Abstract
Cardiac resynchronization therapy (CRT) is an effective treatment for patients with congestive heart failure and ventricular dyssynchrony. Despite the overall efficacy of CRT, approximately 30% of patients receiving CRT do not improve. One of the main technical problems related to the CRT procedure is inadequate visualisation in X-ray fluoroscopy of the venous anatomy in relation to accurate cardiac chamber visualisation. This paper proposes a novel approach for 3D reconstruction of coronary veins from a single contrast enhanced intra-operative fluoroscopy image. For this application, the method uses back-projection geometry and a Euclidean distance/angle-based cost function. The algorithm is validated on a phantom and five patient datasets, comprising six view-angle orientations for the phantom dataset and two view-angle orientations for each of the patient datasets. Median(inter-quartile range) 3D-reconstruction accuracies of 1.41(0.55–3.00) mm and 3.28(2.10–4.89) mm were established for the phantom and patient data, respectively. The technique can facilitate careful advancement of the cannulating guide over a guidewire or a diagnostic catheter positioned in the coronary sinus, and consequently, improve the chances of response to CRT.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Cardiac resynchronization therapy (CRT) has been shown to improve outcomes in a growing subset of patients with congestive heart failure. Although the majority of patients who meet the criteria for CRT under current guidelines derive benefit, approximately one-third of patients do not respond to this pacing modality [15]. Most of these failures are due to difficulty accessing the coronary sinus (CS) ostium or advancing the pacing lead into an adequate, stable position [11]. In order to maintain the accuracy of the guidance information, thereby allowing accurate determination of pacing treatment sites, volumetric coronary vein roadmaps overlaid on X-ray fluoroscopy can be used. Coronary vein anatomy can be provided pre-operatively with multislice computed tomography (CT) [9]. However, CT requires an additional use of ionizing radiation and nephrotoxic contrast agents. Cardiac MR (CMR) imaging is also used to depict the anatomy of the venous system of the heart [3], although a high-spatial resolution and longer scan time is required to adequately depict the relatively small coronary vessels.
The standard visualisation method is a 2D X-ray examination using an injection of contrast material, called a venogram. This uses less radiation than CT and is able to visualise vessels that cannot be seen in CMR [5]. Many methods exist for reconstruction of coronary arteries from venograms [4]. The vascular tree can be reconstructed in 3D by triangulation from venograms if at least two views of the coronary vascular tree are obtained. For reconstruction of the vascular tree, however, corresponding vessels must be identified either manually or by use of the vessel hierarchy [6]. Corresponding points along the vessel centerlines can then be established by means of an epipolar-line technique [13]. Paired images for 3D coronary vein reconstruction can be acquired using a biplane X-ray system [2, 14], although these are less common than a monoplane system in the clinical setting, and involve increased radiation exposure for both the patient and the clinician. Alternatively, 3D reconstruction of coronary veins can be achieved using a monoplane system. This requires either acquisition of a rotational X-ray sequence [1], which involves a long radiation exposure, or two sequences at arbitrary orientations [10]. Such techniques require both cardiac and respiratory phase matching of the images.
In this paper, a novel semi-automatic approach is presented for 3D reconstruction of coronary veins to overcome the limitations of the already proposed techniques. Unlike all previous techniques the proposed technique can reconstruct the coronary vein centrelines from a single contrast enhanced X-ray fluoroscopic image registered to an MR segmentation. This technique reduces radiation dose and simplifies clinical workflow.
2 Methods
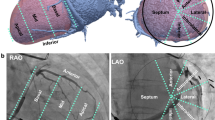
In this section the formation of a 3D model of the coronary veins, reconstructed from a single contrast injected X-ray fluoroscopy image, is described. The workflow of the image analysis framework is illustrated in Fig. 1. Initially, the left ventricle (LV), segmented from preoperative MR, is registered to an intraoperative X-ray fluoroscopic image. The coronary veins are manually annotated on the contrast injected X-ray fluoroscopic image and back-projected to the 3D registered LV mesh. The algorithm makes use of the 3D intersection points between the back-projected rays and the surface of the LV mesh to search for and locate the position of the coronary veins around the LV surface.
2.1 Registration of LV Mesh and Coronary Vein Annotation
The LV is automatically segmented from pre-procedural MRI. A fully-automatic slice-by-slice segmentation and propagation of the epicardial LV borders in long (two-, three- and four-chamber) and short axis images is computed. The system then generates a mesh of the epicardial cavity that follows the epicardial contours at end diastole, using a model-based segmentation algorithm [8].
A single contrast enhanced (end diastolic, end expiration) frame from the X-ray sequence is automatically selected using Masked-principal component analysis motion gating [12]. As part of the proposed workflow, manual annotation of the centrelines of visible vessels is required on the chosen X-ray image (Fig. 1b). The method requires that the first annotated 2D point should correspond to a point on the posterior side of the LV. This can easily be identified, as the CS ostium is always visible in these images and based on the CS cardiac anatomy it lies on the posterior side of the LV mesh at standard angulations. Finally, a clinical expert manually registers the segmented LV mesh to the X-ray fluoroscopic image using a custom-made visualisation software. This is also done intra-procedurally (Fig. 1a).
2.2 Back-Projection of 2D CS Vessel Annotated Points
X-ray fluoroscopy follows the ideal pinhole camera model that describes the relationship between a 3D point and its corresponding 2D projection onto the image plane. The back-projection of a 2D point in the image plane is a line, called the projection line, calculated using the camera parameters of the X-ray fluoroscopy projective modality [7]. The camera parameters are obtained from the DICOM header of the X-ray images. Using projection geometry, each of the 2D coronary venous positions annotated in Fig. 1b is back-projected to form a 3D line, illustrated in red in Fig. 1c.
2.3 3D Reconstruction of Coronary Veins
To reconstruct the coronary veins in 3D (Fig. 1d), the algorithm uses the \(1^{st}\) reconstructed point and a Euclidean distance/angle-based cost function to determine subsequent points along the vessel. The first 3D point, part of the coronary sinus, lies on the 3D line back-projected from the \(1^{st}\) 2D annotated point. This line intersects the mesh at two points and the correct point must be chosen for the reconstruction. The \(1^{st}\) 3D point is known to be the posterior point. Following the determination of the \(1^{st}\) 3D-reconstructed point, subsequent 3D points are defined according to
where \(D(\mathbf{p}_{i}; \mathbf{p}_{i-1})\) is a function that computes the Euclidean distance between the previously defined 3D point, \(\mathbf{p}_{i-1}\), and the candidate points, \(\mathbf{p}_{i}\), as defined in Eq. (2). \(A(\mathbf{p}_{i}; \mathbf{p}_{i-1}, \mathbf{p}_{i-2})\) is a function that computes the angle between \(\mathbf{p}_{i}\) and the two previously defined 3D points, \(\mathbf{p}_{i-1}\) and \(\mathbf{p}_{i-2}\), as defined by Eq. (3) and illustrated in Fig. 2.
where \(\cos (\theta _{i}) = \frac{\overrightarrow{\mathbf{p}_{i-2} \mathbf {p}_{i-1}} \cdot \overrightarrow{\mathbf{p}_{i-1} \mathbf {p}_{i}}}{||\overrightarrow{\mathbf{p}_{i-2} \mathbf {p}_{i-1}}|| ||\overrightarrow{\mathbf{p}_{i-1} \mathbf {p}_{i}}||}\). \(\lambda \) is the weight given to the distance and angle functions. For this algorithm \(\lambda =2\) was found to favour a smoothly curving path of points, which is important at the edges of the projection where the two distances are very similar.
3 Experiments
3.1 Data Acquisition
The proposed algorithm was quantitatively and qualitatively evaluated on a phantom data set and clinical images acquired from 5 different patients undergoing CRT; these comprised a total of 6 view-angle orientations for the phantom dataset and 2 view-angle orientations for each of the clinical datasets. The phantom experiments were performed to evaluate the proposed approach in a clinical imaging environment with a known ground truth registration. The LV epicardial surface (segmented from an MR image) was 3D printed and wires were attached to model the vascular tree. Intra-operative data were then obtained by acquiring a cone beam CT, which provided the registered LV mesh and 6 X-ray images.
Imaging of three of the clinical datasets was carried out using a monoplane 25 cm flat panel cardiac X-ray system (Philips Allura Xper FD10, Philips Healthcare, Best, The Netherlands) while imaging of the phantom dataset and the remaining two clinical datasets was carried out using a biplane cardiac X-ray system (Artis, syngo X Workplace VC10N, Siemens Healthcare GmbH). This study was approved by our Local Ethics Committee.
3.2 Gold Standard 3D Reconstruction of Coronary Veins
Multiple view-angle orientations were used to obtain a manual ground truth coronary vein reconstruction for each of the tested datasets. Using projection geometry, each of the 2D coronary vein positions was carefully annotated (Fig. 3a). Each annotated point was back-projected to form a 3D line, which was then forward projected to generate a 2D epipolar line (Fig. 3b) in a \(2^{nd}\) view that contains the corresponding 2D coronary vein position. For each epipolar line generated, the corresponding coronary vein position was manually detected. Each pair of matching points from the two projection planes was then back-projected (Fig. 3c) to reconstruct the coronary veins in 3D (Fig. 3d).
Illustration of ground truth 3D-reconstruction workflow. (a) 2D manual coronary vein annotation. (b) Use of epipolar line to manually annotate corresponding points in the second view. (c) Back-projection of annotated points from each view angle. (d) 3D coronary vein reconstruction from closest points of intersection between the back-projected rays from each view angle.
3.3 Success Rate and Accuracy of Reconstruction
A successfully reconstructed vessel was defined as one that was reconstructed on the correct side of the LV mesh, following a path similar to the gold standard reconstruction. In cases where the wrong path of a vessel was chosen by the algorithm the reconstruction of the specific vessel was considered a failure. Percentage success rates were computed as the proportion of vessels that were successfully reconstructed, for the phantom and patient datasets. The accuracy of the successfully reconstructed vessels, for both the phantom and the patient datasets, was calculated as the mean of the mm distance from each 3D reconstructed point and the nearest point on the gold standard reconstruction.
(a) Registration of the LV printed mesh to the X-ray image for three view-angle orientations. (b) 3D-reconstructed coronary veins (red) and gold standard (blue). The black arrows illustrate the vessels that were unsuccessfully reconstructed by the algorithm. (c) 3D-reconstructed coronary veins overlaid onto the 16 segment colour coded LV epicardium mesh. (Color figure online)
Example reconstructions of patient data. See the caption to Fig. 4 for the meaning of each image.
4 Results
For both the phantom and patient datasets the algorithm was applied on all available view-angle orientations and the coronary veins were reconstructed from each view. Example results of the reconstructions are shown in Figs. 4 and 5. For the phantom dataset, the gold standard registration of the segmented LV mesh to the X-ray fluoroscopic image is illustrated in Fig. 4a, for the three view angles that included failed reconstructions. Figure 4b illustrates the 3D-reconstructed coronary veins. As part of the planning stage of the CRT procedure the LV surface is divided into 16 segments using the standard 16-segment American Heart Association (AHA) model of the LV for regional analysis. Since clinical decisions for the optimal pacing site are made per segment, Fig. 4c illustrates the 3D-reconstructed coronary veins overlaid on the 16-segment colour coded LV mesh. Figure 5 is a similar illustration, with manual registration of the LV mesh, for one of the patient datasets.
The success rates of the phantom and patient datasets were computed to be \(86\%\) and \(100\%\), respectively. The accuracy of the successfully reconstructed vessels is 1.41 mm (inter-quartile range 1.15–2.01 mm) for the phantom and 3.28 mm (2.2–3.72 mm) for patient data. As shown in Fig. 4, the accuracy of the algorithm is reduced for vessels at the edge of the LV projection, and this is where all of the failures occurred. This is because the distance between the two possible reconstructed points is considerably smaller at the edges, and consequently the angle constraint increases in importance resulting in the algorithm failing to choose the correct 3D point. The accuracy of the unsuccessfully reconstructed vessels varies between 3.5–8.19 mm. Even though these vessels are considered failures of the algorithm when compared to the gold standard vessels, they will usually reach the same segments when overlaid onto the 16-segment colour coded mesh given that the average segment width is around 5cm. As a result, this will not negatively affect the guidance during the CRT procedure, as the clinicians only need to know the segments through which the coronary veins pass.
5 Conclusions
This paper presents a novel and clinically useful algorithm for 3D-reconstruction of the coronary veins from a single contrast enhanced intra-procedural X-ray image. Unlike all previously developed techniques, this technique does not disrupt the CRT clinical workflow, it does not require any additional radiation dose to the patient and staff, and there is no requirement to phase match X-ray images or to find the correspondences between points along the vessels in different projections. This technique enables a superior single shot 3D visualisation of the coronary venous system in relation to the regions of the LV. This may enable placement of the LV lead in the optimal location and therefore improve response rates to CRT. It could also be applicable to other procedures, such as percutaneous coronary intervention for chronic total occlusions and radio frequency ablation procedures. A limitation of the method is that the 3D reconstruction may be inaccurate in cases where a vessel is found at the edges of the LV registered mesh. The accuracy is also very dependent on an accurate registration of the mesh to the X-ray. Future work will focus on automating the procedure by replacing the manual centreline annotation step with an automatic coronary vein detection using a deep learning technique, and by using an automatic registration method.
References
Blondel, C., Malandain, G., Vaillant, R., Ayache, N.: Reconstruction of coronary arteries from a single rotational x-ray projection sequence. IEEE Trans. Med. Imaging 25(5), 653–663 (2006)
Chen, S.Y.J., Carroll, J., Metz, C., Hoffmann, K.: Method and apparatus for three-dimensional reconstruction of coronary vessels from angiographic images (2000(b))
Chiribiri, A., Kelle, S., Götze, S., Kriatselis, C., Thouet, T., Tangcharoen, T., Paetsch, I., Schnackenburg, B., Fleck, E., Nagel, E.: Visualization of the cardiac venous system using cardiac magnetic resonance. American J. Cardiol. 101(3), 407–412 (2008)
Çimen, S., Gooya, A., Grass, M., Frangi, A.: Reconstruction of coronary arteries from x-ray angiography: a review. Med. Image Anal. 32, 46–68 (2016)
Duckett, S., Chiribiri, A., Ginks, M., Sinclair, S., Knowles, B., Botnar, R., Carr White, G., Rinaldi, C., Nagel, E., Razavi, R.: Cardiac MRI to investigate myocardial scar and coronary venous anatomy using a slow infusion of dimeglumine gadobenate in patients undergoing assessment for cardiac resynchronization therapy. J. Magn. Reson. Imaging 33(1), 87–95 (2011)
Guggenheim, N., Doriot, P., Dorsaz, P., Descouts, P., Rutishauser, W.: Spatial reconstruction of coronary arteries from angiographic images. Phys. Med. Biol. 36(1), 99 (1991)
Hartley, R., Zisserman, A.: Multiple View Geometry in Computer Vision, 2nd edn. Cambridge University Press, Cambridge (2004). ISBN: 0521540518
Jolly, M.-P., Guetter, C., Lu, X., Xue, H., Guehring, J.: Automatic segmentation of the myocardium in cine MR images using deformable registration. In: Camara, O., Konukoglu, E., Pop, M., Rhode, K., Sermesant, M., Young, A. (eds.) STACOM 2011. LNCS, vol. 7085, pp. 98–108. Springer, Heidelberg (2012). doi:10.1007/978-3-642-28326-0_10
Jongbloed, M.R., Lamb, H.J., Bax, J.J., Schuijf, J.D., de Roos, A., van der Wall, E.E., Schalij, M.J.: Noninvasive visualization of the cardiac venous system using multislice computed tomography. J. Am. Coll. Cardiol. 45(5), 749–753 (2005)
Messenger, J., Chen, S., Carroll, J., Burchenal, J., Kioussopoulos, K., Groves, B.: 3D coronary reconstruction from routine single-plane coronary angiograms: clinical validation and quantitative analysis of the right coronary artery in 100 patients. Int. J. Card. Imaging 16(6), 413–427 (2000)
Moss, A., Hall, W., Cannom, D., Klein, H., Brown, M., Daubert, J., Estes III, N.M., Foster, E., Greenberg, H., Higgins, S.: Cardiac-resynchronization therapy for the prevention of heart-failure events. N. Engl. J. Med. 361(14), 1329–1338 (2009)
Panayiotou, M., King, A., Housden, R., Ma, Y., Cooklin, M., O’Neill, M., Gill, J., Rinaldi, C., Rhode, K.: A statistical method for retrospective cardiac and respiratory motion gating of interventional cardiac x-ray images. Med. phys. 41(7), 071901 (2014)
Parker, D., Pope, D., Van Bree, R., Marshall, H.: Three-dimensional reconstruction of moving arterial beds from digital subtraction angiography. Comput. Biomed. Res. 20(2), 166–185 (1987)
Rivero-Ayerza, M., Jessurun, E., Ramcharitar, S., van Belle, Y., Serruys, P., Jordaens, L.: Magnetically guided left ventricular lead implantation based on a virtual three-dimensional reconstructed image of the coronary sinus. Europace 10(9), 1042–1047 (2008)
Ypenburg, C.E.: Noninvasive imaging in cardiac resynchronization therapy-part 1: selection of patients. Pacing Clin. Electrophysiol. 31(11), 1475–1499 (2008)
Acknowledgements and Disclaimer
We acknowledge financial support from the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London, King’s College Hospital NHS Foundation Trust and Innovate UK. This work was supported by the Engineering and Physical Sciences Research Council [grant number EP/L505328/1] and Innovate UK. Concepts and information presented are based on research and are not commercially available.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Panayiotou, M. et al. (2017). 3D Reconstruction of Coronary Veins from a Single X-Ray Fluoroscopic Image and Pre-operative MR. In: Mansi, T., McLeod, K., Pop, M., Rhode, K., Sermesant, M., Young, A. (eds) Statistical Atlases and Computational Models of the Heart. Imaging and Modelling Challenges. STACOM 2016. Lecture Notes in Computer Science(), vol 10124. Springer, Cham. https://doi.org/10.1007/978-3-319-52718-5_8
Download citation
DOI: https://doi.org/10.1007/978-3-319-52718-5_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-52717-8
Online ISBN: 978-3-319-52718-5
eBook Packages: Computer ScienceComputer Science (R0)