Abstract
The discovery of RNA interference (RNAi) as an alternate mode of gene regulation utilized by eukaryotes has greatly expanded the arsenal of molecular tools that a biomedical researcher can employ to target tumors. Thus, it will be natural to extend this methodology from bench to the bedside as a novel mode of clinical intervention against intracranial tumors, as these are frequently refractive to conventional therapies, i.e. surgery, radiotherapy, and chemotherapy. Induction of RNAi in the targeted tumor can be multifaceted, with two primary modes of therapeutic intervention postulated to date; delivery of in silico designed and in vitro synthesized non-coding RNA molecules to the tumor, or the pharmacological perturbation of non-coding RNA molecules that are endogenously expressed by the tumor. Both modes silence gene expression, but via somewhat different molecular pathways. Two key non-coding RNA molecules that are known to impart RNAi are small interfering RNA and microRNA. Both are currently under investigation in pre-clinical and clinical settings for their potential utility against tumors, with small interfering RNA already proving their efficacy in several small-scale clinical trials. However, despite demonstration of excellent potential as an anti-cancer therapy at the bench, the current inability to deliver therapeutically significant levels of RNAi into the diseased tissue remains a primary hurdle that needs to be overcome for widespread utility at the bedside. This chapter presents an update on both the latest methodologies that have been utilized for induction of RNAi in brain tumors, both in pre-clinical and clinical settings, as well as descriptions of the molecular pathways that have been targeted via RNAi. The pioneering RNAi-based clinical trials that have been tested against brain tumors are also described to illustrate both the utility and the impact of this novel mode of brain tumor therapy at the bedside.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
22.1 Introduction
Malignant brain tumors are frequently refractory to conventional clinical treatments, i.e. surgical intervention followed by combined radiation therapy and chemotherapy. Despite aggressive multimodal therapy, the prognosis still remains quite poor for these patients. Thus, translation of any novel molecular tool used in brain tumor research to the bedside as a clinical therapy will be of great significance for improving the clinical outcome for such patients.
Brain tumors are classified according to their cellular origin and are graded based on their histopathology [World Health Organization (WHO) grading system]. The grading takes into account atypical nuclear and cellular morphology, mitotic activity, endothelial proliferation, necrosis, and proliferation index (Louis et al. 2007). Brain tumors can be broadly divided into primary brain tumors (those with intracranial origin) and brain metastases (those that metastasize from elsewhere in the body). Approximately 40% of brain tumors are metastatic with origins in cancers of the lung, breast, melanoma, renal, or colon.
Primary brain tumors can arise from any cell type within the central nervous system (CNS). Of these, gliomas are neuroectodermal in origin and arise from the glia, the supporting cells of the CNS. They are the most common type of primary brain tumors, and harbor distinct clinical, histological, and molecular characteristics. They are grouped into low-grade gliomas (WHO grades I and II) and high-grade gliomas (WHO grades III and IV). Other primary brain tumors include meningiomas, ependymomas, choroid plexus papillomas, pituitary adenomas, and medulloblastomas.
It is these most malignant of these tumors (grades III and IV glioma) that are frequently refractory to conventional clinical treatments, with patients having a median survival of 12–15 months from the time of diagnosis. Thus, it is these patients that urgently need novel modes of clinical intervention, where interference RNA can play a role in identifying adjuvant therapies in support of current clinical interventional regimens.
Worldwide, 13% of all deaths occur due to cancer, which translates into 7.4 million deaths. The age-adjusted worldwide incidence rate for malignant brain tumors is 3.7 per 100,000 person-years for males and 2.6 for females (IARC; International Agency for Research on Cancer; http://www.iarc.fr), with rates being higher in the so-called developed countries than in less-developed areas (IARC). The detection rate of malignant brain tumors has increased over the past two decades, especially among the elderly, most likely due to improved diagnostic imaging regimens.
22.1.1 RNA Interference (RNAi)
RNAi had its origins in horticulture research. The existence of a mechanism for interfering with the cellular transcription machinery was first identified in 1990 by Napoli and co-workers (1990) during genetic engineering experiments on the petunia plant to engineer deeper colored flowers. When extra copies of the chalcone synthase gene (which encodes an enzyme crucial for plant pigment synthesis) were added to the plant’s genetic makeup, white or variegated flowers developed instead of the expected deeply colored flowers. Although the added copies of the gene were being correctly transcribed, the chalcone synthase mRNA were somehow being destroyed, preventing production of the enzyme. This new form of gene suppression was termed post-transcriptional gene silencing (PTGS).
Similar gene-silencing effects were also observed in the nematode Caenorhabditis elegans (Guo and Kemphues 1995), where specific gene transcripts could be selectively silenced or inactivated via application of copies of sense RNA identical to the targeted mRNA sequence. Here too, the introduced sense RNA was acting in a manner similar to a classical antisense RNA strand, although the latter imparts its effect due to its sequence complementarity to the target messenger RNA. The mechanism behind this phenotype was identified in 1998 when Fire and co-workers (1998) showed that the transcriptional interference effect was due to the inadvertent presence of double-stranded RNA (dsRNA) molecules in the synthetic RNA preparations used in these experiments. Thus, application of both sense and antisense RNA had a far more potent silencing effect than either sense or antisense RNA alone. Fire and Mello (who were awarded the Nobel Prize in Physiology or Medicine in 2006 for their discovery) designated this mechanism as RNAi, which was later found to be active in mammals as well (Elbashir et al. 2001).
22.1.2 Non-coding RNAs that Impart RNAi
Since these early reports on RNAi by in vitro engineered RNA on the specific mRNA transcripts, several new classes of non-coding RNAs that impart RNAi on eukaryotic cells (including mammalian cells) have been identified. The exogenously applied RNAi molecules that led to the original discovery (described above) were referred to simply as small interfering RNA (siRNA). However, recent reports indicate that both insect and mammalian cells also generate similar siRNA in an endogenous manner, which has been termed endo-siRNA (Okamura and Lai 2008; Watanabe et al. 2008). Thus, the exogenously applied siRNA will need to be categorized as exo-siRNA to differentiate the in vitro from the in vivo. MicroRNA (miRNA) are another class of non-coding RNAs that are endogenously transcribed (Birchler and Kavi 2008; Kim and Rossi 2007; Liu et al. 2008; Mathupala et al. 2007; Pillai 2005).
While siRNA and miRNA populate most of the RNAi molecules studied to date, additional classes of RNAi molecules have been recently described. Piwi-interacting RNA (piRNA) is a class of non-coding RNAs present in germline and adjacent tissues (Lau et al. 2006). Another two classes have been described that influence the transcription-initiation RNA (tiRNA) or are associated with promoter regions promoter-associated small RNAs (pasRNA) (Kapranov et al. 2007; Taft et al. 2009).
Would RNAi impart only short-term changes to the transcriptome of a tumor cell? To date, studies in both non-mammalian and mammalian systems have indicated that RNAi can influence gene activity at the epigenetic level by additional mechanisms known as RNA-directed DNA methylation (RdDM) and RNAi mediated heterochromatin modification (Malecova and Morris 2010; Moazed 2009; Verdel et al. 2009). In contrast to PTGS commonly mediated by RNAi, this mode of longer term gene silencing, which also encompasses chromatin remodeling, is referred to as TGS or transcriptional gene silencing.
The present chapter will address RNAi studies that have been completed in vivo in model systems, or currently in clinical trials. With regard to miRNA, we will describe miRNA signatures that have been documented in primary glioma resections, as these will be of utility in clinical diagnosis. Thus, for clarity, we will refer to small RNAs of exogenous origins (in vitro synthesized) as siRNAs throughout this chapter, while small RNAs transcribed and processed off nuclear DNA templates will be referred to as miRNA.
22.1.3 MiRNA
As described above, miRNAs are transcribed from intergenic regions of chromosomes as well as non-coding regions within genes, i.e. introns and non-translated regions of protein-coding genes. However, it should be noted that miRNAs of intergenic origin are in fact, encoded by distinct transcription units (i.e. genes) (Kim and Nam 2006; Nelson et al. 2003), despite being non-coding in nature. MiRNAs are now known to mediate expression of target genes by base pairing with regions within the corresponding mRNA, which then result in either inhibition of translation, or cause direct destruction of the mRNA template. The latter is thought to closely parallel the mechanism for RNAi via siRNA. The fate of mRNA template is selected on the basis of perfect or imperfect base-pairing between miRNA and the target mRNA template, with a perfect match targeting the mRNA towards siRNA-type targeted destruction, while mismatches create conditions for inhibition of translation (Krol and Krzyzosiak 2004; Meltzer 2005; Nelson et al. 2003). MiRNAs that were expressed within introns were usually coordinately expressed with their host gene’s mRNA, indicating that they were derived from introns of the same pre-mRNA transcript.
22.2 Mechanisms of Interference
Experimentally, the siRNA-mediated mRNA degradation cascade begins with cleavage of exogenously introduced synthetic dsRNA by an enzyme complex referred to as Dicer. The resultant, approximately 21 base pair (bp) dsRNA cleavage products (siRNA) are then incorporated into a protein multimer composed of two proteins referred to as Argonaute-2 (Ago-2; in the case of mammalian systems) (Kim and Rossi 2007; Liu et al. 2004) and RNA-induced silencing complex (RISC). When the pair of oligonucleotide strands of the dsRNA template loaded onto RISC will have perfect sequence complementarity as in the above situation, Ago-2 will cleave and discard the sense RNA strand to produce an active RISC complex containing the antisense strand only. The resultant siRNA/RISC/Ago-2 complex scans the mRNA milieu for target sites with perfect complementarity. Upon binding, the target mRNA is cleaved by the complex.
In contrast, the miRNA-based mRNA degradation cascade begins with endogenously (nuclear) encoded primary miRNA transcripts referred to as pri-miRNAs. As mentioned before, these may be transcribed from non-coding genes, or be components of introns of pre-mRNA templates of coding genes. The miRNA coding genes are also transcribed by RNA polymerase II, in a manner similar to conventional protein coding genes (Kim and Rossi 2007). During transcription, the pri-miRNA template is also spliced and polyadenylated and then processed by an enzyme complex referred to as Drosha to yield precursor miRNAs (pre-miRNAs). These are then exported to the cytoplasm across the nuclear envelop via the protein exportin-5 to be bound to the Dicer complex, which in turn processes the pre-miRNA template for loading onto the Ago-2/RISC complex in a manner similar to that for siRNA. The mature miRNA strand may, or may not have perfect sequence complementarity with target mRNA. When there is full complementarity, the target mRNA is directed for cleavage via the Ago-2/RISC in a mechanism similar to that for siRNA. When the complementarity is partial, translation off the targeted mRNA is first inhibited due to miRNA/RISC binding, or this step may be followed by degradation of the target mRNA in cytoplasmic vesicles referred to as processing bodies (P-bodies).
22.3 Rational Design of siRNA for Pre-clinical and Clinical Studies
Design algorithms for siRNA have evolved from design of 21-mer siRNA to 27-mer longer siRNA templates (Khvorova et al. 2003; Kim et al. 2005; Reynolds et al. 2004; Siolas et al. 2005). The design changes are due to greater understanding of the mechanisms behind the RNAi, with studies indicating that 21-mers are processed by Dicer (an RNAse III type endonuclease) from longer dsRNA templates, which may be necessary both for loading siRNA onto the RISC complex and for the assembly of RISC itself (Amarzguioui et al. 2006; Preall et al. 2006; Rose et al. 2005). Thus, most current algorithms outline the use of 27-mer or longer templates (known as 27-mer Dicer substrates) when designing siRNA.
siRNA can sometimes down-regulate non-targeted mRNA (Cullen 2006a) voiding experimental data interpretation. However, when RNAi experiments are conducted in vivo, this response is not observed (Heidel et al. 2004). More importantly, in the single clinical trial reported for RNAi against glioma (Zukiel et al. 2006), the authors report the absence of local inflammatory responses at the siRNA site-of-injection.
The 5′ region of the antisense strand (also called the guide strand) of the dsRNA loaded on the RISC complex is referred to as the seed region, due to its importance in targeting a specific mRNA for cleavage. This seed region consists of 6 nucleotides between positions 2–7 of the guide strand of the siRNA duplex. Recent studies indicate that off-target siRNA effects may occur due to partial complementarity between this seed region with the 3′ untranslated regions (3′UTR) of non-specific mRNA targets (Jackson et al. 2006). Such partial matches can initiate down-regulation of non-specific mRNA due to siRNA mimicking the action of a miRNA (explained below). Thus, current algorithms analyze siRNA templates for global mRNA 3′UTR – seed region matches to minimize the potential for off-target effects.
With greater understanding of the mechanism employed by Dicer to process long dsRNA molecules to select the guide strand (referred to as functional polarity of the dsRNA template), and the method employed to load the RISC complex in mammalian model systems, 29 bp dsRNA templates with 2-nucleotide 3′ overhangs are now known to display better efficacy in inducing the siRNA response in comparison to shorter 21 bp dsRNA templates. Also, current algorithms design the two strands in a manner that directs Dicer to preferentially process the guide RNA strand for loading onto the RISC complex for optimal efficiency of targeting (Amarzguioui et al. 2006; Rose et al. 2005).
22.4 Pre-clinical Experimental Strategies
Three experimental methods are commonly employed for introduction of siRNA into a cell; (i) direct transfection of gene specific 21 bp RNA duplexes (dsRNA) (which may be chemically modified for better intracellular longevity) to directly load RISC complexes for short term induction of siRNA (Bumcrot et al. 2006); (ii) pre digestion of long dsRNA (that correspond to a full length mRNA template) in vitro with Dicer to produce a pool of 21-mer siRNA templates against a particular mRNA target, followed by transfection of the entire pool into cells; (iii) transfection of cells with an expression plasmid or PCR based expression cassette that can transcribe a short symmetric RNA strand that folds to form a 21-mer dsRNA (denoted a short hairpin RNA or shRNA) for loading the RISC complex (Brummelkamp et al. 2002; Cullen 2006a, b; Myslinski et al. 2001).
In contrast to siRNA, design and development of experimental strategies with miRNA poses a dilemma. First and foremost, miRNA are cellular products specific to a particular cell line or tissue. One can design synthetic miRNA or miRNA templates for in vitro synthesis in a manner similar to that for siRNA, with the caveat that imperfect complementarity needs to be built into the designed miRNA template. In silico analysis of miRNA sequences and the transcriptome hint at a given miRNA sometimes influencing multiple gene targets, sometimes into the hundreds. If so, it would throw a cell’s intricate network of signaling and metabolic cascades into disarray. Thus, further studies are needed to understand the in vivo selectivity and efficacy displayed by miRNA on the cellular transcriptome.
Recent laboratory strategies have utilized miRNA-adapted shRNA hybrids (shRNAmiR), where 30-mer duplex shRNA are designed to mimic naturally transcribed miRNA transcripts in organism of interest (Chang et al. 2006). When transfected, the shRNAmiR will be processed first by the nuclear Drosha complex and then by the cytoplasmic Dicer and loaded onto the RISC complex. Since the method closely follows the endogenous miRNA processing pathways it has been shown to induce more effective suppression of translation from target mRNA and/or cleavage (Silva et al. 2005).
However, recent reports indicate that endogenous miRNA may, at most, regulate their target mRNA messages by approximately two-fold (Seitz 2009). Thus, while miRNA may be of use as a diagnostic or prognostic tool in the clinic, they may be of limited utility as a therapeutic tool.
22.5 siRNA Targets in Glioma – Pre-clinical Studies
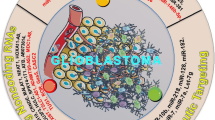
Since the recognition of RNAi as a primary mode of post-transcriptional gene regulation in mammalian cells, over to 300 research reports have appeared in literature on targeting glioma via RNAi. In fact, the first publication on glioma and dsRNA was in 1991, where the authors investigated the anti-proliferative effect of mismatched dsRNA in a human glioma cell line (Hubbell et al. 1991). Since then, RNAi has been utilized to target glioma metabolism, signal transduction, cell-cycle progression, proliferation, invasion, angiogenesis, etc, both in vitro and in vivo (Fig. 22.1).
Primary signaling pathways and extracellular matrix components that have been targeted by RNA interference (RNAi) in pre-clinical and clinical studies. Naked or encapsulated RNAi have been used to target glioma in vivo. Both the transferrin receptor and the epidermal growth factor receptor (EGFR) have been frequently used for ligand coupled homing of siRNA loaded vesicles. Both key signal transduction pathways and extracellular matrix proteins have been targeted to promote glioma apoptosis and growth inhibition, and to inhibit invasiveness. The only clinical trial on using RNAi in glioma therapy has targeted tenascin-C, an extracellular glycoprotein
Since the current chapter addresses the possible routes for using RNAi as a clinical tool, we will summarize published in vivo studies where rodent models were used to test the efficacy of select siRNA or miRNA molecules against implanted glioma cells (of both human and animal origin). The single clinical trial underway on using RNAi against glioma will also be described. Since miRNA may be of utility as a diagnostic tool, the miRNA signatures that have been identified to date from primary glioma tissues or other brain tumor types will be also described. In vitro studies (over 300 at present) that have used RNAi to target glioma, or to elucidate various cellular pathways, will not be discussed for sake of brevity as these in vitro studies have been reviewed elsewhere (Guo et al. 2010; Mathupala et al. 2006, 2007).
22.5.1 Targeting Cell Surface Receptors
Several receptors that are known to be highly expressed in glioma have been studied to either use as a therapeutic target [epidermal growth factor receptor (EGFR) or insulin-like growth factor-1 receptor (IGF-1 R)] or as a tool for targeted delivery of RNAi (transferrin receptor) (Cardoso et al. 2007).
Most malignant gliomas over-express mutant (EGFRvIII) or wild-type EGFR (wtEGFR), crucial for their proliferation and malignancy. In the first in vivo study (in an orthotopic brain tumor nude mouse model) on receptor-mediated targeting of glioma via RNAi, Zhang and co-workers (2004) administered ligand-coupled immuno-liposomes containing an expression vector (shRNA) that targeted EGFR. Two ligands were used, one to target the insulin receptor on the blood-brain barrier (BBB), and the second to target the transferrin receptor on glioma. Others have used HSV-1 based amplicons or siRNA to target EGFR in vivo (Kang et al. 2006; Saydam et al. 2005).
A key signaling pathway activated by EGFR is the PI3kinase (PI3K) pathway, also activated by other members of the receptor tyrosine kinase family, most of which are over-expressed in glioma. Thus, PI3K is a more focused downstream target, after EGFR. This has been targeted in a subcutaneous murine model to show that glioma growth was significantly inhibited (Pu et al. 2006).
Others have targeted the IGF1-R or the IGF-1, as well as basic fibroblast growth factor-2 (FGF-2) as these are highly expressed in human gliomas and activate downstream signaling pathways crucial for tumor growth and proliferation. Rat C6 glioma were implanted subcutaneously in nude mice with intra-tumoral injection of siRNA resulting in regression of tumor growth and reduced tumorigenicity (Dong et al. 2009).
Other cell-surface proteins that have been targeted by siRNA have been: (i) secreted protein acidic and rich in cysteine (SPARC), an extracellular glycoprotein switched-on in malignant gliomas and promote their invasion and cell-survival via Akt pathway activation. Intracranial glioma transfected with siRNA against SPARC were shown to form non-invasive tumors (Seno et al. 2009); (ii) protein tyrosine phosphatase zeta/receptor-type protein tyrosine phosphatase beta (PTPzeta/RPTPbeta) and its ligand pleiotrophin (PTN), which are normally involved in neuronal cell migration, but over-expressed in gliomas; tumor growth was abrogated when glioma engineered to express shRNA against above were implanted in an orthotopic brain tumor model (Ulbricht et al. 2006).
Notch receptors are normally involved in cellular homeostasis, including embryonic development and stem cell maintenance. It is also over-expressed in most gliomas. Thus, Notch-1 was targeted in a nude mouse model which resulted in not only cell-cycle arrest, but reduction in invasiveness and induction of apoptosis of the implanted glioma (Xu et al. 2010).
22.5.2 Targeting Invasive Nature of Glioma
Infiltration of surrounding normal brain tissue by malignant glioma is facilitated by proteolysis of extra-cellular matrix components, initiated by the urokinase plasminogen activator receptor (uPAR). This receptor, along with its agonist urokinase plasminogen activator (uPA) and matrix metalloproteinase-2 (MMP-2, a metalloproteinase over-expressed in glioma) have been targeted to affect the glioma infiltrative process via RNAi in vivo, to show that down-regulation these resulted in decreased cell invasion, angiogenesis, tumor growth, and in a meningioma model, increased radiosensitivity (Gogineni et al. 2010; Gondi et al. 2007, 2009; Kargiotis et al. 2008).
Other proteins that have been targeted in vivo in glioma via RNAi to investigate their effects on migration/invasiveness as well as proliferation and angiogenesis are (i) telomerase (hTERT) (Zhao et al. 2007); (ii) S100 calcium-binding protein A4 (S100A4), a member of the EF-hand family of calcium-binding proteins, which is up-regulated in glioma in contrast to white matter, and known to be involved in both invasion and metastasis. S100A4 was targeted via RNAi in C6 glioma cells (Takenaga et al. 2007) and shown to inhibit migration; (iii) Akt2, a serine/threonine kinase that regulates cell survival and proliferation. The expression of Akt2 was shown to closely correlate with malignancy of primary human glioma. Silencing of Akt2 resulted in decreased levels of MMP-9 and suppression of invasiveness (Zhang et al. 2009a).
22.5.3 Induction of Glioma Apoptosis
RNAi has been used to induce apoptosis in glioma by targeting survivin, an anti-apoptotic protein that belongs to the family of inhibitors of apoptosis (IAP). Survivin is highly expressed in glioma and inhibits caspase activation. Thus, when survivin was targeted in vivo with RNAi, it leads to glioma apoptosis (Uchida et al. 2004).
Another signaling molecule targeted for induction of apoptosis is Wnt, a molecule involved in embryogenesis as well as cancer. Several members of Wnt family are over-expressed in glioma and have been targeted by siRNA to show that silencing of Wnt proteins is associated with decreases in PI3-kinase/Akt signaling, with inhibition of tumor growth (Pu et al. 2009).
22.5.4 Targeting Hypoxia-induced Glioma Angiogenesis Cascades via RNAi
Hypoxia-inducible factor-1 (HIF-1) and vascular endothelial growth factor (VEGF) are both known to be over-expressed in the hypoxic regions of malignant glioma, and promote invasion and radioresistance. Both have been targeted (Gillespie et al. 2007) in vivo to show that silencing of either attenuates glioma migration and invasion.
22.5.5 Modulation of Immuno-activity Against Glioma
Malignant tumors contain microglia, which are known to promote glioma invasion. STAT-3 has been targeted in vivo via RNAi to activate anti-glioma immunity, where microglia and macrophage activation were studied. The latter resulted in glioma growth inhibition indicating that STAT-3 may serve as an adjuvant for immunotherapy against glioma (Zhang et al. 2009b).
22.6 MiRNA Targets in Glioma
An increasing number of investigators have turned to testing miRNA as a molecular tool for glioma therapy. However, in contrast to the extensive list of literature on targeting glioma via siRNA, only a few RNAi studies have been reported that harness the power of miRNA to target glioma even in vitro.
Oncogenic miRNA are classified as oncomirs (He et al. 2005), while those that act as tumor suppressors are termed anti-oncomirs (Goga and Benz 2007). Thus, miRNA therapies that will target oncomirs are referred to as antagomirs (Krutzfeldt et al. 2005). However, due to the homeostatic mechanisms active in mammalian cells, including tumor cells, it is becoming increasingly apparent that perturbing the miRNA to impart significant changes in the tumor proteome will be a difficult task at best. It is highly likely that miRNA will be of significant utility as a diagnostic tool, which has already been proven clinically.
In one expression profiling study, the miRNAs miR-9, -15a, -16, -17, 19a, -20a, -21, -25, -28, -130b, -140, -210 were found to be over-expressed in glioma, while miR-184 and -328 were found to be under-expressed (Malzkorn et al. 2010). Others have located miR-183, -367, -371 to be up-regulated in glioma (Lavon et al. 2010), similar to the profiles seen in neural precursor cells. In another study, miR-10b was found to be up-regulated with increasing grade of glioma, via RT-PCR analysis. The increase closely correlated with uPAR levels which is one of the receptors thought to be regulated by miR-10b (Sasayama et al. 2009). MiR-221 expression was also found to be closely correlated with glioma malignancy in several other RT-PCR screens (Conti et al. 2009; Lu et al. 2009).
In the few in vivo studies that have examined the utility of miRNA, miR-221 and -222 have been shown to promote malignant progression by influencing the Akt signaling cascades (Zhang et al. 2010). Inhibition of miR-21 was shown to suppress tumor growth in vivo, along with inhibition of EGFR, activation of Akt, cyclin D and Bcl-2 (Zhou et al. 2010). In another study miR-21 was targeted for cytotoxic therapy with S-TRAIL, a pro-apoptotic ligand, where synergistic activity was seen between the two molecules (Corsten et al. 2007). MiR-21 is also implicated in cell proliferation, apoptosis, and migration/invasion (Chan et al. 2005; Corsten et al. 2007; Gabriely et al. 2008) as well as regulation of inhibitors of MMP. Inhibition of miR-21 was shown to reduce MMP activity in a mouse model and suppress glioma migration and invasion, indicating that miR-21 contributes to glioma malignancy by down-regulation of MMP inhibitors leading to activation of MMPs, and thus promoting invasiveness of glioma. A detailed review of miRNA in glioma and the potential therapeutic efficacy is provided elsewhere (Mathupala et al. 2007).
22.7 Systemic Delivery of RNAi to Brain Tumors
Brain is a privileged organ, where the tissue is separated from systemic circulation and protected by the BBB, which forms a physical barrier to protect the brain tissue from invading pathogens and other toxins. Tight-junctions between the endothelial cells that line the cerebral capillaries ensure that the BBB will essentially be impenetrable to RNA molecules used in potential RNAi therapies (where the average molecular mass will be around 14 kDa) and to larger shRNA generating DNA vectors, or potential miRNA therapeutics.
Thus, the BBB needs to be traversed for any systemic application of RNAi to succeed. Both basic and potential clinical approaches for systemic application of RNAi have been reviewed in detail elsewhere (Boado 2007; Fountaine et al. 2005; Lesniak 2005; Pardridge 2004; 2007; Pirollo and Chang 2008; Xie et al. 2006).
Several strategies have been tested to deliver RNAi to the brain via the systemic route, by harnessing receptor-mediated transport across the BBB. As described previously, RNAi to target glioma EGFR have been encapsulated in pegylated immunoliposomes (poly-ethylene-glycol encapsulated liposomes) and tested in an orthotopic murine model. The liposomes were surface modified with monoclonal antibodies against the insulin receptor and against the transferrin receptor (Boado 2005; Zhang et al. 2004). The same group has also utilized direct conjugation of transferrin receptor-targeting antibodies to the siRNA, where the antibody was coupled to the RNA molecules via a biotin-streptavidin linker, again in an orthotopic brain tumor model (Xia et al. 2007).
Yet others have utilized coated nanoparticles for delivery of encapsulated siRNA or siRNA generating expression vectors through the BBB (Jain 2007). These nanoparticles are thought to mimic low-density lipoproteins (LDL) and thus interact with the LDL receptors on the endothelium resulting in their uptake across the BBB (Lesniak 2005). However, since the BBB is commonly compromised in patients with late-stage glioma, traversing the vascular endothelium may not be difficult in a systemic siRNA delivery strategy.
The most straightforward, non-obstructive clinical route to brain tumor may be direct application of RNAi therapeutics to the tumor, or the surgical cavity following tumor resection. In fact, the single clinical trial reported to date for glioma therapy with RNAi has followed this route (see Section 22.8), where siRNA was injected directly to infiltrative (inoperable) regions of glioma (Zukiel et al. 2006).
Others have used multifunctional carriers that are pH sensitive for peptide-targeted systemic delivery of siRNA in vivo, where stable nanoparticles were formed to administer HIF-1 targeting complexes in glioma bearing nude mice to show significant tumor inhibition (Wang et al. 2009).
22.8 RNAi-mediated Glioma Therapy Clinical Trials
Tenascin-C is an extracellular matrix glycoprotein that is also expressed during early development. It is also over-expressed in glioma. Tenascin-C expression in the stroma of tumors is known to be associated with poor prognosis. Most importantly, tenascin-C was targeted in the first (and only) clinical trial conducted thus far utilizing RNAi, where a significantly improved outcome was noted in a subset of patients with malignant gliomas (Zukiel et al. 2006).
In the initial study, a cohort of 11 low- and high-grade patients were treated after tumor resection, with at least two of the patients indicating absence of tumor recurrence. Tumor recurrence was observed in at least three other patients, but only at sites distal to the RNAi application point. Currently 46 patients have been treated in this clinical trial, with a median survival of 106.6 weeks for the cohort, whereas the median survival for patients with glioblastoma (grade IV) and anaplastic astrocytoma (grade III) was 48.2 weeks (Rolle et al. 2010; Wyszko et al. 2008). The success in this initial trial points to the efficacy of targeted RNAi-based therapy against glioma.
With regard to clinical trials based on interference RNA strategies, to date, over twenty trials have been initiated with some being completed or terminated, per descriptions outlined in the online database ClinicalTrials.gov. Of these, only five involve strategies against cancer (excluding the above described clinical trial against glioma). Of these, one (NCT00689065) has targeted the M2 subunit of ribonucleotide reductase, where siRNA was systemically delivered, embedded in nanoparticles for a prolonged half-life. In fact, initial results from this study on melanoma patients have provided the first proof-of-principle data to indicate the efficacy of systemic therapy via nanoparticle-mediated application of RNAi, in homing-in on the solid tumor to deliver the siRNA payload (Davis et al. 2010). Another formulation is being tested for advanced solid cancers, again by systemic delivery (NCT00938574). With liver being one of the first-pass organs during systemic siRNA delivery, a third trial targets hepatoma or those tumors with liver metastasis (NCT00882180). Two other trials are targeting melanoma via immuno-therapeutic approaches, where siRNA-transfected (in one case, mRNA) dendritic cells are being utilized (NCT 00672542 and NCT00929019).
22.9 Conclusions
Given the hurdles faced by gene therapy in the past as an adjuvant therapeutic avenue against tumors, a somewhat different set of obstacles need to be overcome by RNAi drugs, primarily targeted delivery to the tumor (in a systemic application mode). However, the pre-clinical studies in animal models and results from the ongoing clinical trials (listed above) indicate the promise of RNAi as a potential adjuvant therapy against both malignant brain tumors as well as against other solid tumors. These initial success stories with systemic delivery clearly indicate that both research and clinical communities have bypassed the threshold problem of targeted delivery of therapeutic doses of RNAi to the tumor site. Studies with miRNA are already indicating their utility as a molecular tool for diagnostic screening and staging of tumors in the pathology laboratory, which are currently supplementing the traditional immune-histological methods. Thus, after a long pause after the discovery at the bench, these miniscule RNA entities are finally displaying their usefulness as a clinical tool for both diagnostic and therapeutic purposes.
References
Amarzguioui M, Lundberg P, Cantin E, et al. Rational design and in vitro and in vivo delivery of Dicer substrate siRNA. Nat Protoc. 2006;1:508–17.
Birchler JA, Kavi HH. Molecular biology. Slicing and dicing for small RNAs. Science. 2008;320:1023–4.
Boado RJ. RNA interference and nonviral targeted gene therapy of experimental brain cancer. NeuroRx. 2005;2:139–50.
Boado RJ. Blood-brain barrier transport of non-viral gene and RNAi therapeutics. Pharm Res. 2007;24:1772–87.
Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–3.
Bumcrot D, Manoharan M, Koteliansky V, et al. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol. 2006;2:711–9.
Cardoso ALC, Simoes S, de Almeida LP, et al. SiRNA detivery by a transferrin-associated lipid-based vector: a non-viral strategy to mediate gene silencing. J Gene Med. 2007;9:170–83.
Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33.
Chang K, Elledge SJ, Hannon GJ. Lessons from Nature: microRNA-based shRNA libraries. Nat Methods. 2006;3:707–14.
Conti A, Aguennouz M, La Torre D, et al. MiR-21 and 221 upregulation and miR-181b downregulation in human grade II-IV astrocytic tumors. J Neurooncol. 2009;93:325–32.
Corsten MF, Miranda R, Kasmieh R, et al. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994–9000.
Cullen BR. Enhancing and confirming the specificity of RNAi experiments. Nat Methods. 2006a;3:677–81.
Cullen BR. Induction of stable RNA interference in mammalian cells. Gene Ther. 2006b;13:503–8.
Davis ME, Zuckerman JE, Choi CH, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–70.
Dong WL, Hu J, Hu SY, et al. RNA interference affects tumorigenicity and expression of insulin-like growth factor-1, insulin-like growth factor-1 receptor, and basic fibroblast growth factor-2 in rat C6 glioma cells. Neural Regen Res. 2009;4:597–605.
Elbashir SM, Harborth J, Lendeckel W, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–8.
Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11.
Fountaine TM, Wood MJ, Wade-Martins R. Delivering RNA interference to the mammalian brain. Curr Gene Ther. 2005;5:399–410.
Gabriely G, Wurdinger T, Kesari S, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–80.
Gillespie DL, Whang K, Ragel BT, et al. Silencing of hypoxia inducible factor-1alpha by RNA interference attenuates human glioma cell growth in vivo. Clin Cancer Res. 2007;13:2441–8.
Goga A, Benz C. Anti-oncomir suppression of tumor phenotypes. Mol Interv. 2007;7:199–202.
Gogineni VR, Nalla AK, Gupta R, et al. Radiation-inducible silencing of uPA and uPAR in vitro and in vivo in meningioma. Int J Oncol. 2010;36:809–16.
Gondi CS, Dinh DH, Klopfenstein JD, et al. MMP-2 downregulation mediates differential regulation of cell death via ErbB-2 in glioma xenografts. Int J Oncol. 2009;35:257–63.
Gondi CS, Lakka SS, Dinh DH, et al. Intraperitoneal injection of a hairpin RNA-expressing plasmid targeting urokinase-type plasminogen activator (uPA) receptor and uPA retards angiogenesis and inhibits intracranial tumor growth in nude mice. Clin Cancer Res. 2007;13:4051–60.
Guo S, Kemphues KJ. Par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–20.
Guo D, Wang B, Han F, et al. RNA interference therapy for glioblastoma. Expert Opin Biol Ther. 2010;10:927–36.
He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33.
Heidel JD, Hu S, Liu XF, et al. Lack of interferon response in animals to naked siRNAs. Nat Biotechnol. 2004;22:1579–82.
Hubbell HR, Boyer JE, Roane P, et al. Cyclic AMP mediates the direct antiproliferative action of mismatched double-stranded RNA. Proc Natl Acad Sci USA. 1991;88:906–10.
Jackson AL, Burchard J, Schelter J, et al. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–87.
Jain KK. Use of nanoparticles for drug delivery in glioblastoma multiforme. Expert Rev Neurother. 2007;7:363–72.
Kang CS, Zhang ZY, Jia ZF, et al. Suppression of EGFR expression by antisense or small interference RNA inhibits U251 glioma cell growth in vitro and in vivo. Cancer Gene Ther. 2006;13:530–8.
Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8.
Kargiotis O, Chetty C, Gondi CS, et al. Adenovirus-mediated transfer of siRNA against MMP-2 mRNA results in impaired invasion and tumor-induced angiogenesis, induces apoptosis in vitro and inhibits tumor growth in vivo in glioblastoma. Oncogene. 2008;27:4830–40.
Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–16.
Kim DH, Behlke MA, Rose SD, et al. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–6.
Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165–73.
Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173–84.
Krol J, Krzyzosiak WJ. Structural aspects of microRNA biogenesis. IUBMB Life. 2004;56:95–100.
Krutzfeldt J, Rajewsky N, Braich R, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9.
Lau NC, Seto AG, Kim J, et al. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–7.
Lavon I, Zrihan D, Granit A, et al. Gliomas display a microRNA expression profile reminiscent of neural precursor cells. Neuro Oncol. 2010;12:422–33.
Lesniak MS. Novel advances in drug delivery to brain cancer. Technol Cancer Res Treat. 2005;4:417–28.
Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–41.
Liu X, Fortin K, Mourelatos Z. MicroRNAs: biogenesis and molecular functions. Brain Pathol. 2008;18:113–21.
Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109.
Lu XM, Zhao P, Zhang CZ, et al. Analysis of miR-221 and p27 expression in human gliomas. Mol Med Rep. 2009;2:651–6.
Malecova B, Morris KV. Transcriptional gene silencing through epigenetic changes mediated by non-coding RNAs. Curr Opin Mol Ther. 2010;12:214–22.
Malzkorn B, Wolter M, Liesenberg F, et al. Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 2010;20:539–50.
Mathupala SP, Guthikonda M, Sloan AE. RNAi based approaches to the treatment of malignant glioma. Technol Cancer Res Treat. 2006;5:261–9.
Mathupala SP, Mittal S, Guthikonda M, et al. MicroRNA and brain tumors: a cause and a cure? DNA Cell Biol. 2007;26:301–10.
Meltzer PS. Cancer genomics: small RNAs with big impacts. Nature. 2005;435:745–6.
Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–20.
Myslinski E, Ame JC, Krol A, et al. An unusually compact external promoter for RNA polymerase III transcription of the human H1RNA gene. Nucleic Acids Res. 2001;29:2502–9.
Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2:279–89.
Nelson P, Kiriakidou M, Sharma A, et al. The microRNA world: small is mighty. Trends Biochem Sci. 2003;28:534–40.
Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673–8.
Pardridge WM. Intravenous, non-viral RNAi gene therapy of brain cancer. Expert Opin Biol Ther. 2004;4:1103–13.
Pardridge WM. shRNA and siRNA delivery to the brain. Adv Drug Deliv Rev. 2007;59:141–52.
Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA. 2005;11:1753–61.
Pirollo KF, Chang EH. Targeted delivery of small interfering RNA: approaching effective cancer therapies. Cancer Res. 2008;68:1247–50.
Preall JB, He Z, Gorra JM, et al. Short interfering RNA strand selection is independent of dsRNA processing polarity during RNAi in Drosophila. Curr Biol. 2006;16:530–5.
Pu PY, Kang CS, Zhang ZY, et al. Downregulation of PIK3CB by siRNA suppresses malignant glioma cell growth in vitro and in vivo. Technol Cancer Res. 2006;5:271–80.
Pu P, Zhang Z, Kang C, et al. Downregulation of Wnt2 and beta-catenin by siRNA suppresses malignant glioma cell growth. Cancer Gene Ther. 2009;16:351–61.
Reynolds A, Leake D, Boese Q, et al. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–30.
Rolle K, Nowak S, Wyszko E, et al. Promising human brain tumors therapy with interference RNA intervention (iRNAi). Cancer Biol Ther. 2010;9:396–406.
Rose SD, Kim DH, Amarzguioui M, et al. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33:4140–56.
Sasayama T, Nishihara M, Kondoh T, et al. MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer. 2009;125:1407–13.
Saydam O, Glauser DL, Heid I, et al. Herpes simplex virus 1 amplicon vector-mediated siRNA targeting epidermal growth factor receptor inhibits growth of human glioma cells in vivo. Mol Ther. 2005;12:803–12.
Seitz H. Redefining microRNA targets. Curr Biol. 2009;19:870–3.
Seno T, Harada H, Kohno S, et al. Downregulation of SPARE expression inhibits cell migration and invasion in malignant gliomas. Int J Oncol. 2009;34:707–15.
Silva JM, Li MZ, Chang K, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–8.
Siolas D, Lerner C, Burchard J, et al. Synthetic shRNAs as potent RNAi triggers. Nat Biotechnol. 2005;23:227–31.
Taft RJ, Glazov EA, Cloonan N, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41:572–8.
Takenaga K, Nygren J, Zelenina M, et al. Modified expression of Mts1/S100A4 protein in C6 glioma cells or surrounding astrocytes affects migration of tumor cells in vitro and in vivo. Neurobiol Dis. 2007;25:455–63.
Uchida H, Tanaka T, Sasaki K, et al. Adenovirus-mediated transfer of siRNA against survivin induced apoptosis and attenuated tumor cell growth in vitro and in vivo. Mol Ther. 2004;10:162–71.
Ulbricht U, Eckerich C, Fillbrandt R, et al. RNA interference targeting protein tyrosine phosphatase zeta/receptor-type protein tyrosine phosphatase beta suppresses glioblastoma growth in vitro and in vivo. J Neurochem. 2006;98:1497–506.
Verdel A, Vavasseur A, Le Gorrec M, et al. Common themes in siRNA-mediated epigenetic silencing pathways. Int J Dev Biol. 2009;53:245–57.
Wang XL, Xu RZ, Wu XM, et al. Targeted systemic delivery of a therapeutic siRNA with a multifunctional carrier controls tumor proliferation in mice. Mol Pharmaceut. 2009;6:738–46.
Watanabe T, Totoki Y, Toyoda A, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–43.
Wyszko E, Rolle K, Nowak S, et al. A multivariate analysis of patients with brain tumors treated with atn-RNA. Acta Pol Pharm. 2008;65:677–84.
Xia CF, Zhang Y, Boado RJ, et al. Intravenous siRNA of brain cancer with receptor targeting and avidin-biotin technology. Pharm Res. 2007;24:2309–16.
Xie FY, Woodle MC, Lu PY. Harnessing in vivo siRNA delivery for drug discovery and therapeutic development. Drug Discov Today. 2006;11:67–73.
Xu P, Qiu MZ, Zhang ZY, et al. The oncogenic roles of Notch1 in astrocytic gliomas in vitro and in vivo. J Neurooncol. 2010;97:41–51.
Zhang LY, Alizadeh D, Van Handel M, et al. Stat3 Inhibition Activates Tumor Macrophages and Abrogates Glioma Growth in Mice. Glia. 2009b;57:1458–67.
Zhang BB, Gu F, She CH, et al. Reduction of Akt2 inhibits migration and invasion of glioma cells. Int J Cancer. 2009a;125:585–95.
Zhang JX, Han L, Ge YL, et al. MiR-221/222 promote malignant progression of glioma through activation of the Akt pathway. Int J Oncol. 2010;36:913–20.
Zhang Y, Zhang YF, Bryant J, et al. Intravenous RNA interference gene therapy targeting the human epidermal growth factor receptor prolongs survival in intracranial brain cancer. Clin Cancer Res. 2004;10:3667–77.
Zhao P, Wang C, Fu Z, et al. Lentiviral vector mediated siRNA knock-down of hTERT results in diminished capacity in invasiveness and in vivo growth of human glioma cells in a telomere length-independent manner. Int J Oncol. 2007;31:361–8.
Zhou X, Ren Y, Moore L, et al. Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest. 2010;90:144–55.
Zukiel R, Nowak S, Wyszko E, et al. Suppression of human brain tumor with interference RNA specific for tenascin-C. Cancer Biol Ther. 2006;5:1002–7.
Acknowledgments
Research support for S.P. Mathupala was provided by a grant from the National Cancer Institute/National Institute of Health (CA 116257), the Fund for Medical Research and Education (FMRE), Wayne State University, and a gift from the Marvin E. Klein, M.D., Charitable Trust. A.E. Sloan is supported by grants from the National Cancer Institute/National Institute of Health (KO8 101954) and the Case Western Reserve University School of Medicine.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer Netherlands
About this chapter
Cite this chapter
Mathupala, S.P., Mittal, S., Guthikonda, M., Sloan, A.E. (2011). RNAi-based Approaches to the Treatment of Brain Tumors. In: Cho, W. (eds) MicroRNAs in Cancer Translational Research. Springer, Dordrecht. https://doi.org/10.1007/978-94-007-0298-1_22
Download citation
DOI: https://doi.org/10.1007/978-94-007-0298-1_22
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-007-0297-4
Online ISBN: 978-94-007-0298-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)