Abstract
l-Ascorbic acid (AsA, ascorbate or vitamin C) is the most abundant water-soluble antioxidant found in plants. Ascorbate is synthesized via four pathways involving d-mannose/l-galactose, d-galacturonate, l-gulose, and myo-inositol as main precursors. In addition to protecting plant tissues from damage caused by reactive oxygen species produced through normal oxygenic metabolism or those generated from biotic and abiotic stresses, ascorbate is also an enzyme cofactor and a modulator of cell division, cell expansion, flowering time, and gene regulation. Plants that are deficient in ascorbate are affected in multiple ways including alterations in cell division, cell expansion, seed germination, growth, floral induction, and photosynthesis. Additionally, elevated ascorbate content in plants leads to an increase in their nutritive value, lengthening of their shelf life, enhancement on their growth rate and biomass accumulation, and to an increased tolerance to multiple abiotic stresses including salt, cold, heat, and water deficit. Increasing the productivity of crops is imperative to satisfy the growing demand for food, feed, and fuels in the world, and biotechnology can lead to the development of plants with higher yields capable of thriving under adverse conditions. To this end, phenotype screening and characterization of a large number of plants experimentally obtained is time consuming and requires a significant amount of resources, skills, and expertise. In this chapter, we will present an overview of how the use of high-throughput phenotyping or phenomics is revolutionizing the way plant phenotypes are characterized and will illustrate the power of digital phenotyping in the characterization of plants overexpressing enzymes in the inositol pathway to ascorbate.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
l-Ascorbic acid (ascorbate, AsA) is the most abundant water-soluble antioxidant found in plants. Ascorbate content varies widely among species but in general AsA concentration in plants is in the mM range (Noctor and Foyer 1998). Ascorbate has a wide variety of physiological roles. It functions as an enzyme cofactor, as a modulator of the cell cycle, and as a scavenger protecting tissues against damage caused by reactive oxygen species (ROS) produced from a multitude of environmental stresses including water deficit, soil salinity, cold, and heat, among others. Alteration of the redox balance resulting in accumulation of ROS (e.g., singlet oxygen, superoxide, and hydrogen peroxide) is a feature common to multiple environmental stresses. When the intensity of those stresses exceeds the antioxidant and repair capacity of cells, ROS can accumulate and cause oxidative damage to all macromolecules, promoting apoptosis and senescence. On the other hand, there is growing evidence of the important role of low levels or ROS as signaling molecules in pathways that stimulate adaptive hormonal and metabolic responses. Therefore whether ROS induce oxidative damage or trigger acclimation depends on the rate and site of production and is controlled by the antioxidant system, of which ascorbate and glutathione are central components (Foyer et al. 1994; Foyer and Noctor 2011).
A better understanding about how ascorbate is made in plants and the factors that regulate its content is of chief importance because this molecule provides the dietary source of vitamin C to humans, primates, and other animals. Mutations in the terminal enzyme of the animal vitamin C pathway (l-gulono-1,4-lactone oxidase) are what render its synthesis impossible for humans and make this an essential vitamin that we must acquire through diet (Linster and Van Schaftingen 2007). Deficiency of ascorbic acid is often associated with human health problems such as anemia, infections, bleeding gums, scurvy, poor wound healing, capillary hemorrhage, muscle degradation, atherosclerotic plaques, and neurotic disturbances (Chambial et al. 2013). Vitamin C deficiency is a common issue for malnourished people in developing countries, and also for various subpopulation groups in developed countries including those who are incarcerated, smokers, widows, and people who suffer various eating and nutrient absorption disorders (Beal et al. 2017).
Vitamin C is involved in essential physiological processes that impact the agronomical value of crops and its physiology. The growth and the world population present unique challenges to agriculture. We need to find ways to make crops more productive with less inputs. One of the potential solutions to this great challenge is the development of improved crops with enhanced nutritional value, higher tolerance to environmental insults, and superior yields.
There are five routes for the production of AsA, one in animals and four in plants (Fig. 1). The biosynthetic pathway for AsA in animals was elucidated in the early 1950s (Ishikawa et al. 2006). On the other hand, plants possess four possible ways to make this key molecule. These are the d-mannose/l-galactose (Wheeler et al. 1998), l-gulose (Wolucka and Van Montagu 2003), d-galacturonate (Agius et al. 2003), and myo-inositol (Lorence et al. 2004) routes (Fig. 1). The best characterized of these pathways is the d-mannose/l-galactose route. The degree of which the so called “alternative routes” is operational in each plant species and the tissue and temporal specificity of these alternative routes is currently unclear.
The ascorbate metabolic network. There is a single biosynthetic pathway for l-ascorbic acid (ascorbate, vitamin C) in some animals, while four distinct routes lead to its formation in plants: MI (myo-inositol) pathway, Gul (l-gulose) pathway, Man/Gal (d-mannose/l-galactose) pathway, and GaIU (d-galacturonic acid) pathway. The enzymes involved in the four different routes are: MI pathway: myo-inositol oxygenase (MIOX); glucuronate reductase (GlcUR); gluconolactonase (GNL); and l-gulono-1,4-lactone oxidase (GuILO). Man/Gal pathway: phosphomannose isomerase (PMI); phosphomannose mutase (PMM); GDP-mannose pyrophosphorylase (VTC1); GDP-3′5′-epimerase (GME); l-galactose guanyltransferase (VTC2); l-galactose-1-phosphate phosphatase (VTC4); l-galactose dehydrogenase (GalDH); l-galactono-1,4-lactone dehydrogenase (GLDH). GaIU pathway: d-galacturonate reductase (GaIUR). The enzymes in the recycling pathway are monodehydroascorbate reductase (MDHAR) and dehydroascorbate reductase (DHAR). Adapted from Suza et al. 2010
In this chapter, we present an overview of the successful strategies to metabolically engineer elevated AsA content in model plants and crops and summarize the studies in which the stress tolerance of those high ascorbate plants has been analyzed. We also discuss how the use of high-throughput phenotyping or phenomics is revolutionizing the way plant phenotypes are characterized and illustrate the power of digital phenotyping in the development of model plants with elevated AsA and enhanced ability to withstand stresses.
2 Successful Metabolic Engineering Strategies to Enhance Ascorbate Content in Plants
During the last 20 years, we have witnessed an unprecedented progress in the understanding of the multiple pathways leading to AsA formation in plants. Table 1 summarizes the successful strategies that have been reported in the engineering of elevated AsA content in plants.
All genes in the d-mannose/l-galactose pathway have been cloned and characterized, and several of those have been used to increase AsA content in more than one species. Overexpression of those genes has led to a 1.2-fold to 6-fold AsA increase.
Phosphomannose mutase (PMM) is one of the key enzymes in the regulation of the initial steps of the AsA synthesis via the d-mannose/l-galactose pathway (Fig. 1). Badejo et al. (2009) developed transgenic tobacco (Nicotiana tabacum) overexpressing PMM from acerola (Malpighia glabra), leading to a 2-fold AsA increase compared to controls.
Overexpression of kiwifruit GDP-l-galactose phosphorylase increased AsA content 6-fold in tomato fruits, while overexpression of this gene in potato increased AsA content in the tubers (Bulley et al. 2012). This gene has also been expressed in Arabidopsis (Zhou et al. 2012) and rice (Zhang et al. 2015).
The importance of GDP-galactose guanylyl transferase (GGT) as a rate-limiting step in the production of AsA production was confirmed with the overexpression of the GGT gene in Arabidopsis resulting in a 2.9-fold increase in AsA content (Zhou et al. 2012).
The overexpression of tomato GME leads to an increase of 1.2-fold in the AsA content of leaves and a boost of 1.6-fold in fruits (Zhang et al. 2011). This gene has also been successfully expressed in Arabidopsis leaves (Zhou et al. 2012; Huang et al. 2014) and alfalfa shoots (Ma et al. 2014).
GDP-d-Mannose pyrophosphorylase has been widely studied in a number of plants including tobacco (Badejo et al. 2008), tomato (Cronje et al. 2012), and Arabidopsis (Zhou et al. 2012; Sawake et al. 2015). The AsA content of transgenic tobacco plants expressing acerola GMP was 2-fold higher compared to wild type (Badejo et al. 2008). Overexpression of l-galactose dehydrogenase (GalDH) in Arabidopsis resulted in a modest increase (1.2-fold) in AsA content (Zhou et al. 2012).
l-Galactono-1,4-lactone dehydrogenase (GLDH) catalyzes the last step in the d-mannose/l-galactose pathway to AsA (Fig. 1). Liu et al. (2011) showed that suppression of GLDH expression resulted in a loss of chlorophyll, lower ribulose-1,5-bisphosphate carboxylase (RUBISCO) protein content, and a lower rate of CO2 assimilation. As a consequence, a slower rate of plant growth and lower seed set were observed. However, overexpression of this enzyme maintained high levels of chlorophyll, RUBISCO protein, and a higher rate of net photosynthesis, resulting in higher seed set. Also increased GLDH expression correlated with the reduced lipid peroxidation, indicating that this enzyme functions protecting the plant against ROS (Liu et al. 2011, 2013). GLDH has been overexpressed in multiple plants including rice (Liu et al. 2011; Zhang et al. 2015), Arabidopsis (Zhou et al. 2012), tobacco (Liu et al. 2013), and lettuce (Landi et al. 2015) leading to 1.3-fold to 2.1-fold AsA content compared to controls.
Engineering increased AsA content using a combination of genes has been more successful than using a single enzyme. Zhou et al. (2012) overexpressed the combination of GGT + GPP and GGT + GLDH separately in Arabidopsis. Their results indicate that the GGT + GPP transgenic lines performed best leading to a 4.1-fold AsA content.
Ripening strawberry has been reported to synthesize AsA from d-galacturonic acid and its methyl ester (Mapson and Isherwood 1956; Loewus and Kelly 1961). In 2003, Agius et al., discovery of a d-galacturonic acid reductase (GaIUR) gene from strawberry, open the possibility to engineer AsA content using this new gene. GalUR is the only enzyme identified in this pathway, which has been shown to be cytosolic (Agius et al. 2003). GalUR overexpression has led to a 2- to 3-fold AsA increase in Arabidopsis (Agius et al. 2003). In plant storage organs, possible sources of AsA include in situ synthesis and long-distance transport of AsA synthesized in other tissues. Hemavathi et al. (2009) developed transgenic potatoes via overexpression of GalUR. These resulting potato tubers had two times more AsA than controls. Transgenic tomato hairy roots overexpressing GalUR had 2.5-fold AsA (Wevar Oller et al. 2009) while overexpression of GalUR in fruits led to 1.4- to 2.5-fold AsA increases (Amaya et al. 2014; Cai et al. 2014; Lim et al. 2016).
Biochemical and molecular data indicate that myo-inositol can also be a precursor for the biosynthesis of AsA in Arabidopsis (Lorence et al. 2004; Zhang et al. 2008). This pathway involves four enzymes, starting from the oxidation of myo-inositol to d-glucuronic acid and further reduction to l-gulonic acid and to l-gulono-1,4-lactone, and further conversion to AsA. These reactions are catalyzed by myo-inositol oxygenase (MIOX) , glucuronate reductase (GlcUR) , gluconolactonase (GNL) , and l-gulono-1,4-lactone oxidase (GuILO , a.k.a. GLOase), respectively. These four enzymes have been already characterized by the Lorence Laboratory (Table 1). Arabidopsis lines overexpressing MIOX contained up to 3-fold ascorbate in leaves and presented increased biomass accumulation, growth rate compared to controls (Lorence et al. 2004). These results are similar with the findings by Kulkarni (2012) that showed a 3-fold increase in AsA content after transient AtMIOX4 expression in tomato fruits. Glucuronate reductase (GlcUR) is the second enzyme in the inositol pathway to AsA, and an isoform from Arabidopsis has been already characterized (Lorence and Nessler 2007; Yactayo-Chang 2011). The overexpression of GlcUR in Arabidopsis led to a 2-fold ascorbate content in leaves compared to the wild-type controls (Yactayo-Chang 2011). The first two enzymes in the inositol pathway to AsA seem to be cytosolic. The Lorence Laboratory has evidence indicating that some isoforms of the last two enzymes in this pathway reside in the chloroplast (Yactayo-Chang 2016) and the endoplasmic reticulum (ER) (Aboobucker 2014; Aboobucker et al. 2017), respectively. Our group recently characterized a GNL isoform that is localized in the plastids. When overexpressed in Arabidopsis, transgenics GNL over-expressers have a 5-fold AsA content in leaves (Yactayo-Chang 2016; Yactayo-Chang and Lorence 2016). The last enzyme in the inositol pathway to AsA is l-gulono-1,4-lactone oxidase (GuILO) . GuILO belongs to the aldonolactone oxidoreductases protein family. GuILO participates in both the l-gulose and inositol pathways to ascorbate. Transgenic tobacco and lettuce plants expressing rat GuILO accumulated up to 7-fold more ascorbate than untransformed plants (Jain and Nessler 2000), while Arabidopsis lines overexpressing the rat isoform were reported to contain a more modest AsA increase (Lisko et al. 2013).
Heick et al. (1972) reported that AsA has been found in a number of yeasts including Saccharomyces cerevisiae. The arabino-1,4-lactone oxidase (ALO) enzyme catalyzes the last step in the formation of d-erythroascorbate in yeast. This enzyme uses l-galactono-1,4-lactone as efficiently as d-arabino-1,4-lactone to produce AsA. Overexpression of ALO resulted in elevated ascorbate (1.3-fold) in tomato fruits and stylo leaves (3.1-fold) (Cronje et al. 2012; Bao et al. 2016).
Once oxidized, AsA can also be recycled back to its reduced form. The reduced AsA is ready for its role again. Overexpression of dehydroascorbate reductase (DHAR) and monodehydro ascorbate reductase (MDHAR) has been implemented successfully to engineer plants with elevated AsA. Overexpression of DHAR has led to 1.3-fold to 6-fold AsA increase in various plant species (Table 1). Constitutive expression of MDHAR isoforms has also been successfully implemented to increase AsA content in plants (Table 1).
The existence of four different pathways leading to AsA formation in plants indicates the need of a complex regulatory mechanism for this network. Regulation within and between pathways in the network is largely uncharacterized. Six master regulators for AsA have been described in the literature. The ascorbic acid mannose pathway regulator 1 (AMR1) is one of these regulators. AMR1 negatively regulates the d-Man/l-Gal pathway (Zhang et al. 2009).
Another regulator of this network is the three KNOTTED-like homeobox gene (HBK3) . Homeobox genes encode a typical DNA-binding domain of 60 amino acids, known as homeodomain that characterizes a large family of transcription factors. These genes encode transcription factors which regulate important events in plant growth and development. The HBK3 gene from Norway spruce regulates somatic embryo yield through alterations in glutathione and AsA metabolism, which have been previously implicated in controlling development and maturation both in vivo and in vitro (Belmonte and Stasolla 2009).
The size of the AsA pool in plants is controlled by a combination of its biosynthesis, recycling, and transport and translocation. Some factors that regulate the AsA pool size by modulating biosynthesis are light, development, and environmental stress conditions. The ethylene response factor (ERF98) is considered a positive regulator of AsA. This regulator enhances tolerance to salt stress through direct activation of AsA synthesis in Arabidopsis (Zhang et al. 2012). Arabidopsis ERF98 transcriptionally activates gene expression of VTC1 to improve AsA content and tolerance to salt stress.
3 Effect of Ascorbate in the Ability of Plants to Withstand Abiotic Stresses
3.1 Low AsA Plants Are Sensitive to Abiotic Stresses
Ascorbate plays vital role in maintaining the plant health status. Mutation of genes involved in the d-mannose/l-galactose pathway led to the characterization of vitamin C defective (vtc) mutants. The vtc 1-1, vtc 2-1, and vtc 3 mutants have 50–75% lower AsA content than wild type. Plants with low AsA are sensitive to heat and light stress compared to control plants (Pavet et al. 2005; Conklin et al. 2013). Interestingly, the vtc 1-1 and vtc 2-1 lines are resistant to bacterial pathogen, Pseudomonas syringe (Tóth et al. 2011). The vtc mutant plants are sensitive to osmotic stress and oxidative stress (Cho et al. 2016) as shown in Table 2. This table illustrates that lowering AsA content makes plants more vulnerable to abiotic stresses.
3.2 High AsA Lines Are Tolerant to Abiotic Stresses
Table 3 presents a summary of the studies in which diverse research groups have evaluated the tolerance to stresses of plants with enhanced AsA content. High AsA plants overexpressing genes involved in the d-mannose/l-galactose pathway have been found to be tolerant to salt stress (up to 100 mM NaCl), herbicide stress (up to 75 μM methyl viologen), cold stress (4 °C), and oxidative stress (wounding). Plant biomass, plant height, shoot length, root length, leaf color, survival rate, and seed germination rate are the most common readouts used to assess the tolerance to abiotic stresses in plants (Zhang et al. 2015, Zhang et al. 2011; Landi et al. 2015; Li et al. 2012).
Similarly plants overexpressing d-galacturonate reductase were found to be tolerant to salt stress (up to 600 mM NaCl), herbicide stress (up to 75 μM methyl viologen), cold stress (4 °C), and osmotic stress (up to 300 mM mannitol). Chlorophyll content, root length/biomass and shoot length/biomass, germination response, and malondialdehyde content have been the main readouts used to assess the tolerance to abiotic stresses in these plants (Hemavathi et al. 2009; Cai et al. 2014; Lim et al. 2016).
Plants overexpressing l-gulono-1,4-lactone oxidase (GulLO) , the enzyme involved in the l-gulose and the inositol pathway of AsA, have elevated AsA. When subjected to abiotic stresses, those plants were tolerant to herbicide stress (up to 20 μM methyl viologen), osmotic stress (up to 300 mM mannitol), and salt stress (up to 600 mM NaCl). Phenotypic changes, chlorophyll content, and malondialdehyde content were the variables used to assess the tolerance to stresses in these plants (Hemavathi et al. 2009; Lim et al. 2016).
Our group and others have characterized the response to stresses of high AsA Arabidopsis lines overexpressing MIOX4, one of the enzymes involved in the myo-inositol pathway to AsA. Increased level of AsA in those plants correlates with tolerance to salt stress (up to 200 mM NaCl), cold stress (16 °C for 3 weeks), heat stress (29 °C for 10 days/40 °C for 15 min), environmental pollutant (up to 200 ppb pyrene), and light stress (up to 300 μmol m−2 s−1). Photosynthetic efficiency, root growth, and shoot growth are the main readouts used to assess abiotic stress tolerance (Tóth et al. 2011; Lisko et al. 2013).
Arabidopsis plants overexpressing a recently characterized GNL containing 5-fold AsA content were found to display enhanced growth rate, tolerance to light stress, and improved photosynthetic efficiency and seed yield (Yactayo-Chang 2016; Yactayo-Chang and Lorence 2016).
Monodehydroascorbate reductase (MDHAR) and dehydroascorbate reductase (DHAR) are the recycling enzymes for AsA in plants. MDHAR and DHAR help to maintain redox status of AsA (Haroldsen et al. 2011). Overexpression of MDHAR from Lycopersicum spp. and Malpighia spp. resulted in 1.2–1.8-fold elevated AsA level in leaves compared to wild-type plants. Elevated AsA level correlated with tolerance to herbicide stress (up to 100 μM methyl viologen), heat stress (40 °C for 24 h), cold stress (4 °C for 24 h), and salt stress (up to 300 mM NaCl) (Li et al. 2010; Eltelib et al. 2012). Similarly, overexpression of DHAR resulted in 1.2–4.0-fold increase in AsA level in leaves, fruits, and roots compared to controls. High AsA plants were found to be tolerant to ozone stress (up to 0.2 ppm), drought stress (5% polyethylene glycol), herbicide stress (up to 3 μM methyl viologen), aluminum stress (up to 400 μM AlCl3), salt stress (up to 150 mM NaCl), light stress (up to 1000 μmol m−2 s−1), and cold stress (4 °C for 1 week). Photosynthetic efficiency, plant height, number of leaves, plant growth, ion leakage, H2O2 accumulation, transpiration, CO2 accumulation, and stomatal conductance were the parameters used to assess the abiotic stress tolerance on these plants (Kwon et al. 2003, Chen and Gallie 2005, Eltayeb et al. 2011, Li et al. 2012, Qin et al. 2015; Liu et al. 2011).
Overexpression of homologues of the plant GulLO enzymes from Saccharomyces cerevisiae, o-arabiono-1,4 lactone oxidase and Stylosanthes guianensis 9-cis-epoxycarotenoid deoxygenase, resulted in a boost of the AsA content in plants. These transgenics showed tolerance to herbicide stress (up to 0.1 mM methyl viologen), light stress (up to 1200 μmol m−2 s−1), aluminum stress (100 mM AlCl3), cold stress (3 and 6 °C), and water limitation stress (Chen et al. 2005; Bao et al. 2016).
On the other hand, overexpression of master regulators of the AsA metabolic network, ethylene response factor 8, SID 22, SIHZ24 and CSN5B from Arabidopsis thaliana and Solanum lycopersicum resulted in 1.2- to 2.0-fold increase in AsA content in leaves and fruits compared to controls. Plants with elevated AsA were found to be tolerant to salt stress (300 mM NaCl), light stress, herbicide stress (up to 50 mM methyl viologen), and oxidative stress. Shoot biomass, root biomass, aluminum accumulation, relative water content, ion leakage, chlorophyll content, and malondialdehyde content were the parameters used to assess the abiotic stress tolerance in those studies (Zhang et al. 2012; Wang et al. 2013; Cai et al. 2016; Hu et al. 2016).
In summary, increased AsA level in plants is associated with enhanced abiotic stresses tolerance, whereas decreasing AsA level has led to sensitivity to abiotic stresses. Surprisingly, plants with low AsA level (vtc mutants) have been shown to be resistant to some biotic stresses.
To date, the most common readouts of abiotic stress tolerance have been plant size, plant shape, and color and photosynthesis parameters. In all published studies to date research teams have relied on manual phenotyping to make these assessments. Manual phenotyping is time consuming and sensitive to bias and is limited to the resolution of the naked eye. Based on these drawbacks, it is important to implement the use of more advanced tools to analyze the phenotype of high AsA plants to fully realize the potential of these metabolic engineering strategies.
4 Plant Phenomics, a Modern Approach to Characterize Plant Phenotypes
4.1 Closing the Gap Between Genomics and Phenomics
Understanding crop adaptation to abiotic and biotic stress is very important, especially now in the face of climate change and global population growth. Powered by the advances in gene editing (Barrangou and Doudna 2016) and the study of natural genetic variability (Nunes-Nesi et al. 2016), plant genomics has been growing to select and improve crop adaptation and yield. This is why it is important to understand plant plasticity using phenomics. Plant phenomics is defined as the application and development of different methodologies to capture information related with performance, function, and structure of a large number of plants (Houle et al. 2010; Tardieu et al. 2017). The main purpose of plant phenomics is to understand plant behavior under a vast variety of scenarios and how exactly the genotypic traits are expressed through the plant phenotype.
Scientists that have had the opportunity to work with manual plant phenotyping understand how slow this process can be. Measuring plant area, plant height, seed number, panicle number, fruit color, leaf number, and other traits of interest can be time consuming and affected by human bias. Phenomics is a research area that is moving fast due to the progress in the development of new sensors and imaging techniques for several traits, plant organs, and responses (Furbank and Tester 2011). However, a major challenge that remains in this area is data handling and processing when analyzing the sensor information and translating that information into knowledge.
4.2 Plant High-Throughput Phenotyping
Being able to gather massive amounts of information based on high resolution images is very convenient. The capability of eliminating destructive measurements increases the capacity of adjusting the experimental design and allows gathering information throughout the life cycle of plants. Plant high-throughput phenotyping technology consists of taking images with different sensors using hundreds of plants per day (Fahlgren et al. 2015). Data acquisition and analysis have become more feasible due to advances in automation for plant phenotyping, specifically robotic and sensor based technology (Rahaman et al. 2015). Currently, this technology has expanded not only at the laboratory (controlled environment) and greenhouse levels but also to the field scale.
The adaptability of this new technology is overwhelming, and it has made it possible to have platforms such as the Field Scanalyzer System at Rothamsted Research, UK and the TERRA-REF (http://terraref.org/) project in Maricopa, AZ, examples of large field crop analytic robots in the world. These robots take images by moving on a surface of 100–200 m2 using different sensors. This technology can also be used on a smaller scale with options that are less expensive, such as taking images using a digital camera, a tripod, and a light box attached (Chitwood et al. 2014). The data gathered with this technology can then be analyzed using commercial software or open source algorithms. A compilation of software options for plant phenomics can be found in the online database plant-image-analysis.org/ (Lobet et al. 2013).
4.3 Key Sensors in Plant Phenomics
Plant plasticity is very complex. When a genomic trait is adjusted, plants show very different architectures and phenotypic responses depending on the environmental conditions they face. Here is where different sensors come into play to help understand these behaviors. Sensor technology has improved since RGB images were used for the first time to determine plant growth, where top view RGB images were taken and correlated to obtain plant fresh weight (Leister et al. 1999).
The sensors available in the market work at different wavelength ranges of the light spectrum (Fig. 2). Color cameras are restricted to the 400–700 nm range and they have three colored sensors (red, green and blue) that are used to calculate the true color of each pixel (Fahlgren et al. 2015). These sensors use pixel-based maps to analyze several phenotypic characteristics such as biomass, area, diameter, flowering time, root architecture, plant height, and others. Although these sensors are the most commonly used, there is a disadvantage to them since they cannot distinguish between overlapping leaves or soil background unless some segmentation process is applied to the image (Li et al. 2014; Rahaman et al. 2015).
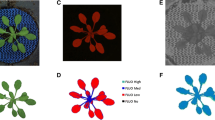
Representative images acquired with the visible (VIS), near infrared (NIR), and fluorescence (FLUO) sensors. (a–c) Raw images. (d–f) Analyzed images where the different parameters were quantified. (d) Shows geometrical parameters and color classification computed from VIS images. (e) In planta water content values computed from NIR images. (f) In planta chlorophyll fluorescence values computed from FLUO images. Values in E and F have been corrected to be relative to plant area
Due to global warming, scientists have focused their research in finding solutions to reduce the negative effect of abiotic/biotic stresses in plant yield. When a plant is affected by high temperature or soil water is depleted, plants tend to close the stomata to avoid water loss. The physiology of plants is also affected due to the increase in leaf temperature, and responses such as leaf tissue and enzyme damage, lower photosynthetic rate, higher respiration rate, and others occur (Schauberger et al. 2017). For this reason, sensors that detect near-infrared (NIR) and infrared (IR) light are very important for plant high-throughput phenotyping. These sensors use pixel-based maps and are used to determine plant water content (900–1700 nm) and canopy/leaf temperature (700–1000 nm), respectively (Fahlgren et al. 2015).
Plants under stress conditions are affected internally, specifically in their photosynthetic machinery, which will affect crop yield. The use of fluorescence (FLUO) sensors allows for the quantification of chlorophyll molecules in photosystem II by artificial excitation, and this information is used to measure how efficient the plant is during photosynthesis (Jansen et al. 2009; Li et al. 2014). The sensor corresponds to a charge-couple device (CCD) camera with sensitive fluorescent signals. These signals occur when the sample is illuminated with visible or UV light (Rahaman et al. 2015). The FLUO sensors are pixel-based maps that are used to detect disease, infestation, photosynthetic status, carbon assimilation, quantum yield, non-photochemical quenching, and total chlorophyll content (Kuhlgert et al. 2016).
Agronomical traits such as tiller, leaf, and panicle number are key for yield estimation. This is why 3D sensors play an important role in plant phenomics. These type of sensors use depth maps and allow acquiring noncontact and nondestructive measurements. By combining a 3D camera with laser scanning, the amount of point clouds detected enables a valid and accurate description of the plant geometry (Paulus et al. 2013). Some of the phenotype parameters that can be detected with this sensor include shoot structure, leaf angle, canopy health status, leaf growth, coverage density, and panicle health (Li et al. 2014).
The latest sensor developed, and the most expensive one, corresponds to imaging spectroscopy, also called hyperspectral imaging. This technology combines features of RGB imaging based on pixel mapping and spectroscopy based on intensity from different spectral bands. The most important feature about this sensor is that it enables measuring plant chemical traits such as water, nutrients, lipids, sugars, and others (Pandey et al. 2017). This sensor is currently the slowest of all, and the image analysis requires additional computational knowledge due to the data dimensions and complexity.
4.4 Phenomics to Understand Plant Abiotic/Biotic Stress Response
The power of phenomics relies on the fact that it is possible to extract a great deal of information from a single image. Figure 3 presents representative images of Arabidopsis plants that have been phenotyped using RGB, FLUO, and NIR sensors (panels A, B, and C). Once the images were analyzed through a commercial software (panels D, E, and F), the information was plotted and analyzed. Each of the sensors quantifies different phenotype parameters (Fig. 3). For example, from the RGB sensor, data such as plant size, architecture, biomass and growth can be obtained. From the NIR, quantification and localization of water can be obtained. More information can be gathered and analyzed, that will help understand in a more detailed manner plant adaptation to the new environment due to climate change. Subtle changes that are not detectable with the naked eye can be measured and quantified using sensors.
Comparison of different cameras used in high-throughput plant phenotyping and the parameters that can be measured with each of them. Each sensor captures signals from the visible and infrared spectrum of the light. The VIS (visible range) camera can quantify plant size, shape, and color. The IR (infrared range) camera can quantify plant stress. The PSII (photosystem II efficiency) sensor can quantify plant photosynthetic activity. The NIR/SWIR (near infrared and short-wave infrared) camera can detect plant relative water content. The TIR/LWIR (thermal infrared and long-wave infrared) sensor can detect plant canopy or leaf temperature. The hyperspectral camera can measure plant stress and chemical composition by detecting a wide variety of spectral bands
Figure 4 illustrates the use of plant phenomics approaches in the characterization of high AsA Arabidopsis lines. As shown in this figure, AtMIOX4 over-expressers are salt tolerant as indicated by their higher growth rate and biomass accumulation compared to wild-type controls. Although not evident to the naked eye, but captured by the RGB camera AtMIOX4 displayed less chlorosis than wild-type plants exposed to NaCl stress.
Characterization of the phenotype of a high ascorbate line compared to controls. Arabidopsis seeds were sterilized, vernalized, and planted on Murashige and Skoog media. After germination vigorous seedlings were transferred to soil and grown in an environment controlled chamber (23 °C, 65% humidity, 16:8 h photoperiod, 200 μmol m−2 s−1 light intensity). After acclimation to soil, plants were subjected to salt stress (150 mM NaCl) 21 days after germination. (a, b) Raw RGB images after segmentation. (c) Projected leaf area of the Arabidopsis rosette; data are means ± SE, n = 15. (d, e) Color classification of the plants analyzed in (b). Values were corrected relative to plant area. This parameter distinguishes yellow color (chlorosis) from green color (healthy tissue). Chlorosis was not apparent by visual inspection, but detected by the RGB camera. MIOX Myo-inositol oxygenase over-expresser, WT wild-type control
4.5 Data Analysis and Handling
Plant high-throughput phenotyping studies generate large-scale, multidimensional data sets that require proper analysis (Klukas et al. 2014). One of the main challenges in the field is standardization of the methodologies and data publication. Storage and data collection needs to be consistent and linked with specific phenotypes and genomic information (Krajewski et al. 2015).
Optimization of the experimental design is key to obtain good quality images and, therefore, reproducible data. First, it is necessary to determine the objective of the experiment before acquiring any information. Then, once the objective is determined, it is necessary to select the sensor/sensors to use. Moreover, having consistency in how the images are acquired is imperative. Improving the quality of the images facilitates the analysis. One simple example of this is adding a blue mesh as a soil cover, in order to improve background/object contrast and to reduce evaporation (Junker et al. 2014).
Plant development research has been positively affected by the advance in phenotyping technologies, which help to determine phenotypic traits that change depending on genotype and environment. Further analysis that integrates several sensors and overlaps the data collected is necessary to better understand plant plasticity and adaptation.
5 Conclusions
During the past two decades, we have significantly advanced our understanding about the different pathways plants use to make AsA, a key molecule involved in the modulation of plant growth and development, plant health, and stress tolerance. Full realization of the potential that this knowledge represents for the development of more nutritious and improved crops must involve the incorporation of novel approaches to engineer crops and to characterize their phenotype. We propose that gene editing (Barrangou and Doudna 2016) and plant phenomics are two approaches that are likely to revolutionize the way high AsA crops are made and characterized before some of them are able to reach widespread adoption.
References
Aboobucker SI (2014) Identification and characterization of a functional L-gulono-1,4-lactone oxidase in Arabidopsis thaliana. PhD dissertation, Arkansas State University, Jonesboro, AR
Aboobucker SI, Suza WP, Lorence A (2017) Characterization of two Arabidopsis L-gulono-1,4-lactone oxidases, AtGulLO3 and AtGulLO5, involved in ascorbate biosynthesis. React Oxyg Spec 4(12):1–29
Agius F, Gonzáles-Lamothe R, Caballero JL, Muňoz-Blanco J, Botella MA, Valpuesta V (2003) Engineering increased vitamin C levels in plants by over-expression of a D-galacturonic acid reductase. Nat Biotechnol 21:177–181
Amaya I, Osorio S, Martinez-Ferri E, Lima-Silva V, Doblas VG, Fernández-Muňoz R, Fernie AR, Botella MA, Valpuesta V (2014) Increased antioxidant capacity in tomato by ectopic expression of the strawberry D-galacturonate reductase gene. Biotechnol J 10(3):490–500
Badejo A, Tanaka N, Esaka M (2008) Analysis of GDP-D-mannose pyrophosphorylase gene promoter from acerola (Malpighia glabra) and increase in ascorbate content of transgenic tobacco expressing the acerola gene. Plant Cell Physiol 49(1):126–132
Badejo A, Eltelib H, Fukunaga K, Fujikawa Y, Esaka M (2009) Increase in ascorbate content of transgenic tobacco plants overexpressing the acerola (Malpighia glabra) phosphomannomutase gene. Plant Cell Physiol 50(2):423–428
Bao G, Zhuo C, Qian C, Xiao T, Guo Z, Lu S (2016) Co-expression of NCED and ALO improves vitamin C level and tolerance to drought and chilling in transgenic tobacco and stylo plants. Plant Biotechnol J 14:206–214
Barrangou R, Doudna J (2016) Applications of CRISPR technologies in research and beyond. Nat Biotechnol 34:933–941
Beal T, Massiot E, Arsenault JE, Smith MR, Hijmans RJ (2017) Global trends in dietary micronutrient supplies and estimated prevalence of inadequate intakes. PLoS One 12(4):e0175554
Belmonte M, Stasolla C (2009) Altered HBK3 expression affects glutathione and ascorbate metabolism during the early phases of Norway spruce (Picea abies) somatic embryogenesis. Plant Physiol Biochem 47:904–911
Bulley S, Wright M, Rommens C, Yan H, Rassam M, Lin-Wang K, Andre C, Brewster D, Karunairetnam S, Allan AC, Laing WA (2012) Enhancing ascorbate in fruits and tubers through over-expression of the L-galactose pathway gene GDP-L-galactose phosphorylase. Plant Biotechnol J 10:390–397
Cai X, Zhang C, Ye J, Hu T, Ye Z, Li H, Zhang Y (2014) Ectopic expression of FaGalUR leads to ascorbate accumulation with enhanced oxidative stress, cold, and salt tolerance in tomato. Plant Growth Regul 76(2):187–197
Cai X, Zhang C, Shu W, Ye Z, Li H, Zhang Y (2016) The transcription factor SlDof22 involved in ascorbate accumulation and salinity stress in tomato. Biochem Biophys Res Commun 474:736–741
Chambial S, Dwivedi S, Kant Shukla K, John P, Sharma P (2013) Vitamin C in disease prevention and cure: an overview. Indian J Clin Biochem 4:314–328
Chen Z, Gallie D (2005) Increasing tolerance to ozone by elevating foliar ascorbic acid confers greater protection against ozone than increasing avoidance. Plant Physiol 138:1673–1689
Chen Z, Young T, Ling J, Chang S, Gallie D (2003) Increasing vitamin C content of plants through enhanced ascorbate recycling. Proc Natl Acad Sci U S A 100(6):3525–3530
Chitwood D, Ranjan A, Martinez C, Headland L, Thiem T, Kumar R, Covington M, Hatcher T, Naylor D, Zimmerman S, Downs N, Raymundo N, Buckler E, Maloof J, Aradhya M, Prins B, Li L, Myles S, Sinha N (2014) A modern ampelography: a genetic basis for leaf shape and venation patterning in grape. Plant Physiol 164:259–272
Cho K, Nguyen H, Kim S, Shin J, Cho D, Hong S, Shin J, Ok S (2016) CML10, a variant of calmodulin, modulates ascorbic acid synthesis. New Phytol 209:664–678
Conklin P, De Paolo D, Wintle B, Schatz C, Buckenmeyre G (2013) Identification of Arabidopsis VTC3 as a putative and unique dual function protein kinase: protein phosphatase involved in the regulation of the ascorbic acid pool in plants. J Exp Bot 64:2793–2804
Cronje C, George GM, Fernie AR, Bekker J, Kossmann J, Bauer R (2012) Manipulation of L-ascorbic acid biosynthesis pathway in Solanum lycopersicum: elevated GDP-mannose pyrophosphorylase activity enhanced L-ascorbate levels in red fruit. Planta 235:553–564
Eltayeb A, Kawano N, Badawi G, Kaminaka H, Sanekata T, Morishima I, Shibahara T, Inanaga S, Tanaka K (2006) Enhanced tolerance to ozone and drought stresses in transgenic tobacco overexpressing dehydroascorbate reductase in cytosol. Physiol Plant 127:57–65
Eltayeb A, Kawano N, Badawi GH, Kaminaka H, Sanekata T, Shibahara T, Inanaga S, Tanaka K (2007) Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta 225:1255–1264
Eltayeb A, Yamamoto S, Eltayeb Habora M, Yin L, Tsujimoto H, Tanaka K (2011) Transgenic potato overexpressing Arabidopsis cytosolic AtDHAR1 showed higher tolerance to herbicide, drought and salt stresses. Breed Sci 61:3–10
Eltelib H, Fujikawa Y, Esaka M (2012) Overexpression of the acerola (Malpighia glabra) monodehydroascorbate reductase gene in transgenic tobacco plants results in increased ascorbate levels and enhanced tolerance to salt stress. S Afr J Bot 78:295–301
Fahlgren N, Gehan M, Baxter I (2015) Lights, camera, action: high-throughput plant phenotyping is ready for a close-up. Curr Opin Plant Biol 24:93–99
Foyer CH, Noctor G (2011) Ascorbate and glutathione: the heart of the redox hub. Plant Physiol 155:2–18
Foyer C, Descourvieres P, Kunert KJ (1994) Protection against oxygen radicals: an important defense mechanism studies in transgenic plants. Plant Cell Environ 17:507–523
Furbank R, Tester M (2011) Phenomics—technologies to relieve the phenotyping bottleneck. Trends Plant Sci 16:635–644
Gest N, Garchery C, Gautier H, Jiménez A, Stevens R (2013) Light-dependent regulation of ascorbate in tomato by a monodehydroascorbate reductase localized in peroxisomes and the cytosol. Plant Biotechnol J 11:344–354
Goo Y, Chun H, Kim T, Lee C, Ahn M, Bae S, Cho K, Chun J, Chung C, Lee S (2008) Expressional characterization of dehydroascorbate reductase cDNA in transgenic potato plants. J Plant Biol 51:35–41
Haroldsen V, Chi-Ham C, Kulkarni S, Lorence A, Bennett A (2011) Constitutively expressed DHAR and MDHAR influence fruit, but not foliar ascorbate levels in tomato. Plant Physiol Biochem 49(10):1244–1249
Heick H, Graff G, Humpers J (1972) The occurrence of ascorbic acid among the yeast. Can J Microbiol 18(5):597–600
Hemavathi, Upadhyaya C, Young K, Akula N, Kim H, Heung J, Oh O, Aswath C, Chun S, Kim D, Park S (2009) Over-expression of strawberry D-galacturonic acid reductase in potato leads to accumulation of vitamin C with enhanced abiotic stress tolerance. Plant Sci 177:659–667
Hemavathi, Upadhyava C, Akula N, Young K, Chun S, Kim D, Park S (2010) Enhanced ascorbic acid accumulation in transgenic potato confers tolerance to various abiotic stresses. Biotechnol Lett 32:321–330
Houle D, Govindaraju DR, Omholt S (2010) Phenomics: the next challenge. Nat Rev Genet 11:855–866
Hu T, Ye J, Tao P, Li H, Zhang J, Zhang Y, Ye Z (2016) The tomato HD-Zip I transcription factor SlHZ24 modulates ascorbate accumulation through positive regulation of the D-mannose/L-galactose pathway. Plant J 85:16–29
Huang M, Xu Q, Deng X (2014) L-ascorbic acid metabolism during fruit development in an ascorbate-rich fruit crop chestnut rose (Rosa roxburghii Tratt). J Plant Physiol 171(14):1205–1216
Imai T, Ban Y, Yamamoto T, Moriguchi T (2012) Ectopic overexpression of peach GDP-D-mannose pyrophosphorylase and GDP-D-mannose-3’5’-epimerase in transgenic tobacco. Plant Cell Tissue Organ Cult 111:1–13
Ishikawa T, Dowdle J, Smirnoff N (2006) Progress in manipulating ascorbic acid biosynthesis and accumulation in plants. Physiol Plant 126:343–355
Jain A, Nessler C (2000) Metabolic engineering of an alternative pathway for ascorbic acid biosynthesis in plants. Mol Breed 6:73–78
Jansen M, Gilmer F, Biskup B, Nagel K, Rascher U, Fischbach A, Briem S, Dreissen G, Tittmann S, Braun S, De Jaeger I, Metzlaff M, Schurr U, Scharr H, Achim W (2009) Simultaneous phenotyping of leaf growth and chlorophyll fluorescence via GROWSCREEN FLUORO allows detection of stress tolerance in Arabidopsis thaliana and other rosette plants. Funct Plant Biol 36:902–914
Junker A, Muraya M, Weigelt-Fischer K, Arana-Ceballos F, Klukas C, Melchinger A, Meyer R, Riewe D, Altmann T (2014) Optimizing experimental procedures for quantitative evaluation of crop plant performance in high throughput phenotyping systems. Front Plant Sci 5:770
Klukas C, Chen D, Pape J (2014) Integrated analysis platform: an open-source information system for high-throughput plant phenotyping. Plant Physiol 165:506–518
Krajewski P, Chen D, Ćwiek H, Dijk A, Fiorani F, Kersey P, Klukas C, Lange M, Markiewicz A, Nap J, Oeveren J, Pommier C, Scholz U, Schriek M, Usadel B, Weise S (2015) Towards recommendations for metadata and data handling in plant phenotyping. J Exp Bot 66:5417–5427
Kuhlgert S, Austic G, Zegarac R, Osei-Bonsu I, Hoh D, Chilvers M, Roth M, Bi K, TerAvest D, Weebadde P, Kramer D (2016) MultispeQ Beta: a tool for large-scale plant phenotyping connected to the open PhotosynQ network. R Soc Open Sci 3:160592
Kulkarni S (2012) Elevating ascorbate content in tomato and studying the role of jasmonates in modulating ascorbate in Arabidopsis. MS Thesis, Arkansas State University, Jonesboro, AR
Kwon S, Choi S, Ahn Y, Lee H, Lee H, Park Y, Kwak S (2003) Enhanced stress-tolerance of transgenic tobacco plants expressing a human dehydroascorbate reductase gene. J Plant Physiol 160:347–353
Laing W, Wright M, Cooney J, Bulley S (2007) The missing step of the L-galactose pathway of ascorbate biosynthesis in plants, and L-galactose guanyltransferase, increases leaf ascorbate content. Proc Natl Acad Sci U S A 104(22):9534–9539
Landi M, Fambrini M, Basile A, Salvini M, Guidi L, Pugliesi C (2015) Overexpression of L-galactono-1,4-lactone dehydrogenase (L-GalLDH) gene correlates with increased ascorbate concentration and reduced browning in leaves of Lactuca sativa L. after cutting. Plant Cell Tissue Organ Cult 123:109–120
Leister D, Varotto C, Pesaresi P, Niwergall A, Salamini F (1999) Large-scale evaluation of plant growth in Arabidopsis thaliana by non-invasive image analysis. Plant Physiol Biochem 37:671–678
Li F, Wu QY, Sun YL, Wang LY, Yang XH, Meng QW (2010) Overexpression of chloroplastic monodehydroascorbate reductase enhanced tolerance to temperature and methyl viologen-mediated oxidative stresses. Physiol Plant 139:421–434
Li Q, Li Y, Li C, Yu X (2012) Enhanced ascorbic acid accumulation through overexpression of dehydroascorbate reductase confers tolerance to methyl viologen and salt stresses in tomato. Czech J Genet Plant Breed 48(2):74–86
Li L, Zhang Q, Huang D (2014) A review of imaging techniques for plant phenotyping. Sensors 14:20078–20111
Lim M, Jeong B, Jung M, Harn C (2016) Transgenic tomato plants expressing strawberry D-galacturonic acid reductase gene display enhanced tolerance to abiotic stresses. Plant Biotechnol Rep 10:105–116
Linster CL, Van Schaftingen E (2007) Vitamin C: biosynthesis, recycling and degradation in mammals. FEBS J 274:1–22
Lisko K, Torres R, Harris R, Belisle M, Vaughan M, Julian B, Chevone B, Mendes P, Nessler C, Lorence A (2013) Elevating vitamin C content via overexpression of myo-inositol oxygenase and L-gulono-1,4-lactone oxidase in Arabidopsis leads to enhanced biomass and tolerance to abiotic stresses. In Vitro Cell Dev Biol Plant 49(6):643–655
Liu Y, Yu L, Wang R (2011) Level of ascorbic acid in transgenic rice for L-galactono-1,4-lactone dehydrogenase overexpressing or suppressed is associated with plant growth and seed set. Acta Physiol Plant 33:1353–1363
Liu W, An H, Yang M (2013) Overexpression of Rosa roxburghii L-galactono1,4-lactone dehydrogenase in tobacco plant enhances ascorbate accumulation and abiotic stress tolerance. Acta Physiol Plant 35:1617–1624
Liu F, Guo X, Yao Y, Tang W, Zhang W, Cao S, Han Y, Liu Y (2015) Cloning and function characterization of two dehydroascorbate reductases from kiwifruit (Actinidia chinensis L). Plant Mol Biol Rep 34(4):815–826
Lobet G, Draye X, Périlleux C (2013) An online database for plant image analysis software tools. Plant Methods 9(38):1–8
Loewus F, Kelly S (1961) The metabolism of D-galacturonic acid and its methyl ester in the detached ripening strawberry. Arch Biochem Biophys 95:483–493
Lorence A, Nessler C (2007) Pathway engineering of the plant vitamin C metabolic network. In: Verpoorte R, Alfermann AW, Johnson TS (eds) Applications of plant metabolic engineering. Springer, Dordrecht, pp 197–217
Lorence A, Chevone B, Mendes P, Nessler C (2004) myo-Inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol 134:1200–1205
Ma L, Wang Y, Liu W, Liu Z (2014) Overexpression of an alfalfa GDP-mannose 3,5-epimerase gene enhances acid, drought and salt tolerance in transgenic Arabidopsis by increasing ascorbate accumulation. Biotechnol Lett 36(11):2331–2341
Mapson L, Isherwood F (1956) Biological synthesis of ascorbic acid: the conversion of derivatives of D-galacturonic acid into L-ascorbic acid by plant extract. Biochem J 64:13–22
Naqvi S, Zhu C, Farre G, Ramessar K, Bassie L, Breitenbach J, Perez Conesa D, Ros G, Sandmann G, Capell T, Christou P (2009) Transgenic multivitamin corn through biofortification of endosperm with three vitamins representing three distinct metabolic pathways. Proc Natl Acad Sci U S A 106(19):7762–7767
Noctor G, Foyer H (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49:249–279
Nunes-Nesi A, Carrari F, Lytovchenko A, Smith A, Loureiro M, Ratcliffe R, Sweeetlove L, Fernie A (2005) Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol 137:611–622
Nunes-Nesi A, Nascimento Vde L, de Oliveira Silva FM, Zsogon A, Araujo W, Sulpice R (2016) Natural genetic variation for morphological and molecular determinants of plant growth and yield. J Exp Bot 67:2989–3001
Pandey P, Ge Y, Stoerger V, Schnable J (2017) High throughput in vivo analysis of plant leaf chemical properties using hyperspectral imaging. Front Plant Sci 8:1–12
Paulus S, Dupuis J, Mahlein A, Kuhlmann H (2013) Surface feature based classification of plant organs from 3D laser scanned point clouds for plant phenotyping. BMC Bioinformatics 14:1–12
Pavet V, Olmos E, Kiddle G, Mowla S, Kumar S, Antoniw J, Alvarez M, Foyer C (2005) Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiol 139:1291–1303
Qin A, Huang X, Zhang H, Wu J, Yang J, Zhang S (2015) Overexpression of PbDHAR2 from Pyrus sinkiangensis in transgenic tomato confers enhanced tolerance to salt and chilling stress. Hortscience 50:789–796
Radzio J, Lorence A, Chevone B, Nessler C (2003) L-Gulono-1,4-lactone oxidase expression rescues vitamin C-deficient Arabidopsis (vtc) mutants. Plant Mol Biol 53:837–844
Rahaman M, Chen D, Gillani Z, Klukas C, Chen M (2015) Advanced phenotyping and phenotype data analysis for the study of plant growth and development. Front Plant Sci 6:619
Sawake S, Tajima N, Mortimer JC, Lao J, Ishikawa T, Yu X, Yamanashi Y, Yoshimi Y, Kawai-Yamada M, Dupree P, Tsumuraya Y, Kotake T (2015) KONJAC1 and 2 are key factors for GDP-mannose generation and affect L-ascorbic acid and glucomannan biosynthesis in Arabidopsis. Plant Cell 27:3397–3409
Schauberger B, Archontoulis S, Arneth A, Balkovic J, Ciais P, Deryng D, Elliott J, Folberth C, Khabarov N, Muller C, Pugh T, Rolinski S, Schaphoff S, Schmid E, Wang X, Schlenker W, Frieler K (2017) Consistent negative response of US crops to high temperatures in observations and crop models. Nat Commun 8:1–9
Suza W, Avila C, Carruthers K, Kulkarni S, Gogging F, Lorence A (2010) Exploring the impact of wounding and jasmonates on ascorbate metabolism. Plant Physiol Biochem 48:337–350
Tardieu F, Cabrera-Bosquet L, Pridmore T, Bennett M (2017) Plant phenomics, from sensors to knowledge. Curr Biol 27:R770–R783
Tóth S, Nagy V, Puthur J, Kovács L, Garab G (2011) The physiological role of ascorbate as photosystem II electron donor: protection against photo-inactivation in heat-stressed leaves. Plant Physiol 156:382–392
Ushimaru T, Nakagawa T, Fujioka Y, Daicho K, Naito M, Yamauchi Y, Nonaka H, Amako K, Yamawaki K, Muratad N (2006) Transgenic Arabidopsis plants expressing the rice dehydroascorbate reductase gene are resistant to salt stress. J Plant Physiol 163:1179–1184
Wang Z, Xiao Y, Chen W, Tang K, Zhang L (2010) Increased vitamin C content accompanied by an enhanced recycling pathway confers oxidative stress tolerance in Arabidopsis. J Integr Plant Biol 52(4):400–409
Wang J, Yu Y, Zhang Z, Quan R, Zhang H, Ma L, Deng XW, Huang R (2013) Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis. Plant Cell 25:625–636
Wevar Oller A, Agostini E, Milrad S, Medina M (2009) In situ and de novo biosynthesis of vitamin C in wild type and transgenic tomato hairy roots: a precursor feeding study. Plant Sci 177:28–34
Wheeler G, Jones M, Smirnoff N (1998) The biosynthetic pathway of vitamin C in higher plants. Nature 393:365–369
Wolucka B, Van Montagu M (2003) GDP-mannose3′5′-epimerase forms GDP-L-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. J Biol Chem 278(48):47483–47490
Yactayo-Chang J (2011) Stable co-expression of vitamin C enhancing genes for improved production of a recombinant therapeutic protein, hIL-12, in Arabidopsis thaliana. MS thesis, Arkansas State University, Jonesboro, AR
Yactayo-Chang J (2016) The role of the chloroplastic and endoplasmic reticulum ascorbate subcellular pools in plant physiology. PhD thesis, Arkansas State University, Jonesboro, AR
Yactayo-Chang J, Lorence A (2016) Method of improving chloroplast function. US Patent pending
Yin L, Wang S, Eltayeb A, Uddin I, Yamamoto Y, Tsuji W, Takeuchi Y, Tanaka K (2010) Overexpression of dehydroascorbate reductase, but not monodehydroascorbate reductase, confers tolerance to aluminum stress in transgenic tobacco. Planta 231:609–621
Zhang W, Gruszewski H, Chevone B, Nessler C (2008) An Arabidopsis purple acid phosphatase with phytase activity increases foliar ascorbate. Plant Physiol 146:431–440
Zhang W, Lorence A, Gruszewski H, Chevone B, Nessler C (2009) AMR1, an Arabidopsis gene that coordinately and negatively regulates the mannose/L-galactose ascorbic acid biosynthetic pathway. Plant Physiol 150:942–950
Zhang C, Liu J, Zhang Y, Cai X, Gong P, Zhang J, Wang T, Li H, Ye Z (2011) Overexpression of SlGMEs leads to ascorbate accumulation with enhanced oxidative stress, cold, and salt tolerance in tomato. Plant Cell Rep 30:389–398
Zhang Z, Wang J, Zhang R, Huang R (2012) The ethylene response factor AtERF98 enhances tolerance to salt through the transcriptional activation of ascorbic acid synthesis in Arabidopsis. Plant J 71:273–287
Zhang G, Liu R, Zhang C, Tang K, Sun M, Yan G, Liu Q (2015) Manipulation of the rice L-galactose pathway: evaluation of the effects of transgene overexpression on ascorbate accumulation and abiotic stress tolerance. PLoS One 10(5):1–14
Zhou Y, Tao QC, Wang ZN, Fan R, Li Y, Sun XF, Tang KX (2012) Engineering ascorbic acid biosynthetic pathway in Arabidopsis leaves by single and double gene transformation. Biol Plant 56(3):451–457
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG, part of Springer Nature
About this chapter
Cite this chapter
Yactayo-Chang, J.P., Acosta-Gamboa, L.M., Nepal, N., Lorence, A. (2017). The Role of Plant High-Throughput Phenotyping in the Characterization of the Response of High Ascorbate Plants to Abiotic Stresses. In: Hossain, M., Munné-Bosch, S., Burritt, D., Diaz-Vivancos, P., Fujita, M., Lorence, A. (eds) Ascorbic Acid in Plant Growth, Development and Stress Tolerance. Springer, Cham. https://doi.org/10.1007/978-3-319-74057-7_13
Download citation
DOI: https://doi.org/10.1007/978-3-319-74057-7_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-74056-0
Online ISBN: 978-3-319-74057-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)