Abstract
Recently, multi-task feature selection methods have been applied to jointly identify the disease-related brain regions for fusing information from multiple modalities of neuroimaging data. However, most of those approaches ignore the complementary label information across modalities. To address this issue, in this paper, we present a novel label-alignment-based multi-task feature selection method to jointly select the most discriminative features from multi-modality data. Specifically, the feature selection procedure of each modality is treated as a task and a group sparsity regularizer (i.e., \(\ell _{2,1}\) norm) is adopted to ensure that only a small number of features to be selected jointly. In addition, we introduce a new regularization term to preserve label relatedness. The function of the proposed regularization term is to align paired within-class subjects from multiple modalities, i.e., to minimize their distance in corresponding low-dimensional feature space. The experimental results on the magnetic resonance imaging (MRI) and fluorodeoxyglucose positron emission tomography (FDG-PET) data of Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset demonstrate that our proposed method can achieve better performances over state-of-the-art methods on multimodal classification of Alzheimer’s disease (AD) and mild cognitive impairment (MCI).
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Alzheimer’s disease (AD) is the most common form of dementia in people over 65 years of age. It is reported that there are 26.6 million AD sufferers worldwide, and 1 in 85 people will be affected by 2050 [1]. Thus, effective and accurate diagnosis of AD and its prodromal stage (i.e., mild cognitive impairment, MCI), is very important for possible delay and early treatment of the brain disease. Lots of efforts have been made for possible identification of such changes at the early stage by leveraging neuroimaging data [3, 13]. At present, several modalities of biomarkers have been proved to be sensitive to AD and MCI, such as the brain atrophy measured in magnetic resonance imaging (MRI) [7] and the cerebral metabolic rates of glucose measured in fluorodeoxyglucose positron emission tomography (FDG-PET) [8].
As multiple features are extracted from different imaging modalities, there may exist some irrelevant or redundant features. So, feature selection, which can be considered as the biomarker identification for AD and MCI, is commonly used to remove these redundant and irrelevant features. Some feature selection methods based on multi-modality data have been proposed for jointly selecting the most discriminative features relevant to disease. For example, Zhang et al. [12] proposed a multi-modal multi-task learning for joint feature selection for AD classification and regression. Liu et al. [5] proposed inter-modality relationship constrained multi-task feature selection for AD/MCI classification. Jie et al. [4] presented a manifold regularized multi-task feature selection method for classification of AD, and achieved the state-of-the-art performance on Alzheimer’s Disease Neuroimaging Initiative (ADNI) database. However, those methods ignore the label information of data from multiple modalities, i.e., the subjects from the same class across multiple modalities should be closer in the low-dimensional feature space.
In this paper, to address this issue, we propose a novel label-alignment-based multi-task feature selection method that considers the intrinsic label relatedness among multi-modality data and preserves the complementary information conveyed by different modalities. We formulate the classification of multi-modality data as a multi-task learning (MTL) problem, where each task focuses on the classification of each modality. Specifically, two regularization items are included in the proposed model. The first item is a group Lasso regularizer [11], which ensures only a small number of features to be jointly selected across different tasks. The second item is a label-alignment regularization term, which can minimize the distance of within-class subjects from multiple modalities after projection to low-dimensional feature space leading to the selection of more discriminative features. Then, we use a multi-kernel support vector machine to fuse the above-selected features from each individual modality. The proposed method has been evaluated on ADNI dataset and obtained promising results.
The rest of this paper is organized as follows. In Sect. 2, we present the proposed label-alignment-based multi-task feature selection method in detail. Experimental results on ADNI dataset using MRI and FDG-PET biomarkers are given in Sect. 3. Finally, Sect. 4 concludes this paper and indicates points for future work.
2 Methods
2.1 Label-Alignment-Based Multi-Task Feature Selection
In this paper, we treat feature selection as a multi-task regression problem that incorporates the relationship between different modalities. Suppose we have M supervised learning tasks (i.e., the number of modalities). Denote \(\varvec{X}^m=[\varvec{x}^m_1,\varvec{x}^m_2,\dots ,\varvec{x}^m_N]^T\in \mathbb {R}^{N \times d}\) as a \(N \times d\) matrix that represents d features of N training samples on the m-th task (i.e., m-th modality), and \(\varvec{Y}=[y_1, y_2, \dots , y_N]^T \in \mathbb {R}^N\) as the response vector from these training subjects, where \(\varvec{x}_i^m\) represents feature vector of the i-th subjects of the m-th modality, and \(y_i\) is the corresponding class label (i.e., patient or normal control). Suppose \(\varvec{w}^m \in \mathbb {R}^d\) is the regression coefficient vector of the m-th task. Then the multi-task feature selection (MTFS) model is to solve the following objective function:
where \(\varvec{W}=[\varvec{w}^1,\varvec{w}^2,\dots ,\varvec{w}^M] \in \mathbb {R}^{d\times M}\) is the weight matrix whose row \(\varvec{w}_j\) is the vector of coefficients associated with the j-th feature across different tasks. \(||\varvec{W} ||_{2,1}\) is the \(\ell _{2, 1}\)-norm of matrix \(\varvec{W}\) defined as \(||\varvec{W} ||_{2,1}=\sum _{j=1}^d||\varvec{w}_j ||_2\) which is the sum of the \(\ell _2\)-norms of the rows of matrix \(\varvec{W}\) [11]. The first term of Eq. (1) measures the empirical error on the training data while the \(\ell _{2,1}\)-norm encourages matrix with many zero rows. So the \(\ell _{2,1}\)-norm combines multiple tasks and ensures that a small number of common features will be selected across different tasks. \(\lambda _1\) is a regularization parameter which balances the relative contributions of the two terms.
The MTFS model using a linear mapping function transforms the data from the original high-dimensional space to one-dimensional space. The limitation of the model is that only the relationship between data and class label for each task is considered, while the mutual dependence among data and the complementary information conveyed by different modalities are ignored, which may result in large deviations even for very similar data after mapping. To address this problem, we introduce a new regularization term called label-alignment regularization term which minimizes the distance between feature vectors of multiple modalities of the within-class subjects after feature projection:
where \(\varvec{x}_i^p\) and \(\varvec{x}_j^q\) are the feature vectors of the i-th and the j-th subjects in the p-th and q-th modalities respectively. \(S_{ij}\) denotes the element of the similarity matrix \(\varvec{S}\) across different subjects. Here, the similarity matrix can be defined as:
The regularization term Eq. (2) can be explained as follows. \(||(\varvec{w}^p)^T\varvec{x}_i^p - (\varvec{w}^q)^T\varvec{x}_j^q ||_2^2 S_{ij}\) measures the distance between \(\varvec{x}_i^p\) and \(\varvec{x}_j^q\) in the projected space. It implies that, if \(\varvec{x}_i^p\) and \(\varvec{x}_j^q\) are from the same class, the distance between them should be as small as possible. Otherwise, the distance between them should be as large as possible. When \(p = q\), the local geometric structrue of the same modality data are preserved during the mapping and when \(p < q\), the complementary information provided from different modalities are preserved after projection of feature vectors onto the one-dimensional feature space. By incorporating the regularizer Eq. (2) into Eq. (1), we can obtain the objective function of our label-alignment-based multi-task feature selection model:
where \(\lambda _1\) and \(\lambda _2\) are the two positive constants that control the sparseness and the degree of preserving the distance between subjects, respectively. To optimize the problem in Eq. (4), we use Accelerated Proximal Gradient (APG) method [6] and only those features with non-zero regression coefficients are used for final classification.
2.2 Multi-modality Data Fusion and Classification
In this paper, we adopt a multi-kernel based support vector machine (SVM) method to integrate features from different modalities for classification [13]. Specifically, we calculate the linear kernels based on the features selected by the above-proposed feature selection method by using multi-modal biomarkers. Then, a combined kernel matrix is constructed by linearly combining kernels from different modalities and used in multi-kernel based SVM. The optimal parameters used for combining different kernels are determined by using a coarse-grid search through cross-validation on the training samples.
We conduct standard 10-fold cross-validation to evaluate classification performance. For each of the 10 trials, within the training data, an internal 10-fold cross-validation is performed to fine tune the parameters, i.e., the regularization parameters \(\lambda _1\), \(\lambda _2\) and the kernel combination parameter. The model that reaches the best performance during the inner cross-validation stage is considered as the optimal model and is adopted to classify unseen testing samples. This process is repeated 10 times independently to avoid any bias introduced by randomly partitioning dataset in the 10-fold cross-validation and the average results are reported.
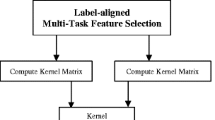
Figure 1 gives a schematic illustration of our multimodal data fusion and classification pipeline, where two modalities of data (e.g., MRI and FDG-PET) are used for jointly selection features corresponding to different tasks.
3 Experiments
In this section, we perform a series of experiments on the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu) to evaluate the effectiveness of the proposed method.
3.1 Subjects and Settings
We use a total of 202 subjects with corresponding baseline MRI and FDG-PET data from ADNI dataset: 51 AD patients, 99 MCI patients (including 43 MCI converters who had converted to AD within 18 months and 56 MCI non-converters), and 52 normal controls (NC). Standard image pre-processing is carried out for all MRI and FDG-PET images, including spatial distortion, skull-stripping, removal of cerebellum. Then for structural MR images, we partition each subject image into 93 manually labeled regions-of-interest (ROIs) [9] with atlas wraping. The gray matter tissue volume of these 93 ROIs is used as features extracted by the FSL package [14]. FDG-PET image of each subject is aligned onto its corresponding MR image using a grid transformation and the average intensity of each ROI in the FDG-PET image is calculated as features. Therefore, we can finally acquire 93 features from MRI image and other 93 features from PET image.
In our experiments, we compare our proposed method with multi-modality multi-kernel method [13] without feature selection (denoted as Baseline), single-modality feature selection with Lasso integrated with multi-modality multi-kernel SVM (denoted as SMFS) and multi-task feature selection method [12] (denoted as MTFS). In addition, we also concatenate all features from MRI and FDG-PET into one feature vector and perform Lasso-based feature selection and then use the standard SVM with linear kernel for classification (denoted as CONFS). For each comparison, different methods are evaluated on multiple binary classification tasks, i.e., AD vs. NC, MCI vs. NC and MCI converters (MCI-C) vs. MCI non-converters (MCI-NC), respectively. To evaluate the performances of different methods, we use four performance measures, including classification accuracy, area under receiver operating characteristic (ROC) curve (AUC), sensitivity (i.e., the proportion of patients that are correctly predicted), and specificity (i.e., the proportion of normal controls that are correctly predicted).
3.2 Results
Table 1 shows the experimental results achieved by five different methods. As can be seen from Table 1, the proposed feature selection method is always superior to other methods on three classification tasks. Specifically, our method obtains the classification accuracy of 95.51%, 82.15% and 70.50% for AD vs. NC, MCI vs. NC and MCI-C vs. MCI-NC, respectively. On the other hand, the best classification accuracy of other methods are 92.25 %, 74.34 % and 61.67 % on three tasks, respectively. Besides, we perform the standard paired t-test on the accuracies of our proposed and those of compared methods. It is shown that our proposed method is significantly better than the comparison methods with p values smaller than 0.05. In addition, Fig. 2 further plots the corresponding ROC curves of different methods for three classification tasks. These results demonstrate that considering the complementary label information of multi-modality data can significantly improve the classification performance, with comparison to traditional methods.
Furthermore, in Table 2, we compare our proposed method with several recent start-of-the-art methods for multimodal AD/MCI classification. Gray et al. got the classification accuracy of 89.0 %, 74.6 % and 58.0 % for AD vs. NC, MCI vs. NC, and MCI-C vs. MCI-NC, respectively with four different modalities (MRI + PET + CSF + genetic) [2]. When using two modalities of features (MRI + PET), Jie et al. [4] achieved the accuracy of 95.0 %, 79.3 % and 68.9 % for classification of AD vs. NC, MCI vs. NC and MCI-C vs. MCI-NC, respectively, which are inferior to our method. These results further validate the efficacy of our proposed method for multimodal AD/MCI classification.
4 Discussion
This paper addresses the problem of integrating the complementary label information to build the multi-task feature selection method for jointly selecting features from multi-modality neuroimaging data to improve AD/MCI classification. Specifically, we formulate the multi-modality classification as a multi-task learning framework and introduce the label-alignment regularization term to seek the optimal features which preserve the discriminative information between within-class subjects across multiple modalities. Experimental results demonstrate that our proposed method can achieve better performance than all conventional methods. In future work, we will extend our method to include more modalities and test other classifiers for further improvement of classification performance.
References
Brookmeyer, R., Johnson, E., Ziegler-Graham, K., Arrighi, H.M.: Forecasting the global burden of alzheimer’s disease. Alzheimer’s Dementia 3(3), 186–191 (2007)
Gray, K.R., Aljabar, P., Heckemann, R.A., Hammers, A., Rueckert, D.: Random forest-based similarity measures for multi-modal classification of alzheimer’s disease. NeuroImage 65, 167–175 (2013)
Huang, S., Li, J., Ye, J., Wu, T., Chen, K., Fleisher, A., Reiman, E.: Identifying alzheimer’s disease-related brain regions from multi-modality neuroimaging data using sparse composite linear discrimination analysis. In: Advances in Neural Information Processing Systems, pp. 1431–1439 (2011)
Jie, B., Zhang, D., Cheng, B., Shen, D.: Manifold regularized multi-task feature selection for multi-modality classification in alzheimer’s disease. In: Mori, K., Sakuma, I., Sato, Y., Barillot, C., Navab, N. (eds.) MICCAI 2013, Part I. LNCS, vol. 8149, pp. 275–283. Springer, Heidelberg (2013)
Liu, F., Wee, C.-Y., Chen, H., Shen, D.: Inter-modality relationship constrained multi-task feature selection for AD/MCI classification. In: Mori, K., Sakuma, I., Sato, Y., Barillot, C., Navab, N. (eds.) MICCAI 2013, Part I. LNCS, vol. 8149, pp. 308–315. Springer, Heidelberg (2013)
Liu, J., Ye, J.: Efficient L1/Lq norm regularization. Technical report, Arizona State University (2009)
McEvoy, L.K., Fennema-Notestine, C., Roddey, J.C., Hagler Jr., D.J., Holland, D., Karow, D.S., Pung, C.J., Brewer, J.B., Dale, A.M.: Alzheimer disease: quantitative structural neuroimaging for detection and prediction of clinical and structural changes in mild cognitive impairment. Radiology 251(1), 195 (2009)
Mosconi, L., Berti, V., Glodzik, L., Pupi, A., De Santi, S., de Leon, M.J.: Pre-clinical detection of alzheimer’s disease using FDG-PET, with or without amyloid imaging. J. Alzheimers Dis. 20(3), 843–854 (2010)
Shen, D., Davatzikos, C.: Hammer: hierarchical attribute matching mechanism for elastic registration. IEEE Trans. Med. Imaging 21(11), 1421–1439 (2002)
Westman, E., Muehlboeck, J., Simmons, A., et al.: Combining MRI and CSF measures for classification of alzheimer’s disease and prediction of mild cognitive impairment conversion. Neuroimage 62(1), 229–238 (2012)
Yuan, M., Lin, Y.: Model selection and estimation in regression with grouped variables. J. Roy. Stat. Soc.: Ser. B (Stat. Methodol.) 68(1), 49–67 (2006)
Zhang, D., Shen, D.: Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in alzheimer’s disease. Neuroimage 59(2), 895–907 (2012)
Zhang, D., Wang, Y., Zhou, L., Yuan, H., Shen, D.: Multimodal classification of alzheimer’s disease and mild cognitive impairment. Neuroimage 55(3), 856–867 (2011)
Zhang, Y., Brady, M., Smith, S.: Segmentation of brain MR images through a hidden markov random field model and the expectation-maximization algorithm. IEEE Trans. Med. Imaging 20(1), 45–57 (2001)
Acknowledegment
This work is supported in part by National Natural Science Foundation of China (Nos. 61422204, 61473149, 61170151), Jiangsu Natural Science Foundation for Distinguished Young Scholar (No. BK20130034), NUAA Fundamental Research Funds (No. NE2013105), the Jiangsu Qinglan Project, Natural Science Foundation of Anhui Province (No. 1508085MF125), the Open Projects Program of National Laboratory of Pattern Recognition (No. 201407361).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing AG
About this paper
Cite this paper
Zu, C., Jie, B., Chen, S., Zhang, D. (2016). Label-Alignment-Based Multi-Task Feature Selection for Multimodal Classification of Brain Disease. In: Rish, I., Langs, G., Wehbe, L., Cecchi, G., Chang, Km., Murphy, B. (eds) Machine Learning and Interpretation in Neuroimaging. MLINI MLINI 2013 2014. Lecture Notes in Computer Science(), vol 9444. Springer, Cham. https://doi.org/10.1007/978-3-319-45174-9_6
Download citation
DOI: https://doi.org/10.1007/978-3-319-45174-9_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-45173-2
Online ISBN: 978-3-319-45174-9
eBook Packages: Computer ScienceComputer Science (R0)