Abstract
Immuno-SERS (iSERS) microscopy is an emerging imaging technique in tissue-based cancer diagnostics, which is based on antibodies labelled with SERS-active noble metal nanoparticles (NPs) in conjunction with Raman microspectroscopy for localizing target proteins. Advantages of SERS over existing labeling approaches include its high capacity for spectral multiplexing (parallel detection of target molecules), quantification (based on the characteristic SERS signatures), high photostability (no or minimal photobleaching), minimization of autofluorescence from biological specimens (via red to near-infrared excitation), and the technical advantage of using only a single laser excitation line. In this book chapter, we will first give a very brief tutorial on the fundamentals of SERS, followed by an introduction into the concept and current designs of target-specific SERS probes based on noble metal NPs. Next, the fast developing applications of iSERS microscopy for tissue-based cancer diagnostics are highlighted, and finally the challenges and future perspectives of this emerging field are presented.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
1 Introduction
Cancer is one of the biggest causes of mortality in the world. Effective diagnostic tools for cancer detection are highly desirable in order to provide early intervention by therapy as well as to reduce costs associated with cancer patient management. Detection and localization of prognostic markers in tissues is of central importance for cancer diagnostics. Currently, the most widely used method to detect protein expression in tissue specimens is immunohistochemistry (IHC), a staining method based on antigen-antibody recognition. In order to visualize the antigen-antibody immunoreaction, the antibody must be labelled, usually by enzymes , fluorophores , radioactive compounds or colloidal gold [1]. Multiplexed immunohistochemistry (mIHC), in which two or more antigens are detected simultaneously, is powerful in tumor diagnosis and classification as it offers greater insights into, spatial relationships and heterogeneity of protein expression in the same tissue specimen [2]. However, current readout methods for mIHC based on dyes or molecular fluorophores are limited because of spectral overlap due to the broad electronic absorption and emission bands, respectively. The introduction of quantum dots [3,4,5], semiconductor nanocrystals with size-dependent fluorescence emission, has been a major improvement in the multiplexing capacity of fluorescence-based detection schemes: approx. 3–10 quantum dots may be detected simultaneously because of their narrower emission peaks (peak width \(\sim \)30 nm compared with molecular fluorophores (typically >50 nm)) [6].
Surface-enhanced Raman scattering (SERS) has experienced significant growth in biomedical research during the past decade [7,8,9]. Numerous applications have demonstrated the potential of SERS for label-free detection of various analytes, especially in human biological samples such as blood, saliva, cells, and serum [10,11,12]. More recently, “SERS labels / nanotags” that combine metallic nanoparticles (NPs) and organic Raman reporter molecules have been designed and used for the selective and sensitive detection of proteins and oligonucleotides [13,14,15]. These SERS-active colloids produce strong, characteristic Raman signals, exhibit optical labeling functions similar to external chromophores like organic dyes, fluorophores, and quantum dots, and additionally, have much narrower emission peaks (<2 nm) [16]. Compared with existing labeling approaches, SERS labels offer advantages such as tremendous spectral multiplexing capabilities for simultaneous target detection (>10 different “colors”), the need of only a single laser excitation wavelength to excite multiple SERS labels simultaneously, no or only minimal photobleaching and improved image contrast by red or near-infrared (NIR) laser excitation [17].
Immuno-SERS (iSERS) microscopy is an emerging imaging method for the selective and sensitive localization of target molecules. Target-specific SERS particles, obtained after the conjugation of SERS labels to target-specific binding molecules, can be localized in cells and tissue and identified by their characteristic signature using Raman microscopy [18]
Immuno-SERS (iSERS) microscopy (Fig. 15.1), which employs target-specific colloidal SERS probes in combination with Raman microspectroscopy, is an emerging imaging method in tissue diagnostics [17]. The concept of protein localization in iSERS is the same as in conventional IHC, i.e., the target antigen is recognized by a labelled antibody (Fig. 15.2). Tissues, either frozen or formalin-fixed and paraffin-embedded (FFPE) specimens, are usually prepared as micrometer-thick sections using a microtome. In order to minimize non-specific binding, blocking agents such as goat serum or bovine serum albumin (BSA) may be employed prior to staining. Then the tissue specimen is incubated with antibodies to form corresponding antigen-antibody complexes, and subsequently unbound antibodies are removed by washing. The characteristic signals of the SERS labels indicate target protein localization because the labels are attached to the antibody, which specifically binds to the target protein.
Principle of immunohistochemistry for target protein localization. For minimizing nonspecific binding, the tissue specimen is first blocked and then incubated with the labelled antibody as the target-specific probe. The antibody selectively recognizes the antigen via the binding sites and forms the corresponding antigen-antibody complex. Finally, washing removes unbound antibodies. The antigen can be localized by the characteristic SERS spectrum of the SERS label using Raman microspectroscopy [18]
2 Brief Tutorial on SERS
In addition to conventional Raman spectroscopy, SERS requires metallic nanostructures that can support localized surface plasmon resonances (the so-called SERS substrate). Figure 15.3 shows for a gold nanoparticle how the oscillating electric field of the incoming laser radiation with angular frequency \(\omega _{inc}\) and amplitude \(E_0\) drives an oscillation of the conduction electrons of the metal, leading to charge separation (dipole). This type of resonance is termed dipolar localized surface plasmon resonance (LSPR) . The sign of the localized induced dipole \(\mu _{ind}\) changes periodically with the angular frequency \(\omega _{inc}\) of the electromagnetic wave as the external driving force. Overall, a Hertzian dipole (see electric field lines in Fig. 15.3a) on the nanoscale has been generated. This nanoantenna can then emit radiation at the same frequency \(\omega _{inc}\) (green color in Fig. 15.3a, b; elastic scattering).
Electromagnetic enhancement in SERS. a A gold nanoparticle acts as a nanoantenna by excitation of a dipolar localized surface plasmon resonance (LSPR); b Both the “incoming” field (\(\omega _{inc}\), green) and the “outgoing” field (\(\omega _{inc}-\omega _{vib}\), orange) are enhanced by elastic light scattering off the LSPR-supporting metal nanostructure [19]
The increased local electric field \(E_{loc}(\omega _{inc})\) in the vicinity of the metal particle can interact with a molecule nearby the surface of the metal sphere (Fig. 15.3b) and induce a molecular dipole (Rayleigh and Raman scattering). The Stokes-Raman scattering component with \(\omega _{inc}-\omega _{vib}\) (left orange arrow in Fig. 15.3b) itself can excite a LSPR of the metal colloid in Fig. 15.3a. Again, elastic light scattering off the metal sphere can take place (right orange arrow in Fig. 15.3b) and the emitted radiation is finally detected in the far field. This physical picture applies to all nanostructures which support LSPRs.
The overall SERS intensity depends on both the “incoming” (\(\omega _{inc}\)) and the “outgoing” (\(\omega _{inc}-\omega _{vib}\)) field: \(I_{SERS} = I_{inc}(\omega _{inc}) I (\omega _{inc}-\omega _{vib}) = |E_{inc}(\omega _{inc})|^2 \cdot |E(\omega _{inc}-\omega _{vib})|^2\). Optimal SERS enhancements therefore require that both the incident radiation at \(\omega _{inc}\) and the Stokes-Raman-shifted radiation at \(\omega _{inc}-\omega _{vib}\) are in resonance with the plasmon peak of the metal nanostructure. For \(\omega _{inc} \approx \omega _{inc}-\omega _{vib}\) the widely used \(|E|^4\) approximation of the SERS intensity results: \(I_{SERS} \approx |E(\omega _{inc})|^4\).
This electromagnetic enhancement is the dominant contribution to SERS since it is responsible for the tremendous signal increase compared to conventional Raman scattering: moderate local field enhancements (\(E_{loc}\)) yield huge SERS enhancements (\(\sim |E|^4\)). SERS is additionally a truly surface-selective effect: only molecules on or nearby such plasmonic nanostructures exhibit huge enhancement due to the SERS distance dependence. The electric field strength of dipolar radiation scales with \(E(r)\sim r^{-3}\). The SERS distance dependence is therefore \(I_{SERS} \sim r^{-12}\) under the \(|E|^4\) approximation, i.e. the SERS intensity drops down drastically with increasing distance r between the molecule and the metal surface. The SERS enhancement factor (EF) is a very central figure in SERS for quantifying the overall signal enhancement. The experimental determination of EFs in SERS requires measurements of the SERS intensity for the adsorbed molecule on the metal surface, relative to the normal Raman intensity of the same, “free” molecule in solution. Both intensities must be normalized to the corresponding number of molecules on the surface (SERS) and in solution (conventional Raman), respectively. Further information on SERS can be retrieved from several review articles [19,20,21,22,23] and monographs [24, 25].
3 SERS Nanoparticle Labels
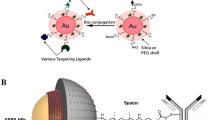
The rational design of SERS nanoparticle labels with defined physicochemical properties is essential for successful applications in bioanalytical assays and cancer diagnostics. For an introduction to the topic of the synthesis and characterization of SERS labels and their application in bioanalysis, recent reviews [13, 14] are recommended. SERS colloids for targeted research contain three basic constituents: a colloidal SERS substrate for signal enhancement, Raman reporter molecules adsorbed onto its surface for identification, and biomolecules attached to the SERS label for target-specific recognition. Figure 15.4a shows an example of a target-specific SERS probe [17, 26], comprising a monolayer of Raman reporter molecules on the surface of a Au/Ag nanoshell , a silica shell , which chemically and mechanically stabilizes the SERS label, and a spacer molecule for bioconjugation of the antibody to the SERS label. The protective encapsulant, such as silica or polymer coatings, is optional but desirable since it enhances the stability and biocompatibility of the colloid. Using a spacer molecule between the antibodies and the SERS label instead of direct conjugation of the antibody to the metal surface introduces conformational flexibility and may be beneficial for proper target recognition via the antigen binding sites.
a Components of a target-specific SERS probe: Colloidal SERS substrate (Au/Ag nanoshell), Raman reporters adsorbed to its surface (organic monolayer), an optional protective encapsulant (glass shell) and a target-specific binding molecule (antibody) attached to the SERS label via a spacer molecule; b Synthesis of SERS-labelled antibodies: The metal colloid is incubated with Raman labels and optionally encapsulated with a protective shell. The resulting encapsulated SERS label is then conjugated to antibodies via spacer units [18]
Figure 15.4b depicts the sequence of chemical modifications by which SERS-labelled antibodies are synthesized from metal colloids. The addition of Raman reporter molecules to the colloid with subsequent encapsulation leads to a stabilized SERS particle. Modification of the shell with spacers and their conjugation to antibodies finally yields SERS-labelled antibodies.
3.1 Choice of Metal Colloids as SERS Substrates
The first step to design a SERS label is to choose and prepare a metal colloid with the desired optical properties, in particular with respect to the position of the LSPR peak and the achievable signal enhancement. Noble metal NPs in the size range 10–100 nm are typically suitable for SERS and in addition to size, other parameters such as shape and chemical composition determine their optical properties. Ideally, the Raman enhancement is so high that single nanoparticles can be detected.
3.1.1 Single Nanoparticle-Based SERS Substrates
Optical excitation of localized surface plasmons is achieved by matching the plasmon peak of the metal colloid with a given laser wavelength for resonant excitation. In biomedical applications, SERS substrates with a LSPR peak in the red or NIR region are often favored, because of minimal autofluorescence of biological specimens and improved image contrast [27, 28]. Silver and gold NPs are the most widely used SERS substrates, however, their extinction maximum occurs in the blue to green region, respectively. Au/Ag nanoshells are suitable substrates in biomedical applications since their LSPR peak is tunable from the red to NIR region (Fig. 15.5a). The optical properties of nanospheres can be calculated by the Lorenz-Mie theory.
a Photographs of different noble metal colloids with a diameter of \(\sim \)55 nm, together with the approximate wavelength of the plasmon peak in water. In contrast to Ag and Au solid nanospheres, the plasmon resonance of Au/Ag nanoshells occurs in the red to near-infrared region and - in addition to the particle diameter - can be controlled by the shell thickness; b Experimental extinction spectra of different noble metal nanoparticles (\(\sim \)60 nm AgNPs, Au NPs and Au/Ag nanoshells) in water [18]
Figure 15.6 illustrates the tunable plasmon band of Au/Ag nanoshells [29] with varying shell thickness based on Mie calculations [30, 31]. All particles have a diameter of 55 nm and the shell thickness d decreases from left to right. The position of the LSPR peak depends on a number of parameters, in particular the size of the NPs and the dielectric function of both the metal and the surrounding medium. The experimental extinction spectra of quasi-spherical 60 nm AgNPs, AuNPs and Au/Ag nanoshells in water are shown in Fig. 15.5b: the corresponding LSPR peaks are observed at \(\sim \)430, \(\sim \)540 and \(\sim \)630 nm, respectively. By using red laser (He-Ne laser, 632.8 nm) excitation, it has been shown experimentally that 60 nm Au/Ag nanoshells yield about 8 times higher SERS intensities compared with gold nanospheres of the same size [26]. Besides the nanoshells with a solvent core and a Au/Ag alloy shell discussed here, several kinds of other nanoshell structures, such as SiO\(_\text {2}\)/Au nanoshells, hollow Au nanoshells, and nanocages, have been developed as SERS substrates [32,33,34,35].
Extinction spectra of single Au/Ag nanoshells with a constant diameter of 55 nm but varying shell thickness, calculated by Lorenz-Mie theory with water as the surrounding medium. The plasmon peak shifts to higher wavelengths as the shell becomes thinner [18]
A new class of multibranched metal NPs such as nanostars or nanoflowers [36, 37] are of particular interest because minor shape modifications enable manipulation of their optical properties. Similar to the nanoshells, the LSPR peak of nanostars is tunable throughout the visible and NIR spectrum. Furthermore, the increased surface area and roughness allows more Raman reporter molecules to be attached on the nanostars than on spherical NPs with a same size and due to their sharp tips, they exhibit high SERS enhancement factors, enabling single-particle SERS detection [38]. In contrast to nanospheres and nanoshells, which exhibit a single LSPR peak, nanorods [39, 40] have two resonances: a weak transverse plasmon peak in the visible region and a strong longitudinal plasmon peak in the longer wavelength region. Au nanorods have also attracted much attention because their longitudinal plasmon resonance as well as the Raman enhancement can be tuned by changing their aspect ratio [41].
3.1.2 Aggregates and Assemblies of Metal Nanoparticles as SERS Substrates
Aggregation of metal NPs (Fig. 15.7a) exhibit great SERS enhancements at the junctions between the NPs, which are usually called “hot spots”.
The electromagnetic field enhancement at hot spots of up to several orders of magnitude enables even single-molecule detection [42]. Particle aggregation is initiated very easily, for example, simply by changing solvents or adding salts. Control over size and shape of NP aggregates is highly desirable for fabricating uniform SERS labels with reproducible signals on a particle-by-particle basis. The biotechnology research group of Intel Corporation claimed to have established a method of Raman label-induced controlled aggregation of silver NPs, the so called Composite Organic-Inorganic Nanoparticles (COINs) [43]. BSA encapsulation is adopted to stabilize the NPs from further aggregation and introduce surface functional groups for bioconjugation [44]. Alternatively, different methods such as electrophoresis, filtration, size exclusion chromatography and density gradient centrifugation can be used to separate small clusters from larger aggregates in crude colloidal mixtures. For example, Salehi and Steinigeweg et al. have established a method to purify small glass-coated clusters (dimers and trimers) of Au NPs by using continuous density gradient centrifugation. The purified clusters exhibit the desired single-particle SERS brightness, even at acquisition times as short as 30 ms per pixel [45].
In addition to small NP aggregates, well defined NP assemblies, which contain controllable hot spots (Fig. 15.7b), are also developed as bright SERS substrates. Dimers of metal NPs are the smallest possible assemblies, which exhibit a single hot spot between the two NPs upon resonant laser illumination. Geometrical configurations that integrate more than two metal NPs into three dimensions represent the next level of complexity beyond dimers and produce multiple hotspots [46]. Silica particles are commonly used as a template for the assembly of many small metal NPs onto its surface, followed by the adsorption of Raman reporter molecules onto the small satellites. The satellite NPs can be either synthesized in situ by adsorbing metal salts and adding reducing agents [47, 48] or be assembled with as-prepared NPs based on covalent linkage or electrostatic interaction [49]. For example, Jeong and co-workers have introduced fluorescent SERS dots, which consist of silica NPs as a core with Raman reporter-labelled silver NPs on their surface and a fluorescent dye-conjugated outer silica shell. The overall size of the assemblies is ca.240 nm and they have been applied for multiplexed molecular diagnostics [48]. Further, also 3D plasmonic nanoassemblies were successfully synthesized via electrostatic self-assembly by Schlücker’s group [46]. Specifically, monodispersed 80 nm gold nanospheres were incubated with Raman reporters and encapsulated with an ultrathin, about 2 to 3 nm thick silica shell and functionalized with amino groups, yielding positively charged glass-encapsulated Au cores. Negatively charged, citrate-stabilized 20 nm AuNPs were then assembled onto the surface of the Au cores via electrostatic attraction. Finite element method (FEM) calculations indicate that the plasmonic coupling between the core and satellite particles as well as between satellites results in large \(|E|^4\) values of up to \({\sim 2} \times 10^{10}\). In order to avoid the relatively large core-satellite gap (>2 nm) due to the silica shell, which reduces the plasmonic enhancement, recently, the same group presented a new route to core-satellite assemblies based on heterobifunctional Raman reporters which serve as molecular linkers for electrostatic conjugation [50]. With a small core-satellite gap (1–2 nm), 3D noble metal NP assemblies using both AgNPs and AuNPs as core particles in combination with smaller AgNPs or AuNPs as satellites were fabricated.
3.2 Labeling with Raman Reporter Molecules
The second step to construct a SERS label is the choice of Raman reporter molecules. A large variety of molecules may be employed as Raman reporters. Ideal Raman reporter molecules exhibit high differential Raman scattering cross sections for bright SERS signals, a minimal number of Raman bands for multiplexing, strong adsorption to the colloid metal surface for stable binding and low or no photobleaching for signal stability. Generally, Raman reporter molecules must be attached directly on or at least near the surface of SERS substrates to experience significant Raman signal enhancement (SERS distance dependence). Positively charged dyes, such as crystal violet , Nile blue , basic fuchsin and cresyl violet may adsorb to the metal surface by electrostatic interactions and delocalized \(\pi \)-electrons. Sulfur containing molecules such as fluorescein isothiocyanate (FITC) and rhodamine B isothiocyanate (RBITC) are also often used because of their high affinity to silver and gold [14]. Recently, Chang’s group reported on the combinatorial synthesis and screening of a triphenylmethane dye library for developing highly sensitive SERS tags, from which they found at least 13 compounds show stronger SERS signal than crystal violet [51]. With a lipoic acid linker , the novel Raman reporters strongly bind to gold NPs and produce strong and stable SERS signals [52]. While most of the commonly used Raman signature molecules are active in the UV/Vis range, endeavors were also made to develop Raman reporters with absorption maxima that are resonant with NIR detection lasers. By synthesizing and screening of an 80-member tricarbocyanine library, Chang and co-workers identified CyNAMLA-381 as a NIR SERS reporter with 12-fold higher sensitivity than the standard 3,3’-diethylthiatricarbocyanine (DTTC), and validated its advantages for the construction of ultrasensitive in vivo SERS probes [53, 54]. More recently, Kircher’s group reported the design and synthesis of a class of NIR absorbing 2-thienyl-substituted chalcogenopyrylium dyes tailored to have high affinity for gold [55]. One notable feature of the pyrylium dyes is the ease in which a broad range of absorptivities can be accessed, and consequently be matched with the NIR light source by careful tuning of the dye’s optical properties. When adsorbed onto gold NPs, these dyes produce biocompatible SERRS probes with attomolar limits of detection amenable to ultrasensitive in vivo multiplexed tumour and disease marker detection [55].
The SERS intensity correlates with the number of Raman reporter molecules on the metal substrates. Porter and co-workers were among the first to employ self-assembled monolayers (SAM) of Raman reporter molecules on noble metal NPs for SERS in immunoassays [56]. They used arylthiols/disulfides as Raman reporters which form a SAM on the metal surface via stable Au-S bonds [57]. The formation of a SAM provides several significant advantages: the uniform orientation of Raman reporter molecules leads to reproducible SERS signatures; the dense packing minimizes co-adsorption of other molecules on the particles surface and thereby reduces or excludes unwanted spectral signals; in addition, a SAM maximizes the number of Raman reporter molecules on the metal surface and creates a maximum SERS intensity. For instance, Au/Ag nanoshells covered with a complete SAM yield ca. 22 times more intense signals compared with the sub-monolayer coverage Fig. 15.8 upon red laser excitation (632.8 nm) [26].
Influence of surface coverage on the brightness of SERS labels: complete SAM compared to submonolayer coverage with Raman labels. The concentration of the colloid with submonolayer coverage is two times larger than in the colloid with monolayer coverage. The increase in signal brightness is 22 ± 5 (n = 3) [26]
3.3 Protection and Stabilization
In biomedical applications, further stabilization and protection of the SERS labels is generally a prerequisite. The encapsulation of SERS particles has several benefits. For instance, the resulting chemical and mechanical stability of the colloidal particles allows particle storage and prevents particle aggregation. Furthermore, the desorption of Raman labels from the metal surface as well as the adsorption of spectrally interfering molecules from the environment to the surface can be eliminated. Various encapsulants, including proteins, organic polymers, silica and hydrophilic SAMs, have been applied to improve colloid stability and biocompatibility of SERS labels as well as to provide functional groups for further bioconjugation [27, 44, 45, 58].
3.3.1 Stabilization with Proteins or Polymers
BSA and thiolated polyethylene glycol (PEG) are two kinds of commonly used surface-coating molecules. BSA adsorbs on the metal surfaces via electrostatic interactions and produces a protective shell. For instance, in a recent study Ren and co-workers introduced a facile way to prepare BSA-coated SERS-based pH sensors [59]. Modification of the NPs with BSA not only protected the Raman reporter molecules in complex conditions, such as high ionic strength solution or biological systems, which ensures the sensitivity and stability of the sensor, but also prevented further aggregation of the NPs and improved their biocompatibility. Another example is the functionalization of COINs [44] (Fig. 15.9a): BSA and glutaraldehyde were introduced to the COINs to form a ca. 10 nm-thick organic encapsulant around them, and to also provide carboxylic acid groups for subsequent bioconjugation. PEG is a nontoxic and hydrophilic polymer that is commonly used to improve drug compatibility and system circulation. Nie and co-workers have investigated the stability of PEGylated gold NPs (Fig. 15.9b) by measuring their SERS signals under a wide range of conditions. The thiol- PEG -coated NPs were stable in even very harsh conditions including strong acids or bases, concentrated salts, or organic solvents (methanol, ethanol and dimethylsulfoxide) [27].
3.3.2 Silica-Encapsulation
Silica-encapsulated SERS labels are attractive because of their high mechanical stability and the option for long-term storage. The concept of silica encapsulation for SERS labels was first introduced by Natan [60] and Nie [61] in 2003. Co-adsorption of Raman labels and SiO\(_\text {2}\) precursors (typically in a 1:20 stoichiometry) leads to formation of a sub-monolayer coverage with Raman reporter molecules onto the metal surface, followed by silica encapsulation with a modified Stöber method [62]. The glass shell provides chemical and physical stability to the SERS label, as well as storage stability and protection against mechanical deformation.
As mentioned in Sect. 15.3.2, in contrast to the sub-monolayer coverage with Raman reporter molecules, a complete SAM with full surface coverage leads to a higher sensitivity. Silica-encapsulated SERS particles comprising a complete SAM combine the chemical stability of a glass shell with the spectroscopic advantages resulting from the maximum and dense surface coverage with Raman reporters in a uniform orientation. Figure 15.10 shows two different synthesis routes towards silica-encapsulated SERS labels comprising a full SAM.
Two different routes to SERS labels with a glass shell as an encapsulant and a self-assembled monolayer (SAM) of Raman reporters on the surface of the metal nanoparticle. Left: Three-step synthesis, starting from a SAM 1a of Raman reporter molecules (Ra) on the surface of Au/Ag nanoshells, subsequent layer-by-layer deposition of the polyelectrolytes PAH (1b) and PVP (1c) and finally the glass encapsulation using tetraethoxyorthosilicate (TEOS) to 1d [26]. Right: One-step approach via a SAM 2a of Raman reporter molecules containing terminal SiO\(_\text {2}\) precursors (Ra-SiO\(_\text {2}\)) and its direct silica encapsulation to 2b using TEOS [63]
In the first route [26] (Fig. 15.10 left), the addition of Raman labels (Ra) to the SERS substrate produces SAM-coated metal nanospheres (1a). Layer-by-layer adsorption of poly(allylamine hydrochloride) (PAH) and polyvinylpyrrolidone (PVP) onto the SAM leads to the polyelectrolyte-coated SAMs 1b and 1c, respectively. The silica shell in 1d is then formed upon addition, hydrolysis and finally condensation of tetraethoxy orthosilicate (TEOS). This route involves different solvents exchange and multiple centrifugations, it is therefore labor and time intensive. A simpler, faster and generally applicable route to silica-encapsulated SERS labels is achievable via a SAM containing terminal SiO\(_\text {2}\)-precursors [63] (Fig. 15.10 right): Ra and terminal SiO\(_\text {2}\)-precursor are covalently bound to each other (Ra-SiO\(_\text {2}\)), i.e. both functions are merged into a single molecular unit. This strategy is independent of the SAM’s surface charge, and is therefore universally applicable. However, it requires an additional synthesis step for the Ra-SiO\(_\text {2}\) precursor conjugate. The transmission electron microscopy (TEM) images in Fig. 15.11 demonstrate the monodispersity of the resulting glass-encapsulated SERS NPs [26]. The thickness of the glass shell can be controlled by the amount of TEOS. The 60 nm Au/Ag nanoshells in Fig. 15.11 have a \(\sim \)10 nm thick (top left) and \(\sim \)25 nm thick (bottom left, right) silica shell. In addition to Au NPs and Au/Ag nanoshells, the silica encapsulation has also been demonstrated for a variety of other plasmonic NPs, including gold nanostars [64], dimers of gold NPs [65], aggregates [45], and core-satellite assemblies [46, 50], which highlights the potential of this approach.
Transmission Electron Microscopy (TEM) images of silica-encapsulated SERS labels with \(\sim \)10 nm (top left) and \(\sim \)25 nm (bottom left and right) thick silica shells. The diameter of the gold/silver nanoshell core is \(\sim \)60 nm [26]
3.3.3 Hydrophilic Stabilization of SAMs
Instead of coating an additional polymer or glass layer on the SERS labels as introduced above, another SERS label design is based on relatively short terminal hydrophilic spacers attached to the SAM [58, 66]. In this rational design of SERS labels depicted in Fig. 15.12, the stabilization of the SAM is achieved by attaching hydrophilic monoethylene glycol (MEG) units with terminal OH groups to the Raman reporters. A small portion of Raman reporter molecules is conjugated to longer triethylene glycol (TEG) units with terminal COOH moieties for bioconjugation. Four advantages result from this strategy: (i) the colloidal surface is completely covered exclusively by Raman reporters for maximum sensitivity; (ii) the entire SERS label is hydrophilic and water soluble due to the MEG/TEG units, i.e. independent of a particular Raman reporter molecule; (iii) increased sterical accessibility of the SAM for bioconjugation via the longer TEG spacers with terminal COOH groups, and (iv) the option for controlled bioconjugation by varying the ratio of the two spacer units (TEG-OH : MEG-COOH).
Hydrophilic SERS labels for controlled bioconjugation . A complete SAM of arylthiols as Raman reporters is adsorbed onto the surface of the metal colloid. Hydrophilic mono- and triethylene glycols (MEG and TEG, respectively) are conjugated to the Raman reporters and stabilize the SAM. The water soluble SERS label can be conjugated to biomolecules via the terminal carboxy moieties of the longer TEG spacers [58]
3.4 Bioconjugation with Antibodies
For iSERS applications, SERS labels must be conjugated to target-specific ligands such as antibodies for antigen recognition. A comparison of different conjugation protocols is summarized in a recent review [67], including physical adsorption, covalent binding and use of adaptor biomolecules. The antibodies can be attached either directly to the NP surface, to the Raman labels, or to the encapsulant such as a PEG or silica shell. Different conjugation strategies are discussed in this section.
3.4.1 Conjugation of Antibodies to SERS Substrates or to Raman Reporter Molecules
Adsorption of proteins, especially of antibodies to colloidal gold (immunogold) based on electrostatic forces, has been explored for decades. An initial design of SERS labels for immunoassays by Porter and co-workers was based on the co-adsorption of antibodies and Raman labels on the surface of gold NPs [56]. In this approach, Raman reporter molecules (arylthiols) and antibodies are directly co-adsorbed to the surface of Au NPs. By using different labels with little spectral overlap, two different antigenic species can be detected simultaneously. However, this design suffers from non-specific binding and “cross-talk” between different SERS labels [57]. An alternative approach based on covalent binding of the antibodies to the Raman reporter molecules improved particle stability and lowered the detection limit, but the steric accessibility of the COOH moieties reduced due to the densely packed SAM [68]. Recently, the same research group presented an improved design using a mixture of two different thiols: an aromatic thiol for the generation of intense and characteristic Raman signals and an alkylthiol with a terminal functional group (e.g. succinimidyl group) for bioconjugation [57]. Schlücker and co-workers have presented an approach (Sect. 15.3.3.3) to synthesize hydrophilic SERS labels in which the colloidal particle was stabilized by two different ethylene glycols (MEG-OH and TEG-COOH) attached to the SAM. This hydrophilically stabilized SAM design guarantees maximum SERS signal intensity and enables efficient conjugation of antibodies, while the number of antibodies can be controlled via the stoichiometric ratio of the two spacers [58].
3.4.2 Conjugation of Antibodies to Protecting Shells
Proteins such as BSA contain a large number of amino and carboxyl groups, which can be used for conjugation with targeting molecules. For example, the functionalization of an encapsulated COIN is based on the activation of the carboxylic acid groups of BSA by N-(3-(dimethylamino)-propyl)-N’-ethylcarbodiimide (EDC); the resulting O-acylisourea intermediates are reacted with amino groups from the antibodies to form amide bonds between the encapsulation layer and the antibodies [69]. Polymer-encapsulated SERS labels usually provide terminal functional groups from the polymer shell. For instance, Nie and co-workers introduced AuNPs coated with thiolated PEG containing terminal carboxyl groups, to which single-chain variable fragments (scFv) were covalently linked [27].
Silica encapsulation provides chemical and physical stability to the SERS labels. Bioconjugation of silica-encapsulated SERS labels usually requires the introduction of functional groups via silane chemistry, e.g. APTMS (NH\(_\text {2}\)) or MPTMS (SH) [13]. The presence of functional moieties on the silica shell then allows bioconjugation of antibodies by established protocols, i.e. amino groups (via succinimides, isothiocyanates or hydrazines), carboxyl groups (via carbodiimides), thiol groups (via maleimides or acetyl bromides) and azides (via click chemistry) [70].
Silica-encapsulated clusters of AuNPs coated with a SAM of Raman reporter molecules (left) are bright SERS labels with single-particle SERS brightness. a Uncontrolled binding of the antibody onto the glass surface of the SERS NP clusters. Due to the uncontrolled orientation of the antibodies, antigen binding sites may be blocked. b Controlled binding and directed orientation of the antibodies onto the silica surface of the SERS NP cluster via coating with protein A/G (blue), which exhibits multiple binding sites for the Fc fragment of the antibody (red). In this case, all antigen binding sites are accessible [71]
3.4.3 Conjugation of Antibodies by Chimeric Proteins
Molecular recognition of the target antigen by the antigen recognition sites of the antibody is highly important for obtaining molecular specificity and reliable diagnostic results in IHC. However, conventional bioconjugation schemes (Fig. 15.13a) through covalent binding do not provide site-specificity, i.e. there is no control over reaction with a particular amino acid residue: the amino acid residues might be close to the Fc part of the receptor (preferred) or within the antigen recognition sites (nonpreferred). Overall, this uncontrolled binding in terms of missing site-specificity may result in a “scrambled” orientation of the antibodies on the surface of the SERS labels. In contrast, the approach (Fig. 15.13b) employed by Salehi et al. avoids this problem by using the chimeric protein A/G, which exhibits multiple bindings to the Fc domain of the antibody [71]. Coating the silica surface of the SERS labels with protein A/G leads to controlled binding and directed orientation of the antibodies onto the silica surface, and is therefore desirable in iSERS staining since all antigen recognition sites of the antibody are accessible for binding to the antigen.
3.5 Single-Particle Brightness and Homogeneity of SERS Labels
iSERS microscopy is a novel imaging technique in tissue diagnostics based on the selective staining of multiple target proteins in tissues. The quality of the IHC staining results strongly depends on the quality of the SERS NPs. Brightness, ideally a detectable SERS signal at the single-particle level, and homogeneity, ideally every SERS particle is equally bright, are the two key issues related to (quantitative) iSERS. The term “single-particle brightness” may be defined in a pragmatic way: a SERS label with single-particle brightness per definitionem exhibits a detectable SERS signal within the defined acquisition time per point (pixel), usually milliseconds [45] or at most 1 s. In a recent study [45], two central questions in iSERS microscopy were addressed: (i) What is the role of SERS labels with single-particle brightness in IHC in terms of false-positive results? and (ii) What are the resulting requirements on the purity of the SERS colloid at the ensemble level?
The role of single-particle brightness in IHC is schematically illustrated in Fig. 15.14. Silica-encapsulated dimers of gold NPs have been demonstrated to exhibit single-particle SERS brightness (1 sec acquisition time) in correlative single-particle AFM/SERS experiments [65]. Figure 15.14a shows the situation of “binding plus detectable SERS signal” when such a dimer, conjugated to the corresponding antibody, binds to the antigen presented on the tissue section. Due to the single-particle SERS brightness of this type of highly active SERS label, a Raman signal is detectable within the defined acquisition time of the Raman mapping experiment. Overall, the presence of the antigen is correctly indicated by the Raman spectrum of the SERS label (positive result). In contrast to dimers, single gold NPs do not exhibit a notable plasmonic enhancement. In previous experiments on single 60 nm gold NPs, no SERS signal could be detected within 1 s acquisiton time [65]. A recent experimental study also demonstrated that 80 nm gold and silver NP monomers were both not SERS-active [72]. Figure 15.14b shows the situation of “binding but no detectable SERS signal” for gold NPs monomers conjugated to antibodies for target recognition. Although the antibody correctly recognizes its antigen, no SERS signal is detectable due to the lack of plasmonic activity of the single gold NP used as a SERS label. Overall, a false-negative result is obtained.
The role of single-particle brightness in IHC via iSERS microscopy. a Clusters (dimers and trimers) of gold NPs with single-particle brightness yield positive results, while b monomers without single-particle brightness give false negative results [45]
The second question on the role of using homogenous SERS colloids at the ensemble level is directly related to the first one. Again, antibody-monomer conjugates yield false-negative results (Fig. 15.14b) and the corresponding antigen is not accessible anymore for detection by an antibody-dimer conjugate. Therefore, a colloidal mixture containing both NPs with and without single-nanoparticle brightness is not optimal for iSERS microscopic experiments; instead, only highly sensitive SERS labels with single-particle brightness should be employed. In summary, in a population of SERS labels, every particle exhibits single-particle brightness. Every individual SERS NP that selectively binds to the target should be detectable within the given (short) acquisition time (per pixel). Otherwise, false-negative results may be generated.
Correlative SEM/SERS experiment of single particles provide the most direct and unambiguous way for establishing structure-activity correlations [46]. Determining the structure of a single plasmonic nanostructure as well as its optical activity requires electron and optical microscopic experiments on the same particle. First, it is necessary to ensure that exactly the same particle is characterized by both electron and optical microscopy. Second, this particle must be spatially isolated from other particles in order to avoid disturbing spectral interferences in the optical experiments. Salehi et al. have compared SERS signals from a glass-coated monomer, dimer, and trimer of gold NPs by correlative SEM/SERS experiments [45] Fig. 15.15. Colloids were deposited on a framed silicon waver and high resolution scanning electron microscopy (HR-SEM) was employed to identify single NPs/clusters. With 30 ms integration time, the glass-coated trimer (red) and dimer (blue) yielded detectable SERS signals, while the glass-coated monomer (black) did not. The SERS signal from the single trimer is about two to three times stronger than that from the single dimer. This is presumably due to orientation effects since the dimer response is highly anisotropic while that of the trimer is not. Also, the trimer has a larger number of hot spots than the dimer. Multiple hotspots occurs at the tips of nanostars and at the junctions between two nanospheres in 3D plasmonic superstructures. Thus, they exhibit a quasi polarization-independent SERS response at the single-particle level [38, 46, 50].
Correlative dark-field/SERS/SEM single-particle experiments on glass-coated monomers and clusters of gold NPs. a Dark-field microscopy, b SERS microscopy and c HR-SEM images. Only the glass-coated gold trimer (yellow arrow) and dimer (green arrow) exhibit single-particle SERS brightness (30 ms integration time), while the glass-coated monomer (blue arrow) does not. d Normalized single-particle SERS spectra of the glass-coated monomer, dimer, and trimer from (b) [45]
4 Application of iSERS Microscopy for Tissue-Based Cancer Diagnostics
iSERS microscopy is the combination of target-specific SERS probes and Raman microspectroscopy. Various methodologies in Raman microspectroscopy are available [73]. Mapping approaches with point or line focus illumination, providing full spectral information in combination with a xy-translation stage are most commonly applied.
4.1 Localization of Single Proteins by iSERS Microscopy
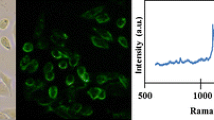
The proof of principle for iSERS microscopy using a SERS-labelled primary antibody for protein localization in tissue was demonstrated by Schlücker and coworkers in 2006 [74]. Prostate-specific antigen (PSA) was chosen as a target protein because of its high expression levels in prostate tissue and its selective histological abundance in the epithelium of the prostate gland . By using Au/Ag nanoshells as colloidal SERS substrates, aromatic thiols as Raman reporters, the localization of PSA in tissue specimens of biopsies from patients undergoing prostatectomy was achieved with SERS-labelled primary anti-PSA antibodies. SERS signals were detected in the PSA-(+) epithelium of the incubated prostate tissue section (Fig. 15.16A–C), while locations in the PSA-(-) stroma and lumen served as negative controls (Fig. 15.16D, E).
Proof of principle for iSERS microscopy using SERS-labelled primary antibodies for tissue diagnostics. Prostate-specific antigen (PSA) was localized in biopsies from patients with prostate cancer. Left: White light image of the prostate tissue section. Three different histological classes can be observed: epithelium, stroma or connective tissue and lumen, in which PSA is either abundant (+) or not (−). Right: Spectra obtained at the locations (A) to (E) indicated by arrows in the white light image. SERS-labelled anti-PSA antibodies are detected in the PSA-(+) epithelium (locations A-C), exhibiting the characteristic Raman signals of the SERS label. Locations in the PSA-(-) stroma (D) and lumen (E) serve as negative controls where no spectral contributions of the SERS-labelled antibody are detected [74]
Further applications of iSERS microscopy for tissue diagnostics with various types of SERS probes appeared soon after this initial study. For example, Sun and coworkers used BSA-coated aggregated silver clusters (COINs) as SERS NPs for PSA localization in prostate tissue sections [44]. In order to illustrate the multiplexing capabilities of iSERS in tissue staining, a two color-COIN staining against PSA was performed: SERS labels with two distinct Raman signatures were conjugated to anti-PSA antibodies, and simultaneous applied to the tissue. The characteristic Raman signatures from both COINs were detected at almost every location in the epithelium, suggesting that steric hindrance from the SERS probes does not represent a major problem. In a subsequent study, the same group presented a detailed comparison of the staining performance with SERS- and fluorophore-PSA antibody conjugates on adjacent tissue sections [69]. Staining with COIN- and Alexa-fluorophore labelled antibodies yielded similar results, with a lower staining accuracy for COINs, which is attributed to an elevated false negative rate, but signal intensities were comparable for COINs and Alexa-fluorophore, supporting the further development of SERS NP labels for iSERS.
Application of Au/Ag nanoshells with a hydrophilic-spacer SAM on selective imaging of PSA by SERS microscopy is illustrated in Fig. 15.17 [58]. A bright field image is shown in Fig. 15.17a, the region used for Raman point mapping is indicated with a white box. Three different histological classes can be identified: epithelium (E), stroma (S) and lumen (L). Epithelium is PSA-(+), while (S) and (L) are PSA-(-). The false-color image in Fig. 15.17b is based on the intensity of the Raman marker band at 1336 rel. cm\(^{-1}\), which shows the selective abundance of PSA in the epithelium. Representative SERS spectra recorded at selected locations in the (E), (S) and (L) are shown in Fig. 15.17c. In negative control experiments with either bare SERS labels or SERS labels conjugated to BSA, no binding to the tissue was observed. This additionally confirms the specific recognition capacity of the SERS-labelled anti-PSA antibody in the epithelium.
iSERS imaging for tissue diagnostics. a Bright field microscope image of a prostate tissue section. Different histological classes are indicated: lumen (L), epithelium (E) and stroma (S). The rectangular box shows the region in which spectra were acquired by point mapping. b The false-color SERS image. c Representative SERS spectra recorded at selected locations in the (E), (L) and (S). d Overlaid bright field image with the false-color SERS image shows that the characteristic signal of the SERS-labelled antibody is observed selectively in the epithelium. Low nonspecific binding is attributed to the hydrophilic ethylene glycols of the SERS probes [58]
Localization of the basal cell marker p63 is a clear test whether iSERS microscopy provides the necessary specificity for selectively staining the nuclei of the basal cells. In a recent study [66], hydrophilically stabilized gold nanostars served as SERS labels for imaging of p63 in benign prostate biopsies. The comparison of the SERS false-color image and the white light image in Fig. 15.18a reveals that p63 is only abundant in the basal epithelium of benign prostate tissue, but not in the lumen or other histological classes and subclasses such as connective tissue (stroma) and secretory epithelium. Five representative SERS spectra from different regions in Fig. 15.18a are displayed in Fig. 15.18b: (1–3) basal epithelium, (4) lumen and (5) stroma. The corresponding locations are marked with white crosses in Fig. 15.18a (1–5: from top to bottom). The SERS mapping experiment clearly demonstrates the specific binding of the SERS-labelled anti-p63 antibody to its target molecule p63 in the basal epithelium. Negative control experiments with SERS-labelled BSA did not exhibit detectable Raman signals.
a Bright field image of a prostate tissue specimen with an overlaid SERS false-color image based on the intensity of the 1340 rel. cm\(^{-1}\) Raman marker band of the SERS label. b Five representative SERS spectra from different locations in (a), indicated by white crosses in the SERS false-color image (from top to bottom) [66]
Epstein-Barr virus (EBV) -encoded latent membrane protein 1 (LMP1) is closely associated with the occurrence and development of nasopharyngeal carcinoma and can be used as a tumor marker in screening for the disease [75]. Chen and coworkers fabricated LMP1-functionalized and 4-mercaptobenzoic acid (4-MBA)-labelled Au/Ag core-shell bimetallic NPs. They applied them for analyzing LMP1 in nasopharyngeal tissue FFPE sections obtained from 34 cancer patients and 20 healthy controls. Data from iSERS microscopy showed that this kind of SERS NP conjugates detected LMP1 in nasopharyngeal tissue sections with high sensitivity and specificity. The results are superior to those of conventional IHC staining for LMP1 and in excellent agreement with those of in situ hybridization for EBV-encoded small RNA [75].
The development of increasingly sensitive SERS NP labels with single-particle brightness enables faster imaging experiments. Earlier reports of iSERS microscopy used integration times of 100 ms [44, 66] or even 1 sec [74]. In a recent study presented by Salehi et al. a proof of concept for rapid iSERS microscopy with 30 ms acquisition time per pixel for selective imaging of p63 in prostate tissue sections was demonstrated [45]. Purified glass-coated clusters (dimers and trimers) of gold NPs were applied as SERS labels and conjugated to anti-P63 antibodies. With 30 ms acquisition time per pixel, the selective staining of p63 in the basal cells of the epithelium is clearly observable. Negative control experiments (100 ms per pixel) with SERS-labelled BSA indicate no or only minimal non-specific binding. Time efficiency is critically important in particular when large or even very large areas in histopathological studies must be investigated, not necessarily with high spatial resolution, but with a large number of points.
SERS signatures of two different SERS labels comprising two different aromatic thiols present as a SAM on the surface of AuNPs: 4-nitrothiobenzoic acid (4-NTB) and 4-mercaptobenzoic acid (4-MBA). The colloidal SERS particles were dispersed in ethanol [71]
4.2 Co-localization of Multiple Proteins by iSERS Microscopy
A central advantage of iSERS is its enormous spectral multiplexing capacity for multi-color IHC. Generally, the term multiplexing refers to the parallel determination of several parameters within a single experiment. In the context of SERS as a labeling strategy for targeted research, multiplexing addresses the issue of simultaneously detecting and identifying the spectral fingerprint of several distinct SERS nanoparticle labels conjugated to different target-specific ligands, i.e. antibodies in the case of IHC.
Fig. 15.19 shows the SERS spectra of two different SERS labels comprising aromatic thiols present as a SAM on the surface of AuNPs [71]. The two Raman reporter molecules are 4-nitrothiobenzoic acid (4-NTB, red solid line) and 4-mercaptobenzoic acid (4-MBA, green solid line), respectively. Since unique and spectrally separated Raman marker bands for both SERS labels are available, a univariate approach based on the integrated Raman intensities of single peaks - here around 1340 rel. cm\(^{-1}\) (4-NTB, red) and around 1590 rel. cm\(^{-1}\) (4-MBA, green) - is sufficient for spectral discrimination in a two-color SERS microscopic experiment employing 4-NTB- and 4-MBA-based SERS labels.
Using highly sensitive, silica encapsulated gold nanoparticle clusters as SERS labels, two-color iSERS microscopy for co-localization of PSA and p63 in non-neoplastic prostate tissue FFPE specimens was demonstrated [71]. The tissue sections were incubated with both SERS-labelled PSA antibodies (Raman reporter: 4-NTB) and SERS-labelled p63 antibodies (Raman reporter: 4-MBA). The false-color SERS image in Fig. 15.20 confirms the binding specificity and signal brightness of the two SERS probes: p63 is selectively observed in the nuclei of the basal cells, while PSA is abundant in the entire epithelium, but not in the stroma.
Two-color immuno-SERS microscopy for the co-localization of p63 (green) and PSA (red). a White light image of a healthy prostate sample. b Overlay with a p63 false-color SERS image; Raman reporter: 4-MBA. c Overlay with a PSA false-color SERS image; Raman reporter: 4-NTB. d Co-localization by overlay with p63/PSA false-color images. Acquisition time: 100 ms per pixel [71]
For multi-color (>2) SERS experiments, data analysis based on single Raman peaks is usually not feasible because of spectral overlap. For decomposition of the overlapping spectral contributions of the individual SERS probes, signal processing algorithms must be applied. Knudsen and coworkers reported a multiplex spectral fitting method that exploits the entire fingerprint to separate and quantify individual SERS probe signals [76]. They first tested four COINs conjugated to antibodies in a quadruplex plate binding assay detecting PSA in solution. Figure 15.21a shows the spectral decomposition for one sample. The best-fit spectrum (black) is overlaid on the measured spectrum (gray, largely covered by the black) and contains signatures of all four antibody-COIN conjugates. Isolated peaks arising from a single COIN are indicated by colored arrows, while peaks arising from multiple COINs are indicated by colored boxes. The typical single peak method cannot be used for quantification due to spectral overlap. The spectral fitting method exploits the entire spectral signature to extract individual component spectra (Fig. 15.21a, colored) and quantitative signals for each antibody-COIN conjugate. The calculated intensity of individual antibody-COIN conjugates in Fig. 15.21b is proportional to the coated PSA concentration and comparable to the linear antibody-COIN dose-response in singlet experiments. Then the authors applied the spectral fitting method to image the expression of PSA, cytokeratin-18, and DNA in FFPE prostate tissue sections (Fig. 15.21c). The spectral analysis method effectively removes tissue autofluorescence and other unknown background, allowing accurate and reproducible imaging at subcellular spatial resolution.
a Spectral deconvolution of the quadruplex assay. The upper traces represent the measured spectrum (gray, largely covered by the black spectrum) and best-fit (black) spectrum. The colored lines represent the pure COIN spectra extracted with the spectral deconvolution method. Peaks that primarily arise from a single COIN are shown by colored arrows, and peaks arising from multiple COINs are color-coded by boxes. b Dose-response curves for the multiplex plate binding assay using four COIN preparations conjugated to anti-PSA antibody. c Spectral deconvolution in a multiplex FFPE tissue assay. BFU-COIN was conjugated to anti-CK18 antibody (BFU-CK18); AOH-COIN was conjugated to anti-PSA antibody (AOH-PSA), and a fluorescent dye targeted DNA (YOYO) [76]
4.3 Non-specific Binding
Rapid advances in nanotechnology have led to an increasing number of nanomaterials for biomedical applications in the areas of cancer diagnostics and therapeutics. For both in vitro and in vivo applications of NPs conjugated to targeting molecules, one of the major challenges is the reduced targeting efficiency caused by non-specific binding. The main reasons for non-specific binding using functionalized NPs are: the increased size, weight and surface area of NPs compared to traditional labeling with dyes or molecular fluorophores; conjugation of the targeting molecules onto NPs usually requires the introduction of reactive functional groups on the NPs, while excess or unreacted groups often lead to increased nonspecific protein interaction; aggregation of NPs during the preparation process or in physiological media. Numerous efforts have been made to analyse and reduce non-specific binding of bioconjugated NPs, including: comparing the effect of size and shape of NPs on their targeting efficiency; surface modification of NPs; blocking the surface of NPs with proteins or hydrophilic molecules. A new strategy to minimize the effect of non-specific binding in tissue staining is the simultaneous detection and ratiometric quantification of targeted and non-targeted NPs by staining tissues with an equimolar mixture of targeted NPs and non-targeted NPs that account for nonspecific effects [77].
4.3.1 Size and Shape of NPs
Immunogold labeling, in which colloidal gold in the size range from few nm up to 30 nm is attached to antibodies, has been broadly applied for identification and localization of proteins at the ultrastructural level. Colloidal gold particles with different diameters (5 and 20 nm) were compared with respect to non-specific binding. The results indicate that small gold NPs have a greater tendency to non-specific binding although they present higher labeling densities resulting from the greater accessibility to binding sites and lower steric interference [78]. This is probably due to a larger percentage of uncoated surface area with proteins on 5 nm NPs than the larger NPs [79]. Recently, Mitragotri and coworkers reported a surprising finding that particle shape enhances avidity as well as specificity of interactions of antibodies with their targets. Using spherical, rod-, and disk-shaped polystyrene NPs and trastuzumab as the targeting antibody, they studied specific and nonspecific binding and uptake in breast cancer cells. The conclusion is that, rod-shaped NPs exhibit higher specific and lower nonspecific accumulation compared with their quasi-spherical counterparts [80].
4.3.2 Surface Modification and Blocking of NPs
Covalent conjugation of target-specific binding molecules to the SERS nanoparticle labels requires the presence of functional groups on the surface of the particles. However, following conjugation, excess active functional groups, which are capable of binding to or reacting with various chemical and biological species, might induce aggregation of NPs or non-specific adsorptions, and lead to false positive/negative results. Therefore, nanoparticle surface design involves an optimum balance of the use of inert and active surface functional groups [81]. Tan and coworkers produced silica NPs with different functional groups, including carboxylate, amine, amine/phosphonate, poly(ethylene glycol), octadecyl, and carboxylate/octadecyl groups, and compared their non-specific binding behavior using confocal imaging of amine-modified glass slides. The results indicate that silica NPs modified with a combination of carboxylate and octadecyl groups have less agglomeration and non-specific binding to the glass slide as compared to NPs having only carboxylate, octadecyl, or PEG groups on the surface [81]. In another immunoassay performed by Arslanoglu et al., carboxyl-terminated SERS tags show strong Raman signal, specificity for the target protein, robust response in the presence of various inorganic pigments, and reduced aggregation on sample surfaces compared to amino-terminated or commercial SERS tags [82]. An elegant strategy established by Nie’s group for modifying SERS labels is to functionalize the NPs with a mixed layer of 85% thiolated PEG (HS-PEG), which ensures minimal non-specific binding and stabilizes the NPs against aggregation, and 15% HS-PEG-COOH containing an additional carboxyl group for conjugation with targeting molecules [27, 83]. The dual-SAM design reported by Schlücker’s group is a similar approach to synthesize hydrophilic SERS probes. Two different ethylene glycol spacers are covalently bound to the same type of Raman reporter molecule: a short monoethylene glycol (MEG-OH) and a longer triethylene glycol (TEG-COOH) moiety. The terminal of hydroxyl group of the MEG spacer guarantees water solubility and reduces non-specific interactions, while the terminal carboxyl group of the TEG spacer allows subsequent conjugation to biomolecules such as antibodies. Controlled bioconjugation is possible by varying the stoichiometric ratio of both spacers [58, 66].
An alternative approach to reduce non-specific binding is to block the NPs with proteins like BSA or protein/PEG mixtures [84]. BSA is a blocking agent widely used in enzyme-linked immunosorbent assay (ELISA), western blot and IHC, which blocks potential sites of nonspecific interaction, eliminates background without altering or obscuring the epitope for antibody binding. It has also been reported that BSA-conjugated NPs show improved stability against flocculation and low toxicity, while simultaneously providing functional groups for further bioconjugation [85]. In recent studies on NP-based cell imaging, intracellular sensing or tissue staining, it was found that an additional blocking step with BSA on the NPs effectively reduces non-specific binding and minimizes cross talk [59, 85, 86].
4.3.3 Ratiometric Quantification of Specific and Non-specific Binding
In the approaches discussed above, efforts are mainly made to minimize non-specific binding through optimizing the preparation or modification process of the SERS NPs. Recently, the dual-reporter ratiometric detection with multiplexed positive- and negative-control contrast agents has become a valuable new strategy for accurately quantifying specific versus non-specific signals [77, 87]. Distinguishing between specific and non-specific probe accumulation on tissues is critical. Current designs of the negative control to detect non-specific binding are normally performed in two ways: i) staining two adjacent sections with target antibody-modified SERS NPs and non-targeted isotype control antibody-modified SERS NPs, respectively; ii) compare accumulation of target antibody-modified SERS probes on normal tissue sections and cancerous tissue sections. However, variable tissue optical and mechanical properties (e.g. diffusion, porosity, interstitial pressure), uneven rinse removal of unbound probes, and uneven detection working distance can all lead to non-specific and misleading sources of contrast [88]. For example, Liu and coworkers observed that non-targeted NPs which are topically applied on tissues show higher retention in normal tissues versus xenograft tumors [77]. Instead of using FFPE tissue sections, they stained fresh tissues with an equimolar mixture of targeted NPs and non-targeted NPs. A calibrated ratiometric analysis method was used to investigate the performance of SERS probes under clinically relevant conditions, demonstrating the ability to accurately quantify molecularly specific versus nonspecific NP accumulation on tumor and normal tissues with a spectral integration time of 0.1 s [77]. The authors claimed that, SERS NPs are particularly well-suited for ratiometric detection, not only because of their high multiplexing capability but also due to the ability to excite multiple SERS NP colors with a single laser wavelength, ensuring that all SERS NPs are interrogated identically in terms of illumination intensity, spot size and effective excitation depth [77]. Using a cocktail of many targeted SERS NPs and one non-targeted NP to account for nonspecific effects, this approach will offer increased accuracy for multiplexed-tumor detection in spite of the large degree of molecular heterogeneity amongst tumors [88].
5 Summary and Outlook
iSERS microscopy is a novel and promising imaging approach for tissue-based cancer diagnosis, in particular because of its immense multiplexing capacity for simultaneous target localization in combination with quantification (see Fig. 15.22) [17]. First applications of iSERS microscopy in ex-vivo tissue imaging, i.e. the combination of target-specific antibody-SERS NP conjugates in conjunction with Raman microspectroscopy, have been demonstrated within the last 10 years. These studies were mainly performed on prostate tissue, which was also used in the proof of concept experiment as a model, but multiplexed detection of prognostic markers in various kinds of tissues are expected to appear within the next years.
SERS microscopy has the potential for the detection of multiple targets (multiplexing) and their quantification, two important aspects for tissue diagnostics [17]
As emerging optical nanodiagnostic probes, SERS NP labels have been developed as an attractive class of biological labeling agents with unique advantages and applications that are not achievable with organic dyes or molecular fluorophores. Advantages include spectral multiplexing, quantitation in combination with high sensitivity and photostability. High quality data, in particular the ability to clearly distinguish the SERS signals from background autofluorescence, is also an important aspect. A disadvantage may be the relatively large size of SERS labels compared with molecular fluorophores and quantum dots. However, in a recent study using SERS NPs with two distinct spectral signatures for staining the same protein in tissue, both particles were detected in all spots, suggesting that steric hindrance from the SERS probes does not represent a major problem [44].
Two most essential issues in the preparation of colloidal SERS labels are their brightness, ideally at the single-particle level, and their homogeneity/uniformity at the ensemble level. Several groups worldwide are actively working on improved designs and syntheses of SERS labels with high brightness and reproducible signals. Target-specific binding molecules, including antibodies, peptides, engineered proteins and nucleic acid aptamers, are “smart molecules” which recognize and selectively bind to the corresponding targets. Antibodies are the most widely used targeting molecules in current applications, however, in future studies, smaller targeting molecules like peptides or aptamers may be increasingly used, in particular because of their lower cost.
Advances in laser, filter, spectrometer and detector technology contributed to the fact that Raman including SERS spectroscopy is now a mature analytical technique. Easy to use commercial equipment permits to perform experiments without an in-depth knowledge of optics and lasers. For Raman microscopy, extremely fast Raman systems equipped with motorized xy-stages are commercially available, enabling data acquisition down to only few milliseconds or even sub-milliseconds per pixel. In order to enable a direct comparison and correlation of Raman images to other optical microscopic images, like fluorescence and dark-field, an integration of different optical techniques on the same microscope setup is favorable.
References
D.E. Carvajal-Hausdorf, K.A. Schalper, V.M. Neumeister, D.L. Rimm, Lab. Invest. (2014)
E.C. Stack, C. Wang, K.A. Roman, C.C. Hoyt, Methods 70(1), 46 (2014)
J. Liu, S.K. Lau, V.A. Varma, R.A. Moffitt, M. Caldwell, T. Liu, A.N. Young, J.A. Petros, A.O. Osunkoya, T. Krogstad et al., ACS nano 4(5), 2755 (2010)
L.W. Wang, C.W. Peng, C. Chen, Y. Li, Breast Cancer Res. Treat. 151(1), 7 (2015)
T.Q. Vu, W.Y. Lam, E.W. Hatch, D.S. Lidke, Cell Tissue Res. 360(1), 71 (2015)
R. Lankowicz, Joseph, Principles of Fluorescence Spectroscopy, 3rd edn. (Springer, USA, 2006)
A. Matschulat, D. Drescher, J. Kneipp, ACS Nano 4(6), 3259 (2010)
M. Vendrell, K.K. Maiti, K. Dhaliwal, Y.T. Chang, Trends Biotechnol. 31(4), 249 (2013)
W. Xie, S. Schlücker, Phys. Chem. Chem. Phy. 15(15), 5329 (2013)
S. Feng, S. Huang, D. Lin, G. Chen, Y. Xu, Y. Li, Z. Huang, J. Pan, R. Chen, H. Zeng, Int. J. Nanomedicine 10, 537 (2015)
S. Feng, D. Lin, J. Lin, B. Li, Z. Huang, G. Chen, W. Zhang, L. Wang, J. Pan, R. Chen et al., Analyst 138(14), 3967 (2013)
K. Kong, C. Kendall, N. Stone, I. Notingher, Adv. Drug Delivery Rev. (2015)
Y. Wang, S. Schlücker, Analyst 138(8), 2224 (2013)
Y. Wang, B. Yan, L. Chen, Chem. Rev. 113(3), 1391 (2012)
W.E. Doering, M.E. Piotti, M.J. Natan, R.G. Freeman, Adv. Mater. 19(20), 3100 (2007)
R.L. McCreery, Raman Spectroscopy for Chemical Analysis, vol. 225 (Wiley, New York, 2005)
S. Schlücker, Chem. Phys. Chem. 10(9–10), 1344 (2009)
S. Schlücker (ed.), Surface Enhanced Raman Spectroscopy: Analytical, Biophysical and Life Science Applications (Wiley, New York, 2011)
S. Schlücker, Angew. Chem. Int. Ed. 53(19), 4756 (2014)
M. Moskovits, Rev. Mod. Phy. 57(3), 783 (1985)
A. Otto, J. Raman Spectrosc. 22(12), 743 (1991)
A. Campion, P. Kambhampati, Chem. Soc. Rev. 27(4), 241 (1998)
M. Moskovits, J. Raman Spectrosc. 36(6–7), 485 (2005)
P.L. Stiles, J.A. Dieringer, N.C. Shah, R.P. Van Duyne, Annu. Rev. Anal. Chem. 1, 601 (2008)
R. Aroca, Surface-Enhanced Vibrational Spectroscopy (Wiley, New Yark, 2006)
B. Küstner, M. Gellner, M. Schütz, F. Schöppler, A. Marx, P. Ströbel, P. Adam, C. Schmuck, S. Schlücker, Angew. Chem. Int. Ed. 48(11), 1950 (2009)
X. Qian, X.H. Peng, D.O. Ansari, Q. Yin-Goen, G.Z. Chen, D.M. Shin, L. Yang, A.N. Young, M.D. Wang, S. Nie, Nat. Biotechnol. 26(1), 83 (2008)
G. von Maltzahn, A. Centrone, J.H. Park, R. Ramanathan, M.J. Sailor, T.A. Hatton, S.N. Bhatia, Adv. Mater. 21(31), 3175 (2009)
M. Gellner, B. Küstner, S. Schlücker, Vib. Spectrosc. 50(1), 43 (2009)
G. Mie, Ann. der Phys. 330(3), 377 (1908)
C.F. Bohren, D.R. Huffman, Absorption and Scattering of Light by Small Particles (Wiley, New Yark, 2008)
Y. Cui, B. Ren, J.L. Yao, R.A. Gu, Z.Q. Tian, J. Phy. Chem. B 110(9), 4002 (2006)
C. Sauerbeck, M. Haderlein, B. Schürer, B. Braunschweig, W. Peukert, R.N. Klupp, Taylor. ACS Nano 8(3), 3088 (2014)
W. Shi, Y. Sahoo, M.T. Swihart, P. Prasad, Langmuir 21(4), 1610 (2005)
M. Rycenga, K.K. Hou, C.M. Cobley, A.G. Schwartz, P.H. Camargo, Y. Xia, Physical Chemistry Chemical Physics 11(28), 5903 (2009)
A.M. Fales, H. Yuan, T. Vo-Dinh, J. Phy. Chem. C 118(7), 3708 (2014)
Y. Liu, H. Yuan, F.R. Kersey, J.K. Register, M.C. Parrott, T. Vo-Dinh, Sensors 15(2), 3706 (2015)
C. Hrelescu, T.K. Sau, A.L. Rogach, F. Jäckel, J. Feldmann, Appl. Phys. Lett. 94(15), 153113 (2009)
P. Negri, R.A. Dluhy, J. Biophotonics 6(1), 20 (2013)
S.H. Seo, B.M. Kim, A. Joe, H.W. Han, X. Chen, Z. Cheng, E.S. Jang, Biomaterials 35(10), 3309 (2014)
C.J. Orendorff, L. Gearheart, N.R. Jana, C.J. Murphy, Phys. Chem. Chem. Phys. 8(1), 165 (2006)
N.J. Halas, S. Lal, W.S. Chang, S. Link, P. Nordlander, Chem. Rev. 111(6), 3913 (2011)
X. Su, J. Zhang, L. Sun, T.W. Koo, S. Chan, N. Sundararajan, M. Yamakawa, A.A. Berlin, Nano lett. 5(1), 49 (2005)
L. Sun, K.B. Sung, C. Dentinger, B. Lutz, L. Nguyen, J. Zhang, H. Qin, M. Yamakawa, M. Cao, Y. Lu et al., Nano lett. 7(2), 351 (2007)
M. Salehi, D. Steinigeweg, P. Ströbel, A. Marx, J. Packeisen, S. Schlücker, J. Biophotonics 6(10), 785 (2013)
M. Gellner, D. Steinigeweg, S. Ichilmann, M. Salehi, M. Schütz, K. Kömpe, M. Haase, S. Schlücker, Small 7(24), 3445 (2011)
J.H. Kim, J.S. Kim, H. Choi, S.M. Lee, B.H. Jun, K.N. Yu, E. Kuk, Y.K. Kim, D.H. Jeong, M.H. Cho et al., Anal. Chem. 78(19), 6967 (2006)
S. Jeong, Y.i. Kim, H. Kang, G. Kim, M.G. Cha, H. Chang, K.O. Jung, Y.H. Kim, B.H. Jun, Y.S. Lee, et al., Sci. Rep. 5 (2015)
C. Wang, Y. Chen, T. Wang, Z. Ma, Z. Su, Adv. Funct. Mater. 18(2), 355 (2008)
M. Schütz, S. Schlücker, Phys. Chem. Chem. Phy. 17(37), 24356 (2015)
S.J. Cho, Y.H. Ahn, K.K. Maiti, U. Dinish, C.Y. Fu, P. Thoniyot, M. Olivo, Y.T. Chang, Chem. Commun. 46(5), 722 (2010)
K.K. Maiti, A. Samanta, M. Vendrell, K.S. Soh, M. Olivo, Y.T. Chang, Chem. Commun. 47(12), 3514 (2011)
A. Samanta, K.K. Maiti, K.S. Soh, X. Liao, M. Vendrell, U. Dinish, S.W. Yun, R. Bhuvaneswari, H. Kim, S. Rautela et al., Angew. Chem. Int. Ed. 50(27), 6089 (2011)
K.K. Maiti, U. Dinish, A. Samanta, M. Vendrell, K.S. Soh, S.J. Park, M. Olivo, Y.T. Chang, Nano Today 7(2), 85 (2012)
S. Harmsen, M.A. Bedics, M.A. Wall, R. Huang, M.R. Detty, M.F. Kircher, Nature communications 6 (2015)
J. Ni, R.J. Lipert, G.B. Dawson, M.D. Porter, Anal. Chem. 71(21), 4903 (1999)
M.D. Porter, R.J. Lipert, L.M. Siperko, G. Wang, R. Narayanan, Chem. Soc. Rev. 37(5), 1001 (2008)
C. Jehn, B. Küstner, P. Adam, A. Marx, P. Ströbel, C. Schmuck, S. Schlücker, Phys. Chem. Chem. Phys. 11(34), 7499 (2009)
X.S. Zheng, P. Hu, Y. Cui, C. Zong, J.M. Feng, X. Wang, B. Ren, Anal. Chem. 86(24), 12250 (2014)
S.P. Mulvaney, M.D. Musick, C.D. Keating, M.J. Natan, Langmuir 19(11), 4784 (2003)
W.E. Doering, S. Nie, Anal. Chem. 75(22), 6171 (2003)
W. Stöber, A. Fink, E. Bohn, J. Colloid Interface Sci. 26(1), 62 (1968)
M. Schütz, B. Küstner, M. Bauer, C. Schmuck, S. Schlücker, Small 6(6), 733 (2010)
M. Li, J. Zhang, S. Suri, L.J. Sooter, D. Ma, N. Wu, Anal. Chem. 84(6), 2837 (2012)
D. Steinigeweg, M. Schütz, M. Salehi, S. Schlücker, Small 7(17), 2443 (2011)
M. Schütz, D. Steinigeweg, M. Salehi, K. Kömpe, S. Schlücker, Chem. Commun. 47(14), 4216 (2011)
J.M. Montenegro, V. Grazu, A. Sukhanova, S. Agarwal, M. Jesus, I. Nabiev, A. Greiner, W.J. Parak, Adv. Drug Delivery Rev. 65(5), 677 (2013)
D.S. Grubisha, R.J. Lipert, H.Y. Park, J. Driskell, M.D. Porter, Anal. Chem. 75(21), 5936 (2003)
B. Lutz, C. Dentinger, L. Sun, L. Nguyen, J. Zhang, A. Chmura, A. Allen, S. Chan, B. Knudsen, Journal of Histochemistry & Cytochemistry 56(4), 371 (2008)
G.T. Hermanson, Bioconjugate Techniques (Academic Press, Cambridge, 2008)
M. Salehi, L. Schneider, P. Ströbel, A. Marx, J. Packeisen, S. Schlücker, Nanoscale 6(4), 2361 (2014)
Y. Zhang, B. Walkenfort, J.H. Yoon, S. Schlücker, W. Xie, Phys. Chem. Chem. Phys. (2015)
S. Schlücker, M.D. Schaeberle, S.W. Huffman, I.W. Levin, Anal. Chem. 75(16), 4312 (2003)
S. Schlücker, B. Küstner, A. Punge, R. Bonfig, A. Marx, P. Ströbel, J. Raman Spectrosc. 37(7), 719 (2006)
Y. Chen, X. Zheng, G. Chen, C. He, W. Zhu, S. Feng, G. Xi, R. Chen, F. Lan, H. Zeng, Int. J. Nanomedicine 7, 73 (2012)
B.R. Lutz, C.E. Dentinger, L.N. Nguyen, L. Sun, J. Zhang, A.N. Allen, S. Chan, B.S. Knudsen, ACS Nano 2(11), 2306 (2008)
Y.W. Wang, A. Khan, M. Som, D. Wang, Y. Chen, S.Y. Leigh, D. Meza, P.Z. McVeigh, B.C. Wilson, J.T. Liu, Technology 2(02), 118 (2014)
P. Lackie, R. Hennessy, G. Hacker, J. Polak, Histochemistry 83(6), 545 (1985)
M. Hayat, Colloidal Gold: Principles, Methods and Applications (San Diego, 1989)
S. Barua, J.W. Yoo, P. Kolhar, A. Wakankar, Y.R. Gokarn, S. Mitragotri, Proc. National Acad. Sci. 110(9), 3270 (2013)
R.P. Bagwe, L.R. Hilliard, W. Tan, Langmuir 22(9), 4357 (2006)
E. Perets, A. Indrasekara, A. Kurmis, N. Atlasevich, L. Fabris, J. Arslanoglu, Analyst 140(17), 5971 (2015)
X. Wang, X. Qian, J.J. Beitler, Z.G. Chen, F.R. Khuri, M.M. Lewis, H.J.C. Shin, S. Nie, D.M. Shin, Cancer Res. 71(5), 1526 (2011)
A. Housni, M. Ahmed, S. Liu, R. Narain, J. Phy. Chem. C 112(32), 12282 (2008)
B. Zhang, X. Wang, F. Liu, Y. Cheng, D. Shi, Langmuir 28(48), 16605 (2012)
H. Xu, J. Xu, X. Wang, D. Wu, Z.G. Chen, A.Y. Wang, ACS Appl. Mater. Interfaces 5(8), 2901 (2013)
K.M. Tichauer, K.S. Samkoe, K.J. Sexton, J.R. Gunn, T. Hasan, B.W. Pogue, J. Biomed. Opt. 17(6), 0660011 (2012)
L. Sinha, Y. Wang, C. Yang, A. Khan, J.G. Brankov, J.T. Liu, K.M. Tichauer, Sci. Rep. 5 (2015)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG
About this chapter
Cite this chapter
Zhang, Y., Schlücker, S. (2018). ISERS Microscopy for Tissue-Based Cancer Diagnostics with SERS Nanotags. In: Toporski, J., Dieing, T., Hollricher, O. (eds) Confocal Raman Microscopy. Springer Series in Surface Sciences, vol 66. Springer, Cham. https://doi.org/10.1007/978-3-319-75380-5_15
Download citation
DOI: https://doi.org/10.1007/978-3-319-75380-5_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-75378-2
Online ISBN: 978-3-319-75380-5
eBook Packages: Physics and AstronomyPhysics and Astronomy (R0)