Abstract
We describe nanotags suitable for both surface enhanced Raman scattering (SERS) and fluorescence detection and imaging. A fluorescently-labeled aptamer conjugated to gold-silver nanorods used for specific and sensitive detection of cervical cancer. NRs with different Au-Ag ratios were synthesized. The Raman reporter 4-aminothiophenol and fluorescently-labeled aptamers were assembled on the surface of NRs via a layer-by-layer process. The fluorescence and SERS signals can be generated independently using different excitation wavelengths, which can avoid the disturbance from each other. The nanotags were proven to be specific to the human protein tyrosine kinase-7 (PTK-7) expressed on Hela (cervical cancer) cells through aptamer-protein interaction. The binding of aptamers towards their targets induced the assembly of nanotags on the cell surface, resulting in strong fluorescence and SERS signals. However, the controls, randomized sequence oligonucleotide conjugated NRs, showed no detectable signal. Fluorescence and SERS mapping images were also performed to confirm targeting ability of the nanotags on the target cell membrane. The success of this method extends the feasibility of the dual mode nanotags for highly sensitive and specific cancer diagnostic.

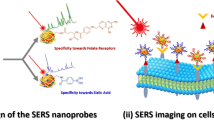
A dual mode SERS-fluorescence nanotag was fabricated using layer-by-layer process. The nanotags were proven to be specific to cervical cancer cells through aptamer-protein interaction. By labeling the target cells with nanotags, strong SERS and fluorescence signal is observed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The demand for specific and accurate cancer screening has driven the development of novel diagnostic probes having high selectivity and sensitivity. In the design of such a probe, one single system that combines precise molecular recognition units, providing selectivity, and the signal transduction moieties, reflecting sensitivity, is very desirable. Recently, ligands termed aptamers, which are single-stranded oligonucleotides, have been isolated and identified for such specific cancer cell recognition [1]. They are generated from an in vitro selection process called Systematic Evolution of Ligands by Exponential enrichment (SELEX) [2, 3]. Aptamers rival antibodies for molecular recognition due to their reproducible synthesis, easy manipulation, and good stability against biodegradation, making them promising candidates as targeting moieties for biosensor applications.
The sensitivity of the targeting probe is also directly related to the transduction of the target-probe interaction into the reporting signal. During the past decades, several optical measurements including fluorescence, surface plasmon resonance (SPR), and colorimetry have been employed for performing molecular detection. Among these, fluorescence based technique has attracted most attention for cancer cells targeting and imaging due to advantages in fast readout, easy operating, and real-time tracking. As a consequence, various types of probes, especially fluorescent nanoparticles and materials, have been continuously used as signal transduction tools for biomedical research and disease diagnostic [4]. Alternatively, Surface enhanced Raman scattering (SERS) has been recently explored as a new platform for highly sensitive molecular targeting and imaging due to advantages in richness of spectroscopic information together with high spatial resolution [5]. The unique “finger print” Raman spectra of particular molecules not only provide the identification and quantitation of specific targets but also allow the multiplex detection [6–8]. In has been reported that a remarkable surface enhance effect can be achieved when Raman molecules are in close proximity to roughened metallic substrate in nanometer-scale. To date, new metallic nanoparticles including nanostars [9], Au/Ag core shell NPs [10], and gold nanorods (GNRs) [11, 12] have been reported as material used for SERS enhancement. GNRs provide a novel type of SERS substrate due to their strong light absorption and scattering [13, 14] with two surface plasmon absorption bands: a short wavelength due to transverse electron oscillation, and a long wavelength in NIR region derived from longitudinal electron oscillation. The long wavelength band, tuned by changing the aspect ratio, can be tailored to overlap with the excitation source wavelength to obtain maximum SERS enhancement for the detection of cancer biomarkers [15, 16]. Particularly, core-shell and alloyed nanomaterial providing superior optical, and electronic properties compare to their monometallic counterpart. It was reported that Au-Ag bimetallic nanoparticles have provided stronger Raman signal than that of pure Au and Ag nanoparticles as SERS substrate [17–19]. Hence, Au-Ag NRs appear to be a new class of nanomaterial as a SERS active substrate.
Although fluorescence and SERS have made impressive progress in bioanalysis, they still suffer from some intrinsic advantages. For example, the spectral overlapping of fluorescence has limited applications in multiplex analysis. While, SERS requires long acquisition time and provides relatively low spatial resolution for large area imaging, such as whole cell analysis. We envisioned a dual mode detection system for cellular analysis by integrating the fast imaging capability of fluorescence with the multiplexing capability of SERS. In the present work, we demonstrated the dual mode SERS and fluorescence nanotags for specific targeting and imaging of the target cancer cells. The nanotags consist of three main compartments; including 4-aminothiophenol (4-ATP) labeled Au-Ag NRs designed for SERS signal generation, aptamers served as the targeting ligands, and fluorophore labeled aptamer for fluorescence imaging. Human cervical cells (Hela cells) with overexpressed protein tyrosine kinase-7 (PTK-7) were used as the model of target cells. With the molecular recognition between the aptamers and the target proteins on the cell membrane, specific targeting ability of the nanotags was realized. The experimental results revealed that these dual mode probes are promising for use in specific cell targeting and imaging. In addition, this multifunctional platform may be further explored for perspective multiplex detection.
Experimental
Reagents
Cetyltrimetylammonuim bromide (CTAB), gold (III) chloride trihydrate (HAuCl4 · 3H2O, 99 %), sodium borohydride (NaBH4, 98 %), silver nitrate (AgNO3, 99 %), ascorbic acid, glycine, sodium hydroxide (NaOH), magnesium chloride (MgCl2), D-(+)-glucose (99.5 %), and 4-aminothiophenol (4-ATP) were obtained from Sigma-Aldrich (MO, USA, http://www.sigmaaldrich.com). Dulbecco’s Phosphate-Buffered Saline (DPBS) was purchased from Invitrogen (NY, USA, http://www.invitrogen.com).
The oligonucleotides with high-pressure liquid chromatography (HPLC) purification were purchased from Integrate DNA technologies (Singapore, http://sg.idtdna.com). The aptamer: sgc8c, 5′- ATC TAA CTG CTG CGC CGC CGG GAA AAT ACT GTA CGG TTA GA-3′ was selected for targeting Hela cell, while a library containing randomized sequence of 41 oligonucleotide was used as a control. Both of the oligonucleotides were labeled with 3′-thiol modifier containing 10 thymine bases as a spacer and coupled with fluorescein at the 5′ end.
Preparation of dual mode nanotags
Synthesis of gold-silver nanorods
Gold-silver nanorods (Au-Ag NRs) were synthesized according to a slightly modified seed-mediated growth method described previously [13, 20]. In aqueous media, CTAB solution (0.2 M, 5.0 mL) was mixed with 5.0 mL of 0.5 mM HAuCl4. The mixture solution was reduced at room temperature by freshly-prepared 0.01 M NaBH4 (0.6 mL) under sonication for 3 min, resulting in a brownish-yellow solution. This seed solution was kept at room temperature for the next step.
The growth solution was prepared by mixing CTAB aqueous solution (0.2 M, 50.0 mL) with 1 mM HAuCl4 (50.0 mL) and AgNO3 (4 mM, 0.25 mL). After gentle mixing, 0.0788 M ascorbic acid (0.7 mL) was added to the solution as a mild reducing agent. The color of the solution was then changed from yellow to colorless, indicating the formation of AuCl2 − ions. The seed solution (0.12 mL) prepared previously was added to the growth solution. The solution color gradually changed to dark-pink over 30 min, indicating the formation of AuNR seeds. For various aspect ratios of AuNR seeds, different amount of AgNO3 (0.075 mL or 0.125 mL) was used for the preparation of growth solution. The AuNR seeds were further used for the next step without purification; noted that the AuNR seed solutions still contained Ag and Au ions, as well as ascorbic acid.
The AuNR seed solution (50.0 mL) was mixed with 0.2 M glycine solution (50.0 mL, pH 8.0). The mixture was then incubated at room temperature overnight without stirring to form Au-Ag NRs. The Au-Ag NRs were recovered by centrifugation at 11,000 g for 10 min. The Au-Ag NRs were washed three times using 0.5 mM CTAB to remove unreacted reagents, and then resuspended in 0.5 mM CTAB. The transverse and longitudinal absorption bands of NRs were verified by a microplate reader (Power wave XS2, Bio-Tek, USA) in a range of 400 to 900 nm with a step size of 2 nm. The sizes and energy dispersive X-ray (EDX) spectra of the prepared Au-Ag NRs were determined through transmission electron microscopy (TEM) analysis (TEM, JEM-2010, JEOL, Japan).
Conjugation of raman reporters and fluorescently-labeled aptamers
The nanotags were prepared by embedding Raman reporters, 4-ATP, on Au-Ag NRs surface. Briefly, the ten-fold dilution of Au-Ag NR solution (100.0 mL) was gently mixed with 1 mM 4-ATP (8.0 mL) for 3 h at room temperature. The coated NRs were then separated from the solution by centrifugation at 9,000 g for 10 min and resuspended in 0.5 mM CTAB (100.0 mL) and stored at room temperature. The solutions of 4-ATP coated NRs (1.0 mL) were washed by centrifugation at 9,000 g for 10 min to remove excess CTAB before aptamer conjugation, and resuspended in the final CTAB concentration of 50.0 μM (1.0 mL). In order to stabilize and functionalize the coated NRs, freshly-prepared mPEG-SH solution (2.0 mM, 0.1 mL) was added to the coated NRs suspension (1.0 mL). Subsequently, 10.0 μL of 2.5 μM fluorescently-labeled aptamers were added and the mixture was then incubated for 12 h at room temperature. The prepared nanotags were purified by centrifugation at 9,000 g for 5 min.
Cell culture and validation of specific targeting by the nanotags
Hela cells were maintained in RPMI supplemented with 10 % fetal bovine serum (FBS) and 100 IU/mL penicillin-streptomycin under a humidified atmosphere (5 % CO2 and 37 °C). The cells were harvested using trypsin-EDTA and washed with DPBS prior to usage. The cell density was then determined using hemacytometer before performing any experiments.
For the fluorescence imaging experiments, 5.0 × 104 Hela cells were placed on each well of a 12-well plate. After 24 h of incubation, the cells were washed with DPBS (pH 7.4) twice. After washing, the nanotags (0.5 mL), which was diluted 1:5 in DPBS containing 0.1 % BSA was added, and the cells were incubated on ice for 1 h. After incubation, the cells were rinsed in triplicated with DPBS prior to fluorescence imaging.
For SERS spectral measurement and SERS mapping images, initially 5.0 × 105 Hela cells were seeded on a 22 × 22 mm glass slip placed in a 6-well plate. After 24 h, the cells were then rinsed with DPBS and immersed in nanotags solution, which was diluted 1:5 in DPBS with 0.1 % BSA for 1 h. After washing with DPBS to remove the unbound nanotags, the cells were left for air-dry before the SERS measurement.
Fluorescence and SERS measurement
Fluorescence experiments were performed using a fluorescence microscope (Olympus IX71, Olympus, USA) with a mercury lamp and a charge couple device (CCD) detector. The cellular images were taken with a 20× objective and the fluorescence were excited at 488 nm (λex for fluorescein), and the emission was detected at 525 nm (λem for fluorescein).
SERS spectral measurements were performed using a Raman spectrometer (NT-MDT, Russia) equipped with an inverted confocal microscope (Olympus IX71, Olympus, USA). The system was connected with a CCD detector cooled at −60 °C. A He-Ne laser operating at λ = 633 nm was used as an excitation source with a power of 3.3 mW. The laser light was coupled through 100× objective lens with a laser spot size at 333 nm. The system was calibrated with Raman signal from a standard silicon wafer at 520 cm−1. The SERS spectra were obtained in a range of 500–2000 cm−1 with the exposure time of 10 s and 12 accumulations. The Raman images of the cells were obtained using Raman point-mapping method. SERS mapping images were collected with a 0.15 μm step size over the specific area of 25 × 25 μm with an integration time of 1 s.
Results and discussion
Herein, we demonstrated the use of dual mode nanotags for the target cells detection, which the outcome signals, fluorescence and SERS, can be generated by different excited wavelengths. The schematic illustration of the nanotags preparation and detection of the target cell was depicted in Fig. 1. Initially, the nanotags were prepared by embedding the Raman reporters, 4-ATP, on NRs surface followed by the conjugation of fluorescently-labeled aptamers specific to the target cancer cells. By using a NR as a scaffold, multiple fluorescein-labeled aptamers can be attached on the nanotags. Consequently, the strong fluorescence signal can be generated, which is much higher than that of individual fluorescence probe [21]. Moreover, when multiple nanotags bind to their target cells through the specific interaction between the aptamers and receptors on the cell membrane, they act cooperatively to form clusters, thereby inducing the coupling of electromagnetic fields. Such strong collective electromagnetic fields lead to the enhancement of SERS signals of the Raman reporter embedded on the NRs indicating the binding event between the nanotags and target molecules.
Choices of materials
To achieve strong detection signal for SERS biosensor, it is necessary to design metallic substrate that can lead to maximum optical field, resulting in maximum Raman signal enhancement. Anisotropic geometries like triangles, cubes, stars, and rods, provide advantages in producing concentrated field at the tips; therefore, strong SERS signal can be generated [22]. Nanorods (NRs) are one of materials that have been extensively used as SERS active substrate. Due to their anisotropic geometries, NRs exhibit two plasmon bands: transverse band at the lower wavelength, and tunable longitudinal band at near-infrared region. It is known that gold nanoparticles are chemically stable, and their size including shape are easily to be controlled during the synthesis. However, gold exhibits low SERS activity owing to their plasmon damping induced by gold interband transition. Silver based-materials can provide high SERS enhancement, conversely, the SERS performances suffer from fast oxidation in ambient conditions. In order to combine distinct properties of both materials, gold-silver nanorods (Au-Ag NRs) have been synthesized and applied in SERS based-sensing systems. It was previously demonstrated that Au-Ag NRs structures can provided significant higher SERS signal enhancement compared to that of pure AuNRs when both attached with similar Raman reporter molecules [23]. Therefore, Au-Ag NRs have been selected as a choice of material to fabricate nanotags for this study in order to achieve intense SERS signal which can be translated into high sensitivity.
Characterization of gold-silver NRs and the dual mode nanotags
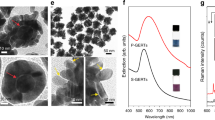
The Au-Ag NRs were synthesized by a modified seed-mediated growth method using a surfactant, CTAB, as a building block. It was previously reported that by changing the amount of AgNO3 solution used in the preparation of the growth solution, the aspect ratio of NRs can be tuned [13]. Figure 2 demonstrated characteristic extinction spectra and TEM images of NRs with three different aspect ratios. The NRs were fabricated with the average width of 15 ± 3 nm, and the length ranging from 20 to 55 ± 6 nm by considering at least 50 NRs for the individual aspect ratio. The increasing of AgNO3 amount used in the preparation of AuNR seeds induced the rising of NRs length resulting in the adjustment of the aspect ratios from 1.5 to 3.5 and shifting of longitudinal bands to the redshift. As seen in Fig. 2, the NRs with the aspect ratio of 2.5 and 3.5 have a weak plasmon band at 510 nm and a strong longitudinal band at 690, and 780 nm, respectively. It is interesting to note that when the aspect ratio was reduced to 1.5, the NRs possess only one plasmon band similar to spherical gold nanoparticles. In order to confirm the presence of Au and Ag in the NRs, the EDX analysis was performed. The results showed that all of the synthesized NRs comprised of both Au and Ag (Fig. S1) with the amplified Ag content when the aspect ratio of NRs increased corresponded to the previous study [20, 24].
After Au-Ag NRs synthesis, the Raman reporters, 4-ATP, were absorbed on NRs surface through thiol-gold interaction defined as 4-ATP-NRs, and the SERS spectra were then investigated. In the experiments, the laser at 633 nm was used as an excitation light with a power of 3.3 mW at the samples, and the integration time of each measurement was 10 s. Figure 3 showed the SERS spectra of 4-ATP which were coated on the surface of NRs. The observed spectra were dominated with the a1 vibrational modes (in-plane, in-phase modes) at 1077 (ν(CS)), 1161 (δ(CH)), and 1587 (ν(CC)) cm−1 and the b2 modes (in-plane, out-of-phase modes) at 1381 and 1420 cm−1 (ν(CC + δ(CH)) [10, 25, 26]. The strong Raman signals are ascribed to the enhancement from the localized electromagnetic fields on metallic surface and charge transfer between the metal to absorbed molecules. Here, the SERS signal of 4-ATP-NRs with an aspect ratio of 2.5 was much stronger than those of 4-ATP-NRs with aspect ratios of 1.5 and 3.5, respectively (Fig. S2). The larger surface enhancement was attributed to a stronger electromagnetic enhancement due to the overlapping between plasmon resonance of the NRs and the excitation source. As a result, the NRs with an aspect ratio of 2.5, which has a longitudinal band overlapping with the laser excitation wavelength at 633 nm, produces the highest SERS signal. Similar phenomena were also observed previously for 4-mercaptopyridine and methylene blue on NRs with different aspect ratios [16, 27]. Therefore, NRs with the aspect ratio of 2.5 were further employed for the aptamer conjugation and target cell detection.
In this study, fluorescein-labeled sgc8c aptamers specific to the human protein tyrosine kinase-7 (PTK-7) expressed on Hela cells [28] were conjugated to the NRs with the aspect ratios of 2.5 after embedded with the Raman reporters, 4-ATP, and defined as the nanotags, while random sequences which had an equal number of bases and similar fluorescein labeling were used as a control. The assembly of aptamers on NRs surface was performed simply through thiol-gold interaction. The thiol-tethered aptamer consists of four important compartments: (1) the thiolalkane hydrocarbon, (2) the poly-Thymine (T) bases as a spacer, (3) the recognition unit of aptamer, and (4) the fluorescent entity. The first part is the hydrocarbon containing a thiol unit that promotes the self-assembly of aptamers onto NRs. The second part is a spacer that extends the aptamers from the surface of NRs in order to avoid steric hindrance of the target proteins interacting with the aptamers. The third one is the ligand; in this case, the aptamer sgc8c, which is used for the specific targeting the membrane proteins of Hela cells. The last part is the fluorophore, fluorescein, which uses to generate fluorescence signal. Furthermore, a monolayer of additional PEG onto the NRs surface minimizes aggregation of the nanotags and resists nonspecific binding in physiological environments.
Figure 4 demonstrated the Raman spectrum of the 4-ATP molecules and SERS spectra of layer-by-layer process of the assembled-system. According to previous reports [15, 29], CTAB and DNA have weak Raman signal intensity, however; the Raman reporter, 4-ATP has a relatively strong signal. The Raman spectrum of 10 mM 4-ATP in ethanol (i) is dominated at 1077, and 1587 cm−1 from the a1 vibrational modes (in-plane, in-phase modes), however; the other vibrational modes were not observed. The weak SERS signal of CTAB on NRs (ii) around 1265 cm−1 was detected [15]. After embedding of 4-ATP on NRs surface (iii), the strong and rich SERS signal of 4-ATP-NRs was exhibited. The small peak shift compared to the Raman spectrum of 4-ATP was noticed, which derived from the interactions of 4-ATP with NRs or neighboring molecules. After subsequent attachment of aptamers to NRs coated with 4-ATP (iv), a similar pattern with relatively slight shifts were observed with additional Raman shifts detected at 668, 722, and 1330 cm−1 corresponding to DNA SERS signal [29]. Nevertheless, the final nanotags still showed a strong Raman intensity which is suitable for the further cellular studies.
Target cell detection by the dual mode nanotags
In order to demonstrate the application of such the dual mode nanotags for specific targeting, Hela (cervical cancer) cell which has high expression of PTK-7 on their surface was chosen as a model. Fluorescein amidite (FAM)-labeled sgc8c-nanotags were used as a target and random sequences-nanotags with similar labeling were used as a control. It is well known that gold nanoparticles (AuNPs) are often considered as efficient quenchers when the fluorophore is directly in contact with a metal surface due to nonradiative energy transfer from the excited state of the fluorophore to the metal. However, in our case, the aptamers with 41 bases and poly T-bases placed a distance of more than 10 nm between the fluorophore and NRs. In addition, a full coverage of NRs with well-packed PEG was also helpful for stretching out the fluorophores away from the metallic surface. Therefore, the distance extended beyond the distance for efficient energy transfer, and the undesired fluorescence quenching in these NR-aptamer hybrids was minimized. Since our aptamers were labeled with fluorescein, the wavelength of 488 nm was used as an excitation source with a 20× objective lens. The binding between sgc8c-nanotags and Hela cells was observed by a bright fluorescence signal especially at the cell membrane indicating the binding capability of the nanotags (Fig. 5a). However, the control showed only minimal fluorescence signal as a background (Fig. 5c). This result suggested that affinity of the aptamer probe is maintained well after the conjugation with NRs and such the nanotags can provide fluorescence mode of detection.
Afterward, the SERS detection modality of the nanotags was investigated. In the present nanotags, Au-Ag NRs were applied as the SERS substrates, and 4-ATP molecules were absorbed onto the NRs to provide SERS signal. The sgc8c aptamers on the nanotags can specifically bind to the PTK-7, which is overexpressed on Hela cell surface with the density of 550 ± 90 receptor μm−2 based on the previous fluorescence correlation spectroscopy (FCS) study [30]. The high density of PTK-7 induced the accumulation of the nanotags into close proximity on the cell surface. It is known that the aggregate of metal nanoparticles induces SERS enhancement due to the coupling of electromagnetic field generated at the junctions of nanoparticles, defined as “hot spots”, thereby gaining strong Raman signals. To evaluate the SERS functionality, the nanotags were incubated with the target Hela cells, and the excitation wavelength was switched to 633 nm. Then, the spectra of the labeled Hela cells after incubation with either the nanotags (Fig. 5b) or controls (Fig. 5d) were demonstrated. More than 10 Hela cells with several spots for each cell labeled with both the nanotags and control were measured. For the nanotags labeling, over 85 % of the measurements showed strong Raman signals (Fig. S3), especially on the cell surface (Fig. S5). In contrast, most of the controls showed no Raman spectra and a few cells gave relatively weak signal at some positions of the cells (Fig. S4).
SERS mapping image
To confirm the binding of nanotags toward the target cell surface, the SERS mapping images of Hela cells were measured and analyzed. Figure 6 demonstrated the SERS images of Hela cells labeled with the nanotags (Fig. 6a) and controls (Fig. 6b) using the excitation wavelength at 633 nm. The laser beam was focusing on the surface of the single cell by adjusting the z-axis. The Raman peak of 4-ATP at 1587 cm−1 which is the most distinctive peak was selected and displayed by a color-coding. The brighter color of the bar demonstrated the higher SERS signal ascribed from the attachment of the nanotags on the cell surface. Here, the mapping image of the target cell using the nanotags (Fig. 6a) was much brighter than that of the controls (Fig. 6b). These results agreed with our fluorescence assays and SERS spectra measurement, as described above, and confirmed the specific recognition of the nanotags, making this a viable and practical technique for the sensitive detection of the target cancer cells.
Conclusion
In summary, we demonstrated the system of functionalized Au-Ag NRs for targeting and imaging of specific cancer markers expressed on the membrane of cervical cancer cells. A procedure of the dual mode nanotags assembled-system using layer-by-layer process was developed. The nanotags were found to maintain their specific recognition and provide a multivalent effect, resulting in strong interaction with their target cells. Significantly, strong detection signal derived from both high affinities of the aptamers conjugated NRs towards their target cells and the collective electromagnetic fields ascribed to the strong longitudinal surface plasmon oscillation of Au-Ag NRs coupled with the overlapping with the excitation wavelength. Consequently, these nanotags with fluorescence labeled aptamers enabled the fluorescence-SERS dual-mode sensor system. Notably, the implementations of SERS technique in bio-analysis extends their important uses in cancer diagnostic and bioimaging.
References
Brody EN, Gold L (2000) Aptamers as therapeutic and diagnostic agents. Rev Mol Biotechnol 74:5–13
Ellington AD, Szostak JW (1990) In vitro selection of RNA molecules that bind specific ligands. Nature 346:818–822
Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249:505–510
Wolfbeis OS (2015) An overview of nanoparticles commonly used in fluorescent bioimaging. Chem Soc Rev 44:4743–4768
Cialla D, März A, Böhme R, Theil F, Weber K, Schmitt M, Popp J (2012) Surface-enhanced raman spectroscopy (SERS): progress and trends. Anal Bioanal Chem 403:27–54
Kneipp J, Kneipp H, Kneipp K (2008) SERS-a single-molecule and nanoscale tool for bioanalysis. Chem Soc Rev 37:1052–1060
Qian XM, Nie SM (2008) Single-molecule and single-nanoparticle SERS: from fundamental mechanisms to biomedical applications. Chem Soc Rev 37:912–920
Doering WE, Piotti ME, Natan MJ, Freeman RG (2007) SERS as a foundation for nanoscale, optically detected biological labels. Adv Mater 19:3100–3108
Ma W, Sun M, Xu L, Wang L, Kuang H, Xu C (2013) A SERS active gold nanostar dimer for mercury ion detection. Chem Commun 49:4989–4991
Luo Z, Chen K, Lu D, Han H, Zou M (2011) Synthesis of p-amiothiophenol-embedded gold/silver core-shell nanostructures as novel SERS tags for biosensing applications. Microchim Acta 173:149–156
Wang Y, Lee K, Irudayaraj J (2010) SERS aptasensor from nanorod-nanoparticle junction for protein detection. Chem Commun 46:613–615
Wang Z, Zong S, Yang J, Li J, Cui Y (2011) Dual-mode probe based on mesoporous silica coated gold nanorods for targeting cancer cells. Biosens Bioelectron 26:2883–2889
Nikoobakht B, El-Sayed MA (2003) Preparation and growth mechanism of gold nanorods (NRs) using seed-mediated growth method. Chem Mater 15:1957–1962
Link S, Mohamed MB, El-Sayed MA (1999) Simulation of the optical absorption spectra of gold nanorods as a function of their aspect ratio and the effect of the medium dielectric constant. J Phys Chem B 103:3073–3077
Huang X, El-Sayed IH, Qian W, El-Sayed MA (2007) Cancer cells assembled and align gold nanorods conjugated to antibodies to produce highly enhanced, sharp, and polarized surface raman spectra: a potential cancer diagnostic marker. Nano Lett 7:1591–1597
Park H, Lee S, Chen L, Lee EK, Shin SY, Lee YH, Son SW, Oh CH, Song JM, Kang SH, Choo J (2009) SERS imaging of HER2-overexpressed MCF7 cells using antibody-conjugated gold nanorods. Phys Chem Chem Phys 11:7444–7449
Kim K, Kim KL, Lee SJ (2005) Surface enrichment of Ag atoms in Au/Ag alloy nanoparticles revealed by surface enhanced raman scattering spectroscopy. Chem Phys Lett 403:77–82
Yang Y, Shi J, Kawamura G, Nogami M (2008) Preparation of Au–Ag, Ag–Au core–shell bimetallic nanoparticles for surface-enhanced raman scattering. Scr Mater 585:862–865
Pande S, Ghosh SK, Praharaj S, Panigrahi S, Basu S, Jana S, Pal A, Tsukuda T, Pal T (2007) Synthesis of normal and inverted gold-silver core-shell architectures in β-cyclodextrin and their applications in SERS. J Phys Chem C 111:10806–10813
Huang YF, Lin YW, Chang HT (2006) Growth of various Au–Ag nanocomposites from gold seeds in amino acid solutions. Nanotechnology 17:4885–4894
Huang YF, Chang HT, Tan W (2008) Cancer cell targeting using multiple aptamers conjugated on nanorods. Anal Chem 80:567–572
Kumar GVP (2012) Plasmonic nano-architectures for surface enhanced raman scattering: a review. J Nanophotonics 6(1–20):64503
Nima ZA, Mahmood M, Xu Y, Mustafa T, Watanabe F, Nedosekin DA, Juratli MA, Fahmi T, Galanzha EI, Nolan JP, Basnakian AG, Zharov VP, Biris AS (2014) Circulating tumor cell identification by functionalized silver-gold nanorods with multicolor, super-enhanced SERS and photothermal resonances. Sci Rep 4(1–8):4752
Huang YF, Huang KM, Chang HT (2006) Synthesis and characterization of Au core-Au-Ag shell nanoparticles from gold seed: impacts of glycine concentration and pH. J Colloid Interface Sci 301:145–154
Hu X, Wang T, Wang L, Dong S (2007) Surface-enhanced raman scattering of 4-aminothiophenol self-assembled monolayers in sand with structure with nanoparticle shape dependence: Off-surface plasmon resonance condition. J Phys Chem C 111:6962–6969
Zheng J, Zhou Y, Li X, Ji Y, Lu T, Gu R (2003) Surface-enhanced raman scattering of 4-aminothiophenol in assemblies of nanosized particles and the macroscopic surface of silver. Langmuir 19:632–636
Sivapalan ST, DeVetter BM, Yang TK, Dijk TV, Achulmerich MV, Carney PS, Bhargava R, Murphy CJ (2013) Off-resonance surface-enhanced raman spectroscopy from gold nanorod suspensions as a function of aspect ratio: not what we thought. ACS Nano 7:2099–2105
Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W (2006) Aptamers evolved from live cells as effective molecular probes for cancer study. PNAS 103:11838–11843
Barthoumi A, Zhang D, Tam F, Halas NJ (2008) Surface-enhanced raman spectroscopy of DNA. J Am Chem Soc 130:5523–5529
Chen Y, Munteanu AC, Huang YF, Phillips J, Zhu Z, Mavros M, Tan W (2009) Mapping receptor density on live cells by using fluorescence correlation spectroscopy. Chem Eur J 15:5327–5336
Acknowledgments
This work was supported by grant from the Development and Promotion of Science and Technology Talents project (DPST), and the National Nanotechnology Center (NANOTEC), Thailand.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 1185 kb)
Rights and permissions
About this article
Cite this article
Bamrungsap, S., Treetong, A., Apiwat, C. et al. SERS-fluorescence dual mode nanotags for cervical cancer detection using aptamers conjugated to gold-silver nanorods. Microchim Acta 183, 249–256 (2016). https://doi.org/10.1007/s00604-015-1639-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-015-1639-9