2.6 NMR Spectroscopy
The lyophilized RNAs are typically dissolved into 250 ul Buffer containing 10 mM sodium phosphate at pH = 6.0 containing 10% 2H2O with RNA concentrations of 40–500 μM.
6 Conclusions and Outlook
Although almost half of the RNA structures deposited in the PDB database were determined by NMR spectroscopy only ten of them are >20 kDa and only 4 >30 kDa (April 2012). The limitations in isotope labeling of RNA are a main factor that prevented solving more RNA structures of larger molecular weight. The severe spectral overlap of RNA resonances and the strong relaxation of the sugar or the H5-H6 protons make RNA structure determination of larger size difficult or impossible. Recent improvements in strategies for selective deuteration/protonation of nucleotides that can be incorporated into RNAs have helped to reduce the spectral overlap but also to decrease broadening of the NMR resonances due to proton-proton relaxation. On the other hand, segmental isotope labeling of RNA is decreasing the spectral overlap providing the basis for studying RNAs of larger size. Specially, isotope labeling of RNA will gain in importance in verifying if a small RNA domain has the same structure in isolation as in the context of the larger biologically relevant RNA. Using a modular approach, it will allow in future to target structures of large RNAs or protein-RNA complexes. Initially, the structures of small RNA domains will be solved by conventional NMR methods or X-ray crystallography. Then, the chemical shifts of the small domains will be compared to the chemical shifts of the domains in context of the large RNA or protein-RNA complexes using segmental isotope labeling of the large RNA. The small RNA domains with identical chemical shifts will then be taken as building blocks for the large RNA. Finally, the relative positions of the different domains with respect to each other will be determined using residual couplings (RDCs), paramagnetic relaxation enhancement (PRE) and other non NMR methods. As the measurement of RDCs and PREs relies on the observation of non-overlapped, well-separated resonances, both segmental isotope labeling of RNA and the incorporation of selectively deuterated NTPs will be necessary to study larger RNAs. Using segmental isotope labeling of RNA in combination with measurements of RDCs and measurements of long-range distances by electron paramagnetic resonance, allowed us to solve the solution structure of a 70 kDa tetra-molecular complex containing a 72 nucleotides RNA bound to three protein dimers (Duss and Allain, unpublished results). This would not have been possible without a highly efficient method for multiple segmental isotope labeling of RNA [37]. We expect that more structures in solution of high molecular weight RNA and protein-RNA complexes will be determined in the future following the same approach.
References
Sharp PA (2009) The centrality of RNA. Cell 136:577–580
Mattick JS (2011) The central role of RNA in human development and cognition. FEBS Lett 585:1600–1616
Varani G, Aboulela F, Allain FHT (1996) NMR investigation of RNA structure. Prog Nucl Magn Reson Spectrosc 29:51–127
Wijmenga SS, van Buuren BNM (1998) The use of NMR methods for conformational studies of nucleic acids. Prog Nucl Magn Reson Spectrosc 32:287–387
Dominguez C, Schubert M, Duss O, Ravindranathan S, Allain FHT (2011) Structure determination and dynamics of protein-RNA complexes by NMR spectroscopy. Prog Nucl Magn Reson Spectrosc 58:1–61
Lukavsky PJ, Puglisi JD (2005) Structure determination of large biological RNAs. Methods Enzymol 394:399–416
Tzakos AG, Grace CRR, Lukavsky PJ, Riek R (2006) NMR techniques for very large proteins and RNAs in solution. Annu Rev Biophys Biomol Struct 35:319–342
Zuo XB, Wang JB, Foster TR, Schwieters CD, Tiede DM, Butcher SE, Wang YX (2008) Global molecular structure and interfaces: refining an RNA: RNA complex structure using solution X-ray scattering data. J Am Chem Soc 130:3292–3293
Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE (2004) Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol 11:257–264
Perez-Canadillas JM (2006) Grabbing the message: structural basis of mRNA 3′UTR recognition by Hrp1. EMBO J 25:3167–3178
Ohtsuki T, Vinayak R, Watanabe Y, Kita K, Kawai G, Watanabe K (1996) Automated chemical synthesis of biologically active tRNA having a sequence corresponding to Ascaris suum mitochondrial tRNA(Met) toward NMR measurements. J Biochem 120:1070–1073
Scaringe SA, Wincott FE, Caruthers MH (1998) Novel RNA synthesis method using 5′-O-silyl-2′-O-orthoester protecting groups. J Am Chem Soc 120:11820–11821
Pitsch S, Weiss PA (2002) Chemical synthesis of RNA sequences with 2′-O-[(triisopropylsilyl)oxy]methyl-protected ribonucleoside phosphoramidites. Curr Protoc Nucleic Acid Chem Chapter 3:Unit 3.8
Ramos A, Varani G (1998) A new method to detect long-range protein-RNA contacts: NMR detection of electron-proton relaxation induced by nitroxide spin-labeled RNA. J Am Chem Soc 120:10992–10993
Wenter P, Reymond L, Auweter SD, Allain FH, Pitsch S (2006) Short, synthetic and selectively 13C-labeled RNA sequences for the NMR structure determination of protein-RNA complexes. Nucleic Acids Res 34:e79
Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, Allain FH (2005) Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science 309:2054–2057
Auweter SD, Fasan R, Reymond L, Underwood JG, Black DL, Pitsch S, Allain FH (2006) Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J 25:163–173
Oberstrass FC, Lee A, Stefl R, Janis M, Chanfreau G, Allain FH (2006) Shape-specific recognition in the structure of the Vts1p SAM domain with RNA. Nat Struct Mol Biol 13:160–167
Skrisovska L, Bourgeois CF, Stefl R, Grellscheid SN, Kister L, Wenter P, Elliott DJ, Stevenin J, Allain FH (2007) The testis-specific human protein RBMY recognizes RNA through a novel mode of interaction. EMBO Rep 8:372–379
Dominguez C, Fisette JF, Chabot B, Allain FH-T (2010) Structural basis of G-tract recognition and encaging by hnRNP F quasi RRMs. Nat Struct Mol Biol 17:853–861
Hobartner C, Rieder R, Kreutz C, Puffer B, Lang K, Polonskaia A, Serganov A, Micura R (2005) Syntheses of RNAs with up to 100 nucleotides containing site-specific 2′-methylseleno labels for use in X-ray crystallography. J Am Chem Soc 127:12035–12045
Rieder R, Lang K, Graber D, Micura R (2007) Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. Chembiochem 8:896–902
Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC (1987) Oligoribonucleotide synthesis using T7 RNA-polymerase and synthetic DNA templates. Nucleic Acids Res 15:8783–8798
Gurevich VV, Pokrovskaya ID, Obukhova TA, Zozulya SA (1991) Preparative in vitro messenger-RNA synthesis using Sp6 and T7 RNA-polymerases. Anal Biochem 195:207–213
Pokrovskaya ID, Gurevich VV (1994) In-vitro transcription – preparative RNA yields in analytical scale reactions. Anal Biochem 220:420–423
Price SR, Ito N, Oubridge C, Avis JM, Nagai K (1995) Crystallization of RNA-protein complexes.1. Methods for the large-scale preparation of RNA suitable for crystallographic studies. J Mol Biol 249:398–408
Nikonowicz EP, Sirr A, Legault P, Jucker FM, Baer LM, Pardi A (1992) Preparation of C-13 and N-15 labeled RNAs for heteronuclear multidimensional NMR-studies. Nucleic Acids Res 20:4507–4513
Batey RT, Battiste JL, Williamson JR (1995) Preparation of isotopically enriched RNAs for heteronuclear NMR. Methods Enzymol 261:300–322
Scott LG, Tolbert TJ, Williamson JR (2000) Preparation of specifically H-2- and C-13-labeled ribonucleotides. Methods Enzymol 317:18–38
Cromsigt J, Schleucher J, Gustafsson T, Kihlberg J, Wijmenga S (2002) Preparation of partially H-2/C-13-labelled RNA for NMR studies. Stereo-specific deuteration of the H5″ in nucleotides. Nucleic Acids Res 30:1639–1645
Lu K, Miyazaki Y, Summers MF (2010) Isotope labeling strategies for NMR studies of RNA. J Biomol NMR 46:113–125
Price RP, Oubridge C, Varani G, Nagai K (1998) Preparation of RNA: protein complexes for X-ray cristallography and NMR. In: Smith CWJ (ed) RNA-protein interactions: a practical approach. Oxford University Press, Oxford, pp 37–74
Gallo S, Furler M, Sigel RKO (2005) In vitro transcription and purification of RNAs of different size. Chimia 59:812–816
Pleiss JA, Derrick ML, Uhlenbeck OC (1998) T7 RNA polymerase produces 5′ end heterogeneity during in vitro transcription from certain templates. RNA 4:1313–1317
Dayie KT (2008) Key labeling technologies to tackle sizeable problems in RNA structural biology. Int J Mol Sci 9:1214–1240
Ferre-D’Amare AR, Doudna JA (1996) Use of cis- and trans-ribozymes to remove 5′ and 3′ heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res 24:977–978
Duss O, Maris C, von Schroetter C, Allain FHT (2010) A fast, efficient and sequence-independent method for flexible multiple segmental isotope labeling of RNA using ribozyme and RNase H cleavage. Nucleic Acids Res 38:e188
Schurer H, Lang K, Schuster J, Morl M (2002) A universal method to produce in vitro transcripts with homogeneous 3′ ends. Nucleic Acids Res 30:e56
Walker SC, Avis JM, Conn GL (2003) General plasmids for producing RNA in vitro transcripts with homogeneous ends. Nucleic Acids Res 31:e82
Guo HC, Collins RA (1995) Efficient trans-cleavage of a stem-loop RNA substrate by a ribozyme derived from neurospora VS RNA. EMBO J 14:368–376
Shields TP, Mollova E, Marie LS, Hansen MR, Pardi A (1999) High-performance liquid chromatography purification of homogenous-length RNA produced by trans cleavage with a hammerhead ribozyme. RNA 5:1259–1267
Santoro SW, Joyce GF (1997) A general purpose RNA-cleaving DNA enzyme. Proc Natl Acad Sci USA 94:4262–4266
Santoro SW, Joyce GF (1998) Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry 37:13330–13342
Pyle AM, Chu VT, Jankowsky E, Boudvillain M (2000) Using DNAzymes to cut, process, and map RNA molecules for structural studies or modification. Methods Enzymol 317:140–146
Inoue H, Hayase Y, Iwai S, Ohtsuka E (1987) Sequence-dependent hydrolysis of RNA using modified oligonucleotide splints and RNase H. FEBS Lett 215:327–330
Lapham J, Crothers DM (1996) RNase H cleavage for processing of in vitro transcribed RNA for NMR studies and RNA ligation. RNA 2:289–296
Kao C, Zheng M, Rudisser S (1999) A simple and efficient method to reduce nontemplated nucleotide addition at the 3′ terminus of RNAs transcribed by T7 RNA polymerase. RNA 5:1268–1272
Ponchon L, Dardel F (2007) Recombinant RNA technology: the tRNA scaffold. Nat Methods 4:571–576
Ponchon L, Beauvais G, Nonin-Lecomte S, Dardel F (2009) A generic protocol for the expression and purification of recombinant RNA in Escherichia coliusing a tRNA scaffold. Nat Protoc 4:947–959
Kim I, Mckenna SA, Puglisi EV, Puglisi JD (2007) Rapid purification of RNAs using fast performance liquid chromatography (FPLC). RNA 13:289–294
Lukavsky PJ, Puglisi JD (2004) Large-scale preparation and purification of polyacrylamide-free RNA oligonucleotides. RNA 10:889–893
Anderson AC, Scaringe SA, Earp BE, Frederick CA (1996) HPLC purification of RNA for crystallography and NMR. RNA 2:110–117
Easton LE, Shibata Y, Lukavsky PJ (2010) Rapid, nondenaturing RNA purification using weak anion-exchange fast performance liquid chromatography. RNA 16:647–653
Cathala G, Brunel C (1990) Use of n-butanol for efficient recovery of minute amounts of small RNA fragments and branched nucleotides from dilute solutions. Nucleic Acids Res 18:201
McKenna SA, Kim I, Puglisi EV, Lindhout DA, Aitken CE, Marshall RA, Puglisi JD (2007) Purification and characterization of transcribed RNAs using gel filtration chromatography. Nat Protoc 2:3270–3277
Murray JB, Collier AK, Arnold JRP (1994) A general purification procedure for chemically synthesized oligoribonucleotides. Anal Biochem 218:177–184
Cheong HK, Hwang E, Lee C, Choi BS, Cheong C (2004) Rapid preparation of RNA samples for NMR spectroscopy and X-ray crystallography. Nucleic Acids Res 32:e84
Kieft JS, Batey RT (2004) A general method for rapid and nondenaturing purification of RNAs. RNA 10:988–995
Batey RT, Kieft JS (2007) Improved native affinity purification of RNA. RNA 13:1384–1389
Walker SC, Scott FH, Srisawat C, Engelke DR (2008) RNA affinity tags for the rapid purification and investigation of RNAs and RNA-protein complexes. Methods Mol Biol 488:23–40
Allain FH, Varani G (1997) How accurately and precisely can RNA structure be determined by NMR? J Mol Biol 267:338–351
Tugarinov V, Muhandiram R, Ayed A, Kay LE (2002) Four-dimensional NMR spectroscopy of a 723-residue protein: chemical shift assignments and secondary structure of malate synthase g. J Am Chem Soc 124:10025–10035
Marino JP, Diener JL, Moore PB, Griesinger C (1997) Multiple-quantum coherence dramatically enhances the sensitivity of CH and CH2 correlations in uniformly C-13-labeled RNA. J Am Chem Soc 119:7361–7366
Lukavsky PJ, Puglisi JD (2001) RNAPack: an integrated NMR approach to RNA structure determination. Methods 25:316–332
Tolbert TJ, Williamson JR (1997) Preparation of specifically deuterated and C-13-labeled RNA for NMR studies using enzymatic synthesis. J Am Chem Soc 119:12100–12108
Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE (2006) RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex (vol 351, pg 371, 2005). J Mol Biol 360:742
Varani G, Tinoco I (1991) RNA structure and NMR-spectroscopy. Q Rev Biophys 24:479–532
Nikonowicz EP (2001) Preparation and use of H-2-labeled RNA oligonucleotides in nuclear magnetic resonance studies. Nucl Magn Reson Biol Macromol Pt A 338:320–341
Bullock SL, Ringel I, Ish-Horowicz D, Lukavsky PJ (2010) A′-form RNA helices are required for cytoplasmic mRNA transport in Drosophila. Nat Struct Mol Biol 17:703–709
D’Souza V, Dey A, Habib D, Summers MF (2004) NMR structure of the 101-nucleotide core encapsidation signal of the Moloney murine leukemia virus. J Mol Biol 337:427–442
D’Souza V, Summers MF (2004) Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature 431:586–590
Miyazaki Y, Irobalieva RN, Tolbert BS, Smalls-Mantey A, Iyalla K, Loeliger K, D’Souza V, Khant H, Schmid MF, Garcia EL, Telesnitsky A, Chiu W, Summers MF (2010) Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography. J Mol Biol 404:751–772
Kim I, Lukavsky PJ, Puglisi JD (2002) NMR study of 100 kDa HCV IRES RNA using segmental isotope labeling. J Am Chem Soc 124:9338–9339
Lukavsky PJ, Kim I, Otto GA, Puglisi JD (2003) Structure of HCV IRES domain II determined by NMR. Nat Struct Biol 10:1033–1038
Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, Divakaruni SS, LaCotti C, Barton S, Tummillo D, Hosic A, Edme K, Albrecht S, Telesnitsky A, Summers MF (2011) NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science 334:242–245
Serganov A, Keiper S, Malinina L, Tereshko V, Skripkin E, Hobartner C, Polonskaia A, Phan AT, Wombacher R, Micura R, Dauter Z, Jaschke A, Patel DJ (2005) Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation. Nat Struct Mol Biol 12:218–224
Akiyama BM, Stone MD (2009) Assembly of complex RNAs by splinted ligation. Methods Enzymol 469:27–46
Moore MJ, Query CC (2000) Joining of RNAs by splinted ligation. Methods Enzymol 317:109–123
Frilander MJ, Turunen JJ (2005) RNA ligation using T4 DNA ligase. In: Hartmann RK, Bindereif A, Schön A, Westhof E (eds) Handbook of RNA biochemistry. WILEY-VCH Verlag GmBH & Co, Weinheim, pp 36–52
Persson T, Willkomm DK, Hartmann RK (2005) T4 RNA ligase. In: Hartmann RK, Bindereif A, Schön A, Westhof E (eds) Handbook of RNA biochemistry. WILEY-VCH Verlag GmBH & Co, Weinheim, pp 53–74
Purtha WE, Coppins RL, Smalley MK, Silverman SK (2005) General deoxyribozyme-catalyzed synthesis of native 3′-5′ RNA linkages. J Am Chem Soc 127:13124–13125
Lang K, Micura R (2008) The preparation of site-specifically modified riboswitch domains as an example for enzymatic ligation of chemically synthesized RNA fragments. Nat Protoc 3:1457–1466
Stark MR, Pleiss JA, Deras M, Scaringe SA, Rader SD (2006) An RNA ligase-mediated method for the efficient creation of large, synthetic RNAs. RNA 12:2014–2019
Ohtsuki T, Kawai G, Watanabe Y, Kita K, Nishikawa K, Watanabe K (1996) Preparation of biologically active Ascaris suum mitochondrial tRNAMet with a TV-replacement loop by ligation of chemically synthesized RNA fragments. Nucleic Acids Res 24:662–667
Ohtsuki T, Kawai G, Watanabe K (1998) Stable isotope-edited NMR analysis of Ascaris suum mitochondrial tRNAMet having a TV-replacement loop. J Biochem 124:28–34
Xu J, Lapham J, Crothers DM (1996) Determining RNA solution structure by segmental isotopic labeling and NMR: application to Caenorhabditis elegans spliced leader RNA 1. Proc Natl Acad Sci USA 93:44–48
Tzakos AG, Easton LE, Lukavsky PJ (2006) Complementary segmental labeling of large RNAs: economic preparation and simplified NMR spectra for measurement of more RDCs. J Am Chem Soc 128:13344–13345
Tzakos AG, Easton LE, Lukavsky PJ (2007) Preparation of large RNA oligonucleotides with complementary isotope-labeled segments for NMR structural studies. Nat Protoc 2:2139–2147
Nelissen FH, van Gammeren AJ, Tessari M, Girard FC, Heus HA, Wijmenga SS (2008) Multiple segmental and selective isotope labeling of large RNA for NMR structural studies. Nucleic Acids Res 36:e89
Kawahara I, Haruta K, Ashihara Y, Yamanaka D, Kuriyama M, Toki N, Kondo Y, Teruya K, Ishikawa J, Furuta H, Ikawa Y, Kojima C, Tanaka Y (2012) Site-specific isotope labeling of long RNA for structural and mechanistic studies. Nucleic Acids Res 40:e7
Schubert M, Lapouge K, Duss O, Oberstrass FC, Jelesarov I, Haas D, Allain FH (2007) Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA. Nat Struct Mol Biol 14:807–813
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Hayase Y, Inoue H, Ohtsuka E (1990) Secondary structure in formylmethionine tRNA influences the site-directed cleavage of ribonuclease H using chimeric 2′-O-methyl oligodeoxyribonucleotides. Biochemistry 29:8793–8797
Acknowledgements
We thank Christophe Maris for setting up the HPLC and Christine von Schroetter for the production of isotopically labeled NTPs. This work was supported by the SNF-NCCR Iso-lab, the SNF grant Nr. 3100A0-118118 (to F.A.) and the grant from HFSP [RGP0024/2008] (to P.J.L.).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer Science+Business Media Dordrecht
About this chapter
Cite this chapter
Duss, O., Lukavsky, P.J., Allain, F.HT. (2012). Isotope Labeling and Segmental Labeling of Larger RNAs for NMR Structural Studies. In: Atreya, H. (eds) Isotope labeling in Biomolecular NMR. Advances in Experimental Medicine and Biology, vol 992. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-4954-2_7
Download citation
DOI: https://doi.org/10.1007/978-94-007-4954-2_7
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-4953-5
Online ISBN: 978-94-007-4954-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)