Abstract
NMR spectroscopy has become an indispensable tool for high-resolution structure determination of biomolecules at physiological conditions both in solutions and solids. Currently, NMR is routinely used to study the structure and dynamics of high molecular weight biomolecules in sizes ranging up to ~ 50–100 kDa and to evaluate complexes as large as 500–1 MDa. The latest advances in spectrometer technology, methodologies and advents in newer and highly innovative NMR active isotope-labeling strategies now enable us to overcome an earlier speculated size barrier of ~ 20 kDa for de novo structure determination. Of these, developments in NMR active isotope-labeling strategies are of great significance as they allow reduction in spectral crowding and yield selective spin correlations. Moreover, NMR isotope enrichment schemes permit exploitation of heteronuclear magnetization transfer pathways for enhanced sensitivity and selectivity. Functionally relevant sites or domains in very large complexes can also be selectively evaluated by specific labeling strategies in which other regions are masked. Further, labeling schemes can be effectively used to favourably overcome deleterious relaxation effects. Recently evolved labeling strategies include uniform labeling, perdeuteration, specific labeling of an amino acid or a side chain, selective deuteration or protonation, segmental labeling and biosynthesis of biomolecules in various organisms, cell lines and cell-free systems. The present review is aimed at introducing various NMR isotope labeling strategies and discusses their impact in widening the scope of biomolecular NMR spectroscopy driven structural biology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Nuclear magnetic resonance spectroscopy has emerged as an important tool to derive high-resolution three-dimensional structure of biomolecules, study motions and conformational exchange processes at various time scales and amplitudes, as well as residue-specific interactions with the ligand. NMR turns out to be advantageous over its contemporary techniques such as X-ray crystallography or cryo-electron microscopy due to the feasibility of studies near physiological conditions, yielding spatiotemporal information as NMR can probe both structure and dynamics. Nonetheless, high-resolution structure determination of large biomolecules (> 20 kDa) by NMR is quite cumbersome because of various factors such as spectral overlap, inherent low sensitivity of NMR active nuclei leading to poor signal to noise ratio and resonance broadening due to exchange processes, etc.
Biomolecular NMR spectroscopy has undergone significant changes since the advent of first solution structure of the bull seminal protease inhibitor (BUSI) and α-amylase inhibitor Tendamistat1, 2. Early structure determination projects primarily relied on exploiting 1H-1H interactions network manifested in 2D experiments such as NOESY, TOCSY and COSY. It was soon realized that the proton-based approach could only yield structures of small sized biomolecules (~ 100 aa) as spectral overlap in larger proteins would not allow unambiguous resonance assignments and subsequent structure determination. Alternate approaches utilizing heteronuclei (e.g., 13C and 15N) were partially successful due to an inherently low natural abundance of 13C and 15N. At this juncture, developments in molecular biology and biochemistry opened new avenues to produce uniformly 13C and 15N labeled NMR samples3,4,–5.
Combinations of various NMR active nuclei such as 13C, 15N and 2H can be used as the sample labeling scheme for different types of multi-dimensional and multi-resonance experiments. Introduction of labels in biological macromolecules has come a long way since the early 1990s. Isotope labeling in general can be categorized in four classes viz uniform labeling, fractional labeling, residue selective labeling and site-specific labeling.
In the present review, we have discussed various labeling schemes in biomolecules and their significance in NMR based experiments. Further, we describe recent developments in eukaryotic and cell-free expression systems, which facilitate native-like protein expression and labeling necessary for large, as well as multi-domain proteins and their complexes.
2 Uniform Isotope Labeling
Heteronuclear isotope labeling facilitated evolution of triple resonance experimentsFootnote 1 for sequential backbone chemical shift assignments and subsequently led to the study of protein structure and dynamics up to ~ 20 kDa molecular weight. Bacterial systems are the most widely used expression systems due to their ease in handling, faster biomass accumulation and ease of genetic manipulation. In these systems, a recombinant gene is cloned, often in an E. coli strain (such as BL21(DE3), codon plus as in Rosetta, RIL, etc.), followed by its over-expression in minimal medium and purification using an affinity tag. To achieve uniform 13C and/or 15N enrichment, proteins are overexpressed in minimal medium, supplemented with U-13C glucose/Glycerol/Sodium acetate and/or U-15N ammonium chloride/ammonium sulphate as the sole source of carbon and nitrogen, respectively3, 4. The resultant protein so obtained would be uniformly 13C and/or 15N labeled. The simplicity in producing uniformly labeled samples and NMR methodology developments have resulted in determining over 10,000 three-dimensional solution structures till date as seen from the PDB data. However, poor cell growth in minimal medium, sub-optimal induction of heterologous proteins, lethality of the foreign protein to the cells, codon usage biasFootnote 2 and, most importantly, non-native folding of the recombinant protein, and lack of post-translational modifications specific to eukaryotic proteins remain some of the serious issues with bacterial expression. To circumvent this issue, media containing isotopically enriched algal or microbial hydrolysates have been used5. To support the cell growth and protein induction, trace elements and cofactors like biotin can be supplemented.
Incorporation of 13C and 15N in polypeptide chain also paved way for the development of triple resonance experiments enabling sequential chemical shift assignments (such as HNCACB, HN(CO)CACB, HNCO, HN(CA)CO, HCCH-TOCSY) and three-dimensional 13C/15N-edited NOESY-HSQC experiments6. Heteronuclear triple resonance experiments exploit heteronuclear 1J and 2J scalar coupling for magnetization transfer (e.g., 1H–13C = 125–160 Hz, and 1H–15N = 87–95 Hz; 2JHNCα = 4–9 Hz) than 3J 1H-1H homonuclear coupling, which is often 0–10 Hz. Efficiency of magnetization transfer from proton to covalently connected 13C or 15N extends up to 50–90%, which makes 2D-heteronuclear correlation spectroscopy a very sensitive technique6, 7. The enhancement in the heteronuclear magnetization for 13C and 15N is significant as they have relatively lower gyromagnetic ratios, i.e., ~ 1/4th (10.71 MHz/T) and ~ 1/10th (− 4.316 MHz/T) to that of proton (42.58 MHz/T), respectively. In these cases, the 1H (I) magnetization is transferred to the coupled heteronuclei—13C or 15N (S)—via a selective coherence transfer pathway using a tailored pulse sequence containing radio-frequency pulses and time delays that are tuned to the coupling evolution. After the propagation of magnetization through desired magnetization transfer pathway, it is transferred back to 1H (I) for detection as the sensitivity of 1H (I) is higher leading to increased signal to noise ratio. Overall sensitivity in heteronuclear correlation is proportional to S/N ∝ γex γ 3/2det [1-exp(− R1exT)], where γex and γdet are gyromagnetic ratios of spin excited and detected, respectively, R1ex is spin–lattice relaxation of spin excited and T is recycling time of experiment. Further, the gain in the sensitivity of signals from magnetization transfer from I to S and subsequent detection on S is in order of n(γI/γS)3/2, where γI and γS are gyromagnetic ratios of I and S spin, and n is number of protons bonded with the heteronucleus. Thus, increase in sensitivity would be ~ 24 for methyl, ~ 16 for methylene and ~ 8 for methine proton spins. Similarly, the sensitivity enhancement for H–N pair would be in the order of ~ 30. Larger spin–lattice relaxation rate constant (R1) of proton compared to heteronuclei gives an additional sensitivity advantage for experiments starting with 1H magnetization because of the [1−exp(− R1exT)] factor8.
While this strategy works well with biomolecules < 20 kDa, the increased number of hydrogen atoms in large MW systems leads to undesirable magnetization redistribution occurring due to a dense network of neighboring spins through a process known as spin diffusion9. Moreover, due to slow tumbling of the molecule, the transverse relaxation rate constant (R2) increases with increasing molecular weight and results in short-lived spin magnetization leading to broadening of resonances. Additionally, in large MW systems, significant spectral overlap hinders the unambiguous assignment of individual peaks. Further, peaks arising from side-chains, especially methylene protons and carbons, become degenerate due to narrow spectral dispersion. The spectral degeneracy leads to redundant and misassigned peaks and subsequent structural inaccuracy10, 11. Hence, the quantification of cross-peak intensities to determine inter-atomic distances becomes complicated in NOE-based experiments due to effective spin-diffusion pathways provided by side-chain protons12.
3 Perdeuteration
Perdeuteration is the substitution of almost all non-exchangeable protons with deuterium in a polypeptide chain. Perdeuteration enhances the signal to noise ratio by reducing loss of magnetization occurring through the process of spin diffusion to the neighbouring spins. Furthermore, perdeuteration also confers advantage of reduction in dipolar couplingFootnote 3 between 13C and 15N and covalently bonded protons13. Smaller gyromagnetic ratio of deuterium compared to hydrogen (γD:γH::1:6.5) decreases the relaxation rates in the proportion of (γD:γH)2 ~ 0.02. Therefore, the relaxation time for 13C or 15N is greatly increased leading to smaller linewidths or enhanced signal to noise ratio14. Moreover, perdeuteration paired with transverse relaxation optimized spectroscopy (TROSY) further improves the spectral resolution.
Non-exchangeable protons in a protein sample can be deuterated uniformly or fractionally in a random or selective (residue selective, stereo-selective or regio-selective) manner. One of most useful methods is random and fractional deuteration of a sample along with complete 13C and 15N labeling, where the percentage of deuteration can be standardized for optimum cell growth and NMR spectra. To optimize deuteration percentage and isotope labeling, transformed bacterial cells are grown with U–13C and/or U–15N sources in the minimal medium with a desired H2O:D2O ratio. As metabolism of D2O is quite a bit slower, the doubling time in the conventional bacterial growth cycle is often seen to be ~ 2 h. The issue of slow growth can be circumvented by initially growing cell culture in a rich media such as LB, harvesting uninduced culture at the desired O. D. and suspending it into minimal media containing D2O and other desired labels. Alternatively, gradual increase in the D2O level per growth cycle is also performed so that bacteria gets accustomed to the slow growth environment in D2O media.
Advantages of perdeuteration are demonstrated in studies on Crc (~ 32 kDa) from P. syringae Lz4W. The protonated [1H–15N] HSQC shows significant spectral overlap and resonance broadening, particularly in the central region, due to faster relaxation rates (Fig. 1a). The [1H–15N] HSQC spectrum shows significant improvement upon perdeuteration (Fig. 1b). The spectral resolution and dispersion are further improved upon utilization of [1H–15N] TROSY–HSQC, where the TROSY component neutralizes 1H–15N dipole–dipole relaxation and chemical shift anisotropy to further enhance the signals (Fig. 1c).
Perdeuteration and effect of transverse relaxation optimized spectroscopy (TROSY) in Crc (~ 32 kDa). a [1H–15N] HSQC spectrum of U–15N Crc (protonated). b [1H–15N] HSQC spectrum of U–15N Crc with perdeuteration. c The spectrum represents [1H–15N] TROSY–HSQC of U–15N perdeuterated Crc. Well resolved and sharp cross-peaks in c aid in seamless backbone chemical shift assignment. All three spectra were collected using near identical conditions of the sample, data acquisition and processing.
Exchangeable amide protons residing in the impenetrable hydrophobic core remain a major issue with perdeuteration as these resonances display significant broadening in the HSQC and TROSY. Moreover, the presence of large numbers of similar amino acids in identical environment often leads to spectral overlap. Specific 15N/13C labeling of a single amino acid in an otherwise unlabeled environment can be used to simplify the spectrum and to aid the chemical shift assignment process.
4 Selective Amino Acid Labeling in Proteins
Addition of completely protonated amino acids in M9 media prepared in D2O was utilized in early NMR studies for specific labeling. In 1968, Crespi and Katz showed that the unlabeled Leu can be added to proteins expressed in uniformly deuterated medium15. Subsequently, various permutations and combinations for most of the 20 amino acids were exploited10, 16, 17. The residue-specific protonation resulted in the retention of side-chain protons on these specific residues of protein in otherwise deuterated background, thus increasing the sensitivity of many proton-detected experiments. However, this approach significantly suffers from amino acid scrambling and high-cost involvements.
Residue specific selective 15N-labeling of a protein often enables rapid and precise sequential assignments in larger proteins with crowded and complex spectra. Specific amino acid labeling schemes have been successfully used for few amino acids such as His, Lys, Arg and Met. Selective enrichment can be achieved biosynthetically, by over-expressing the protein of interest in culture medium supplemented with the corresponding U–15N labeled amino acids (Fig. 2a–c). A cost effective method to identify long chain amino acids (such as Arg) is by reverse labeling in which unlabeled amino acid is added in minimal media supplemented with U–15N salts. The strategy would allow intended unlabeling of amino acids in otherwise labeled background (Fig. 2b). However, the utility of these approaches is restricted because of isotope dilution and metabolic scrambling, particularly with Asp and Glu. Dilution of the supplemented isotope occurs by endogenous amino acid biosynthesis3. Moreover, the labeled amino acids can be scrambled to other amino acid residues by specific metabolic conversion or due to aminotransferase (transaminase) activity18. The residues that are end products in the amino acid biosynthetic pathway and are not acted upon by general aminotransferases (such as Arg, Cys, His, Gln, Lys, Met, Pro, and Thr) are less susceptible to metabolic scrambling. Nonetheless, under prolonged growth periods, isotopic dilution and scrambling have been observed even for these residues. Amino acid biosynthesis is controlled by feedback inhibition, so isotopic dilution and scrambling can be mitigated to some extent using a high concentration of all 20 amino acids for supplementing the growth medium3.
[1H–15N] TROSY–HSQC for specifically labeled residues in Crc. a [1H–15N] TROSY–HSQC of U–15N, 2H labeled Crc. b The spectrum is an overlay of reverse labeled Arg (red) and U–15N, 2H labeled Crc (blue). The missing cross-peaks in reverse labeled spectrum (blue) represent Arg. We could identify 20 out of 22 Arg resonances. c Overlay spectrum displays specific labeling of Lys (blue) where 15N-Lys was supplemented in otherwise unlabeled cell culture. Eight out of 8 Lys were traced using this sample. d–f An overlay of specific amino acid labeling of Ile, Leu and Val-Ala by 15N (blue) and [1H–15N] TROSY–HSQC of U–15N, 2H labeled Crc (red). The specific labeling of a single amino acid type was obtained by home-made auxotrophic strain of BL21(DE3). We were successful in identifying 11 out 11 Ile, 29 out of 29 Leu, 25 out of 25 Ala and 12 out of 12 Val using this strategy. The missing peaks in Arg may be attributed to the spectral overlap and/or intermediate scale exchange processes causing resonance broadening.
A more specific, efficient and less expensive approach is to employ bacterial strains that have been altered to contain the necessary genetic lesions to regulate amino acid biosynthesis19. These auxotrophsFootnote 4 are routinely used for selective labeling in NMR studies. Auxotrophs are generated using genetic lesions that are imposed at specific sites in the bacterial genome, which results in inhibiting the metabolism of exogenously added amino acid as the cells are impaired in its synthesis. Bacterial strains with the desired genotype are then constructed by defective genes transfer from chromosome of one strain to another. Generalized transduction, using bacteriophage P1 is often utilized as a vehicle for delivering DNA to the recipient bacteria and, if necessary, co-transduction with a selection marker. The process involves use of transposable genetic elements as they insert in the bacterial chromosome at various locations and deliver a selectable phenotype. Auxotrophs can be further selected using an antibiotic marker, as well as the auxotrophy of the constructed strain. E. coli strains with transposon insertions at sites adjacent to defective alleles of amino acid metabolism enzymes can be procured from E. coli Genetic Stock Center (Keio Collection) and subsequently, required auxotrophs can be created19, 20.
For selective incorporation of amino acids Arg, Cys, Gln, Gly, Lys, His, Ile, Met, Ser, Pro and Thr, a single lesion is required to construct the required genotype. Furthermore, aforementioned residues lie at the end of biosynthetic pathways and with the exception of Ile and Thr, are not substrates for the aminotransferases. In the case of the remaining residues, a combination of genetic lesions is required to nullify dilution and scrambling of the label. Most of the amino acids in this category are either substrates of the aminotransferases or are metabolic precursors of other residues. Genetic lesions corresponding to four known general aminotransferases in E. coli have been reported and are the products of ilvE, aspC, tyrB and avtA genes. Construction of mutants for Phe, Val, Leu, Tyr, Asp and possibly Trp requires genetic lesions in the genes (one or all) pertaining to general aminotransferases concomitant with the genes directly involved in their biosynthetic pathways. Asn and Ser require two (asnA, asnB) and four (serB, glyA, cysE, tyrB) genetic lesions in the wild type bacteria, respectively for ideal auxotrophic genotype. Ala and Glu auxotrophs are not effective due to involvement of these residues in multiple pathways19.
To achieve the selective labeling using auxotrophy of multiple amino acids, various strains have been developed, a few of which are discussed below. Strain DL39 (lesions in aspC, ilvE, tyrB); is a general transaminase-deficient strain auxotrophic for isoleucine, leucine, valine, aspartate, phenylalanine and tyrosine20, and has been utilized for 15N- labeling of Ala + Val, Asp + Asn (Asx), Ile, Leu, Phe and Tyr. Similarly, AB1255 (metB, argH, hisG, ilvA) is auxotrophic for Arg, His, Ile and Met, PC0950 (thr, argF, argL serB, purA) exhibits auxotrophy for Arg, Ser, Thr and adenine, AT2457 (glyA) for Gly, PA340 (gdh, gltB); for Glu and strain JE5811 (lys) is deficient in Lys21.
Although these strains provide an efficient tool for selective labeling of amino acids, they exhibit slow growth due to lesions imposed on the genome. Moreover, the protein yield in these strains is far lesser than that obtained from usual BL21(DE3) expression system. To address this issue, auxotrophs such as DL39(DE3), CT8 and CT19 were created with T7 expression system for enhanced expression of proteins22. A detailed table elaborating amino acid auxotrophs and the lesion required is given by David Waugh in mid-90’s19 and is reproduced with permission in this review as Table 1.
In our laboratory, due to significant reduction in protein yields in the DL39 strain compared to the BL21(DE3) expression system, a similar approach was adopted in the creation of an auxotrophic strain for assistance in backbone assignment of Crc. Lesions in ilvE, aspC and tyrB were introduced in the BL21(DE3) strain by transposon mutagenesis. Keio mutants for ilvE, aspC and tyrB were obtained, and respective P1 lysates were prepared. Transduction was performed in a stepwise manner to incorporate these lesions in BL21(DE3). Selection was done on the basis of an antibiotic marker in the transposon, as well as the auxotrophy introduced in the strain. This modified strain can be utilized for assistance in the backbone and side-chain chemical shift assignments of Ile, Leu, Val and Ala. For selective labeling of these residues, we have added U–15N Ile, U–15N Leu and U–15N Val (100 mg each, 30 min prior to induction) and have obtained a [1H–15N] TROSY–HSQC spectrum exhibiting well-defined resonances for each residue (Fig. 2d–f). As we have not introduced the avtA mutation, it was expected that the incorporation of labeled Val will yield both Val and Ala resonances (Fig. 2f).
These auxotrophic strains can be further utilized for residue and stereo-specific 13C methyl labeling of Leu, Val and Thr residues of proteins.
5 Methyl Sidechain Labeling of Amino Acids
In large MW systems, perdeuteration significantly eliminates 1H–1H dipolar relaxation network and hence enhances longevity of NMR resonances. However, it is associated with a severe decrease in the inter-proton NOE network mostly between amides and sidechains, which are crucial for distance constraints in structure determination. Various labeling schemes for site-specific protonation in a highly deuterated environment have been devised to overcome these issues.
Hydrophobic amino acids containing methyl groups such as Ile, Leu and Val are abundantly present in the core of proteins (~ 21–25% of all residues). Specific labeling of methyl groups has emerged as an effective approach as methyl groups are ideal probes for NMR studies of high molecular weight systems because of sensitivity and sharper line widths due to rapid rotation about the three-fold methyl symmetry axis and multiplicity of protons. Hence, labeling strategies involving these residues are widely used and enable efficient assignments. It also facilitates detection of long-range amide-methyl and methyl–methyl NOEs, which aided in determining the global folds of large proteins. These residues also serve as excellent reporters of dynamics in proteins. Additionally, methyl protons have distinct chemical shifts (− 1.5 to 2.5 ppm) that enable their identification in crowded spectra.
For specific labeling of an amino acid containing methyl group (Ala, Met, Thr, Ile, Leu and Val), specifically labeled precursors can be chosen by evaluating their ability to enter in an anabolic pathway without any complications from 1H to 2H exchange, ease of preparation and their utilization by E. coli. A simplified biosynthetic pathway for methyl group containing amino acids in E. coli has been shown in Fig. 3, which can be manipulated for specific site-specific 13C/1H or 2H enrichment.
Simplified amino acid biosynthetic pathway in E. coli. Carbon atoms derived from pyruvate or alanine are coded in green and that derived from aspartate are shown in red. Various enzymes involved in the pathway are represented in numerals. AHAS: α-hydroxy acid synthase; AlaA, AlaC and AvtA: E. coli alanine transaminases; BACT: branched-chain amino acid aminotransferase; DHAD: dihydroxy-acid dehydratase; KARI: ketol-acid reductoisomerase; TD: biosynthetic threonine deaminase. Figure adapted and modified with permission from44.
The methyl group of pyruvate acts as the precursor for methyl groups in Ala, Val, Leu, and Ile (γ2)23. Use of protonated and 13C enriched pyruvate as a carbon source in deuterated media ensures the incorporation of 13C, 1H labeled methyl groups in Ala, Val, Leu, and Ile (γ2) in an otherwise deuterated protein23, 24. It was shown that the level of protonation in these methyl groups varies from 40% (Ala) to 60% (Val and Ile) to 80% (Leu)23. Specific protonation at Ala, Ile (γ2), Val and Leu methyl groups along with deuteration ensures enhanced sensitivity for triple resonance experiments for backbone and side-chain chemical shift assignments. A major disadvantage involving use of pyruvate is formation of methyl isotopomers (CH3, CH2D, and CHD2), which leads to reduced sensitivity and resolution. Moreover, the protein yields in pyruvate-based media is halved in comparison to glucose-based media in E. coli. To overcome these problems, glucose media supplemented with amino acid precursors and amino acids was introduced for over-expression of proteins in bacteria24.
Earlier attempts for achieving methyl-specific labeling schemes employed the use of 2-keto-3-[D2], 4-[13C]-butyrate as sole source of protons in perdeuterated media to yield [U-D], Ile-δ1—[13CH3]-labeled protein25. 2-keto-3-[d]-[13CH3, 13CH3]-isovalerate was utilized as a precursor for labeling both pro-chiral methyl groups of Ile and Val26. Combinations of [13CH3]-methyl-labeling schemes for aid in chemical shift assignments have been extensively discussed in the literature. These comprise of ILV (Ile-δ1/Leu-δ/Val-γ)27,28,–29 and include use of precursors 2-keto-3-[13CH3, 13CH3]-isovalerate (α-ketoisovalerate) for labeling Leu/Val and 2-keto-3-,4-[13C]-butyrate (α-ketoisobutyrate) for labeling Ile. Use of selective 13C, 1H labeling for only methyl groups or U–13C, 1H labeling in α-ketoisovalerate and α-ketoisobutyrate further gave options to use 13C methyl only samples for NOE measurements and U–13C methyl labeled samples for side-chain assignments using H(CCO)NH–TOCSY and (H)C(CCO)NH–TOCSY30, 31.
Figure 4 represents stereo-selective 13C, 1H methyl chemical shift assignments of Crc (~ 32 kDa) in which samples were prepared using specifically as well as uniformly labeled α-ketoisovalerate and α-ketoisobutyrate and yielded over 85% of Ile (δ1), Leu (δ1/δ2) and Val (γ1/γ2) unambiguous assignments.
[1H–13C] HSQC spectrum of Crc. Spectrum represents cross-peaks from stereospecific methyl groups of Leu(δ1 and δ2), Val(γ1 and γ2) and Ile(δ1). Figure adapted with permission from130.
As amino acids like Leu and Val contain more than one methyl group at the side-chain terminus, exact stereo-specific discrimination of pro-chiral methyl groups needed further evolution of labeling strategies.
6 Stereo-Specific and Other Recent Advances in Methyl Group Labeling
Initial attempts to resolve the stereo-specific peaks of pro-chiral methyl groups in Leu and Val relied on preparation of a sample containing 10% of U-[13C] glucose in 90% unlabeled glucose as sole carbon source32. In the partial carbon labeling scheme, 13CH3 Leu-δ2/Val-γ2 (pro-S) remain isolated and 13CH3 Leu-δ1/Val-γ1 groups (pro-R) couple with 13Cγ and 13Cβ, respectively, that leads to a doublet separated by ~ 35 Hz, which can be easily detected with [1H–13C] HSQC.
Further, selective labeling in pro-chiral groups in Leu and Val were obtained by using 2-keto-3-[d]-[13CH3, 13CD3] isovalerate, which allowed labeling of only one of the pro-chiral group33. Use of 2-acetolactate34 or addition of labeled Val35 in the media along with deuterated Leu was shown to yield labeled methyl groups of Val exclusively. Similarly, 2-ketoisocaproate29 or [13CH3]—Leu35 have been suggested for non-stereo-specific or stereo-specific labeling of Leu. The ε-methyl group of methionine can be isotopically enriched by adding labeled Met residue in the growth medium36 and 2-hydroxy-2-ethyl-3-keto-butanoic acid can be utilized to label the Ile-γ2 methyl group37, 38.
Despite their ability to achieve stereo-specific discrimination, the partial carbon labeling schemes suffered from poor NMR sensitivity due to fractional 13C labeling. To surmount the challenge of low spectral sensitivity and stereo-specific discrimination of pro-chiral methyl groups of Val and Leu, a novel synthetic route for the production of specifically methyl-labeled acetolactate (or 2-hydroxy-2-[13C]methyl-3-oxo-4-[2H3]butanoic acid) was introduced39. This approach was used to demonstrate the stereo-specific protonation of Leu and Val methyl groups in recombinant perdeuterated proteins. The strategy relied on the stereo-specific rearrangement of methyl groups in (S)-2-acetolactate (in vivo) in the early steps of Leu and Val biogenesis. This labeling scheme was applied to 82 kDa Malate Synthase G, for which Methyl TROSY and inter-methyl NOE cross-peaks of enhanced pro-S were obtained. Cross-peaks for pro-R methyls were eliminated in the process. In a nutshell, combinatorial approaches for specific labeling of methyl groups for MILV (Met-ε/Ile-δ1/Leu-δ/Val-γ)36, AILV (Ala-β/Ile-δ1/Leu-δ/Val-γ)40, and MILVT (Met-ε/Ile-δ1/Leu-δ/Val-γ/Thr-γ2)41 have been reported.
The aforesaid methyl labeling schemes are appropriate for assemblies with symmetrical, lower molecular weight subunits. For larger proteins/assemblies, overlap between the methyls of Val and Leu preclude the proper NMR based spectral analysis. To alleviate this challenge, a straightforward labeling scheme was introduced to incorporate stereospecific 13CH3 isotopomers into Val residues without labeling the corresponding Leu groups34. Introduction of 13CH3 is based on the simultaneous incorporation of 13CH3 acetolactate and 12C,2H L-Leu in the culture medium, yielded specific labeling of 13CH3 methyl groups of Val resulting in a simplified [1H–13C]-Methyl TROSY spectra of 468 kDa homododecameric peptidase TET2. Thirty-two out of 37 Val in TET2 and [1H–13C] HMQC–NOESY derived methyl proton NOEs separated up to 7–8 Å could be assigned using this labeling strategy by combining mutagenesis, innovative labeling and adapted triple resonance experiments.
Jerome Boisbouvier and coworkers proposed an improved AILV methyl labeling scheme with stereo-specificity for methyl groups of Val and Leu42, 43. A ready-reckoner in the form of a table describing the strategies for methyl labeling schemes has been detailed by Boisbouvier and co-workers44. Despite significant developments in obtaining assignments for Ile-γ2, Val and Leu, a specific analogue for selectively labeling Ile-δ1 was not available. A robust and cost effective enzymatic synthesis of precursor for Ile, 2-hydroxy-2-(1′-[2H2], 2′–[13C])ethyl-3-keto-4-[2H3]butanoic acid, was proposed in order to stereo-specifically assign 13CH3 in the Ile δ1 position in the backbone via a linear 13C spin system since the Ile-γ2 methyl group remains 12C and deuterated. As the method is metabolically leak-proof, isotope scrambling was eliminated. The labeling scheme was applied to 82 kDa Malate Synthase G and 1H–1H NOE crosspeaks between methyls separated by 10 Å.
Moreover, previously mentioned auxotrophic strains developed in our group and elsewhere can also be used for specific methyl labeling of any of Val-γ1/γ2, Leu-δ1/δ2, Val- γ1/Leu- δ1, Val γ2/Leu- δ2, Val- γ1, Val- γ2, Leu- δ1 or Leu- δ2 by providing appropriately labeled amino acids in the culture media45. Available options for 13C methyl labeling are listed in Table 2.
Along with the development of methyl labeling, Kay and co-workers have demonstrated that the 2D [1H–13C] HMQC (called Methyl TROSY) is already optimized to exploit destructive interference between the multiple 1H–13C and 1H–1H dipolar interactions30. The utility of Methyl TROSY in accessing methyl 1H and 13C chemical shifts paved the way to study significantly larger macromolecular assemblies. For example, Methyl TROSY has been used to decipher functionally relevant motions and interactions in ~ 670 kDa 20S proteasomal assembly46, the interface between heptameric rings in 300 kDa, cylindrical protease ClpP and its exchange between two conformations47, and ~ 450 kDa chromatin remodeling complex where buried Ile, Leu and Val in H4 displayed dynamics during the nucleosome interaction with SNF2H48. Methyl TROSY was proved to be instrumental in characterizing the oligomerization process and folding intermediates of half a megadalton, homododecameric tetrahedral (TET) aminopeptidase49.
7 Isotope Labeling Using Yeast Cell Lines
Prokaryotic cells (bacteria, especially E. coli) have proven to be ideal expression systems and are widely used due to the low cost carbon source requirement for growth, rapid biomass accumulation, and simple scale up process. However, expression of eukaryotic proteins in the prokaryotic system could be a challenge due to codon bias, non-native folding and lack of post-translational modifications. Use of eukaryotic expression systems would most likely circumvent several of these problems and yeast expression systems provide a better alternative. Yeast as an expression system has many advantages such as low isotope labeling cost, high expression yields and easy genetic manipulation. It can be easily grown in deuterated media and deliver yields comparable to bacterial systems. Yeast system allows the native folding of protein along with post-translational modifications such as proteolytic truncation, formation of disulfide bonds, glycosylation, phosphorylation and acylation. Moreover, it also allows expression of both cellular and secretory proteins precluding the chances of cytotoxicity.
The two most used yeast strains are Saccharomyces cerevisiae and the methylotrophic yeast Pichia pastoris. Isotope labeling using Pichia pastoris is well established and widely used as it provides higher yields of recombinant proteins and more native glycosylation pattern50. Expression of an isotopically labeled eukaryotic protein, tick anticoagulant, was already established in the mid-1990s using P. pastoris51.
P. pastoris grows well in minimal medium where 15N-labeled ammonium salts are added as a nitrogen source. For carbon labeling, 13C-glucose or glycerol can be supplemented before induction. As the protein expression is performed under a strong alcohol oxidase promoter, 13C-methanol is used for induction, which is a primary carbon source during the expression phase. Cell growth is also consonant with deuterated medium52. Further, methods for specific amino acid labeling have been also developed for Cys, Lys, Leu and Met53, 54.
Despite being a powerful expression system with post-translational modifications in the recombinant proteins, yeasts have imitations of hyperglycosylation, uncertainty in disulphide bond formation and often encounter poor or no yields55,56,–57.
8 Isotope Labeling Using Insect Cell Lines
Baculovirus-mediated expression (BvE) in insect cells offers an advanced expression system with superior post-translational modification machinery. As baculovirus genome is considered too big for direct incorporation of foreign genetic material, the gene is first cloned in transfer vector containing regions flanking polyhedron gene in virus genome. Further, the viral genome is co-transfected with transfer vector inside insect cells allowing the incorporation of the gene in the viral genome via homologous recombination. Commonly used cell lines for Baculovirus-mediated amplification and recombinant protein expression are SF9 and SF21 (derived from fall armyworm). Trichoplusia ni (cabbage looper moth) and BTI5B1-4 (High Five™) cells are used for secreted recombinant proteins58, 59.
Isotope enrichment of proteins using insect cell lines is a cost ineffective process as it requires supplementation of labeled amino acids. Alternatively, commercially available media kits such as Bioexpress 2000 (CIL) provide options to produce U–13C,15N labeled samples with up to 90% labeling. Using BvE system, sample for U–13C,15N Abelson Kinase domain was prepared and subsequently used for NMR driven structural studies60.
Because of the high-mannose and paucimannose types of glycosylation, expression of therapeutic proteins (e.g. insulin, recombinant monoclonal antibodies) using insect cell lines is limited as it leads to compromised bioactivity and acts as potential allergens of the recombinant proteins61, 62.
9 Isotope Labeling Using Mammalian Cell Lines
Mammalian cells are preferred for expression of therapeutic recombinant proteins as they provide more native-like fold with appropriate, human-like post-translational modification. Nevertheless, mammalian cells are considered difficult to handle and protein yields are low compared to bacterial and yeast expression systems. Regardless, in the past decade most eukaryotic membrane proteins, including several drug targets that could not be produced in prokaryotic systems in sufficient functional quantity or quality were successfully expressed in animal cell lines. One major hurdle in protein overexpression in mammalian cell lines pertaining to NMR driven structural biology is high-cost involvement for isotope labeling.
In early 1990’s, U–15N and U–15N, 13C urokinase was expressed in Sp2/0 and CHO cells using culture media containing amino acids isolated from E. coli and lyophilized algae with Cys and Glu supplements63, 64. During this experiment, ~ 5% of heat-inactivated serum was added to culture medium, which did not affect the isotope enrichment. Currently, efforts are in place to reconstitute a cost-effective stable medium against commercially available expensive mammalian cell culture medium (CIL)65. Uniform isotope labeling in mammalian cells is achieved by novel serum-free medium, which includes stable isotope labeled autolysate and lipids from algae, yeast and bacteria. These microorganisms are relatively easy to label with commercially available metabolic precursors and lead to reduction in cost by sixfold. The medium was used for expressing recombinant proteins in Chinese hamster ovary (CHO) cells and human embryonic kidney (HEK293) cell lines65. Recombinant human chronic gonadotropin and human IgG were expressed in CHO cells and enriched with 13C and 15N using labeled algal hydrolysate to conduct in situ structural-conformational analysis66, 67. Recently, mouse hybridoma cells were used to specifically label IgG2b glycoprotein that was metabolically labeled using [δ2–13C; Hα, Hβ, Hγ, Hδ1–2H7] Leu and its non-deuterated counterpart68.
Despite advanced protein synthesis machinery, cell lines often suffer from low or no expression of the desired protein. Also, recombinant proteins are toxic to the host cells that make the whole process impractical for structural biology. Another drawback of cell-based expression systems is their inability to introduce stereo-specifically labeled methyl probes as of now.
10 Cell-Free Expression and Isotope Labeling
A cell-free protein expression system is an in vitro protein synthesis reaction, which comprises cell extract from different living organisms including all the cellular machinery pertaining to protein expression. Cell-free systems provide opportunity for expression of higher molecular mass proteins and depending on the protein of interest host organism can be chosen like microorganism, plant, insect or mammalian cells (Fig. 5).
Model for the cell-free protein expression system. The cell-free expression system is divided into reaction and feeding chambers. Cell extract from bacteria, plant cell or mammalian cells is prepared and set up in reaction chamber that is separated by a semi-permeable membrane. Energy source (ATP), amino acids, rNTPs, etc. are added into the feeding chamber (inlet) and metabolic wastes and by-products are removed from it (outlet). The membrane allows the influx and efflux of small molecules like amino acids, ATP, metabolic wastes and so on. In reaction chamber necessary DNA template or mRNA is added for transcription and subsequent translation.
In 1988, a continuous flow cell-free translation system was introduced with MS2 phage RNA or brome mosaic virus RNA 4 as templates and small substrates such as ATP, GTP and amino acids69. The system was tested for prokaryotic (E. coli) and eukaryotic (wheat embryo) cell lysates. Subsequently, a dialysis-based cell-free expression system was utilized to obtain 15N-Ser/15N-Asp Ras protein with increased yields i.e., 0.1 mg/mL70 followed by production of 6 mg/mL 13C/15N labeled Ras protein using algal labeled amino acids71. Recently, an E. coli cell-free system was used for scalable characterization of CRISPR technology72. Recombinant proteins, yeast Ubiquitin and RbpA1 are expressed in a wheat germ extract (WGE) cell-free system in much higher quantities (200–400 ng/μL) compared to E. coli extract73. Expression of recombinant eukaryotic protein in bacterial cell-free extract often results in non-functional sample and to circumvent the issue other eukaryotic cell lysates were tested. Further, insect cell line extract provides increased protein yields (71 μg/mL)74. Insect cell (Sf21) lysates are readily used to express many G-protein coupled receptors ranging from 40 to 133 kDa in a detergent-free manner75, 76. Insect cell lysates are suitable for GPCRs as many of them require post-translational modifications such as phosphorylation, palmitoylation, glycosylation and disulfide bond formation to stabilize their active state and correct folding77. Use of extracts derived from mammalian cell lines like Rabbit reticulocyte lysates (RRL), Ehrlich ascites cells, HeLa cells, CHO cells and mouse L cells have further expanded the scope of cell-free expression systems78. Recently, a cell-free system based on rabbit reticulocyte (RLL) lysate is developed to express HBV capsid proteins79.
Apart from the aforementioned hosts, plant cells provided suitable alternative for higher molecular weight, well-folded recombinant proteins with higher yields. For example, wheat germ cell-lysate is used extensively for high-throughput immuno-screening of P. falciparum proteins in search of novel anti-malarial druggable targets80. However, WGE lysate preparation is time consuming and RLL lysate suffers low yields. To circumvent this issue, a novel cell-free system from tobacco bright yellow 2 (BY 2) cells is developed81. BY-2 lysate (BYL) can be prepared rapidly (in about 4–5 h) compared to WGE, which usually takes about 4–5 days. Further, yields from BYL are much higher compared to that of WGE, e.g., BYL had a maximum yield of 80 μg/mL of eYFP and 100 μg/mL of luciferase, compared to only 45 μg/mL of eYFP and 35 μg/mL of luciferase in WGEs81.
Arabidopsis cell-free extract (ACE) is another alternative for BYL and WGE, and the lysate is prepared from callus culture derived from seedlings followed by evacuolation of protoplast82. Yields from ACE medium is akin to that of WGE and extracts from 5′ to 3′ exoribonuclease-deficient mutants of Arabidopsis, xrn4-5, exhibited increased stability of an uncapped mRNA as compared with that from wild-type Arabidopsis. However, usage of ACE in stable isotope labeling is yet to be tested. Although, cell-free expression systems provide a wider scope for selective labeling of recombinant proteins with higher yields and no apparent metabolic scrambling or other expression system based issue, its laboratory usage is limited as E. coli, WGE and BYL are the only commercially available options.
Even though WGE, BYL and ACE media appear to be lucrative alternatives for expression of eukaryotic proteins, the NMR isotope labeling strategies necessary for NMR studies have not been established so far.
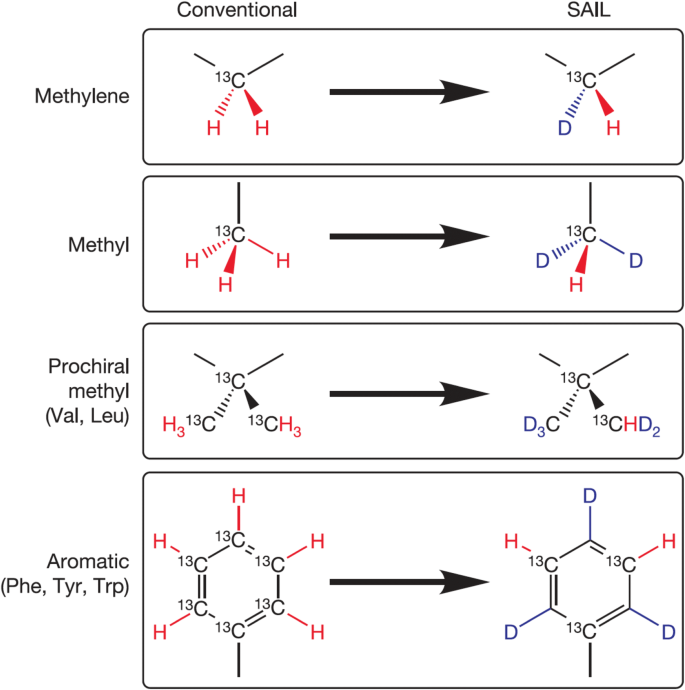
For stable NMR isotope labeling, the cell-free system provides an excellent opportunity to incorporate site- and regio-specific labels. A newly designed stereo-array isotope labeling, or SAIL, provides opportunity for 2H and 13C labeling in U–15N-labeled protein in a controlled manner and is depicted in Fig. 683. Signal to noise ratio and sensitivity in SAIL is better than conventional uniform labeling as the number of observable protons is reduced without sacrificing relevant structural information. Replacement of 1H by 2H decreases the transverse relaxation during the magnetization transfer during experiments such as [1H–13C] constant time-HSQC that enhances signal to noise ratio. Reduction in long range coupling further sharpens the signals. The signals for methylene group increases by three to seven times in SAIL, compared to uniformly labeled sample under same experimental conditions. The SAIL approach is, however, limited by feasibility of a small number of stereospecific assignments. SAIL (stereo-array isotope labeling) uses a cell-free expression system for high-quality structure determination of proteins ~ 40–50 kDa with ease of smaller proteins. For efficient incorporation of stereo-specifically labeled amino acids, the E. coli based cell-free system was employed to express 17 kDa calmodulin (CaM) from X. laevis and 41 kDa maltodextrose-binding protein (MBP)83. Final yields obtained post-purification were 5.5 mg for CaM and 5.3 mg for MBP and the samples were further used for solution structure determination by NMR. Improvement in signal intensity was more pronounced in MBP compared to CaM with straightforward aromatic 13C assignments. Structures so derived for SAIL–CaM and SAIL–MBP were in good agreement with their previously known crystal structures79. Later, SAIL-Phe and SAIL-Tyr were incorporated in 18.2 kDa protein, E. coli peptidyl-prolyl cis–trans isomerase b (EPPIb) using an E. coli based cell-free system to yield δ-, ε- or ζ-13C/1H assignments84.
Stereo-array isotope labeling (SAIL). The figure represents amino acid design for stereo-specific and regio-specific labeling used in cell-free systems. Protons are selectively deuterated in methylene groups, pro-chiral methyl groups in Val and Leu, and alternating C–H groups in six membered aromatic rings. Figure adapted with permission from83.
11 Segmental Labeling
As discussed in earlier sections, increasing molecular weight of biomolecules makes structural studies of functionally relevant sites by NMR extremely challenging. Another strategy to characterize large multi-domain proteins utilizes isotopic labeling of defined segments/single domains with NMR active nuclei, whereby remaining domains are expressed using NMR inactive nuclei. Since only some of the multi-domain complex is NMR visible, this technique drastically reduces peak overlap and spectral complexity. This approach involves production of samples with selectively labeled domains/segments followed by their ligation. However, the feasibility of this technique is limited due to decreased efficiency of the ligation step. Several options have been suggested to facilitate ligation of protein segments such as native chemical ligation (NCL), expressed protein ligation (EPL), protein trans-splicing (PTS) and sortase-mediated ligation (SML)85,86,–87.
NCL involves the ligation of two synthetic unprotected peptides, one possessing an N-terminal cysteine residue (α-cysteine) and the other containing a C-terminal thioester (α-thioester), which leads to the formation of a peptide bond in aqueous conditions. Peptides or protein segments with specific termini can be synthesized by solid-phase peptide synthesis (SPPS)87. NCL allows for incorporation of all types of site-specific label or any type of modification (phosphorylation, methylation, glycosylation, etc.) in the peptides. Since SPPS can accurately generate peptides only up to approximately ~ 50 amino acids; it cannot be utilized for synthesis of larger protein segments or will require ligation of more than two peptides. Another disadvantage of this method is the cost ineffectiveness of the overall process.
Inteins are a class of self-splicing proteins, which cleave themselves from larger polypeptide chains leading to formation of peptide bond between the leftover protein fragments (exteins). Inteins lack any function in intended protein sequence and undergo self-cleavage upon translation while remaining N- and C-exteins form a new peptide bond to fold into the native structure. Two related approaches that exploit the process of protein splicing based on intein properties are routinely used for segmental labeling of proteins are expressed protein ligation (EPL) and protein trans-splicing (PTS)88,89,–90.
In expressed protein ligation (EPL), the NCL approach is combined with recombinant protein production to overcome the size limit posed by SPPS (Fig. 7a). Here, either or both of the peptide fragments for ligation are produced by recombinant bacterial expression. The reaction requires protein fragments containing an α-thioester and an α-cysteine. Hence, the presence of a native cysteine is necessary at the ligation site and if absent, it needs to engineered. Peptide α-thioesters are prepared synthetically by SPPS or biosynthetically using intein-fusion strategies. N-extein is fused with modified intein that lacks the ability of trans-thioesterification88. A thiol group is exogenously added to generate N-extein α-thioester intermediate and cleaved intein. An N-extein α-thioester intermediate is then attacked by Cys of C-extein and undergoes an S → N-acyl transfer to form a native peptide bond, and the resulting peptide product is obtained. N-Cys peptides are synthesized by routine SPPS.
Segmental labelling schemes to produce recombinant protein. a Schematic representation depicts expressed protein ligation (EPL) where the protein of interest is expressed in two segments or exteins (red and blue) separately with intein fused at the C-terminus of N-extein (green) and the N-terminus of C-extein (yellow). Intein in EPL is mutated and does not undergo self-cleavage. Thus, the thio- group is exogenously added to N-extein to generate an α-thioester intermediate. Cys of C-extein attacks the N-extein thioeaster to undergo transthiolesterification, which results in peptide bond formation between the two exteins. b In intein-mediated protein trans-splicing (PTS) two exteins (red and blue) are fused with N- and C-fragments of Intein (green). Individually, Intein fragments do not have cleavage activity; upon fusion, a complete intein is formed followed by trans-splicing or self-excision of intein to give a ligated recombinant protein. c The figure is a pictorial representation of sortase A (green)-based protein ligation where the N-terminal protein segment (red) has a conserved LPXTG motif and the C-terminal fragment (blue) starts with a Gly. The LPXTG motif is recognized by sortase A and end Gly is cleaved off to generate an N-terminal fragment added with sortase A. Gly of the C-terminal fragment attacks the previous ligated product leading to the removal of sortase A and the formation of a ligated product of two protein fragments.
EPL is regularly utilized for segmental isotope labeling of proteins. Mostly two protein fragments are ligated; however, ligation of three or more fragments can also be performed to study large proteins. Cotton et al. performed an experiment where three-piece protein ligation was achieved by the regioselective incorporation of CK(Dns)G, a fluorescent peptide label between the recombinant SH3 and SH2 domains of Abl, Abelson nonreceptor protein tyrosine kinase91. A 50 kDa protein C-terminal Src Kinase (Csk) has been successfully studied using intein-based expressed protein ligation92.
Protein trans-splicing (Fig. 7b) (PTS) involves fusion of N-terminal fragment of intein to C-terminus of first segment and C-terminal fragment to N-terminus of another segment of protein of interest90. As it involves the functional reconstitution of a split intein, the ligation step in PTS is done under conditions suitable for protein folding. Upon fusion of both fragments of intein, an N → O/S acyl rearrangement is facilitated at its N-terminal Cys or Ser residue resulting in formation of an ester or thioester bond, respectively between the side-chain and the peptide backbone of the N-extein. After self-cleavage of intein, exteins form an amide bond that is indistinguishable from a ribosome-assembled fusion protein. Protein trans-splicing can be achieved in vivo by co-transforming the two fragments, but under different promoters or in vitro to yield a domain-specific labeled 140 kDa dimeric multi-domain protein CheA with 2H, 15N enrichment93, 94.
Though these segmental labeling techniques have been used to express many proteins, they have limited success as they are time consuming and necessitate more reagents than conventional labeling. If the protein of interest is a single polypeptide chain, then refolding to the native conformation remains a challenging step. Further, unligated precursors should be removed by an extra purification step. In PTS ligation, significant cross-labeling is observed due to leaky expression. In EPL, reducing agents are used for generation of thioester, thus preventing the utilization of this method for proteins containing disulfide linkages. Moreover, a large concentration of the cysteine containing cargo is required for efficient EPL, which makes this strategy costly.
To overcome the shortcomings of the aforementioned techniques in segmental labeling, recently Sortase A (SrtA, a cysteine transpeptidase that anchors virulent surface proteins to cell wall in gram positive bacteria) has been employed to ligate two differently expressed protein fragments95,96,–97. Staphylococcus aureus sortase, SrtA catalyzes the transpeptidation reaction between a C-terminal LPXTG recognition motif in the proteins and poly-glycine bridge in the cell wall. The enzyme cleaves the amide bond between Thr and Gly of LPXTG to form an acyl-enzyme intermediate. This is followed by nucleophilic attack on the carboxyl group of Thr of the thioester intermediate by the amino group of the tris-Gly moiety resulting in the formation of an LPXT–GGG bond between protein and the peptidoglycan wall, and release of the free enzyme (Fig. 7c).
Sortase A has exhibited applications in protein conjugation to ligate model peptides/proteins together if the reactants harbor − LPXTG-COOH or − NH2-GGG tags95. The tris-Gly moiety functions as a nucleophile even when attached to non-protein species such as polyethylene glycol or to a surface and is not dependent on the presence of a protein terminus in solution98. Several primary amine derivatives such as alkylamines and hydroxylamine can also be used as tris-Gly moiety surrogates; however, the efficiency of these substrates is lesser than oligoglycine derivatives98. The sole requirement of sortase mediated ligation (SML) is the presence of an LPXTG motif on the C-terminus of the N-terminal peptide of the protein. The attachment of this motif does not lead to decrease in solubility or expression as observed in intein-mediated ligation system96, 99. SML is performed under mild conditions, does not require any additional cofactors (ATP, biotin etc.) and artificial modifications in the ligated domains. Furthermore, efficiency of the ligation step can be optimized by biochemical approaches as ligation fragments and sortase are individually produced and then mixed to initiate the reaction.
Mao et al., demonstrated the use of SrtA as a novel protein ligation tool. Recombinant GFP harboring a C-terminus LPETG-His6 sequence was utilized as a model protein for specific modifications with diverse native and non-native peptides95. Freiburger et al., demonstrated efficient and selective labeling of RNA recognition motifs (RRM) of splicing factor T cell-restricted intracellular antigen-1 (TIA-1) and domains of Hsp90 where a released aminoglycine peptide fragment was removed by simple centrifuge filters or dialysis100. Similarly, structural and dynamic studies were conducted on selectively labeled individual bromodomains of BRD4 using this method101.
Another promising candidate for protein ligation is butelase-1 isolated from the plant Clitoria ternatea102, 103. Butelase-1 is the fastest known ligase with catalytic efficiency up to 542,000 M−1 s−1 ; however, it is unavailable in the recombinant form as of now.
12 LEGO–NMR
In theory, macromolecular complexes with more than one subunit can be reconstituted in vitro. However, in multiple instances, individual subunits expressed separately may not be stable or soluble in isolation and require binding partners to retain a stable fold. To circumvent this issue, various subunits of asymmetric protein complexes can be sequentially co-expressed in bacterial cells and reconstituted in vivo. Herein, the method has advantages of in vivo reconstitution and partial isotope labeling. The E. coli cells are transformed with plasmids under different promoters, enabling induction of different sub-units independently. The first set of plasmids (generally under weak promoter) can be induced in NMR active medium followed by induction of another set of plasmids (with strong promoter) in NMR inactive medium. All the labeled and unlabeled subunits get organized in quaternary arrangement in vivo and create a montage of labeled-unlabeled complex. The aforesaid method is known as “label, express and generate oligomers” for NMR (LEGO–NMR)104. Seven sub-units of ~ 75 kDa LSm complexes were selectively labeled with 2H and 15N by LEGO–NMR to map the RNA-binding site104.
13 Isotope Enrichment in Nucleic Acids
Akin to proteins, biological function of RNA is tightly regulated by structure and dynamics. NMR offers suitable means to study nucleic acids in their native state and observe changes under physiological conditions. Nonetheless, NMR based studies of nucleic acids are far more challenging as there are only four nucleotides compared to 20 amino acids in proteins. Thus, the spectral dispersion in case of DNA and RNA is far less compared to that of proteins. Narrower spectral dispersion leads to spectral overlap, which is further augmented by transverse relaxation and inadequate 1H–1H homonuclear long distance restraints.
In addition to the famous Watson–Crick base pairing, double stranded DNA folds into a variety of conformations such as Holliday junction during recombination, G-quadratets in chromosome telomeres and single stranded trinucleotide repeats such as (CNG)n. Dynamic intergenerational expansions in copy number of DNA simple repeats, and hence structural alterations are causes of various hereditary genetic disorders such as Huntington’s syndrome, spinal and bulbar muscular atrophy, several ataxias and Fragile-X syndrome.105. To completely understand the structural–functional diversity of DNA, its structure needs to be exploited further. Similarly, in recent years, a variety of RNAs have emerged as major gene regulatory elements, involved in maintenance of sub-cellular structure, catalysis and propagation of genetic information. Contrary to smaller nucleotide sequences, which can be characterized without any labeling at higher field fully structured long nucleotide sequences require isotope enrichment106, 107.
Nucleic acids can be labeled with stable isotopes uniformly, fractionally and in a site-specific manner by supplementing labeled carbon and nitrogen sources. For production of labeled NTPs, nucleic acid is extracted from microorganisms to enzymatically degraded to NMP followed by their conversion to NTP in vitro108. For synthesis of labeled DNA/RNA inside microorganisms, E. coli or Methylophilus methylotrophus cells are grown in 13C or 15N supplemented minimal medium. Nucleic acids are extracted and digested with nuclease P1 and DNAse I into rNMPs and dNMPs, respectively. rNTPs and dNTPs are separated on HPLC using boronate gel matrix and further phosphorylated to respective NTPs109.
Parallel to advances in isotope enrichment of proteins, specific labeling strategies for nucleic acids were established during the past decades. Deoxynucleotides with specific labels can be synthesized chemically by phosphoramidite-based solid phase synthesis110, 111 and specific labels can be incorporated with ease112, 113. Zimmer and Crothers first demonstrated enzymatic synthesis of labeled DNA where they designed self-priming hairpin ssDNA with modified 3′ terminal ribonucleotide114. The DNA template so provided would be acted upon by Klenow fragment of DNA Polymerase I to make new 13C, 15N labeled ssDNA. 13C, 15N labeled dsDNA can be obtained either by growing bacteria with appropriate plasmid in medium containing isotope labels or by incorporation of labeled deoxynucleoside triphosphate in PCR reaction115, 116. Stable NMR isotope labeling has enabled structural–functional characterization studies of many G-quadruplexes117, 118.
For synthesis of labeled RNA, currently, enzymatic in vitro synthesis by T7 RNA polymerase is the most popular and widely used method to incorporate commercially available 13C, 15N and 2H labeled nucleotides119,120,–121. Apart from the aforementioned in vitro transcription, RNAs smaller than 15 nucleotides are synthesized chemically by using phosphoramidites122. However, phosphoramidites are not commercially available, and hence their laboratory use is restricted. A newer method, PLOR, is designed to incorporate site-specific labels that combine both liquid phase transcription and solid phase chemical synthesis123. PLOR has a DNA template attached to a solid support allowing step-wise buffer and rNTP change. PLOR is initiated by mixture of T7 RNAP, rNTPs and template attached to beads. The mixture is devoid of one or more type of rNTPs that causes transcription stalling. The mixture is then replaced by the one with desired labels and the transcription is resumed. The number of pause/resume cycle depends on the quantity of RNA required. In the termination step, the mixture of all the rNTPs along with T7 RNAP are provided and the transcription reaction is completed123. Segmental labeling in RNA is achieved by a simple cut and paste approach where differently labeled RNA fragments, either chemically generated or in vitro transcribed, are ligated by T4 DNA ligase120, 124 (Fig. 8a). Apart from T4 ligase, segmental labeling of RNA can also be achieved by deoxyribozyme-catalyzed synthesis of 30–50 nt long RNA as RNA ligase always do not provide desirable yields125. Deoxyribozymes (DNA catalysts that mediate reactions involving nucleic acids) provide rapid 3′–5′ linkage without monophosphate requirement at 5′ end donor and has very modest sequence requirements (Fig. 8b)126. 5′ leader sequence of HIV-1 composed of 357 nt was segmentally enriched with 13C in order to elucidate its dimerization and nucleocapsid binding mechanism127. Isotope enrichment of non-coding RNA RsmZ helped in deciphering the mechanism of sequestering RsmE protein dimer by RsmZ128. For conformational and dynamic characterization of Inosone edited RNA, site-specific inosine phosphoramidite was chemically incorporated in Inosine containing 20 mer RNA duplex129.
RNA ligation in segmental labeling in RNA. a DNA ligase mediated b RNA ligase mediated RNA ligation. c Deoxyribozyme mediated RNA ligation. 9DB1 and 7DE5 are examples of deoxyribozyme. Figure adapted and modified with permission from126.
14 Conclusion and Perspective
In the present review, we have highlighted a wide range of conventional and newly designed labeling schemes and expression systems, which enable solution NMR to counter bigger biomolecular structures and complexes. Biomolecules that were earlier intractable are now readily analyzed by advanced labeling techniques such residue-specific and stereo-specific labeling, methyl labeling and relaxation optimized pulse programs such as TROSY (Methyl TROSY). Segmental labeling and LEGO–NMR have further widened the scope of NMR in addressing macromolecular complex structure and dynamics. However, the best labeling scheme used is still case-specific and is subject to protein expressed, spectral quality required, cost and time. The continued interest in devising newer labeling strategies depicts a brighter future for the biomolecular NMR in tackling difficult structural biology problems and deciphering functionally relevant dynamics.
Notes
Experiments involving three nuclei, typically 13C, 15N and 1H.
Organisms prefer a subset of codons during amino acid incorporation in protein translation.
Interaction of dipolar spins through space causing relaxation in solution NMR and broad lines in solid-state NMR.
Auxotrophs are organisms genetically impaired in producing important amino acids or metabolites.
References
Williamson MP, Havel TF, Wüthrich K (1985) Solution conformation of proteinase inhibitor IIA from bull seminal plasma by 1H nuclear magnetic resonance and distance geometry. J Mol Biol 182:295–315
Kline AD, Braun W, Wüthrich K (1986) Studies by 1H nuclear magnetic resonance and distance geometry of the solution conformation of the α-amylase inhibitor Tendamistat. J Mol Biol 189:377–382
Muchmore DC, McIntosh LP, Russell CB, Dahlquist FW (1989) Expression and nitrogen-15 labeling of proteins for proton nitrogen-15 nuclear magnetic resonance. Methods Enzymol 177:44–73
Venters RA, Calderone TL, Spicer LD, Fierke CA (1991) Uniform 13C isotope labeling of proteins with sodium acetate for NMR studies: application to human carbonic anhydrase II. Biochemistry 30:4491–4494
Reilly D, Fairbrother WJ (1994) A novel isotope labeling protocol for bacterially expressed proteins. J Biomol NMR 4:459–462
Bax A, Grzesiek S (1993) Methodological advances in protein NMR. Acc Chem Res 26:131–138
Bax A (1994) Multidimensional nuclear magnetic resonance methods for protein studies. Curr Opin Struct Biol 4:738–744
Cavanagh J, Fairbrother WJ, Palmer AG III, Rance M, Skelton NJ (2007) Heteronuclear NMR experiments, protein NMR spectroscopy, 2nd edn. Academic Press, Cambridge, pp 533–678
Rule GS, Hitchen TK (2006) Dipolar coupling, fundamentals of protein NMR spectroscopy. Focus on structural biology, vol 5. Springer, Dordrecht, pp 357–368
Metzler WJ, Leiting B, Pryor K, Mueller L, Farmer BT (1996) The three-dimensional solution structure of the SH2 domain from p55 blk kinase. Biochemistry 35:6201–6211
Smith BO, Ito Y, Raine A, Teichmann S, Ben-Tovim L, Nietlispach D, Broadhurst RW, Terada T, Kelly M, Oschkinat H, Shibata T, Yokoyama S, Laue ED (1996) An approach to global fold determination using limited NMR data from larger proteins selectively protonated at specific residue types. J Biomol NMR 8:360–368
Pachter R, Arrowsmith CH, Jardetzky O (1992) The effect of selective deuteration on magnetization transfer in larger proteins. J Biomol NMR 2:183–194
Sattler M, Fesik SW (1996) Use of deuterium labeling in NMR: overcoming a sizeable problem. Structure 4:1245–1249
Kushlan DM, LeMaster DM (1993) Resolution and sensitivity enhancement of heteronuclear correlation for methylene resonances via 2H enrichment and decoupling. J Biomol NMR 3:701–708
Crespi HL, Katz JJ (1969) High resolution proton magnetic resonance. Studies of fully deuterated and isotope hybrid proteins, nature 224:560–562
Brodin P, Drakenberg T, Thulin E, Forsén S, Grundström T (1989) Selective proton labeling of amino acids in deuterated bovine calbindin D9 K: a way to simplify1H–NMR spectra. Protein Eng 2:353–357
Oda Y, Nakamura H, Yamazaki T, Nagayama K, Yoshida M, Kanaya S, Ikehara M (1992) 1H NMR studies of deuterated ribonuclease HI selectively labeled with protonated amino acids. J Biomol NMR 2:137–147
McIntosh LP, Griffey RH, Muchmore DC, Nielson CP, Redfield AG, Dahlquist FW (1987) Proton NMR measurements of bacteriophage T4 lysozyme aided by 15N isotopic labeling: structural and dynamic studies of larger proteins. Proc Natl Acad Sci 84:1244–1248
Waugh DS (1996) Genetic tools for selective labeling of proteins with α-15 N-amino acids. J Biomol NMR 8:184–192
LeMaster DM, Richards FM (1988) NMR sequential assignment of Escherichia coli thioredoxin utilizing random fractional deuteriatiod. Biochemistry 27:142–150
Yamasaki K, Muto Y, Ito Y, Wälchli M, Miyazawa T, Nishimura S, Yokoyama S (1992) A 1H–15N NMR study of human c-Ha-ras protein: biosynthetic incorporation of 15N-labeled amino acids. J Biomol NMR 2:71–82
Fiaux J, Bertelsen EB, Horwich AL, Wüthrich K (2004) Uniform and residue-specific 15N-labeling of proteins on a highly deuterated background. J Biomol NMR 29:289–297
Rosen MK, Gardner KH, Willis RC, Parris WE, Pawson T, Kay LE (1996) Selective methyl group protonation of perdeuterated proteins. J Mol Biol 263:627–636
Gardner KH, Kay LE (1998) The Use of 2H, 13C, 15N multidimensional NMR to study the structure and dynamics of proteins. Annu Rev Biophys Biomol Struct 27:357–406
Gardner KH, Kay LE (1997) Production and Incorporation of 15N, 13C, 2H (1H-δ1 Methyl) isoleucine into proteins for multidimensional NMR studies. J Am Chem Soc 119:7599–7600
Goto NK, Gardner KH, Muellerb GA, Willis RC, Kay LE (1999) A robust and cost-effectivemethod for the production of Val, Leu, Ile (δ1) methyl-protonated 15N-, 13C-, 2H-labeled proteins. J Biomol NMR 13:369–374
Gross JD, Gelev VM, Wagner G (2003) A sensitive and robust method for obtaining intermolecular NOEs between side chains in large protein complexes. J Biomol NMR 25:235–242
Lichtenecker RJ, Coudevylle N, Konrat R, Schmid W (2013) Selective isotope labeling of leucine residues by using α-ketoacid precursor compounds. Chem. BioChem. 14:818–821
Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE (2003) Cross-correlated relaxation enhanced 1H–13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J Am Chem Soc 125:10420–10428
Hajduk PJ, Augeri DJ, Mack J, Mendoza R, Yang J, Betz SF, Fesik SW (2000) NMR-based screening of proteins containing 13C-labeled methyl groups. J Am Chem Soc 122:7898–7904
Gardner KH, Konrat R, Rosen MK, Kay LE (1996) An (H)C(CO)NH-TOCSY pulse scheme for sequential assignment of protonated methyl groups in otherwise deuterated 15N, 13C-labeled proteins. J Biomol NMR 8:351–356
Neri D, Szyperski T, Ötting G, Senn H, Wüthrich K (1989) Stereospecific nuclear magnetic resonance Assignments of the methyl groups of Valine and Leucine in the DNA-Binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry 28:7510–7516
Tugarinov V, Kay LE (2004) An isotope labeling strategy for methyl TROSY spectroscopy. J Biomol NMR 28:165–172
Mas G, Crublet E, Hamelin O, Gans P, Boisbouvier J (2013) Specific labeling and assignment strategies of valine methyl groups for NMR studies of high molecular weight proteins. J Biomol NMR 57:251–262
Miyanoiri Y, Takeda M, Okuma K, Ono AM, Terauchi T, Kainosho M (2013) Differential isotope-labeling for Leu and Val residues in a protein by E. coli cellular expression using stereo-specifically methyl labeled amino acids. J Biomol NMR 57:237–249
Gelis I, Bonvin AM, Keramisanou D, Koukaki M, Gouridis G, Karamanou S, Economou A, Kalodimos CG (2007) Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell 131:756–769
Ruschak AM, Velyvis A, Kay LE (2010) A simple strategy for 13C,1H labeling at the Ile-γ2 methyl position in highly deuterated proteins. J Biomol NMR 48:129–135
Ayala I, Hamelin O, Amero C, Pessey O, Plevin MJ, Gans P, Boisbouvier J (2012) An optimized isotopic labeling strategy of isoleucine-γ 2 methyl groups for solution NMR studies of high molecular weight proteins. Chem Commun 48:1434–1436
Gans P, Hamelin O, Sounier R, Ayala I, Durá MA, Amero CD, Noirclerc-Savoye M, Franzetti B, Plevin MJ, Boisbouvier J (2010) Stereospecific isotopic labeling of methyl groups for NMR spectroscopic studies of high-molecular-weight proteins. Angew Chemie Int Ed 49:1958–1962
Godoy-Ruiz R, Guo C, Tugarinov V (2010) Alanine methyl groups as NMR probes of molecular structure and dynamics in high-molecular-weight proteins. J Am Chem Soc 132:18340–18350
Saio T, Guan X, Rossi P, Economou A, Kalodimos CG (2014) Structural basis for protein antiaggregation activity of the trigger factor chaperone. Science 344:597–612
Kerfah R, Hamelin O, Boisbouvier J, Marion D (2015) CH3-specific NMR assignment of alanine, isoleucine, leucine and valine methyl groups in high molecular weight proteins using a single sample. J Biomol NMR 63:389–402
Kerfah R, Plevin MJ, Pessey O, Hamelin O, Gans P, Boisbouvier J (2015) Scrambling free combinatorial labeling of alanine-β, isoleucine-δ1, leucine-proS and valine-proS methyl groups for the detection of long range NOEs. J Biomol NMR 61:73–82
Kerfah R, Plevin MJ, Sounier R, Gans P, Boisbouvier J (2015) Methyl-specific isotopic labeling: a molecular tool box for solution NMR studies of large proteins. Curr Opin Struct Biol 32:113–132
Monneau YR, Ishida Y, Rossi P, Saio T, Tzeng SR, Inouye M, Kalodimos CG (2016) Exploiting E. coli auxotrophs for leucine, valine, and threonine specific methyl labeling of large proteins for NMR applications. J Biomol NMR 65:99–108
Sprangers R, Kay LE (2007) Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature 445:618–622
Sprangers R, Gribun A, Hwang PM, Houry WA, Kay LE (2005) Quantitative NMR spectroscopy of supramolecular complexes: dynamic side pores in ClpP are important for product release. Proc Natl Acad Sci 102:16678–16683
Sinha KK, Gross JD, Narlikar GJ (2017) Distortion of histone octamer core promotes nucleosome mobilization by a chromatin remodeler. Science 355:eaaa3761. https://doi.org/10.1126/science.aaa3761
Macek P, Kerfah R, Erba EB, Crublet E, Moriscot C, Schoehn G, Amero C, Boisbouvier J (2017) Unraveling self-assembly pathways of the 468-kDa proteolytic machine TET2. Sci Adv 3:1–10
Pickford AR, O’Leary JM (2004) Isotopic labeling of recombinant proteins from the methylotrophic yeast Pichia pastoris. Methods Mol Biol 278:17–33
Laroche Y, Storme V, De Meutter J, Messens J, Lauwereys M (1994) High-level secretion and very efficient isotopic labeling of tick anticoagulant peptide (TAP) expressed in the methylotrophic yeast, Pichia pastoris. Nat Biotechnol 12:1119–1124
Morgan WD, Kragt A, Feeney J (2000) Expression of deuterium-isotope-labelled protein in the yeast Pichia pastoris for NMR studies. J Biomol NMR 17:337–347
Whittaker MM, Whittaker JW (2005) Construction and characterization of Pichia pastoris strains for labeling aromatic amino acids in recombinant proteins. Protein Expr Purif 41:266–274
Chen CY, Cheng CH, Chen YC, Lee JC, Chou SH, Huang W, Chuang WJ (2006) Preparation of amino-acid-type selective isotope labeling of protein expressed in Pichia pastoris. Proteins Struct Funct Genet 62:279–287
Romanos MA, Scorer CA, Clare JJ (1992) Foreign gene expression in yeast: a review. Yeast 8:423–488
Cereghino JL, Cregg JM (2000) Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev 24:45–66
Heimo H, Palmu K, Suominen I (1997) Expression in Pichia pastoris and purification of Aspergillus awamori glucoamylase catalytic domain. Protein Expr Purif 10:70–79
Kost TA, Condreay JP, Jarvis DL (2005) Baculovirus as versatile vectors for protein expression in insect and mammalian cells. Nat Biotechnol 23:567–575
Koczka K, Peters P, Ernst W, Himmelbauer H, Nika L, Grabherr R (2018) Comparative transcriptome analysis of a Trichoplusia ni cell line reveals distinct host responses to intracellular and secreted protein products expressed by recombinant baculoviruses. J. Biotech. 270:61–69
Strauss A, Bitsch F, Fendrich G, Graff P, Knecht R, Meyhack B, Jahnke W (2005) Efficient uniform isotope labeling of Abl kinase expressed in Baculovirus-infected insect cells. J Biomol NMR 31:343–349
Harrison RL, Jarvis DL (2006) Protein N-glycosylation in the baculovirus-insect cell expression system and engineering of insect cells to produce “mammalianized” recombinant glycoproteins. Adv Virus Res 68:159–191
Saxena K, Dutta A, Klein-Seetharaman J, Schwalbe H (2012) Isotope labeling in insect cells. Methods Mol Biol 831:37–54
Hansen AP, Petros AM, Mazar AP, Pederson TM, Rueter A, Fesik SW (1992) A practical method for uniform isotopic labeling of recombinant proteins in mammalian cells. Biochemistry 31:12713–12718
Archer SJ, Bax A, Roberts AB, Sporn MB, Ogawa Y, Piez KA, Karl A, Weatherbee JA, Tsang MLS, Lucas R, Zheng BL, Wenker J, Torchia DA (1993) Transforming growth factor beta 1: NMR signal assignments of the recombinant protein expressed and isotopically enriched using Chinese hamster ovary cells. Biochemistry 32:1152–1163
Egorova-Zachernyuk TA, Bosman GJ, Degrip WJ (2011) Uniform stable-isotope labeling in mammalian cells: formulation of a cost-effective culture medium. Appl Microbiol Biotechnol 89:397–406
Weller CT, Lustbader J, Seshadri K, Brown JM, Chadwick CA, Kolthoff CE, Ramnarain S, Pollak S, Canfield R, Homans SW (1996) Structural and conformational analysis of glycan moieties in situ on isotopically 13C, 15N-enriched recombinant human chorionic gonadotropin. Biochemistry 35:8815–8823
Yamaguchi Y, Nishimura M, Nagano M, Yagi H, Sasakawa H, Uchida K, Shitara K, Kato K (2006) Glycoform-dependent conformational alteration of the Fc region of human immunoglobulin G1 as revealed by NMR spectroscopy. Biochim Biophys Acta 1760:693–700
Yanaka S, Yagi H, Yogo R, Yagi-Utsumi M, Kato K (2018) Stable isotope labeling approaches for NMR characterization of glycoproteins using eukaryotic expression systems. J Biomol NMR. https://doi.org/10.1007/s10858-018-0169-2
Spirin AS, Baranov VI, Ryabova LA, Ovodov SY, Alakhov YB (1988) A continuous cell-free translation system capable of producing polypeptides in high yield. Science 242:1162–1164
Kigawa T, Muto Y, Yokoyama S (1995) Cell-free synthesis and amino acid-selective stable isotope labeling of proteins for NMR analysis. J Biomol NMR 6:129–134
Kigawa T, Yabuki T, Yoshida Y, Tsutsui M, Ito Y, Shibata T, Yokoyama S (1999) Cell-free production and stable-isotope labeling of milligram quantities of proteins. FEBS Lett 442:15–19
Marshall R, Maxwell CS, Collins SP, Jacobsen T, Luo ML, Begemann MB, Gray BN, January E, Singer A, He Y, Beisel CL, Noireaux V (2018) Rapid and scalable characterization of CRISPR technologies using an E. coli cell-free transcription-translation system. Mol Cell 69:146–157
Morita EH, Sawasaki T, Tanaka R, Endo Y, Kohno TA (2003) Wheat germ cell-free system is a novel way to screen protein folding and function. Protein Sci 12:1216–1221
Ezure T, Suzuki T, Higashide S, Shintani E, Endo K, Kobayashi S, Shikata M, Ito M, Tanimizu K, Nishimura O (2006) Cell-free protein synthesis system prepared from insect cells by freeze–thawing. Biotechnol Progress 22:1570–1577
Sonnabend A, Spahn V, Stech M, Zemella A, Stein C, Kubick S (2017) Production of G protein-coupled receptors in an insect-based cell-free system. Biotechnol Bioeng 114:2328–2338
Suzuki Y, Ogasawara T, Tanaka Y, Takeda H, Sawasaki T, Mogi M, Liu S, Maeyama K (2018) Functional G-protein coupled receptor (GPCR) synthesis: the pharmacological analysis of human histamine H1 receptor (HRH1) synthesized by a wheat germ cell-free protein synthesis system combined with asolectin glycerosomes. Front Pharmacol 9:38
Merk H, Rues R, Gless C, Beyer K, Dong F, Dötsch V, Gerrits M, Bernhard F (2015) Biosynthesis of membrane dependent proteins in insect cell lysates: identification of limiting parameters for folding and processing. Biol Chem 396:1097–1107
Brödel AK, Kubick S (2014) Developing cell-free protein synthesis systems: a focus on mammalian cells. Pharm Bioprocess 2:339–348
Liu K, Hu J (2018) Host-regulated hepatitis B virus capsid assembly in a mammalian cell-free system. Bio Protoc 8:e2813
Morita M, Takashima E, Ito D, Miura K, Thongkukiatkul A, Diouf A, Fairhurst RM, Diakite M, Long CA, Torii M, Tsuboi T (2017) Immunoscreening of Plasmodium falciparum proteins expressed in a wheat germ cell-free system reveals a novel malaria vaccine candidate. Sci Rep 7:46086
Buntru M, Vogel S, Spiegel H, Schillberg S (2014) Tobacco BY-2 cell-free lysate: an alternative and highly-productive plant-based in vitro translation system. BMC Biotechnol 14:1–11
Murota K, Hagiwara-Komoda Y, Komoda K, Onouchi H, Ishikawa M, Naito S (2011) Arabidopsis cell-free extract, ACE, a new in vitro translation system derived from arabidopsis callus cultures. Plant Cell Physiol 52:1443–1453
Kainosho M, Torizawa T, Iwashita Y, Terauchi T, Mei OA, Güntert P (2006) Optimal isotope labeling for NMR protein structure determinations. Nature 440:52–57
Takeda M, Ono AM, Terauchi T, Kainosho M (2010) Application of SAIL phenylalanine and tyrosine with alternative isotope-labeling patterns for protein structure determination. J Biomol NMR 46:45–49
Dawson PE, Muir TW, Clark-Lewis I, Kent SB (1994) Synthesis of proteins by native chemical ligation. Science 266:776–779
Harmand TJ, Murar CE, Bode JW (2014) New chemistries for chemoselective peptide ligations and the total synthesis of proteins. Curr Opin Chem Biol 22:115–121
Merrifield RB (1973) Solid-phase peptide synthesis. In: Katsoyannis PG (ed) The chemistry of polypeptides. Springer, Boston, MA, pp 335–361
Muir TW, Sondhi D, Cole PA (1998) Expressed protein ligation: a general method for protein engineering. Proc Natl Acad Sci USA 95:6705–6710
Vila-Perelló M, Liu Z, Shah NH, Willis JA, Idoyaga J, Muir TW (2013) Streamlined expressed protein ligation using split inteins. J Am Chem Soc 135:286–292
David Y, Vila-Perelló M, Verma S, Muir TM (2015) Chemical tagging and customizing of cellular chromatin states using ultrafast trans-splicing inteins. Nat Chem 7:394–402
Cotton GJ, Ayers B, Xu R, Muir TW (1999) Insertion of a synthetic peptide into a recombinant protein framework: a protein biosensor. J Am Chem Soc 121:1100–1101
Liu D, Xu R, Cowburn D (2009) Segmental isotopic labeling of proteins for nuclear magnetic resonance. Methods Enzymol 462:151–157
Muona M, Aranko AS, Raulinaitis V, Iwaï H (2010) Segmental isotopic labeling of multi-domain and fusion proteins by protein trans-splicing in vivo and in vitro. Nature protocol 5:574–587
Minato Y, Ueda T, Machiyama A, Shimada I, Iwaï H (2012) Segmental isotopic labeling of a 140 kDa dimeric multi-domain protein CheA from Escherichia coli by expressed protein ligation and protein trans-splicing. J Biomol NMR 53:191–207
Mao H, Hart SA, Schink A, Pollok BA (2004) Sortase-mediated protein ligation: a new method for protein engineering. J Am Chem Soc 126:2670–2671
Kobashigawa Y, Kumeta H, Ogura K, Inagaki F (2009) Attachment of an NMR-invisible solubility enhancement tag using a sortase-mediated protein ligation method. J Biomol NMR 43:145–150
Levary DA, Parthasarathy R, Boder ET, Ackerman ME (2011) Protein-protein fusion catalyzed by sortase A. PLoS One 6:e18342
Parthasarathy R, Subramanian S, Boder ET (2007) Sortase A as a novel molecular “Stapler” for sequence specific protein conjugation. Bioconjugate Chem 18:469–476
Yamazaki T, Otomo T, Oda N, Kyogoku Y, Uegaki K, Ito N, Ishino Y, Nakamura H (1998) Segmental isotope labeling for protein NMR using peptide splicing. J Am Chem Soc 120:5591–5592
Freiburger L, Sonntag M, Hennig J, Li J, Zou P, Sattler M (2015) Efficient segmental isotope labeling of multi-domain proteins using sortase A. J Biomol NMR 63:1–8
Williams FP, Milbradt AG, Embrey KJ, Bobby R (2016) Segmental isotope labeling of an individual bromodomain of a tandem domain BRD4 using sortase A. PLoS One 11:e0154607
Nguyen GK, Wang S, Qiu Y, Hemu X, Lian Y, Tam JP (2014) Butelase 1 is an Asx-specific ligase enabling peptide macrocyclization and synthesis. Nat Chem Biol 10:732–738
Nguyen GKT, Kam A, Loo S, Jansson AE, Pan LX, Tam JP (2015) Butelase 1: a versatile ligase for peptide and protein macrocyclization. J Am Chem Soc 137:15398–15401
Mund M, Overbeck JH, Ullmann J, Sprangers R (2013) LEGO-NMR spectroscopy: a method to visualize individual subunits in large heteromeric complexes. Angew Chemie Int Ed 52:11401–11405
Mirkin SM (2007) Expandable DNA repeats and human disease. Nature 447:932–940
Alláin FH, Varani G (1997) How accurately and precisely can RNA structure be determined by NMR? J Mol Biol 267:338–351
Tolbert BS, Miyazaki Y, Barton S, Kinde B, Starck P, Singh R, Bax A, Case DA, Summers MF (2010) MF, Major groove width variations in RNA structures determined by NMR and impact of 13C residual chemical shift anisotropy and 1H–13C residual dipolar coupling on refinement. J Biomol NMR 47:205–219
Louis JM, Martin RG, Clore GM, Gronenborn AM (1998) Preparation of uniformly isotope-labeled DNA oligonucleotides for NMR spectroscopy. J Biol Chem 273:2374–2378
Nelissen FH, Tessari M, Wijmenga SS, Heus HA (2016) Stable isotope labeling methods for DNA. Prog Nucl Mag Reson Spect 96:89–108
Beaucage SL, Iyer RP (1992) Advances in the synthesis of oligonucleotides by the phosphoramidite approach. Tetrahedron 48:2223–2311
Beaucage SL, Iyer RP (1993) The synthesis of modified oligonucleotides by the phosphoramidite approach and their applications. Tetrahedron 49:6123–6194
Phan AT, Patel DJ (2002) Differentiation between unlabeled and very-low-level fully 15N,13C-labeled nucleotides for resonance assignments in nucleic acids. J Biomol NMR 23:257–262
Phan AT, Patel DJ (2002) A site-specific low-enrichment 15N, 13C isotope-labeling approach to unambiguous NMR spectral assignments in nucleic acids. J Am Chem Soc 124:1160–1161
Zimmer DP, Crothers DM (1995) NMR of enzymatically synthesized uniformly 13C, 15N-labeled DNA oligonucleotides. Proc Natl Acad Sci USA 92:3091–3095
Louis JM, Martin RG, Clore GM, Gronenborn AM (1998) Preparation of uniformly isotope-labeled DNA oligonucleotides for NMR spectroscopy. J Biol Chem 273:2374–2378
Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA (1998) Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487–491
Nguyena SV, Stroevaa E, Germann MW (2018) Simplifying DNA NMR spectroscopy by silencing GH8 and AH8 resonances. J Mol Struct 1166:344–347
Chung WJ, Heddi B, Schmitt E, Lim KW, Mechulam Y, Phan AT (2015) Structure of a left-handed DNA G-quadruplex. Proc Natl Acad Sci USA 112:2729–2733
Milligan JF, Uhlenbeck OC (1989) Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol 180:51–62
Lu K (2010) K., Y. Miyazaki and M. F. Summers, Isotope labeling strategies for NMR studies of RNA. J Biomol NMR 46:113–125
Liu Y, Sousa R, Wang YX (2016) Specific labeling: an effective tool to explore the RNA world. BioEssays 38:192–200
Müller S, Wolf J, Ivanov SA (2004) Current strategies for the synthesis of RNA. Curr Org Synth 1(3):293–307
Liu Y, Yu P, Dyba M, Sousa R, Stagno JR, Wang YX (2016) Applications of PLOR in labeling large RNAs at specific sites. Methods 103:4–10
Duss O, Maris C, von Schroetter C, Alláin FH (2010) A fast, efficient and sequence-independent method for flexible multiple segmental isotope labeling of RNA using ribozyme and RNase H cleavage. Nucl Acids Res 38:e188
Silverman SK, Cech TR (1999) Energetics and cooperativity of tertiary hydrogen bonds in RNA structure. Biochemistry 38:8691–8702
Purtha WE, Coppins RL, Smalley MK, Silverman SK (2005) General deoxyribozyme-catalyzed synthesis of native 3′–5′ RNA linkages. J Am Chem Soc 127:13124–13125
Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, Divakaruni SS, LaCotti C, Barton S, Tummillo D, Hosic A, Edme K, Albrecht S, Telesnitsky A, Summers MF (2011) NMR detection of structures in the HIV-1 5′-Leader RNA that regulate genome packaging. Science 334:242–245
Duss O, Michel E, Yulikov M, Schubert M, Jeschke G, Allain FH (2014) Structural basis of the non-coding RNA RsmZ acting as a protein sponge. Nature 509:588–592
Dallmann A, Beribisky AV, Gnerlich F, Rübbelke M, Schiesser S, Carell T, Sattler M (2016) Site-specific isotope labeling of inosine phosphoramidites and NMR analysis of an inosine containing RNA duplex. Chemistry 22:15350–15359
Sharma R, Sahu B, Ray MK, Deshmukh MV (2015) Backbone and stereospecific 13C methyl Ile (δ1), Leu and Val side-chain chemical shift assignments of Crc. Biomol NMR Assign 9:75–79
Acknowledgements
The authors acknowledge technical support and assistance extended by Dr. Manjula Reddy (CSIR–CCMB) during the preparation of the auxotrophic strain. The authors gratefully acknowledge Dr. Malay K. Ray (CSIR–CCMB) for the collaboration and exciting scientific discussions on the Crc project. The Crc project was financially supported through CSIR network project GenCODE (BSC0123). UR and RS acknowledge their fellowships from University Grants Commission (UGC) and Council of Scientific and Industrial Research (CSIR), respectively.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rai, U., Sharma, R. & Deshmukh, M.V. Accessing Structure, Dynamics and Function of Biological Macromolecules by NMR Through Advances in Isotope Labeling. J Indian Inst Sci 98, 301–323 (2018). https://doi.org/10.1007/s41745-018-0085-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41745-018-0085-1