Abstract
Purpose
We address the automatic segmentation of healthy and cancerous liver tissues (parenchyma, active and necrotic parts of hepatocellular carcinoma (HCC) tumor) on multiphase CT images using a deep learning approach.
Methods
We devise a cascaded convolutional neural network based on the U-Net architecture. Two strategies for dealing with multiphase information are compared: Single-phase images are concatenated in a multi-dimensional features map on the input layer, or output maps are computed independently for each phase before being merged to produce the final segmentation. Each network of the cascade is specialized in the segmentation of a specific tissue. The performances of these networks taken separately and of the cascaded architecture are assessed on both single-phase and on multiphase images.
Results
In terms of Dice coefficients, the proposed method is on par with a state-of-the-art method designed for automatic MR image segmentation and outperforms previously used technique for interactive CT image segmentation. We validate the hypothesis that several cascaded specialized networks have a higher prediction accuracy than a single network addressing all tasks simultaneously. Although the portal venous phase alone seems to provide sufficient contrast for discriminating tumors from healthy parenchyma, the multiphase information brings significant improvement for the segmentation of cancerous tissues (active versus necrotic part).
Conclusion
The proposed cascaded multiphase architecture showed promising performances for the automatic segmentation of liver tissues, allowing to reliably estimate the necrosis rate, a valuable imaging biomarker of the clinical outcome.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Liver cancer is the sixth most common cancer worldwide, and the third most common cause of cancer death according to the Global Cancer Observatory [6]. Among the different types of primary liver cancers, the hepatocellular carcinoma is the most frequent and the most lethal one. Contrast-enhanced computed tomography (CECT) is one of the most informative image modalities for monitoring liver cancer [14, 25]. The main advantage brought by the CECT compared to non-enhanced CT (NECT) is its ability to investigate the dynamics of tissue vascularization and the vessel structure [23]. However, as a prerequisite to a quantitative analysis of such temporal sequences, an efficient deformable registration method is needed to compensate for respiratory motion between the different phases [14, 22].
In the clinical routine, the detection of lesions as well as the estimation of their size and number is still broadly performed using visual inspection of CT and MR images, which can be subjective, operator dependent and prone to diagnosis misinterpretations because of the high inter-tumor variability. The high intra-tumoral heterogeneity should also be considered when characterizing the cancer phenotype [19]. In radiomics studies, this can be achieved by differentiating active from necrotic tissue within the tumor [10, 25] using shape and texture information. This differentiation can also be useful to compute the necrosis rate, which is a valuable imaging biomarker for patients follow-up, since it has been shown to be correlated with survival after treatment [20].
All these observations underline the need for automatic and reliable tools dedicated to tumor segmentation and active and necrotic part discrimination in order to finely characterize liver cancer.
Related work
The segmentation of the liver and of its lesions remains a challenging task, mainly due to the high anatomical shape variability and to the fact that liver may be in contact with neighboring structures of similar intensities. In addition to that, diseases affecting the liver may have a large impact on its textural properties [17].
Intensity-based techniques were proposed either by learning the intensity distribution from training samples in order to pre-segment the region of interest [7], or simply by using thresholding techniques [13]. Machine learning methods were also proposed in combination with the aforementioned techniques to segment the liver [26] and its lesions [8]. Statistical shape models were considered in order to capture the shape of the liver, while coupling them with deformable model [5] or graph cuts [24]. Methods based on probabilistic atlases are an alternative to incorporate a priori knowledge about location and shape of the liver, as well as information about surrounding anatomical structures [15].
More recently, deep learning approaches have led to major breakthroughs in medical image analysis. They have been recognized for their ability to encode complex morphological and texture information in images by directly using raw pixel intensities as inputs, whereas traditional machine learning methods required carefully handcrafted features to achieve descent results [16, 28]. Deep learning networks achieved state-of-the-art results in many medical-related applications, including detection, classification, segmentation, localization and registration [11, 16]. These performances have been reached, even with a small number of training cases, thanks to smart architecture choices, data augmentation or transfer learning [31]. Fully convolutional networks (FCNs) are now widely used for medical applications [9], in particular, for liver and lesions segmentation [2, 3, 27, 30]. They significantly outperform the standard methods mentioned above.
Segmentation using FCNs was popularized by the U-Net network initially introduced to segment cells on histopathological slices [21] and afterward applied to liver and lesion segmentation [2]. More recently, a cascaded version of the U-Net network was used for both liver and lesion segmentation [3]. Only a few studies exploited the multiphase information using FCNs [27], and to our knowledge, only one contribution tackled the delineation of both active and necrotic parts within the lesions, but using MR images [30].
Contributions
In this work, we applied deep learning-based semantic segmentation to delineate the liver and its lesions in contrast-enhanced CT sequences, while discriminating the necrotic from the active parts. This task was previously addressed in the context of MR images [30]. We evaluate the impact of using either single-phase or multiphase CT images on the performance of different FCNs architectures. We showed that portal-phase images alone provide enough contrast for discriminating lesions from healthy parenchyma, while multiphase information brings significant improvement for the segmentation of active and necrotic tumor parts. Finally, our results support the hypothesis that combining several specialized networks in cascade leads to better performance than a single versatile network addressing all tasks simultaneously [3].
Materials and methods
Database
Due to the lack of publicly available datasets providing segmentation of active and necrotic tissues within hepatic lesions, we used an in-house database (Db) containing ground truth 2D annotations for three classes, namely the healthy parenchyma, the active and the necrotic parts of the lesions. A subset of Db was already used in a previous work on the semiautomatic segmentation of liver tissue [4].
The database is composed of data from seven patients suffering from hepatocellular carcinoma (HCC) that underwent from one to three contrast-enhanced computed tomography (CECT) examinations (in plane resolution ranging from 0.66 to 0.97 mm, slice thickness ranging from 0.7 to 1.25 mm), resulting in a total number of 13 CT sequences. Each examination is composed of one image before contrast agent injection (NECT), and two contrast-enhanced images reflecting the arterial (AR) phase (\(\approx 25\)–30 s after injection) and the portal venous (PV) phase (\(\approx 60\)–70 s after injection). For each CT sequence, four experts have manually segmented the three classes on eight slices regularly sampled across the tumor, resulting in a total number of 104 labeled slices. Expert annotations were fused using the STAPLE algorithm to reach a consensus [29]. The manual segmentation task was performed on the PV images, which exhibit the best contrast between the different tissues.
Data pre-processing
As a prerequisite for the use of multiphase information [14], respiratory motion between the different phase images was corrected using a diffeomorphic deformable registration algorithm [1]. The PV image was considered as the reference image in the registration process, following [4].
Images from Db are of heterogeneous sizes and voxel resolutions. Since FCN requires a standardized input size, a scaling transformation was applied to all images to enforce isotropic resolution (\( 0.97 \,\mathrm{mm}^2 \)) and a fixed \(512 \times 512\) size.
An intensity normalization procedure similar to [3] was then applied. We only retained Hounsfield unit (HU) intensities within the range \([-100\, \text {HU}, 400\,\text {HU}]\), corresponding to typical radiodensity values of the liver, to reduce the influence of neighboring organs. Contrary to [3], we did not implement slice-wise histogram equalization since it may be biased by the different proportions of liver present in each slice. Instead, we considered a linear mapping of the retained HU values to the [0, 1] interval.
Cascaded networks
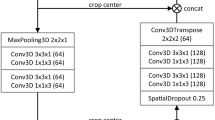
As in [3], we decompose the segmentation task in a hierarchical manner by cascading several networks relying on the U-Net architecture, each of them being specialized in the segmentation of a specific tissue. In the sequel, a given single-phase elementary network will be referred to by the input phase and the target class. For example, PV-Lesion (see Fig. 1) corresponds to a network dedicated to segment lesions using PV images as input, while AR-Necrosis refers to the network dedicated to the segmentation of the active vs necrotic parts of the lesions using AR images as an input.
Two strategies were investigated to exploit multiphase information. The first one, referred as DMP (Dimensional MultiPhase) strategy, consists in concatenating single-phase images in a multi-dimensional feature map used as the input to the network (see Fig. 2). The second one, referred as MPF (MultiPhase Fusion) strategy, consists in processing each phase independently and then merges the output maps (by simple addition) to obtain the final segmentation (see Fig. 3). Only the AR and PV phases were considered in the DMP and MPF networks because NECT does not provide enough inter-tissue contrast.
The cascaded architecture is depicted in Fig. 4 and can be composed of either single-phase or multiphase U-Net sub-networks. Unlike the original cascaded architecture [3], which cropped the original image to the predicted liver area after the first step, we decided to keep the same image size all throughout the networks. This choice was motivated by the fact that cropping the image would require to resample it in order to fit the fixed input size of the network, which would alter its isotropic resolution.
Cascaded network: The first network takes as input a CT image and segments the liver. The resulting segmentation map is used to remove non-liver pixels in the input data of the second network which performs the segmentation of lesions. The last network segments the necrosis within the lesions. The three binary masks are combined in the final segmentation map
To validate the hypothesis suggesting that cascaded specialized networks perform better than a versatile network addressing all tasks simultaneously [3], we consider the original U-Net architecture (denoted {\( \cdot \)}-Full) to simultaneously predict the three classes of liver tissues (see Fig. 5). This network is evaluated for both single-phase and multiphase conditions. To ensure that there was no bias in favor of the cascaded networks in the comparison, the cascaded networks were imposed to have a smaller number of parameters than the versatile one (which had a total of 32 M parameters). 8 M parameters were specified for all cascaded networks, apart from MPF-{\( \cdot \)}, with 16 M parameters (fusion of two networks with 8M parameters each).
Training
FCNs have proven their ability to operate well even when trained on small databases. A data augmentation step is generally required to avoid overfitting and to generalize better on unseen cases. In our case, for each input image, ten new images were randomly generated using the following settings:
-
Rotation (\( 90{^{\circ }} , 180{^{\circ }}, 270{^{\circ }} \)).
-
Horizontal/vertical flip.
-
Horizontal/vertical shift in range [0, 0.1] of the image size.
-
Addition of Gaussian noise (\(\mu = 0, \,\sigma = 3\) HU).
We chose to consider a moderate noise variance since the inter-class difference is generally smaller than 20 HU.
A leave-one-patient-out cross-validation process was used during the training to ensure that no slices from the same patient were present in both training and test sets. The networks were implemented using Keras with a TensorFlow backend and trained on a single NVIDIA GTX 1070 with 8 GB VRAM. We used the Adam optimizer [12] with a learning rate of \( 10^{-4} \) and a decay of \( 10^{-5} \). No additional dropout layers were used since it appeared to decrease the performance, probably due to the small number of images available. The softmax function is considered for the final layer of the different networks. To cope with the imbalanced classes problem, we considered the weighted cross-entropy as the cost function to optimize the weights of the networks, as suggested in [18]. The segmentation quality was evaluated with the Dice similarity coefficient (DSC), which is commonly used for the assessment of semantic segmentation [2,3,4, 30].
Experiments and results
Mean DSCs were computed over all slices for each target class. The different methods were statistically compared using the Wilcoxon signed paired rank tests, since slice-wise DSCs did not follow a normal distribution. We first compared {NECT, AR, PV, DMP, MPF}—Liver networks to evaluate which phase allows better liver segmentation. We then trained {NECT, AR, PV, DMP, MCF}—Lesion and {NECT, AR, PV, DMP, MCF}—Necrosis networks separately on Db by masking all values outside the liver using ground truth annotations in order to assess whether multiphase information is really useful for the segmentation. The results are given in Table 1.
Multiphase performed significantly better than single phase for segmenting the liver (DMP versus PV, \(P=0.001\); DMP versus AR, \(P=0.005\), DMP versus NECT, \(P<0.001\)) and the active part of the lesions (DMP vs PV, \(P<0.001\); DMP versus AR, \(P=0.003\); DMP versus NECT, \(P<0.001\)). When comparing single phase alone, PV achieved significantly better DSCs than AR or NECT for all the segmentation tasks except for the liver segmentation. When comparing multiphase methods, DMP carries out significantly better than MPF for the segmentation of the liver (DMP versus MPF, \(P=0.004\)), the parenchyma (DMP versus MPF, \(P<0.001\)) and the active part of the lesions (DMP versus MPF, \(P=0.005\)).
Since both DMP-Lesion and DMP-Necrosis led to the best results, we combined them in a cascade as explained before, and compared it to both {NECT, AR, PV, DMP, MPF}—Full networks and evaluate them in terms of liver tissue classification performance. (Experiments were conducted on images that were masked with the ground truth liver segmentation.) The mean DSCs are reported in Table 2. Examples of segmentation results are shown in Fig. 6.
The results highlight that the cascaded version performed significantly better than {\(\cdot \)}-Full networks for segmenting the active part of the lesion (Cascaded DMP vs PV-Full, \(P = 0.001\)). From the resulting segmentation maps, we were also able to estimate the necrosis rate, which is commonly used for diagnosis and prognosis of the treatment outcome. Our workflow provided estimates of this valuable biomarker with a mean error rate of 13.0%, which is accurate enough for clinical application.
With our setup, we also achieved slightly better results than those obtained in [30] for the same task, on a different database of MR images. We evaluated our method on the same database used in [4], where a manual expert interaction was required for the segmentation phase, which is not the case in the present deep learning approach. To allow fair comparison, the evaluation was conducted on the areas where all the experts reached an agreement as in [4]. The mean patient-wise segmentation DSCs are depicted in Table 3. Our method enabled a better segmentation of the lesions and both necrotic and active parts. Therefore, we were able to predict the patient-wise necrosis rate with a slightly better precision.
We finally used the complete cascaded network, as depicted in Fig. 4, which combines DMP-Liver, DMP-Lesion and DMP-Necrosis to perform a fully automatic segmentation from the raw (unmasked) CT image. We reached average slice-wise DSCs of \(78.3 \pm 22.1\) for the segmentation of the parenchyma, \(50.6 \pm 24.6\) for the segmentation of the active part and \(68.1 \pm 23.2\) for the necrotic part of the lesions. As a by-product, we also provided a necrosis rate per patient with a mean error of 15.9%. Examples of fully automatic segmentation results are given in Fig. 7.
Conclusion
Following the intuition developed in [3], we validated the hypothesis that FCNs are able to learn the hierarchical structures present in a CT image, even when trained on small databases. It has been shown that several cascaded specialized FCNs yield more accurate results than a single network addressing all tasks simultaneously. For a fair comparison, this has been demonstrated on networks with a comparable number of parameters.
The segmentation accuracy obtained by our method is on par with the one reported in [30], which addresses the same task using a different database of MR images. The accuracy of the FCN-based automatic segmentation method outperforms the results reported in [4] on the same database, using an ensemble classifier and requiring manual expert interaction during the segmentation. As a by-product, we have also shown that the segmentation maps may be used to provide usable estimates of the necrosis rate, which is an important predictor of treatment outcome.
The use of multiphase information has been proven to be beneficial when segmenting active vs necrotic tumor parts. However, this requires an accurate registration of the different phases, which remains a challenging step. As an alternative, the single PV phase can be used, with reasonable results for the lesion segmentation. A previous study has already reported that multiphase information is helpful for lesion segmentation, but that good outcomes may be reached using AR images only [27]. This difference in the most appropriate phase for segmentation may be explained by the fact that our training database was only composed of HCC images, whereas [27] was confronted with different tumor types that respond differently to contrast agents. More interestingly, multiphase information also significantly helped the segmentation of the liver. This may be explained by the high inter-class contrast present between lesion and parenchyma at PV phase, leading PV-Liver to sometimes not consider lesions as part of the liver.
In this work, we have based our 3D segmentation on the segmentations of 2D slices. This choice was motivated by the will to limit the computational cost, but also by the difficulty to cope with the high variability of inter-voxels spacing in the z-direction. In addition, our training database only contained sparse annotations on 2D slices, which prevented a full 3D training. The use of full 3D information during the training, segmentation or in a post-processing phase is a perspective of this work. The benefit of using 3D-CRF in a post-processing step has, for instance, been shown for both liver and lesion segmentation in [3].
To our knowledge, we are the first to provide a method that fully automatically segments liver tissues on CT images. We conclude that automatic segmentation of liver tissue remains a challenging task, but our results prove that it can be tackled using fully convolutional networks. In addition, the resulting lesion segmentations can provide useful clinical information for patients follow-up, such as the necrosis rate.
References
Avants BB, Epstein CL, Grossman M, Gee JC (2008) Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal 12(1):26–41
Ben-Cohen A, Diamant I, Klang E, Amitai M, Greenspan H (2016) Fully convolutional network for liver segmentation and lesions detection. In: Carneiro G et al (eds) Deep learning and data labeling for medical applications, vol 10008. Springer, Cham, pp 77–85. https://doi.org/10.1007/978-3-319-46976-8
Christ PF, Ettlinger F, Grün F, Elshaera MEA, Lipkova J, Schlecht S, Ahmaddy F, Tatavarty S, Bickel M, Bilic P, Rempfler M, Hofmann F, Anastasi MD, Ahmadi SA, Kaissis G, Holch J, Sommer W, Braren R, Heinemann V, Menze B (2017) Automatic liver and tumor segmentation of CT and MRI volumes using cascaded fully convolutional neural networks, pp 1–20. arXiv:1702.05970
Conze PH, Noblet V, Rousseau F, Heitz F, de Blasi V, Memeo R, Pessaux P (2017) Scale-adaptive supervoxel-based random forests for liver tumor segmentation in dynamic contrast-enhanced CT scans. Int J Comput Assist Radiol Surg 12(2):223–233. https://doi.org/10.1007/s11548-016-1493-1
Erdt M, Steger S, Kirschner M, Wesarg S (2010) Fast automatic liver segmentation combining learned shape priors with observed shape deviation. In: Proceedings—IEEE symposium on computer-based medical systems, pp 249–254. https://doi.org/10.1109/CBMS.2010.6042650
Bray F, Ferlay Jacques, Soerjomataram Isabelle, Siegel RL, Torre LA, Jemal A (2018) Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer Journal for Clinicians. https://doi.org/10.3322/caac.21492
Foruzan AH, Aghaeizadeh Zoroofi R, Hori M, Sato Y (2009) Liver segmentation by intensity analysis and anatomical information in multi-slice CT images. Int J Comput Assist Radiol Surg 4(3):287–297. https://doi.org/10.1007/s11548-009-0293-2
Freiman M, Cooper O, Lischinski D, Joskowicz L (2011) Liver tumors segmentation from CTA images using voxels classification and affinity constraint propagation. Int J Comput Assist Radiol Surg 6(2):247–255. https://doi.org/10.1007/s11548-010-0497-5
Hu Z, Tang J, Wang Z, Zhang K, Zhang L, Sun Q (2018) Deep learning for image-based cancer detection and diagnosis : a survey. Pattern Recognit 83:134–149. https://doi.org/10.1016/j.patcog.2018.05.014
Jeong WK, Jamshidi N, Felker ER, Raman SS, Lu DS (2018) Radiomics and radiogenomics of primary liver cancers. Clin Mol Hepatol 25:1–9. https://doi.org/10.3350/cmh.2018.1007
Ker J, Wang L, Rao J, Lim T (2017) Deep learning applications in medical image analysis. IEEE Access 6:9375–9379. https://doi.org/10.1109/ACCESS.2017.2788044
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. In: AIP conference proceedings 1631:58–62. https://doi.org/10.1063/1.4902458. arXiv:1412.6980
Kumar SS, Moni RS, Rajeesh J (2013) Automatic liver and lesion segmentation: a primary step in diagnosis of liver diseases. Signal Image Video Process 7(1):163–172. https://doi.org/10.1007/s11760-011-0223-y
Lee J, Kim KW, Kim SY, Shin J, Park KJ, Won HJ, Shin YM (2015) Automatic detection method of hepatocellular carcinomas using the non-rigid registration method of multi-phase liver CT images. J X-ray Sci Technol 23(3):275–288. https://doi.org/10.3233/XST-150487
Li CY, Wang X, Eberl S, Fulham M, Yin Y, Feng D (2010) Fully automated liver segmentation for low- and high-contrast ct volumes based on probabilistic atlases. In: Proceedings—international conference on image processing ICIP, pp 1733–1736. https://doi.org/10.1109/ICIP.2010.5654434
Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JA, van Ginneken B, Sánchez CI (2017) A survey on deep learning in medical image analysis. Med Image Anal 42(1995):60–88. https://doi.org/10.1016/j.media.2017.07.005
Moghbel M, Mashohor S, Mahmud R, Saripan MIB (2018) Review of liver segmentation and computer assisted detection/diagnosis methods in computed tomography. Artif Intell Rev 50(4):497–537. https://doi.org/10.1007/s10462-017-9550-x
Novikov AA, Lenis D, Major D, Hladuvka J, Wimmer M, Buhler K (2018) Fully convolutional architectures for multi-class segmentation in chest radiographs. IEEE Trans Med Imaging. https://doi.org/10.1109/TMI.2018.2806086
O’Connor JPB, Rose CJ, Waterton JC, Carano RAD, Parker GJM, Jackson A (2015) Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clin Cancer Res 21(2):249–257. https://doi.org/10.1158/1078-0432.CCR-14-0990
Oldhafer KJ, Chavan A, Frühauf NR, Flemming P, Schlitt HJ, Kubicka S, Nashan B, Weimann A, Raab R, Manns MP, Galanski M (1998) Arterial chemoembolization before liver transplantation in patients with hepatocellular carcinoma: Marked tumor necrosis, but no survival benefit? J Hepatol 29(6):953–959. https://doi.org/10.1016/S0168-8278(98)80123-2
Ronneberger O, Fischer P, Brox T (2015) U-Net: convolutional networks for biomedical image segmentation, pp 1–8. arXiv:1505.04597
Ruskó L, Bekes G, Fidrich M (2009) Automatic segmentation of the liver from multi- and single-phase contrast-enhanced CT images. Med Image Anal 13(6):871–882. https://doi.org/10.1016/j.media.2009.07.009
Sadigh G, Applegate KE, Baumgarten DA (2014) Comparative accuracy of intravenous contrast-enhanced CT versus noncontrast CT plus intravenous contrast-enhanced CT in the detection and characterization of patients with hypervascular liver metastases. Critic Apprais Topic Acad Radiol 21(1):113–125. https://doi.org/10.1016/j.acra.2013.08.023
Saito A, Yamamoto S, Nawano S, Shimizu A (2017) Automated liver segmentation from a postmortem CT scan based on a statistical shape model. Int J Comput Assist Radiol Surg 12(2):205–221. https://doi.org/10.1007/s11548-016-1481-5
Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, Chan BK, Matcuk GR, Barry CT, Chang HY, Kuo MD (2007) Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol 25(6):675–680. https://doi.org/10.1038/nbt1306
Shi C, Cheng Y, Liu F, Wang Y, Bai J, Tamura S (2016) A hierarchical local region-based sparse shape composition for liver segmentation in CT scans. Pattern Recognit 50:88–106. https://doi.org/10.1016/j.patcog.2015.09.001
Sun C, Guo S, Zhang H, Li J, Chen M, Ma S, Jin L, Liu X, Li X, Qian X (2017) Automatic segmentation of liver tumors from multiphase contrast-enhanced CT images based on FCNs. Artif Intell Med 83:58–66. https://doi.org/10.1016/j.artmed.2017.03.008
Suzuki K (2017) Overview of deep learning in medical imaging. Radiol Phys Technol 10(3):257–273. https://doi.org/10.1007/s12194-017-0406-5
Warfield SK, Zou KH, Wells WM (2004) Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging 23(7):903–921. https://doi.org/10.1109/TMI.2004.828354
Zhang F, Yang J, Nezami N, Laage-gaupp F, Chapiro J, De Lin M, Duncan J (2018) Liver tissue classification using an auto-context-based deep neural network with a multi-phase training framework. In: Bai W, Sanroma G, Wu G, Munsell BC, Zhan Y, Coupé P (eds) Patch-based techniques in medical imaging. Springer, Cham, pp 59–66
Zheng W, Thorne N, Mckew JC (2015) Deep learning in medical image analysis. Annu Rev Biomed Eng 18:1067–1073. https://doi.org/10.1016/j.drudis.2013.07.001.Phenotypic
Funding
This study was funded by IHU Strasbourg through ANR grant 10-IAHU-0002.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standard
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ouhmich, F., Agnus, V., Noblet, V. et al. Liver tissue segmentation in multiphase CT scans using cascaded convolutional neural networks. Int J CARS 14, 1275–1284 (2019). https://doi.org/10.1007/s11548-019-01989-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-019-01989-z