Abstract
Developmental cues and environmental signals remodel the chromatin structure, thus affecting various processes, including flowering time, imprinting, floral development, and biotic and abiotic stress responses in plants. Chromatin remodeling through histone tail post-translational modifications, DNA methylation, and ATP-dependent nucleosome reorganization represents a ubiquitous mechanism to regulate gene expression. Most of the epigenetic and epigenomic studies for the regulation of gene expression in response to developmental and environmental stimuli have been carried out in Arabidopsis. Although genetic modifications have been used for crop improvement, however, the epigenetic modifications are at their beginning. In this chapter, we summarize the roles of chromatin-remodeling mechanisms in response to environmental stimuli and discuss their potential for crop improvement.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
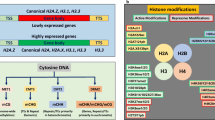
In eukaryotic cell nuclei, genomic DNA is packaged into a highly organized nucleoprotein complex known as chromatin. The fundamental unit of chromatin is the nucleosome, which is composed of ~147 base pairs of DNA wrapped around a core of eight histone molecules (two copies of each of the histones H2A, H2B, H3 and H4). Nucleosomes are not simply static structural units, but are rather dynamic. Nucleosomes can be moved, stabilized/destabilized, and disassembled/reassembled at particular genome locations in response to specific environmental signals or developmental cues. The resulting dynamic of the chromatin structure directly modulates the DNA accessibility, thus regulating all DNA-template processes (i.e., transcription, DNA replication, DNA repair, recombination, transposition, or chromosome segregation) and affecting various processes in plants such as root growth, flowering timing, floral organogenesis, gametophyte or embryo formation, as well as the response to pathogens or environmental changes (Berr et al. 2011). However, not all genes are active at all times. Therefore, cells use several mechanisms along the genome to alter the chromatin structure and the properties of a nucleosome in order to specifically control gene expression. Regulation of gene expression within the chromatin context is controlled by different mechanisms, including nucleosome assembly, ATP-dependent nucleosome reorganization, DNA methylation, and post-translational covalent histone modifications (e.g., acetylation, ubiquitination, methylation, phosphorylation, sumoylation).
Different epigenetic regulators are controlling all the above mechanisms, and the changes in these regulators can influence gene expression of a particular gene or set of genes, while the underlying DNA sequence remains identical (Jablonka and Raz 2009). Most of these changes are reversible developmental effects, and they are part of molecular processes encoding phenotypic plasticity in response to environmental variation (Richards et al. 2010) (Fig. 1). However, inheritable chromatin changes have also been reported (Jablonka and Raz 2009). At this point, it is important to clarify that those modifications which are not inheritable are not included in epigenetics as according to the definition of epigenetics, these modifications must be inheritable (mitotic and/or meiotic). Thus, we can broadly classify these modifications into nonheritable chromatin modifications (chromatin modifications that are the result of processes such as DNA repair or phosphorylation of serine 10 of histone H3, which are observed only at specific times during the cell cycle and are, therefore, unlikely to encode epigenetic information (Springer 2013)) and heritable chromatin modifications. The heritable chromatin modifications can further be classified into mitotically transmissible modifications that are reset in the next generation and meiotically transgenerational chromatin modifications that are inherited/transmitted to the following generations. The mitotically stable epigenetic marks, which accompany development, are mainly histone modification, but there are some examples of involvement of DNA methylation as well (Lauria et al. 2004; Zemach et al. 2010; Khan et al. 2013). However, DNA methylation can exhibit a relatively stable pattern of inheritance even over hundreds of years (Cubas et al. 1999; Manning et al. 2006). Because heritability determines the potential of changes or variations of a trait, it is essential to determine the degree of heritability of epigenetic modifications, their impact on given ecologically important traits (Fisher 1930; Falconer 1996), their role in individual adaptation to changing environment (Visser 2008; Hoffmann and Sgrò 2011) and ultimately in crop improvement.
Gene expression regulation through genetic and epigenetic modifications in natural population in response to environmental stimuli. The genetic and epigenetic changes may act alone or together and regulate the gene expression, which may result in a heritable and nonheritable change and may lead to a survival and/or crop improvement
The heritable epigenetic mutations, i.e., epimutations/epialleles, can be classified into three categories on the basis of relative dependence on the genotype. Pure epialleles constitute the first category, which is solely epigenetic, meaning that they are independent of the genetic variations. The second category is facilitated epialleles, which are not fully dependent on genetic variation, although they are linked and even caused by a genetic variant. The example for this kind of epialleles is DNA methylation spreading into a gene after the insertion of a neighboring transposon where this methylation of the gene is maintained across generations even after the facilitating transposon is excised or segregated away, meaning they could be partly attributable to both genetic and epigenetic differences. The third category is obligate epialleles, which are directly determined by genetic variants and co-segregate with these methylation variants (Woo et al. 2007). For example, methylation of a gene may be dependent on the presence or absence of a nearby transposon. There are many examples of epimutations that provide evidence that the genetic events like transposon insertions, duplications, and other structural rearrangements might trigger the chromatin remodeling which results in epigenetic control for particular haplotypes (Martin et al. 2009; Durand et al. 2012).
Increasing world population and changing climate demand to improve crop species. Although Mendelian-based genetic approaches and DNA sequence variation to select and improve crop varieties capture a substantial portion of heritable variation, dissecting epigenetic mechanisms could lead to more efficient improvement of crops. A crop improvement strategy includes the response to environmental stimuli, and the potential role of chromatin modifications in biotic and abiotic stresses has been recently reported (Chinnusamy and Zhu 2009; Kim et al. 2010; Berr et al. 2012). In view of global climate change, improving our knowledge of epigenetic regulation could have a significant impact on breeding for increased stress tolerance. In this chapter, we summarize the recent advances in epigenetic regulation in response to stress and discuss the potential of epigenetic regulatory mechanisms for crop improvement.
Histone Modifications
The N-terminal tails of histone are subjected to different covalent posttranslational modifications (PTMs) through the addition of acetyl or methyl groups and small peptide such as ubiquitin. Numerous PTMs may occur on one histone or different histones from the same nucleosome. Histone modifications, particularly acetylation/deacetylation and methylation/demethylation, epigenetically regulate the response to various stresses (Table 1). Here below we summarize the current knowledge of the enzymes responsible for histone modifications and involvement to environmental stimuli.
Histone Acetylation
Histone acetylation is linked to transcriptional activation in euchromatin and also related to DNA replication, recombination, and repair (Allfrey et al. 1964; Allis et al. 1985; Unnikrishnan et al. 2010). Acetylation by addition of an acetyl group to histone lysine (K) residues neutralizes the positive charge of lysine and therefore modifies the histone-DNA interaction, relieving DNA from its condensate state and exposing it to the transcriptional machinery. In Arabidopsis, lysine residues of histone H3 (K9, K14, K18, K23, and K27) and H4 (K5, K8, K12, K16, and K20) are subjected to acetylation modifications (Earley et al. 2007; Zhang et al. 2007). Histone acetyltransferases (HATs) are divided into four main classes based on the sequence homology with yeast and mammalian HATs and mode of action: GNAT (GCN5-related N-terminal acetyltransferases), MYST (MOZ, Ybt2, Sas2, Tip60 like), CBP/p300 (CREB-binding protein), and TAF1/TAFII250 families (Sterner and Berger 2000). AtGCN5 (GENERAL CONTROL NON-REPRESSIBLE 5) was shown to acetylate H3 in vitro (Earley et al. 2007). Atgcn5 mutant showed reduced levels of global H3 acetylation (Bertrand et al. 2003), particularly on H3K14 and H3K27 at certain gene loci (Benhamed et al. 2006). AtGCN5 was found to be involved in environmental responses (i.e., cold), along with other development pathways (Vlachonasios et al. 2003). AtGCN5 not only interacts with Arabidopsis Ada2 homologues AtADA2a and AtADA2b in vitro but also acetylates AtADA2a/b (Stockinger et al. 2001; Mao et al. 2006). Atada2b mutants showed a hypersensitive response to salt and abscisic acid (ABA) and altered response to low-temperature stress (Hark et al. 2009). H3 and H4 acetylation was found reduced on COR6.6 (COLD-RESPONSIVE 6.6), RAB18 (RESPONSIVE TO ABA 18), and RD29b (RESPONSIVE TO DESSICATION 29b) genes under salt stress in Atada2b mutants (Kaldis et al. 2011). The cold-induced transcription factor CBF1 (C-repeat/DRE BINDING FACTOR 1) interacts with AtADA2 and AtGCN5 (Mao et al. 2006), and they positively regulate the expression of cold-inducible genes during cold stress (Pavangadkar et al. 2010). This suggests that CBF is recruiting GCN5-containing activator complexes to activate the cold-responsive genes. SGF29 (SAGA-ASSOCIATED FACTOR 29), another component of GCN5-containing complexes in yeast, has two orthologs in Arabidopsis AtSGF29a and AtSGF29b. Atsgf29a mutants showed increased tolerance to salt stress (Kaldis et al. 2011), whereas Atada2b mutants were hypersensitive. This suggests that different components of GCN5-containing HAT complexes may play a different role in plant stress tolerance. Elongator HAT complex is involved in ABA signaling, drought, and oxidative stress responses in Arabidopsis (Chen et al. 2006; Zhou et al. 2009). AtABO1/ELO2 (ABA OVERLAY SENSITIVE 1), an Elp1 homologue of yeast, was identified in a genetic screen of drought-resistant mutant (Chen et al. 2006). Atabo1/elo2/elp1 mutant showed ABA hypersensitivity in germination and seedling growth and also showed drought- and oxidative-resistant phenotype (Chen et al. 2006). Mutation in the genes coding for the core subcomplex subunits AtABO1/ELO2/ELP1 and AtELP2 (ELONGATOR SUBUNIT 2), but not in the genes coding for accessory subcomplex subunits AtELP4 (ELONGATOR SUBUNIT 4) and AtELP6 (ELONGATOR SUBUNIT 6), caused stomatal closing to be hypersensitive to ABA (Zhou et al. 2009). Furthermore, these single mutants showed resistance to oxidative stress and to CsCl compared to the wild type plant (Zhou et al. 2009). AtELP2 and AtELP3 (ELONGATOR SUBUNIT 3) were also required for both basal immunity and effector-triggered immunity (ETI), but not for systemic acquired resistance (SAR) (DeFraia et al. 2010; DeFraia et al. 2013). These results suggest that elongators play crucial roles in ABA signaling pathways and abiotic and biotic stress responses. AtTAF1/HAF2 was shown to be required for light-regulated gene expression (Benhamed et al. 2006). Together, HATs from GNAT family are involved in both biotic and abiotic stresses. However, involvement of HATs from CBP, MYST, and TAF1 classes in biotic and abiotic stresses response is still lacking in Arabidopsis.
Until now, the knowledge of HATs in the field crops is very limited. Eight HATs have been identified in rice and divided into four families: GNAT (OsHAG702, OsHAG703, and OsHAG704), MYST (OsHAM701), CBP/p300 (OsHAC701, OsHAC703, OsHAC704), and TAF1/TAFII250 (OsHAF701) (Liu et al. 2012). Rice HATs respond to ABA, salicylic acid (SA), and various abiotic stresses, i.e., cold, heat, drought, and salt (Liu et al. 2012; Fang et al. 2014). An increase in transcription of OsHAG702, OsHAG703, OsHAC701, OsHAC703, and OsHAM701 was observed with the exogenous application of ABA, whereas OsHAC703 and OsHAC704 transcript levels were reduced with SA application. In addition, OsHAC701, OsHAC703, OsHAC704, and OsHAG703 transcripts were induced by salt and depressed by cold exposure (Liu et al. 2012). Furthermore, H3 (K9, K18, and K27) and H4 (K5) acetylation and transcripts of OsHAG703, OsHAM701, OsHAC703, and OsHAF701 were found increased after drought stress in rice seedlings (Fang et al. 2014). Barley HATs belonging to GNAT (HvGCN5 and HvELP3) and MYST (HvMYST) families respond to ABA (Papaefthimiou et al. 2010). The expression of HvGCN5, HvELP3, and HvMYST was induced with exogenous application of ABA (Papaefthimiou et al. 2010). Together, these studies showed that HATs from all the four families are involved in different stresses in field crops. Therefore, the understanding of molecular mechanism may play an important role to cope with various stresses in field crops. It is hoped that this will eventually lead to a long-term improvement of stress tolerance in field crops, which is important for food security.
Histone Deacetylation
The homeostatic balance of histone acetylation is maintained through the antagonistic action between HATs and histone deacetylases (HDACs). In Arabidopsis, HDACs are classified into three families: the reduced potassium dependency 3 (RPD3/HDA1) superfamily, the HD2-like family, and the silent information regulator 2 (SIR2) family (Imhof et al. 1997; Sterner and Berger 2000; Strahl and Allis 2000). Functional analysis has demonstrated that HDA1 class of HDACs is involved in both biotic and abiotic stresses response in Arabidopsis. Overexpression of AtHDA19 leads to increased expression of a gene that integrates jasmonic acid (JA) and ethylene (ET) signaling pathway, i.e., ERF1 (ETHYLENE RESPONSIVE FACTOR 1) and PR (PATHOGENESIS RELATED) genes. This results in increased plant resistance to Alternaria brassicicola (Zhou et al. 2005). It is also reported that AtHDA19 (HISTONE DEACETYLASE 19) is involved in the repression of SA-mediated defense responses. Athda19 mutant has increased SA contents and the expression of PR genes, resulting in enhanced resistance to Pseudomonas syringae (Choi et al. 2012). AtHDA19 interacts with WRKY38 (WRKY TRANSCRIPTION FACTOR 38) and WRKY62 (WRKY TRANSCRIPTION FACTOR 62) transcriptional activator to regulate plant basal defense responses (Kim et al. 2008). AtHDA6 (HISTONE DEACETYLASE 6), another HDAC, is also involved in JA response, and Ataxe5/hda6 showed reduced expression of JA-responsive genes PDF1.2 (PLANT DEFENSIN 1.2), VSP2 (VEGETATIVE STORAGE PROTEIN 2), JIN1 (JASMONATE INSENSITIVE 1), and ERF1 (Wu et al. 2008). Ataxe5/hda6 mutants also showed reduced freezing tolerance (To et al. 2011), indicating that AtHDA6 has a critical role in freezing tolerance. The expression of ABA and abiotic stress-responsive genes ABI1 (ABA INSENSITIVE 1), ABI2 (ABA INSENSITIVE 2), KAT1 (POTASSIUM CHANNEL IN ARABIDOPSIS THALIANA 1), KAT2 (POTASSIUM CHANNEL IN ARABIDOPSIS THALIANA 2), DREB2A (DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN 2A), RD29A (RESPONSIVE TO DESSICATION 29A), and RD29B was decreased in Ataxe5/hda6 mutant or AtHDA6-RNAi plants (Chen et al. 2010). Similarly, Athda19 mutant also showed a hypersensitive response to ABA and salt stress (Chen and Wu 2010). This suggests that AtHDA19 and AtHDA6 may play a redundant role in modulating ABA and salt stress response. Moreover, AtHDA19 and AtHDA6 play a crucial role in responses to biotic and abiotic stresses. AtHDA2C, an HD2-type HDAC, was also shown to be involved in ABA and salt stress response. Overexpression of AtHD2C in transgenic plants showed enhanced tolerance to salt and drought stress and ABA-insensitive phenotype (Sridha and Wu 2006). Conversely, Athd2c mutant showed a hypersensitive response to ABA and NaCl and decreased tolerance to salt stress (Luo et al. 2012). Furthermore, AtHD2C interacts with AtHDA6 (Luo et al. 2012), suggesting that AtHD2C may functionally associate with AtHDA6 to ABA and salt stress responses and may be a part of HDAC complexes to regulate gene expression through histone modifications. Arabidopsis SIRTUIN 2 (AtSRT2), an SIR2 HDAC expression, is downregulated upon Pseudomonas syringae pv. tomato (PstDC 3000) infection. AtSRT2 suppresses the expression of SA biosynthesis genes PAD4 (PHYTOALEXIN-DEFICIENT 4), EDS5 (ENHANCED DISEASE SUSCEPTIBILITY 5), and SID2 (SPAC24B11.11C), thereby supressing SA production and expression of defense-regulated genes (Wang et al. 2010).
HDAC and its involvement in biotic and abiotic stresses have also been reported in cereals. Rice has 19 genes coding for HDAC (Hu et al. 2009), which may play an important role in regulating various stress responses. OsHDA705, OsHDT701, and OsHDT702 transcripts were found affected by SA, JA, and ABA, whereas OsHDA714, OsSRT701, and OsSRT702 expression is modulated by cold, mannitol, and salt (Fu et al. 2007). In OsSRT1-RNAi transgenic rice, H3K9 acetylation and H3K9 dimethylation (H3K9me2) levels were decreased and increased, respectively, leading to H2O2 production, DNA fragmentation, cell death, and lesion-mimicking plant hypersensitive responses during incompatible interactions with pathogens. In contrast, OsSRT1 overexpression showed an enhanced tolerance to oxidative stress (Huang et al. 2007). Overexpression of OsHDT701, a plant-specific HD2 HDAC, leads to decreased level of H4 acetylation on flowering and defense-related genes and enhanced susceptibility to the Magnaporthe oryzae and Xanthomonas oryzae pv. oryzae (Xoo) pathogens (Li et al. 2011; Ding et al. 2012). In contrast, silencing of OsHDT701 showed increased levels of H4 acetylation and increased transcription of pattern recognition receptor (PRR) and defense-related genes, elevated generation of reactive oxygen species, as well as enhanced resistance to both M. oryzae and Xoo (Ding et al. 2012). HvHDAC2-1 and HvHDAC2-2 genes, HD2-type HDAC from barley, were found to respond to JA, ABA, and SA treatments, implying an association of these barley genes with plant resistance to biotic and abiotic stresses (Demetriou et al. 2009). NtHD2a and NtHD2b genes, HD2-type HDAC from tobacco, were found to work as inhibitors of cryptogein-induced cell death (Bourque et al. 2011). Together, HD2-type HDAC from rice and barley carries the same function, suggesting a common function among species for HDAC homologues but also possible species-specific functional diversification, in response to stress. The involvement of HDACs in biotic and abiotic stresses response in agronomically important crops and their underlying molecular mechanism is of utmost importance for sustainable crop improvement.
Histone Methylation
Histone methylation plays an essential role in diverse biological processes ranging from transcriptional regulation to heterochromatin formation. Methylation of histone can occur on lysine (K) or arginine (R) residues leading to either transcriptional activation or repression. Histone methylation not only occurs at different residues (K and R) and distinct sites (e.g., K4, K9, K27, K36, R2, and R17 of H3 and K20 and R3 of H4, etc.) but also differs in the number of methyl groups added (mono-, di-, and tri-methylated). Methylation of lysine residues does not affect their net charge but elevates the hydrophobicity nature of the side chain and may alter intra- or intermolecular interactions or create new binding surfaces for proteins that bind preferentially to the methylated domains (Liu et al. 2010). Indeed, the arginine residue, after the addition of the methyl group, changes its shape and removes a potential hydrogen bond donor as well (Bedford and Clarke 2009). Mostly the studies have been done on histone modifications only at individual stress-induced plant genes. Very few studies with genome-wide histone methylation analysis have been reported. van Dijk et al. (2010) have studied genome-wide analysis of the histone H3 lysine 4 mono-, di-, and tri-methylation (H3K4me1, H3K4me2, H3K4me3, respectively) patterns in chromatin isolated from Arabidopsis rosette leaves before and after dehydration stress. Genome-wide transcript patterns in watered and dehydration-stressed plants were compared in this study. The presence of the H3K4me1, H3K4me2, and H3K4me3 marks is predominantly located on genes, and the distribution of H3K4me1 and H3K4me2 is higher than H3K4me3. Interestingly, H3K4me1, H3K4me2, and H3K4me3 patterns display different dynamics and specific patterns at upregulated, downregulated, and unaffected genes during the response to dehydration stress. A modest change in H3K4me2 and H3K4me1 levels was found at a subset of known stress response genes, but the H3K4me3 abundance over gene bodies changed more dramatically at genes whose transcript levels increased or decreased during dehydration. The different behaviors of each methylation mark during the response process illustrate that each mark plays a distinct role in the transcriptional response of implicated genes. In a recent study, genome-wide profiling of histone H3K4-tri-methylation of 25-day-old rice plants under dehydration conditions was done. This analysis uncovered a positive correlation between H3K4me3 accumulation and the expression levels of some drought-responsive genes during dehydration. This correlation could be extended to genes involved in stress-related metabolite and hormone signaling pathways (Zong et al. 2012). These genome-wide histone modification studies help broaden our knowledge on whole genome scale and indicate a need to study histone modifications on a genome-wide level in response to other abiotic stresses as well.
Histone Lysine Methylation
Covalent addition of one, two, or three methyl groups (me1, me2, or me3) mainly occurs on H3K4, H3K9, H3K27, H3K36, and H4K20, and this function is exerted through histone methyltransferases (HMTs). All known plant HMTs have a so-called SET [from the initially identified Drosophila HMTs: Suppressor of variegation (Su(var)3-9), Enhancer of Zeste (E(z)), and Trithorax (TRX)] catalytic domain, an evolutionarily conserved sequence of 130–150 amino acids in length. SET Domain Group (SDG) proteins are classified into three subgroups: Su(var)3-9, Enhancer of Zeste (E(z)), and Trithorax (TRX). These subgroups have been shown to establish different chromatin marks, leading to different impacts on transcription. SDGs of the ASH1 and TRX subgroups primarily belong to the Trithorax group (TrxG) and are responsible for methylation on H3K36 and/or H3K4, which are associated with transcriptional activation (Agger et al. 2008; Liu et al. 2010). The E(z) subgroup SDGs catalyze H3K27 methylation associated with transcriptional gene silencing. H3K27 can be mono-, di-, and tri-methylated and seems to be one of the major gene silencing mechanisms in Arabidopsis because ~17 % of the coding genes were marked with H3K27me3 (Turck et al. 2007). Classically and conservatively, the Su(var)3-9 subgroup SDGs potentially show an H3K9 methyltransferase activity and are associated with inactive genes located in a euchromatic region and within highly condensed constitutive heterochromatin (Ng et al. 2007). SDG proteins have been involved in diverse biological processes, including flowering time regulation, floral organogenesis, leaf morphogenesis, parental imprinting, and seed development (Liu et al. 2010; Berr et al. 2011; Shafiq et al. 2014). AtATX1/SDG27, a member of the Trithorax group, is a methyltransferase of H3K4me3. AtATX1 was found to be involved in drought and SA pathway responses (Ding et al. 2011; Berr et al. 2012). Atatx1 mutant displayed larger stomatal apertures, increased transpiration, and decreased tolerance to dehydration stress. AtATX1 is required for the induction of NECD, a gene involved in ABA biosynthesis and deposition of H3K4me3 in response to dehydration stress. AtATX1 can influence gene expression by ABA-dependent as well as ABA-independent pathways (Ding et al. 2011). AtATX1 was described as critical for basal resistance against Pst DC3000, and it regulates the SA-inducible expression of transcriptional factor WRKY70 (Alvarez-Venegas et al. 2006; Berr et al. 2012). AtSDG8, another member of the Trithorax group, is the major H3K36me2/me3 methyltransferase (Xu et al. 2008). AtSDG8 was reported in Pst DC3000-triggered plant defense through the regulation of particular R genes (Palma et al. 2010) and the transcriptional activation of JA/ET signaling-related genes (Berr et al. 2010). Atsdg8 mutant exhibited reduced resistance to Alternaria brassicicola and Botrytis cinerea (Berr et al. 2010). H3K36 methylation on defense-related genes is impaired in Atsdg8 mutant (Berr et al. 2010; Palma et al. 2010), indicating that AtSDG8 mediates the pathogen response by regulating histone methylation of defense-responsive genes. The expression of AtSUVH2, AtSUVH5, AtSUVH6, and AtSUVH8 genes encoding H3K9 methylation decreased in the progenies of salt-stressed plants (Bilichak et al. 2012). In addition, Curly leaf (CLF) gene encoding H3K27 methylation was hypermethylated in the progenies of salt-stressed plants (Bilichak et al. 2012). These results suggest that H3K9 and H3K27 methyltransferases are involved in the plant stress adaptation. Until now, HMT involvement in biotic and abiotic stresses response is very limited in crops. HvTX1, barley TRX-like H3K4 methyltransferase, has been shown to be involved in drought stress. The transcripts of HvTX1 were found increased under drought stress (Shvarts Iu et al. 2010; Papaefthimiou and Tsaftaris 2012b). This suggests that TrxG plays an important role in plant response to environmental stresses. A homologue of polycomb complex subunit from barley HvE(Z) was found to be induced by ABA implying an association with ABA-mediated processes during seed development and stress response (Kapazoglou et al. 2010). Recently, it was shown that 18 genes containing SET domain from maize showed differential expression under salt and drought stress (Qian et al. 2014). Although SET domain proteins are involved in biotic and abiotic stresses in crops, their molecular mechanism is still missing. It is hoped that with emerging new technologies and better understanding of molecular mechanism, the SET domain proteins may have potential for sustainable crop improvement.
Histone Arginine Methylation
Arginine methylation mainly occurs at R2, R8, R17, and R26 of histone H3 and R3 of histone H4 and histone H2A. Arginine methylation can be symmetric or asymmetric and only occurs in mono- and di-methyl states (Aletta et al. 1998; Bedford and Clarke 2009). Arginine methylation is evolutionarily conserved and has been found in fungi, plants, Caenorhabditis elegans, Drosophila melanogaster, and vertebrates (Krause et al. 2007). Arginine methylation is catalyzed by a small group of protein arginine methyltransferases (PRMTs) that harbor a set of four conserved motifs (i.e., I, post-I, II, III) and a THW loop (Katz et al. 2003). Proteins that are arginine methylated play an essential role in transcriptional regulation, DNA repair, signal transduction, nuclear/cytoplasmic shuttling, RNA processing, and formation of silent chromatin (Bedford and Richard 2005; Bedford and Clarke 2009). In mammals, PRMTs are classified into two classes depending on the nature of the modification introduced. Although both type I and type II catalyze arginine monomethylation, they differ in the final type of arginine modification. The type I enzymes result in asymmetrical dimethylarginine, whereas type II enzymes result in symmetrical dimethylarginine (McBride and Silver 2001; Katz et al. 2003; Jelinic et al. 2006). The involvement of arginine methylation in biotic and abiotic stresses is very poorly understood. AtPRMT5/SKB1, a homologue of the human PRMT5 (PROTEIN ARGININE METHYLTRANSFERASE 5), specifically di-methylates symmetrically H4R3 as a type II arginine methyltransferase in Arabidopsis (Deng et al. 2010). Atskb1 mutant displayed salt-hypersensitive phenotype. AtSKB1 suppresses the transcription of stress-responsive genes by increasing the H3R3sm2 (Zhang et al. 2011).
Histone Demethylation
Histone methylation is important for chromatin stability and gene expression and was considered irreversible until the discoveries of demethylases that antagonized or balanced the methylase activities. There are two types of methylases with distinct mechanisms, the lysine-specific demethylases (LSD1) and the Jumonji C (JmjC) domain-containing demethylases. They use different cofactors and act on different substrates to remove methyl groups from methylated lysine residues. LSD1 is catalytically limited to mono- and di-methylated lysine due to the reaction mechanism used to initiate the demethylation (Klose and Zhang 2007). Unlike LSD1, Jmj proteins do not have limitations in their catalytic mechanism and are able to demethylate mono-, di-, and tri-methyl residues (Agger et al. 2008). In Arabidopsis and rice, histone demethylases (HDMs) have been found to be involved in many developmental processes and gene silencing (Noh et al. 2004; Sun and Zhou 2008; Chen et al. 2013; Cui et al. 2013; Shafiq et al. 2014). Although the role of HDMs in stress response is not yet clear, evidences suggest that histone demethylation may be involved in stress responses. Increased level of H3K9/K14ac and H3K4me3 and decreased level of H3K9me2 on ABA-responsive genes (ABI1, ABI2, and RD29B) have been found in Arabidopsis after ABA treatment (Chen et al. 2010), which suggests that some HDMs are working for the demethylation of H3K9 to activate the ABA-responsive genes. Decreased levels of H3K4me1, H3K4me2, and H3K4me3 and downregulation of stress-responsive genes have been reported upon dehydration stress in Arabidopsis (van Dijk et al. 2010). This also suggests that HDMs are modulating the expression of stress-responsive genes. Recent reports describing a putative role of HDMs in stress response are anticipated. Putative plant-specific barley HvPKDM7 histone demethylase was found to be significantly induced by drought stress (Papaefthimiou and Tsaftaris 2012a). Genome-wide analysis of rice showed that a lot of genes were differentially H3K4me3-modified in drought stress (Zong et al. 2012), suggesting that the rice HDMs are involved in stress response.
Other Histone Modifications
Ubiquitination is the covalent attachment of a small (76 amino acids) and highly conserved protein named ubiquitin to the target protein, achieved through the sequential action of the ubiquitin-activating enzyme E1, the ubiquitin-conjugating enzyme E2 (Ubc), and the ubiquitin-protein ligase E3 (Pickart 2001; Smalle and Vierstra 2004). The substrate can remain monoubiquitinated, or the ubiquitin can have several lysine (K) residues that may be substrates themselves for subsequent addition of ubiquitins, resulting in a polyubiquitin chain. H2B monoubiquitination (H2Bub1) in yeast, animals, and Arabidopsis is mainly associated with transcriptional activation (Briggs et al. 2002; Dover et al. 2002; Hwang et al. 2003). AtHUB1, catalyzing H2B monoubiquitination, was reported as a regulatory component of plant defense against necrotrophic fungal pathogens (Dhawan et al. 2009). Athub1 mutant displayed susceptibility to B. cinerea and A. brassicicola. ET and SA but not JA modulate the resistance of Athub1 mutants to necrotrophic fungi. Athub1-6 presents a reduced cell thickness, indicating that HUB1 may regulate resistance by altering plant cell wall-related defense mechanisms (Dhawan et al. 2009). It remains to be explored whether and how H2Bub is involved in plant defense. AtMSI1 (MULTICOPY SUPPRESSOR OF IRAI), a subunit of the Polycomb group (PcG) having H3K27 methylation activity, has been shown to be involved in drought stress (Alexandre et al. 2009). Atmsi1 mutant displayed increased tolerance to drought stress and increased transcripts of stress and ABA-responsive genes, indicating that AtMSI1 suppresses stress-responsive genes in an ABA-dependent manner. Polycomb complex gene homologue from barley HvFIE (FERTILIZATION-INDEPENDENT ENDOSPERM 1) was found to be induced by ABA implying an association with ABA-mediated processes during seed development and stress response (Kapazoglou et al. 2010).
ATP-Dependent Chromatin Remodeling Factors
ATP-dependent chromatin remodeling complexes use the energy of ATP hydrolysis to alter the structure of chromatin for the regulation of gene expression (Vignali et al. 2000). ATP-dependent chromatin remodeling factors have been found to be involved in biotic and abiotic stresses response. ATP-dependent chromatin complexes can be grouped into three classes: the SWI/SNF ATPases, the imitation switch (ISWI) ATPases, and the chromodomain and helicase-like domain (CHD) ATPases. AtCHR12, an SNF/BRAHMA-type (BRM) chromatin remodeling factor, was shown to be involved in plant growth response to adverse environmental conditions (Mlynárová et al. 2007). Under drought, heat, and salinity stress, AtCHR12 overexpressing plants exhibited an arrested growth of normally active primary buds as well as reduced growth of primary stem. Atchr12 mutant plants displayed less growth arrest than the wild type when exposed to stress. However, the molecular mechanism of how the AtCHR12 is involved in growth arrest under adverse environments is not clear. AtBRM, an SWI2/SNF2 chromatin remodeling ATPase, has been demonstrated to be involved in drought stress (Han et al. 2012). Atbrm mutant showed increased drought tolerance and regulated the expression of ABA-responsive gene AB15. Furthermore, nucleosomes were found destabilized upon the loss of BRM activity, indicating that BRM regulates stress responses through the regulation of nucleosome stability of AB15. Using wounding as stimulus, SPLAYED (SYD), a closest homologue of BRM, was shown to be required for the basal as well as stress-induced expression of genes (PDF1.2, VSP2, and MYC2) working downstream of JA/ET signaling pathways (Walley et al. 2008). These results indicate that ATP-dependent chromatin remodeling complexes are playing a crucial role in stress responses, thus influencing plant innate immunity and tolerance. However, the knowledge about the ATP-dependent chromatin remodeling factors in field crops is very limited. After UVB treatment of maize plants, the enrichment of SWI2/SNF2 was found at target genes, implying the involvement of chromatin remodeling factors in abiotic stress responses (Casati et al. 2008). It is expected that by exploiting the rice, maize, and Brachypodium genomes, chromatin remodeling complexes and their association with biotic and abiotic stresses will be studied, thus improving crop production.
DNA Methylation
DNA methylation refers to a chemical modification of genomic DNA by the addition/attachment of a methyl (−CH3) group to specific nucleotide bases, which could be cytosine or adenine. It occurs most commonly on cytosine base leading to a 5-methylcytosine. It is conserved in major eukaryotic groups, i.e., plants, animals, and fungi, with few exceptions (Goll and Bestor 2005; Henderson and Jacobsen 2007). Although methylation at cytosine can be explained in a variety of DNA sequence contexts, mechanistically it can be classified broadly into three contexts, CG, CHG, and CHH (where H denotes A, T, or C) (Law and Jacobsen 2010). The pattern of occurrence of DNA methylation varies, i.e., it mainly occurs at CG sites in mammals, but it can occur in CG, CHG, and CHH contexts in plants (Feng et al. 2010). In Arabidopsis, the genome-wide DNA methylation level is reported to be 24 %, 6.7 %, and 1.7 % for CG, CHG, and CHH contexts, respectively. Alteration in DNA methylation is associated with gene regulation and transposable element silencing in eukaryotes (Law and Jacobsen 2010). It acts differently in different regions of the genome. In transposable elements (TE), where it appears in all three contexts (CG, CHG, and CHH), it is responsible for transcriptional silencing. In genes, DNA methylation is mainly restricted to CG sites (Law and Jacobsen 2010; Zhang et al. 2010), although CHG and CHH methylation has also been reported recently (González et al. 2011). The presence of DNA methylation at the gene promoter region is generally negatively correlated with gene expression (Zhang et al. 2006; Li et al. 2012). Furthermore, DNA methylation can also occur within the gene body (i.e., away from the 5′ and 3′ ends of transcription units), in the so-called bell-shaped CG “gene body methylation” pattern. However, the function of gene body methylation is still not clear (Zhang et al. 2010).
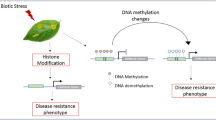
DNA METHYLTRANSFERASE 1 (MET1) primarily maintains CG methylation, which is a homologue of the mammalian DNA METHYLTRANSFERASE1 (DNMT1). Moreover, CHROMOMETHYLASE 3 (CMT3) maintains CHG methylation. Maintenance of CHH methylation is complex because it is asymmetrical; it needs to be reacquired de novo after each replication, through the action of the plant-specific RNA-dependent DNA methylation (RdDM) pathway (Law and Jacobsen 2010) in which small RNAs (24 nucleotides long) target the de novo methyltransferase DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2) to homologous genomic loci to establish DNA methylation. The modifications in DNA methylation in response to environmental stress have been reported in both locus-specific and genome-wide studies (Table 2). Some of such examples are explained below: In tomato, altered DNA methylation levels were observed on Asr1 (ABSCISIC ACID STRESS RIPENING 1) gene under drought stress (González et al. 2011). Asr1 is a non-transposon, protein-coding, water stress-inducible gene of the LEA superfamily in tomato. The expression of Asr1 increases with the longer duration of drought stress. In addition, the CHH methylation was decreased in drought conditions showing the negative correlation with gene expression (González et al. 2011). Furthermore, the existence of all the three contexts of DNA methylation (CG, CHG, CHH) was reported in the regulatory region of Asr2 (ABSCISIC ACID STRESS RIPENING 2). Interestingly, the gene body methylation was restricted to only one context (CG). The site-specific removal of methyl marks from CNN sites in the regulatory region was observed under drought stress. This response of Asr2 is heritable through generations and could have evolutionary importance (González et al. 2013). In maize, genome-wide DNA methylation pattern was studied under cold stress. It led to the identification of a fragment named ZmMI1, which was transcribed only in cold stress conditions (Steward et al. 2002). About 49 transcription factors showed differential expression in soybean on exposure to salinity stress. Moreover, DNA methylation and expression profiles of one MYB (MYELOBLASTOSIS), one b-ZIP (BASIC LEUCINE ZIPPER), and two AP2/DREB transcription factor gene family members were significantly correlated (Song et al. 2012). Choi and Sano (2007) analyzed glycerophospho-diesterase-like protein (NtGPDL) gene in tobacco to understand the effect of various stresses, including aluminum, salt, and cold stress. The increased transcription and CG demethylation in the coding regions of NtGPDL were observed under stress conditions (Choi and Sano 2007). Change in DNA methylation pattern in response to biotic stress was also reported. In tobacco, a pathogen-responsive gene NtAlix1 (ALG-2 INTERACTING PROTEIN X 1) was studied upon the infection of tobacco mosaic virus (TMV). The change of DNA methylation at NtAlix1 was observed after 24 hours of infection, indicating that DNA methylation pattern undergoes alteration in response to biotic stresses which is closely related to the activation of stress-responsive genes (Wada et al. 2004). One interesting example came from the analysis of Arabidopsis, where a putative small RNA target region about 2.6 kb upstream of the ATG start codon of AtHKT1 (HIGH-AFFINITY K + TRANSPORTER 1) gene is normally heavily methylated and its hypomethylation represses the AtHKT1 gene expression, which is crucial for salt tolerance (Baek et al. 2011). In Arabidopsis, the promoter region of AtRMG1 (RESISTANCE METHYLATED GENE 1) gene is targeted by RdDM and ROS1-dependent DNA demethylation as a defense response against the P. syringae pathogen (Yu et al. 2013). Sharma et al. (2009) characterized ten putative DNA methyltransferases in rice. Expression analysis of them was done at different developmental stages and under abiotic stress. High salinity and cold stress induced OsCMT2, but drought stress showed no effect. Drought and salinity stress caused OsCMT3 to exhibit approximately a six- and four fold reduction in mRNA accumulation in rice seedlings subjected to high-salt and dehydration conditions, respectively. In addition to locus-specific stress responses, a good deal of work has been done on the genome-wide level. Genome-wide DNA methylation response to cold stress by MSAP (methylation-sensitive amplification polymorphism) technique in maize revealed global DNA methylation shift. The main part of this shift was attributed to the demethylation of fully methylated fragments (Shan et al. 2013). Lira-Medeiros et al. (2010), in an interesting comparative study of mangrove plants, growing in salt marsh neighborhood and riverside habitat, revealed that riverside plants were much taller and thicker than the plants growing in salt marsh neighborhood. Genome-wide DNA methylation analysis showed considerable hypermethylation in riverside plants in comparison with the plants growing in salt marsh neighborhood suggesting a pivotal role of natural epigenetic variations in a plant population toward environmental adaptation. Similarly, a genome-wide study by MSAP analysis performed in diverse rice genotypes differing in their salt-responsive characteristics highlighted differential methylation and expression of salt stress-related genes, retrotransposons, and chromatin modifier genes (Karan et al. 2012). Another study of genome-wide DNA methylation analysis under drought stress has been reported in rice. In this study, the comparison of two genotypes under drought stress and subsequent recovery revealed the genotype-specific DNA methylation modifications, which were mostly reversed after recovery, but some were maintained even after recovery indicating some sort of stress memory. This study illustrated the importance of these induced epigenetic changes in regulatory mechanisms for adaptation of rice plant to environmental stresses (Wang et al. 2011). Dyachenko et al. (2006) reported hypermethylation of CHG methylation in nuclear genome of Mesembryanthemum crystallinum plants during high-salinity stress imposition. These examples provide a glimpse of the importance of epigenetic mechanisms in the plant response to the environmental variation and their potential involvement in the adaptive strategies devised by the plants. In this respect, the reported data sets of various plant methylomes could provide the basis for the selection of differential epigenetic regions as probable targets for the genetic manipulation for crop improvement.
Epigenetic Outlook for Crop Improvement
One very significant part of the success attained in the field of crop yield improvement is attributed to the plant breeding and genetics. The utilization of desirable available variation has been one of the main roles followed by the scientists for the improvement of crop plants. In the last two decades, the researchers across the globe have accumulated the wealth of knowledge that provides the evidence of prevalence of epigenetic variability (natural as well as generated) and its potential to influence the phenotype (agronomic traits) and large crop improvement. Histone modifications are involved in mitotically stable transcriptional activation or repression and exhibit lower level of transgenerational heritability. So, can the mitotically stable epigenetic information be used for crop improvement? The answer is that very similar to transcriptional factors, chromatin changes also control plant morphology and response to the environment, and a greater control over traits may be achieved by understanding these mechanisms, which is highly important for the breeding point of view. Several cases of naturally occurring epialleles (i.e., DNA methylation alleles that are independent of DNA sequence variation causing a visible phenotype) have been described, such as the Lcyc locus in Linaria vulgaris (Cubas et al. 1999) and an SBP-box gene in tomato (Manning et al. 2006). DNA methylation of natural epialleles has also been described at a larger genomic scale for species such as Arabidopsis (Cervera et al. 2002; Vaughn et al. 2007), Spartina anglica (Salmon et al. 2005), or Populus trichocarpa (Raj et al. 2011). These examples indicate the existence of epigenetic variations in natural populations. Therefore, epigenetic variants could be used in breeding programs for the improvement of crops because breeders select for a particular trait rather than a molecular mechanism.
These genome-specific techniques have important implications on the crop improvement by their potential role in the identification of regions, which show epigenetic modifications under various kinds of stress. This identification could lead to the characterization of these regions of interests in the genome. Further studies of these regions could lead to detection of epialleles which could be incorporated into the breeding programs and play their role in crop improvement. Moreover, recent techniques enabled breeders to generate desired allelic variation through the mutagenesis or transgenic modifications to develop a trait not observed in natural population. Epigenetic regulation affects transgene behavior and could be used to establish novel epialleles for breeding purposes. Different approaches have been proposed, speculated, and/or initiated to use the epigenetic diversity in the breeding programs. One big hurdle in producing or developing epigenetic diversity in crop plants is the lack of availability of genome-wide DNA methylation mutants like met1 and ddm1. In such cases, the usage of chemical inhibitors like 5-azadeoxycytidine is a good alternative. Different studies have reported the transgenerational inheritance of the modifications created by its treatment. (Sano et al. 1990; Akimoto et al. 2007). Akimoto et al. (2007) reported that in progenies of 5-azadeoxycytidine-treated rice, some of the altered phenotypes were stably inherited even after 10 years. Some of these phenotypes were of interest from the breeding point of view like resistance of a bacterial pathogen Xanthomonas oryzae. Another interesting and exciting approach, which is drawing much attention, is the usage of epigenetic- Recombinant Inbred Lines (epiRILs). These lines are created by artificial crossing of DNA methylation mutants, i.e., decreased DNA methylation 1 (ddm1) or methyltransferase 1 (met1) with their wild types (Johannes et al. 2009; Reinders et al. 2009). Since these mutants are deficient in DNA methylation machinery but genetically similar as that of the wild type, the resulting lines (epiRILs) have almost identical DNA sequences but divergent patterns of DNA methylations. These patterns are reported to be stable across many generations through molecular analysis. Analysis of these lines has shown that they have widespread phenotypic variation for morphological or developmental traits, like flowering time, plant height, as well as biotic and abiotic stresses (Johannes et al. 2009; Reinders et al. 2009; Zhang et al. 2013). Although these methylation variants do not necessarily reveal natural variation, they can be very useful in multiple ways. They can serve as a good material to understand the extent and potential role of the epigenetic variations, which are independent of genetic variations. They can also help to understand the extent of the phenotypic variation caused by the random combination of plant-specific epigenome. The above mentioned hypothesis was confirmed from the various recent publications giving further insight into the basic mechanisms which require DNA methylation, like the effect of DNA methylation on crossing over where the results showed that the distribution of crossing over event is sensitive to DNA methylation, but the rate of crossing over is not affected by it (Colomé-Tatché et al. 2012; Mirouze et al. 2012). Similarly, significant heritable variation in growth rate in response to biotic stresses was reported. All these results further support the opinion that considerable heritable variations in economically important traits could be created by variation in the DNA methylation patterns, and these kinds of approaches could be applicable for crop improvement. Utilization of epialleles generated and/or identified by various researchers in diverse plant/crop species should be exploited in different breeding programs. Such kind of program could start with the identification and understanding of epigenetic pattern in individuals of the selected population. This will lead to the identification of specific phenotypes and, finally, the association studies of that inherited phenotype and epigenetic variation. With the advances in the genome editing technologies, the usage of locus-specific epigenetic modifications could also be used for crop improvement (Chen and Gao 2014).
Techniques Used in Epigenetic/Epigenomic
There are different techniques in use for the detection of DNA methylation profile and DNA-protein interaction both at locus-specific level and on genome-wide level. Some examples of these techniques are discussed here. On locus-specific level, the DNA methylation profile can be studied through bisulfite treatment technique and through the use of methyl-sensitive restriction enzymes. On a genome-wide level, DNA methylation profile can be studied through MSAP (methyl-sensitive amplification polymorphism) (Yaish et al. 2014), through the use of HPLC (high-performance liquid chromatography) technique (Friso et al. 2002). A short description of these techniques is given below.
DNA Methylation
Bisulfite Treatment
In bisulfite treatment, the genomic DNA is treated with sodium bisulfite, which converts all the non-methylated cytosines into uracil. This conversion is followed by the PCR through specific primers. All the uracil (non-methylated cytosines before bisulfite treatment) and thymine residues (which were always thymines even before bisulfite treatment) are being amplified by PCR as thymine, whereas only 5-methylcytosine residues are amplified as cytosine. After sequencing of PCR product, the analysis of sequences provides the information about the methylated sites in the amplified region as well as the methylation level in a particular genomic region (Frommer et al. 1992). With the advancement in the sequencing technology, this technique can also be used in genome-wide methylation analysis.
Methylation-Sensitive PCR (MSP)
Methylation-sensitive PCR (MSP) is a modification in the above-described bisulfite treatment technique where the amplification is done with primer pair that is specific for methylated DNA and primer pair specific to unmethylated DNA (Herman et al. 1996). This technique can be used in the development of epigenetic markers to be used in marker-assisted selection.
Methyl-Sensitive Restriction Enzyme Technique
In methyl-sensitive restriction enzyme technique, the genomic DNA is digested with the methylation-sensitive restriction enzyme, and this digested DNA is amplified by primers flanking the restriction site. PCR will work only if the restriction site is not cleaved (due to the methylation at that site) (Singer-Sam et al. 1990). This kind of technique along with its various modified forms paves the way for the development of simple and practical epigenetic marker for the epialleles which could have implications in marker-assisted selection and could have important role in the crop improvement strategies.
Methyl-Sensitive Amplification Polymorphism (MSAP)
Methyl-sensitive amplification polymorphism (MSAP) is a technique for the DNA methylation analysis on a genome-wide level, in which digestion of genomic DNA is done with a methylation-sensitive restriction enzyme like HpaII as a first step. This is followed by the ligation of DNA fragments to adaptors, which facilitate the amplification of these fragments. MspI, a methylation-insensitive isoschizomer of HpaII, is used in parallel for digestion, and this digestion serves as a loading control in the experiment. After that, amplification of these fragments through fluorescently labeled primers is done. Comparison of PCR products from different individuals allows the user to identify the interesting fragments. This leads to the isolation and characterization of that fragment (Yaish et al. 2014).
High-Performance Liquid Chromatography Technique (HPLC)
In high-performance liquid chromatography technique (HPLC), the genomic DNA is enzymatically hydrolyzed, and this hydrolyzed DNA is then separated into its four major DNA bases and 5-methyl-2′-deoxycytidine using HPLC. 5-Methyl-2′-deoxycytidine is obtained. The global DNA methylation status is calculated by comparing the amount of 5-methyl-2′-deoxycytidine per microgram of DNA with percent relative standard deviations (%RSD) (Friso et al. 2002).
Methylated DNA Immunoprecipitation
It is another technique to study genome-wide changes in DNA methylation patterns. In this technique, DNA is isolated from cells and sheared through sonication. By the usage of antibodies specifically targeting methylated DNA fragments, isolation of methylated regions occurs, which then can be identified using high-resolution DNA microarrays or next-generation sequencing techniques. The global changes in the methylation patterns across the varied cells can be detected through this technique.
Chromatin Immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) is a technique used to study the interaction between proteins (e.g., histones) and DNA. The use of highly specific antibodies directed against DNA-binding proteins is required in this technique, and it can be followed by various nucleic acid analysis techniques, including PCR, qPCR, sequencing, and microarray hybridization. It can help to determine whether certain proteins are associated with specific genomic regions and is also useful for identifying regions of the genome associated with specific histone modifications.
Conclusion
Our understanding of epigenetic regulation in plants is rapidly growing. However, up to now, linkage of histone modifications, ATP-dependent chromatin remodeling, and DNA methylation to a specific stress and the origin of this specificity is still unknown. To identify targeted genes and regulatory complexes, genomic binding studies and proteomic analysis will be required, respectively. It is necessary to deepen our investigation of the epigenetic regulators in crops and their underlying molecular mechanism. Understanding the mechanism of epigenetic regulators and their regulatory networks in crops will be a potential tool for further exploitation toward sustainable agriculture. Moreover, it is desirable to design new breeding strategies in which the epigenetic variability should be taken into consideration. This seems even more realistic with the advancement of genomic technologies and cost lowering of next-generation sequencing. Like MAS (marker-assisted selection), epigenetic marker-assisted selection could also be initiated.
References
Agger K, Christensen J, Cloos PA, Helin K (2008) The emerging functions of histone demethylases. Curr Opin Genet Dev 18:159–168
Akimoto K, Katakami H, Kim HJ, Ogawa E, Sano CM, Wada Y et al (2007) Epigenetic inheritance in rice plants. Ann Bot 100:205–217
Aletta JM, Cimato TR, Ettinger MJ (1998) Protein methylation: a signal event in post-translational modification. Trends Biochem Sci 23:89–91
Alexandre C, Moller-Steinbach Y, Schonrock N, Gruissem W, Hennig L (2009) Arabidopsis MSI1 is required for negative regulation of the response to drought stress. Mol Plant 2:675–687
Allfrey V, Faulkner R, Mirsky A (1964) Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci U S A 51:786–794
Allis CD, Chicoine LG, Richman R, Schulman IG (1985) Deposition-related histone acetylation in micronuclei of conjugating Tetrahymena. Proc Natl Acad Sci U S A 82:8048–8052
Alvarez-Venegas R, Sadder M, Hlavacka A, Baluska F, Xia Y, Lu G et al (2006) The Arabidopsis homolog of trithorax, ATX1, binds phosphatidylinositol 5-phosphate, and the two regulate a common set of target genes. Proc Natl Acad Sci U S A 103:6049–6054
Baek D, Jiang J, Chung J-S, Wang B, Chen J, Xin Z et al (2011) Regulated AtHKT1 gene expression by a distal enhancer element and DNA methylation in the promoter plays an important role in salt tolerance. Plant Cell Physiol 52:149–161
Bedford MT, Clarke SG (2009) Protein arginine methylation in mammals: who, what, and why. Mol Cell 33:1–13
Bedford MT, Richard S (2005) Arginine methylation: an emerging regulator of protein function. Mol Cell 18:263–272
Benhamed M, Bertrand C, Servet C, Zhou D-X (2006) Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell Online 18:2893–2903
Berr A, McCallum EJ, Alioua A, Heintz D, Heitz T, Shen WH (2010) Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol 154:1403–1414
Berr A, Shafiq S, Shen WH (2011) Histone modifications in transcriptional activation during plant development. Biochim Biophys Acta 1809:567–576
Berr A, Ménard R, Heitz T, Shen W-H (2012) Chromatin modification and remodelling: a regulatory landscape for the control of Arabidopsis defence responses upon pathogen attack. Cell Microbiol 14:829–839
Bertrand C, Bergounioux C, Domenichini S, Delarue M, Zhou DX (2003) Arabidopsis histone acetyltransferase AtGCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. J Biol Chem 278:28246–28251
Bilichak A, Ilnystkyy Y, Hollunder J, Kovalchuk I (2012) The progeny of Arabidopsis thaliana plants exposed to salt exhibit changes in DNA methylation, histone modifications and gene expression. PLoS ONE 7:e30515
Bourque S, Dutartre A, Hammoudi V, Blanc S, Dahan J, Jeandroz S, Pichereaux C et al (2011) Type-2 histone deacetylases as new regulators of elicitor-induced cell death in plants. New Phytol 192:127–139
Briggs SD, Xiao T, Sun Z-W, Caldwell JA, Shabanowitz J, Hunt DF et al (2002) Gene silencing: trans-histone regulatory pathway in chromatin. Nature 418:498
Casati P, Campi M, Chu F, Suzuki N, Maltby D, Guan S et al (2008) Histone acetylation and chromatin remodeling are required for UV-B-dependent transcriptional activation of regulated genes in maize. Plant Cell 20:827–842
Cervera MT, Ruiz-García L, Martínez-Zapater JM (2002) Analysis of DNA methylation in Arabidopsis thaliana based on methylation-sensitive AFLP markers. Mol Gen Genet 268:543–552
Chen K, Gao C (2014) Targeted genome modification technologies and their applications in crop improvements. Plant Cell Rep 33:575–583
Chen LT, Wu K (2010) Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal Behav 5:1318–1320
Chen Z, Zhang H, Jablonowski D, Zhou X, Ren X, Hong X et al (2006) Mutations in ABO1/ELO2, a subunit of holo-Elongator, increase abscisic acid sensitivity and drought tolerance in Arabidopsis thaliana. Mol Cell Biol 26:6902–6912
Chen LT, Luo M, Wang YY, Wu K (2010) Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J Exp Bot 61:3345–3353
Chen Q, Chen X, Wang Q, Zhang F, Lou Z, Zhang Q et al (2013) Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice. PLoS Genet 9:e1003239
Chinnusamy V, Zhu JK (2009) Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol 12:133–139
Choi CS, Sano H (2007) Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol Genet Genomics 277:589–600
Choi SM, Song HR, Han SK, Han M, Kim CY, Park J et al (2012) HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J 71:135–146
Colomé-Tatché M, Cortijo S, Wardenaar R, Morgado L, Lahouze B, Sarazin A et al (2012) Features of the Arabidopsis recombination landscape resulting from the combined loss of sequence variation and DNA methylation. Proc Natl Acad Sci U S A 109:16240–16245
Cubas P, Vincent C, Coen E (1999) An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401:157–161
Cui X, Jin P, Gu L, Lu Z, Xue Y, Wei L et al (2013) Control of transposon activity by a histone H3K4 demethylase in rice. Proc Natl Acad Sci U S A 110:1953–1958
DeFraia CT, Zhang X, Mou Z (2010) Elongator subunit 2 is an accelerator of immune responses in Arabidopsis thaliana. Plant J 64:511–523
DeFraia CT, Wang Y, Yao J, Mou Z (2013) Elongator subunit 3 positively regulates plant immunity through its histone acetyltransferase and radical S-adenosylmethionine domains. BMC Plant Biol 13:102
Demetriou K, Kapazoglou A, Tondelli A, Francia E, Stanca MA, Bladenopoulos K et al (2009) Epigenetic chromatin modifiers in barley: I. Cloning, mapping and expression analysis of the plant specific HD2 family of histone deacetylases from barley, during seed development and after hormonal treatment. Physiol Plant 136:358–368
Deng X, Gu L, Liu C, Lu T, Lu F, Lu Z et al (2010) Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc Natl Acad Sci U S A A107:19114–19119
Dhawan R, Luo H, Foerster AM, Abuqamar S, Du HN, Briggs SD, Scheid OM et al (2009) HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21:1000–1019
Ding Y, Avramova Z, Fromm M (2011) The Arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways. Plant J 66:735–744
Ding B, Bellizzi Mdel R, Ning Y, Meyers BC, Wang GL (2012) HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell 24:3783–3794
Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M et al (2002) Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem 277:28368–28371
Durand S, Bouché N, Perez Strand E, Loudet O, Camilleri C (2012) Rapid establishment of genetic incompatibility through natural epigenetic variation. Curr Biol 22:326–331
Dyachenko OV, Zakharchenko NS, Shevchuk TV, Bohnert HJ, Cushman JC, Buryanov YI (2006) Effect of hypermethylation of CCWGG sequences in DNA of Mesembryanthemum crystallinum plants on their adaptation to salt stress. Biogeosciences 71:461–465
Earley KW, Shook MS, Brower-Toland B, Hicks L, Pikaard CS (2007) In vitro specificities of Arabidopsis co-activator histone acetyltransferases: implications for histone hyperacetylation in gene activation. Plant J 52:615–626
Falconer DS (1996) Introduction to quantitative genetics. Ronald Press Co., New York, pp 365
Fang H, Liu X, Thorn G, Duan J, Tian L (2014) Expression analysis of histone acetyltransferases in rice under drought stress. Biochem Biophys Res Commun 443:400–405
Feng S, Jacobsen SE, Reik W (2010) Epigenetic reprogramming in plant and animal development. Science 330:622–627
Fisher RA (1930) The genetical theory of natural selection. Clarendon Press, Oxford, pp 272
Friso S, Choi S-W, Dolnikowski GG, Selhub J (2002) A method to assess genomic DNA methylation using high-performance liquid chromatography/electrospray ionization mass spectrometry. Anal Chem 74:4526–4531
Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW et al (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A 89:1827–1831
Fu W, Wu K, Duan J (2007) Sequence and expression analysis of histone deacetylases in rice. Biochem Biophys Res Commun 356:843–850
Goll MG, Bestor TH (2005) Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74:481–514
González RM, Ricardi MM, Iusem ND (2011) Atypical epigenetic mark in an atypical location: cytosine methylation at asymmetric (CNN) sites within the body of a non-repetitive tomato gene. BMC Plant Biol 11:94
González RM, Ricardi MM, Iusem ND (2013) Epigenetic marks in an adaptive water stress-responsive gene in tomato roots under normal and drought conditions. Epigenetics 8:864–872
Han SK, Sang Y, Rodrigues A, Wu MF, Rodriguez PL, Wagner D (2012) The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 24:4892–4906
Hark AT, Vlachonasios KE, Pavangadkar KA, Rao S, Gordon H, Adamakis ID et al (2009) Two Arabidopsis orthologs of the transcriptional coactivator ADA2 have distinct biological functions. Biochim Biophys Acta 1789:117–124
Henderson IR, Jacobsen SE (2007) Epigenetic inheritance in plants. Nature 447:418–424
Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB (1996) Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 93:9821–9826
Hoffmann AA, Sgrò CM (2011) Climate change and evolutionary adaptation. Nature 470:479–485
Hu Y, Qin F, Huang L, Sun Q, Li C, Zhao Y et al (2009) Rice histone deacetylase genes display specific expression patterns and developmental functions. Biochem Biophys Res Commun 388:266–271
Huang L, Sun Q, Qin F, Li C, Zhao Y, Zhou DX (2007) Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol 144:1508–1519
Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD (2003) A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell 11:261–266
Imhof A, Yang X-J, Ogryzko VV, Nakatani Y, Wolffe AP, Ge H (1997) Acetylation of general transcription factors by histone acetyltransferases. Curr Biol 7:689–692
Jablonka E, Raz G (2009) Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q Rev Biol 84:131–176
Jelinic P, Stehle J-C, Shaw P (2006) The testis-specific factor CTCFL cooperates with the protein methyltransferase PRMT7 in H19 imprinting control region methylation. PLoS Biol 4:e355
Johannes F, Porcher E, Teixeira FK, Saliba-Colombani V, Simon M, Agier N et al (2009) Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet 5:e1000530
Kaldis A, Tsementzi D, Tanriverdi O, Vlachonasios KE (2011) Arabidopsis thaliana transcriptional co-activators ADA2b and SGF29a are implicated in salt stress responses. Planta 233:749–762
Kapazoglou A, Tondelli A, Papaefthimiou D, Ampatzidou H, Francia E, Stanca MA et al (2010) Epigenetic chromatin modifiers in barley: IV. The study of barley polycomb group (PcG) genes during seed development and in response to external ABA. BMC Plant Biol 10:73
Karan R, DeLeon T, Biradar H, Subudhi PK (2012) Salt stress induced variation in DNA methylation pattern and its influence on gene expression in contrasting rice genotypes. PLoS ONE 7:e40203
Katz JE, Dlakić M, Clarke S (2003) Automated identification of putative methyltransferases from genomic open reading frames. Mol Cell Proteomics 2:525–540
Khan AR, Enjalbert J, Marsollier A-C, Rousselet A, Goldringer I, Vitte C (2013) Vernalization treatment induces site-specific DNA hypermethylation at the VERNALIZATION-A1 (VRN-A1) locus in hexaploid winter wheat. BMC Plant Biol 13:209
Kim KC, Lai Z, Fan B, Chen Z (2008) Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20:2357–2371
Kim JM, To TK, Nishioka T, Seki M (2010) Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ 33:604–611
Klose RJ, Zhang Y (2007) Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 8:307–318
Krause CD, Yang Z-H, Kim Y-S, Lee J-H, Cook JR, Pestka S (2007) Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol Ther 113:50–87
Lauria M, Rupe M, Guo M, Kranz E, Pirona R, Viotti A et al (2004) Extensive maternal DNA hypomethylation in the endosperm of Zea mays. Plant Cell 16:510–522
Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11:204–220
Li C, Huang L, Xu C, Zhao Y, Zhou D-X (2011) Altered levels of histone deacetylase OsHDT1 affect differential gene expression patterns in hybrid rice. PLoS ONE 6:e21789
Li X, Zhu J, Hu F, Ge S, Ye M, Xiang H et al (2012) Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC Genomics 13:300
Lira-Medeiros CF, Parisod C, Fernandes RA, Mata CS, Cardoso MA, Ferreira PCG (2010) Epigenetic variation in mangrove plants occurring in contrasting natural environment. PLoS ONE 5:e10326
Liu C, Lu F, Cui X, Cao X (2010) Histone methylation in higher plants. Annu Rev Plant Biol 61:395–420
Liu X, Luo M, Zhang W, Zhao J, Zhang J, Wu K et al (2012) Histone acetyltransferases in rice (Oryza sativa L.): phylogenetic analysis, subcellular localization and expression. BMC Plant Biol 12:145
Luo M, Wang YY, Liu X, Yang S, Lu Q, Cui Y et al (2012) HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J Exp Bot 63:3297–3306
Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ et al (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38:948–952
Mao Y, Pavangadkar KA, Thomashow MF, Triezenberg SJ (2006) Physical and functional interactions of Arabidopsis ADA2 transcriptional coactivator proteins with the acetyltransferase GCN5 and with the cold-induced transcription factor CBF1. Biochim Biophys Acta (BBA) Gene Struct Expr 1759:69–79
Martin A, Troadec C, Boualem A, Rajab M, Fernandez R, Morin H et al (2009) A transposon-induced epigenetic change leads to sex determination in melon. Nature 461:1135–1138
McBride AE, Silver PA (2001) State of the arg: protein methylation at arginine comes of age. Cell 106:5–8
Mirouze M, Lieberman-Lazarovich M, Aversano R, Bucher E, Nicolet J, Reinders J et al (2012) Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc Natl Acad Sci U S A 109:5880–5885
Mlynárová L, Nap J, Bisseling T (2007) The SWI/SNF chromatin-remodeling gene AtCHR12 mediates temporary growth arrest in Arabidopsis thaliana upon perceiving environmental stress. Plant J 51:874–885
Ng DW, Wang T, Chandrasekharan MB, Aramayo R, Kertbundit S, Hall TC (2007) Plant SET domain-containing proteins: structure, function and regulation. Biochim Biophys Acta 1769:316–329
Noh B, Lee SH, Kim HJ, Yi G, Shin EA, Lee M et al (2004) Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16:2601–2613
Palma K, Thorgrimsen S, Malinovsky FG, Fiil BK, Nielsen HB, Brodersen P et al (2010) Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog 6:e1001137
Papaefthimiou D, Tsaftaris A (2012a) Significant induction by drought of HvPKDM7-1, a gene encoding a jumonji-like histone demethylase homologue in barley (H. vulgare). Acta Physiol Plant 34:1187–1198
Papaefthimiou D, Tsaftaris AS (2012b) Characterization of a drought inducible trithorax-like H3K4 methyltransferase from barley. Biol Plant 56:683–692
Papaefthimiou D, Likotrafiti E, Kapazoglou A, Bladenopoulos K, Tsaftaris A (2010) Epigenetic chromatin modifiers in barley: III. Isolation and characterization of the barley GNAT-MYST family of histone acetyltransferases and responses to exogenous ABA. Plant Physiol Biochem 48:98–107
Pavangadkar K, Thomashow MF, Triezenberg SJ (2010) Histone dynamics and roles of histone acetyltransferases during cold-induced gene regulation in Arabidopsis. Plant Mol Biol 74:183–200
Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70:503–533
Qian Y, Xi Y, Cheng B, Zhu S, Kan X (2014) Identification and characterization of the SET domain gene family in maize. Mol Biol Rep 41:1341–1354
Raj S, Bräutigam K, Hamanishi ET, Wilkins O, Thomas BR, Schroeder W et al (2011) Clone history shapes Populus drought responses. Proc Natl Acad Sci U S A 108:12521–12526
Reinders J, Wulff BBH, Mirouze M, Marí-Ordóñez A, Dapp M, Rozhon W et al (2009) Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev 23:939–950
Richards CL, Bossdorf O, Verhoeven KJF (2010) Understanding natural epigenetic variation. New Phytol 187:562–564
Salmon A, Ainouche ML, Wendel JF (2005) Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae). Mol Ecol 14:1163–1175
Sano H, Kamada I, Youssefian S, Katsumi M, Wabiko H (1990) A single treatment of rice seedlings with 5-azacytidine induces heritable dwarfism and undermethylation of genomic DNA. Mol Gen Genet MGG 220:441–447
Shafiq S, Berr A, Shen W-H (2014) Combinatorial functions of diverse histone methylations in Arabidopsis thaliana flowering time regulation. New Phytol 201:312–322
Shan X, Wang X, Yang G, Wu Y, Su S, Li S et al (2013) Analysis of the DNA methylation of maize (Zea mays L.) in response to cold stress based on methylation-sensitive amplified polymorphisms. J Plant Biol 56:32–38
Sharma R, Mohan Singh RK, Malik G, Deveshwar P, Tyagi AK, Kapoor S et al (2009) Rice cytosine DNA methyltransferases – gene expression profiling during reproductive development and abiotic stress. FEBSJ 276:6301–6311
Shvarts Iu B, Kahn TG, Pirrotta V (2010) Polycomb and trithorax control genome expression by determining the alternative epigenetic states of chromatin for key developmental regulators. Genetika 46:1413–1416
Singer-Sam J, Grant M, LeBon JM, Okuyama K, Chapman V, Monk M et al (1990) Use of a HpaII-polymerase chain reaction assay to study DNA methylation in the Pgk-1 CpG island of mouse embryos at the time of X-chromosome inactivation. Mol Cell Biol 10:4987–4989
Smalle J, Vierstra RD (2004) The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol 55:555–590
Song Y, Ji D, Li S, Wang P, Li Q, Xiang F (2012) The dynamic changes of DNA methylation and histone modifications of salt responsive transcription factor genes in soybean. PLoS ONE 7:e41274
Springer NM (2013) Epigenetics and crop improvement. Trends Genet 29:241–247
Sridha S, Wu K (2006) Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J 46:124–133
Sterner DE, Berger SL (2000) Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev 64:435–459
Steward N, Ito M, Yamaguchi Y, Koizumi N, Sano H (2002) Periodic DNA methylation in maize nucleosomes and demethylation by environmental stress. J Biol Chem 277:37741–37746
Stockinger EJ, Mao Y, Regier MK, Triezenberg SJ, Thomashow MF (2001) Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res 29:1524–1533
Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403:41–45
Sun Q, Zhou DX (2008) Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci U S A 105:13679
To TK, Nakaminami K, Kim JM, Morosawa T, Ishida J, Tanaka M et al (2011) Arabidopsis HDA6 is required for freezing tolerance. Biochem Biophys Res Commun 406:414–419
Turck F, Roudier F, Farrona S, Martin-Magniette M-L, Guillaume E, Buisine N et al (2007) Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 3:e86
Unnikrishnan A, Gafken PR, Tsukiyama T (2010) Dynamic changes in histone acetylation regulate origins of DNA replication. Nat Struct Mol Biol 17:430–437
van Dijk K, Ding Y, Malkaram S, Riethoven JJ, Liu R, Yang J et al (2010) Dynamic changes in genome-wide histone H3 lysine 4 methylation patterns in response to dehydration stress in Arabidopsis thaliana. BMC Plant Biol 10:238
Vaughn MW, Tanurdzić M, Lippman Z, Jiang H, Carrasquillo R, Rabinowicz PD et al (2007) Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol 5:e174
Vignali M, Hassan AH, Neely KE, Workman JL (2000) ATP-dependent chromatin-remodeling complexes. Mol Cell Biol 20:1899–1910
Visser ME (2008) Keeping up with a warming world; assessing the rate of adaptation to climate change. Proc Biol Sci/R Soc 275:649–659
Vlachonasios KE, Thomashow MF, Triezenberg SJ (2003) Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell Online 15:626–638
Wada Y, Miyamoto K, Kusano T, Sano H (2004) Association between up-regulation of stress-responsive genes and hypomethylation of genomic DNA in tobacco plants. Mol Genet Genomics 271:658–666
Walley JW, Rowe HC, Xiao Y, Chehab EW, Kliebenstein DJ, Wagner D, Dehesh K et al (2008) The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog 4:e1000237
Wang C, Gao F, Wu J, Dai J, Wei C, Li Y (2010) Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression. Plant Cell Physiol 51:1291–1299
Wang WS, Pan YJ, Zhao XQ, Dwivedi D, Zhu LH, Ali J et al (2011) Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza sativa L.). J Exp Bot 62:1951–1960
Woo HR, Pontes O, Pikaard CS, Richards EJ (2007) VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev 21:267
Wu K, Zhang L, Zhou C, Yu CW, Chaikam V (2008) HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J Exp Bot 59:225–234
Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou J-P, Steinmetz A et al (2008) Di- and Tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol Cell Biol 28:1348–1360
Yaish MW, Peng M, Rothstein SJ (2014) Global DNA methylation analysis using methyl-sensitive amplification polymorphism (MSAP). In: Sanchez-Serrano JJ, Salinas J (eds) Arabidopsis protocols, vol 82. Humana Press, Totowa, pp 285–298
Yu A, Lepère G, Jay F, Wang J, Bapaume L, Wang Y et al (2013) Dynamics and biological relevance of DNA demethylation in Arabidopsis antibacterial defense. Proc Natl Acad Sci U S A 110:2389–2394
Zemach A, Kim MY, Silva P, Rodrigues JA, Dotson B, Brooks MD et al (2010) Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci U S A 107:18729–18734
Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SWL, Chen H et al (2006) Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126:1189–1201
Zhang K, Sridhar VV, Zhu J, Kapoor A, Zhu JK (2007) Distinctive core histone post-translational modification patterns in Arabidopsis thaliana. PLoS ONE 2:e1210
Zhang M, Kimatu JN, Xu K, Liu B (2010) DNA cytosine methylation in plant development. J Genet Genomics = Yi chuan xue bao 37:1–12
Zhang Z, Zhang S, Zhang Y, Wang X, Li D, Li Q et al (2011) Arabidopsis floral initiator SKB1 confers high salt tolerance by regulating transcription and pre-mRNA splicing through altering histone H4R3 and small nuclear ribonucleoprotein LSM4 methylation. Plant Cell 23:396–411
Zhang Y-Y, Fischer M, Colot V, Bossdorf O (2013) Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytol 197:314–322
Zhou C, Zhang L, Duan J, Miki B, Wu K (2005) HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell Online 17:1196–1204
Zhou X, Hua D, Chen Z, Zhou Z, Gong Z (2009) Elongator mediates ABA responses, oxidative stress resistance and anthocyanin biosynthesis in Arabidopsis. Plant J 60:79–90
Zong W, Zhong X, You J, Xiong L (2012) Genome-wide profiling of histone H3K4-tri-methylation and gene expression in rice under drought stress. Plant Mol Biol 81:175–188
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer India
About this chapter
Cite this chapter
Shafiq, S., Khan, A.R. (2015). Plant Epigenetics and Crop Improvement. In: Barh, D., Khan, M., Davies, E. (eds) PlantOmics: The Omics of Plant Science. Springer, New Delhi. https://doi.org/10.1007/978-81-322-2172-2_6
Download citation
DOI: https://doi.org/10.1007/978-81-322-2172-2_6
Published:
Publisher Name: Springer, New Delhi
Print ISBN: 978-81-322-2171-5
Online ISBN: 978-81-322-2172-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)