Abstract
In this work we explore a fully convolutional network (FCN) for the task of liver segmentation and liver metastases detection in computed tomography (CT) examinations. FCN has proven to be a very powerful tool for semantic segmentation. We explore the FCN performance on a relatively small dataset and compare it to patch based CNN and sparsity based classification schemes. Our data contains CT examinations from 20 patients with overall 68 lesions and 43 livers marked in one slice and 20 different patients with a full 3D liver segmentation. We ran 3-fold cross-validation and results indicate superiority of the FCN over all other methods tested. Using our fully automatic algorithm we achieved true positive rate of 0.86 and 0.6 false positive per case which are very promising and clinically relevant results.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Liver cancer is among the most frequent types of cancerous diseases, responsible for the deaths of 745,000 patients worldwide in 2012 alone [14]. The liver is one of the most common organs to develop metastases and CT is one of the most common modalities used for detection, diagnosis and follow-up of liver lesions. The images are acquired before and after intravenous injection of a contrast agent with optimal detection of lesions on the portal phase (60–80 s post injection) images. These procedures require information about size, shape and precise location of the lesions. Manual detection and segmentation is a time-consuming task requiring the radiologist to search through a 3D CT scan which may include multiple lesions. The difficulty of this task highlights the need for computerized analysis to assist clinicians in the detection and evaluation of the size of liver metastases in CT examinations. Automatic detection and segmentation is a very challenging task due to different contrast enhancement behavior of liver lesions and parenchyma. Moreover, the image contrast between these tissues can be low due to individual differences in perfusion and scan time. In addition, lesion shape, texture, and size vary considerably from patient to patient. This research problem has attracted a great deal of attention in recent years. The MICCAI 2008 Grand Challenge [3] provided a good overview of possible methods. The winner of the challenge [11] used the AdaBoost technique to separate liver lesions from normal liver based on several local image features. In more recent works we see a variety of additional methods trying to deal with detection and segmentation of liver lesions [1, 9].
In recent years, deep learning has become a dominant research topic in numerous fields. Specially, Convolutional Neural Networks (CNN) have been used for many challenges in computer vision. CNN obtained outstanding performance on different tasks, such as visual object recognition, image classification, handwritten character recognition and more. Deep CNNs introduced by LeCun et al. [5], is a supervised learning model formed by multi-layer neural networks. CNNs are fully data-driven and can retrieve hierarchical features automatically by building high-level features from low-level ones, thus obviating the need to manually customize hand-crafted features. CNN has been used for detection in several medical applications including pulmonary nodule [10], sclerotic metastases, lymph node and colonic polyp [8] and liver tumours [6]. In these works, the CNN was trained using patches taken out of the relevant region of interest (ROI).
In this paper we used a fully convolutional architecture [7] for liver segmentation and detection of liver metastases in CT examinations. The fully convolutional architecture has been recently used for medical purposes in multiple sclerosis lesion segmentation [2]. Fully convolutional networks (FCN) can take input of arbitrary size and produce correspondingly-sized output with efficient inference and learning. Unlike patch based methods, the loss function using this architecture is computed over the entire image segmentation result. Our network processes entire images instead of patches, which removes the need to select representative patches, eliminates redundant calculations where patches overlap, and therefore scales up more efficiently with image resolution. Moreover, there is a fusion of different scales by adding links that combine the final prediction layer with lower layers with finer strides. This fusion helps to combine across different lesion sizes. The output of this method is a lesion heatmap which is used for detection.
Since our dataset is small we use data augmentation by applying scale transformations to the available training images. The variations in slice thickness are large in our data (1.25 to 5 mm) and provides blurred appearance of the lesions for large slice thickness. The scale transformations allows the network to learn alter local texture properties.
We use a fully convolutional architecture for liver segmentation and detection of liver metastases in CT examinations using a small training dataset and compare it to the patch based CNN. To the best of our knowledge, this is the first work that uses fully convolutional neural network for liver segmentation and liver lesions detection.
2 Fully Convolutional Network Architecture
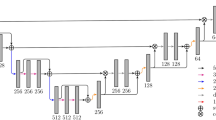
Our network architecture uses the VGG 16- layer net [12]. We decapitate the net by discarding the final classifier layer, and convert all fully connected layers to convolutions. We append a \(1\times 1\) convolution with channel dimension 2 to predict scores for lesion or liver at each of the coarse output locations, followed by a deconvolution layer to upsample the coarse outputs to pixel-dense outputs. The upsampling is performed in-network for end-to-end learning by backpropagation from the pixelwise loss. The FCN-8s DAG net was used as our initial network, which learned to combine coarse, high layer information with fine, low layer information as described in [7]. Our initial network architecture is presented in Fig. 1. We also explored the additional value of adding another lower level linking layer creating an FCN-4s DAG net. This was done by linking the Pool2 layer in a similar way to the linking of the Pool3 and Pool4 layers in Fig. 1.
2.1 3D Information
The input in our task are axial CT slices. In order to use the information from z-axis we modified the input image to have three CT slices, the relevant slice and two adjacent slices (above and below). Due to a very high slice spacing in some of our data we had to interpolate the adjacent slices, using linear interpolation, to be with a fixed spacing of 1 mm.
2.2 Training
Input images and their corresponding segmentation maps are used to train the network with the stochastic gradient descent implementation of MatConvNet [13] with GPU acceleration. Two networks were trained, one for the liver segmentation task and one for the lesion detection tasks. For the lesions detection training the areas surrounding the liver including the different organs and tissues were ignored. Note that one network trained on both lesions and liver was not used in our work since we had two different datasets for each task. The softmax log-loss function was calculated pixel-wise with different weights for each class pixels as in Eq. (1):
Where \(c \in {[1...D]}\) is the ground-truth class and x is the prediction scores matrix (before softmax) and w is the per-pixel weight matrix. As most of the pixels in each image belong to the liver, we balanced the learning process by using fixed weights that are inversely proportional to the population ratios. The learning rate was chosen to be 0.0005 for the first 20 epochs and 0.0001 for the last 30 epochs (total of 50 epochs). The weight decay was chosen to be 0.0005 and the momentum parameter was 0.9.
2.3 Data Augmentation
The lesion detection dataset was much smaller than the liver segmentation dataset since the manual segmentation masks were only in 2D for this dataset so data augmentation was appropriate. Data augmentation is essential to teach the network the desired invariance and robustness properties, when only few training samples are available. We generate different scales from 0.8 to 1.2 as lesion size can change significantly. The scales are sampled using uniform distribution and new images are re-sampled using nearest-neighbour approach. For each image in our dataset, four augmentations were created in different scales.
3 Experiments
3.1 Data
The data used in the current work includes two datasets. The lesion detection dataset includes CT scans from the Sheba medical center, taken during the period from 2009 to 2014. Different CT scanners were used with 0.71–1.17 mm pixel spacing and 1.25–5 mm slice thickness. Each CT image was resized to obtain a fixed pixel spacing of 0.71 mm. The scans were selected and marked by one radiologist. They include 20 patients with 1–3 CT examinations per patient and overall 68 lesion segmentation masks and 43 liver segmentation masks. The data includes various liver metastatic lesions derived from different primary cancers. The liver segmentation dataset includes 20 CT scans with entire liver segmentation masks taken from the SLIVER07 challenge [4] and was used only for training the liver segmentation network.
3.2 Liver Segmentation
We evaluated the liver segmentation network using the lesion detection dataset (43 slices of livers). Two framework variations were used: adding two neighbour slices (above and below), and linking the Pool2 layer for the final prediction (FCN-4s). We evaluated the algorithm’s segmentation performance using the Dice index, and calculated the Sensitivity and Positive predictive values (PPV). The results are shown in Table 1. The best results were obtained using the FCN-8s architecture with the addition of the adjacent slices. We obtained an average Dice index of 0.89, an average sensitivity of 0.86 and an average positive predictive value of 0.95. The fusion of an additional low level layer (FCN-4s) did not improve the results, probably since the liver boundary has a smooth shape and there is no need for a higher resolution. Adding the adjacent slices slightly improved the segmentation performance.
3.3 Detection-Comparative Evaluation
To evaluate the detection performance independent of the liver segmentation results we constrained the training and the testing sets to the liver area circumscribed manually by a radiologist. One of our goals was to examine the behaviour of the lesion detection network compared to the more classical patch based method. We designed a patch-based CNN similar to the one introduced by Li et al. [6]. The liver area is divided into patches of 17X17 pixels which are fed to the CNN for classification into lesion/normal area. We used a CNN model with seven hidden layers which included three convolutional layers, two max-pooling layers, fully connected layers, ReLU and a softmax classifier. We implemented the CNN model with MatConvNet framework [13] with GPU acceleration. We ran 50 epochs with a learning rate of 0.001. In each epoch a batch of 100 examples is processed simultaneously.
Another comparison made was to a recent work using sparsity based learned dictionaries which achieved strong results [1]. In this method each image was clustered into super-pixels and each super-pixel was represented by a feature vector. The super-pixels are classified using a sparsity based classification method that learns a reconstructive and discriminative dictionary.
As in the liver segmentation evaluation we tried the same two variations by adding two neighbour slices (above and below), and linking the Pool2 layer for the final prediction (FCN-4s). The detection performance was visually assessed considering the following two metrics: True positive rate (TPR)- the total number of detected lesions divided by the total number of known lesions; False positive per case (FPC)- the total number of false detections divided by the number of livers. Each lesion was represented by a 5 mm radius disk in its center of mass for the detection evaluation. The same was done for each connected component in our method results. We define a detected lesion when its center of mass overlap with the system lesion candidate center of mass. A 3-fold cross validation was used (each group containing different patients) using the lesion detection dataset. Results are presented in Table 2. To make this clinically interesting the highest TPR is presented with an FPC lower than 2.
The FCN-4s with the addition of neighbour slices performed better than the other methods with better TPR and better FPC. Figure 2 shows example results using the FCN. Seem that for lesions the FCN-4s is more appropriate than the FCN-8s because of the lesions size (smaller objects).
3.4 Fully Automatic Detection results
Finally, we tested the combination of the liver segmentation network (FCN-8s with neighbours) and the lesion detection network (FCN-4s with neighbours) to automatically extract the liver and detect the lesions in the liver segmentation result. We achieved a TPR of 0.86 and an FPC of 0.6 using the fully automatic algorithm. These results are close to the detection results achieved using the manual segmentation of the liver with a lower TPR but a better FPC. The liver automatic segmentation output usually did not include some of the boundary pixels which are darker and less similar to the liver parenchyma. These dark boundary pixels can be falsely detected as a lesion. This was the main reason for the improvement in FPC when using the automatic segmentation in comparison to the manual one.
3.5 Synthetic Data Experiments
Trying to understand the FCN discriminative features we created three synthetic images: (1) The lesion filled with a fixed value equal to its mean Hounsfield Units (HU) value; (2)The boundary of the lesion blurred by giving it a fixed value equal to the mean parenchyma HU; (3) The lesion mean HU value equal to the parenchyma mean HU. By using a fixed value to all of the pixels inside the lesion we eliminate the lesion’s texture. Figure 3 shows that most of the pixels inside the lesion were misclassified as normal liver tissue. The pixels around the lesion’s boundary can look different than normal parenchyma, even if they are not part of the lesion. They are sometimes marked by the experts as part of the lesion and this causes false positives around the boundary of the lesion. The blurred boundary of the lesion reduced the amount of false positives around the lesion’s boundary by making it similar to the liver parenchyma. By making the mean HU value of the lesion equal to that of the mean HU value of the parenchyma we eliminate gray levels difference. In that case the lesion was not detected at all. These results indicate that the network learned the texture of the lesion and mostly relies on the gray level difference between the lesion and the liver parenchyma.
4 Conclusions
To conclude, we showed automated fully convolutional network for liver segmentation and detection of liver metastases in CT examinations. Several approaches were tested including state of the art sparse dictionary classification techniques and patch based CNN. The results indicate that the FCN with data augmentation, addition of neighbour slices, and appropriate class weights provided the best results. Note that we have a small dataset and testing was conducted with 3-fold cross-validation. The detection results are promising. Note that no significant pre-processing or post-processing was implemented in the suggested method. Adding these steps may increase lesion detection accuracy and enable more accurate segmentation as well. Future work entails expanding to 3D analysis on larger datasets.
References
Ben-Cohen, A., Klang, E., Amitai, M., Greenspan, H.: Sparsity-based liver metastases detection using learned dictionaries. In: 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), pp. 1195–1198 (2016)
Brosch, T., Yoo, Y., Tang, L.Y.W., Li, D.K.B., Traboulsee, A., Tam, R.: Deep convolutional encoder networks for multiple sclerosis lesion segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 3–11. Springer, Heidelberg (2015)
Deng, X., Du, G.: Editorial: 3D segmentation in the clinic: a grand challenge II-liver tumor segmentation. In: MICCAI Workshop (2008)
Heimann, T., et al.: Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans. Med. Imaging 28(8), 1251–1265 (2009)
LeCun, Y., Bottou, L., Bengio, Y., Haffner, P.: Gradient-based learning applied to document recognition. Proc. IEEE 86, 2278–2324 (1998)
Li, W., Jia, F., Hu, Q.: Automatic segmentation of liver tumor in CT images with deep convolutional neural networks. J. Comput. Commun. 3(11), 146 (2015)
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431–3440 (2015)
Roth, H., Lu, L., Liu, J., Yao, J., Seff, A., Cherry, K., Kim, L., Summers, R.: Improving computer-aided detection using convolutional neural networks and random view aggregation. IEEE Trans. Med. Imaging, (2015, pre-print)
Rusko, L., Perenyi, A.: Automated liver lesion detection in CT images based on multi-level geometric features. Int. J. Comput. Assist. Radiol. Surg. 9(4), 577–593 (2014)
Setio, A.A., Ciompi, F., Litjens, G., Gerke, P., Jacobs, C., van Riel, S., Wille, M.W., Naqibullah, M., Sanchez, C., van Ginneken, B.: Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans. Med. Imaging, (2016, pre-print)
Shimizu, A., et al.: Ensemble segmentation using AdaBoost with application to liver lesion extraction from a CT volume. In: Proceedings of Medical Imaging Computing Computer Assisted Intervention Workshop on 3D Segmentation in the Clinic: A Grand Challenge II, New York (2008)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014)
Vedaldi, A., Lenc, K.: MatConvNet: convolutional neural networks for matlab. In: Proceedings of the 23rd Annual ACM Conference on Multimedia Conference, pp. 689–692 (2015)
The World Health Report, World Health Organization (2014)
Acknowledgment
Part of this work was funded by the INTEL Collaborative Research Institute for Computational Intelligence (ICRI-CI).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Ben-Cohen, A., Diamant, I., Klang, E., Amitai, M., Greenspan, H. (2016). Fully Convolutional Network for Liver Segmentation and Lesions Detection. In: Carneiro, G., et al. Deep Learning and Data Labeling for Medical Applications. DLMIA LABELS 2016 2016. Lecture Notes in Computer Science(), vol 10008. Springer, Cham. https://doi.org/10.1007/978-3-319-46976-8_9
Download citation
DOI: https://doi.org/10.1007/978-3-319-46976-8_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46975-1
Online ISBN: 978-3-319-46976-8
eBook Packages: Computer ScienceComputer Science (R0)