Abstract
Multimodal classification methods using different modalities of imaging and non-imaging data have recently shown great advantages over traditional single-modality-based ones for diagnosis and prognosis of Alzheimer’s disease (AD), as well as its prodromal stage, i.e., mild cognitive impairment (MCI). However, to the best of our knowledge, most existing methods focus on mining the relationship across multiple modalities of the same subjects, while ignoring the potentially useful relationship across different subjects. Accordingly, in this paper, we propose a novel learning method for multimodal classification of AD/MCI, by fully exploring the relationships across both modalities and subjects. Specifically, our proposed method includes two subsequent components, i.e., label-aligned multi-task feature selection and multimodal classification. In the first step, the feature selection learning from multiple modalities are treated as different learning tasks and a group sparsity regularizer is imposed to jointly select a subset of relevant features. Furthermore, to utilize the discriminative information among labeled subjects, a new label-aligned regularization term is added into the objective function of standard multi-task feature selection, where label-alignment means that all multi-modality subjects with the same class labels should be closer in the new feature-reduced space. In the second step, a multi-kernel support vector machine (SVM) is adopted to fuse the selected features from multi-modality data for final classification. To validate our method, we perform experiments on the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database using baseline MRI and FDG-PET imaging data. The experimental results demonstrate that our proposed method achieves better classification performance compared with several state-of-the-art methods for multimodal classification of AD/MCI.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Alzheimer’s disease (AD) is a physical disease that affects the brain and is the most common cause of dementia. There were more than 26.6 million people worldwide with AD in 2010, and it is predicted that 1 in 85 people will be affected by 2050 (Brookmeyer et al. 2007). So far, there is no treatment for the disease, which worsens as it progresses, and eventually leads to death. Thus, it is very important to accurately identify AD, especially for its early stage also known as mild cognitive impairment (MCI) which has a high risk of progressing to AD (Petersen et al. 1999).
Existing studies have shown that AD is related to the structural atrophy, pathological amyloid depositions, and metabolic alterations in the brain (Jack et al. 2010; Nestor et al. 2004). So far, multiple biomarkers have been shown to be sensitive to the diagnosis of AD and MCI, i.e., structural MR imaging (MRI) for brain atrophy measurement (Leon et al. 2007; Du et al. 2007; Fjell et al. 2010; Mcevoy et al. 2009), functional imaging (e.g., FDG-PET) for hypometabolism quantification (De et al. 2001; Morris et al. 2001), and cerebrospinal fluid (CSF) for quantification of specific proteins (Bouwman et al. 2007; Mattsson et al. 2009; Shaw et al. 2009; Fjell et al. 2010).
In recent years, machine learning and pattern classification methods, which can learn a model from training subjects to predict class label (i.e., patient or normal control) on unseen subject, have been widely applied to studies of AD and MCI based on single modality of biomarkers. For example, researchers have extracted the features from the structural MRI, such as voxel-wise tissue (Desikan et al. 2009; Fan et al. 2007; Magnin et al. 2009), cortical thickness (Desikan et al. 2009; Oliveira et al. 2010) and hippocampal volumes (Gerardin et al. 2009; MJ et al. 2004) for AD and MCI classification. Besides structural MRI, some researchers also used fluorodeoxyglucose positron emission tomography (FDG-PET) (Chételat et al. 2003; Foster et al. 2007; Higdon et al. 2004) for AD or MCI classification.
Different imaging modalities provide different views of brain structure or function. For example, structural MRI reveals patterns of gray matter atrophy, while FDG-PET measures the reduced glucose metabolism in the brain. It is reported that MRI and FDG-PET provide different sensitivity for memory prediction between disease and health (Walhovd et al. 2010). Using multiple biomarkers may reveal hidden information that could be overlooked by using single modality. Researchers have begun to integrate multiple modalities to further improve the accuracy of disease classification (Leon et al. 2007; Fjell et al. 2010; Foster et al. 2007; Walhovd et al. 2010; Apostolova et al. 2010; Dai et al. 2012; Gray et al. 2012; Hinrichs et al. 2011; Huang et al. 2011; Landau et al. 2010; Westman et al. 2012; Yuan et al. 2012; Zhang et al. 2011). For instance, Hinrichs et al. (2011) used two modalities (including MRI and FDG-PET) for AD classification. Zhang et al. (2011) combined MRI, FDG-PET and cerebrospinal fluid (CSF) for classifying patients with AD/MCI from normal controls. Dai et al. (2012) integrated structural MRI (sMRI) and functional MRI (fMRI) for AD classification. Gray et al. (2012) used MRI, FDG-PET, CSF and categorical genetic information for AD/MCI classification.
Although promising results were achieved by existing multimodal classification methods, the problem of small number of subjects and large feature dimensions limits further performance improvement of the above methods. For neuroimaging data, even after feature extraction, the dimension of feature is still relatively high compared to the size of subject. Also, there may exist redundant or irrelevant features for subsequent classification task. Thus, those irrelevant and redundant features need to be removed for reducing feature dimension by feature selection. In the literature, most existing feature selection methods are often performed for each modality individually, which ignores the potential relationship among different modalities. To the best of our knowledge, only a few studies focus on jointly selecting features from multi-modality neuroimaging data for AD/MCI classification. For example, Huang et al. (2011) proposed a sparse composite linear discriminant analysis model (SCLDA) for identification of disease-related brain regions of early AD from multi-modality data. Zhang and Shen (2012) proposed a multi-modal multi-task learning for joint feature selection for AD classification and regression. Liu et al. (2014) proposed inter-modality relationship constrained multi-task feature selection for AD/MCI classification. Jie et al. (2015) presented a manifold regularized multi-task feature selection method for multimodal classification of AD/MCI. However, except for Jie et al.’s work, most of the existing multi-modality feature selection methods focus on using multi-modality information from the same subjects, while ignoring the intrinsic relationship across different subjects, which may also contain useful information for further improving the classification performance. Different from Jie et al.’s method, the proposed approach not only considers the information of each modality, but also regards the relationship across different modalities as extra information. Hence, Jie et al.’s method can be regarded as a special case of our proposed method.
In this paper, we propose a novel learning method that can fully explore the relationships across both modalities and subjects through mining and fusing discriminative features from multi-modality data for AD/MCI classification. Specifically, our proposed learning method includes two major steps: 1) label-aligned multi-task feature selection, and 2) multimodal classification. First, we treat the feature selections from multi-modality data as different learning tasks and adopt a group sparsity regularizer to ensure a subset of relevant features to be jointly selected from multi-modality data. Moreover, to utilize the discriminative information among labeled subjects, we introduce a new label-aligned regularization term into the objective function of standard multi-task feature selection. Here, label-alignment means that all multi-modality subjects with the same class label should be closer in the new feature-reduced space. Then, we use a multi-kernel support vector machine (SVM) to fuse the selected features from multi-modality data for final classification. The proposed method has been evaluated on the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database, demonstrating better results compared to several state-of-the-art multi-modality-based methods.
Method
Neuroimaging data
We use the data obtained from the Alzheimer’s disease Neuroimaging Initiative (ADNI) database (www.loni.usc.edu) in this paper. The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public-private partnership. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials. The initial goal of ADNI was to recruit approximately 200 cognitively normal older individuals to be followed for 3 years, 400 MCI patients to be followed for 3 years, and 200 early AD patients to be followed for 2 years.
We use imaging data from 202 ADNI participants with corresponding baseline MRI and FDG-PET data. In particular, it includes 51 AD patients, 99 MCI patients and 52 normal controls (NC). The MCI patients were divided into 43 MCI converters (MCI-C) who have progressed to AD with 18 months and 56 MCI non-converters (MCI-NC) whose diagnoses have still remain stable within 18 months. Table 1 lists the clinical and demographic information for the study population. A detailed description on acquiring MRI and PET from ADNI as used in this paper can be found in (Zhang et al. 2011). All structural MR scans were acquired from 1.5 T scanners. Raw Digital Imaging and Communications in Medicine (DICOM) MRI scans were downloaded from the public ADNI site (adni.loni.usc.edu), reviewed for quality, and automatically corrected for spatial distortion caused by gradient nonlinearity and B1 field inhomogeneity. PET images were acquired 30–60 min post-injection, averaged, spatially aligned, interpolated to a standard voxel size, intensity normalized, and smoothed to a common resolution of 8 mm full width at half maximum.
Image pre-processing and feature extraction are performed for all MR and PET images by following the same procedures as in (Zhang et al. 2011). First, we do anterior commissure (AC)-posterior commissure (PC) correction on all images, and use the N3 algorithm (Sled et al. 1997) to correct the intensity inhomogeneity. Next, we do skull-stripping on structural MR images using both brain surface extractor (BSE) (Shattuck et al. 2001) and brain extraction tool (BET) (Smith and Stephen 2002), followed by manual edition and intensity inhomogeneity correction. After removal of cerebellum, FAST in the FSL package (Zhang et al. 2001) is used to segment structural MR images into three different tissues: gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF). After registration using HAMMER (Shen and Davatzikos 2002), we obtain the subject-labeled image based on a template with 93 manual labels. Then, we compute the GM tissue volume of each region as a feature. For PET image, we first align it to its respective MR image of the same subject using a rigid transformation, and then compute the average intensity of each ROI in the PET image as a feature. Therefore, for each subject, we totally obtain 93 features from MR image and another 93 features from PET image.
Label-aligned multi-task feature learning
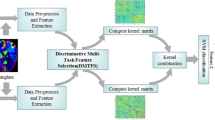
In this section, we will first briefly introduce the conventional multi-task feature selection (Evgeniou and Pontil 2004; Kumar and Daume Iii 2012; Obozinski et al. 2006, 2010; Yuan and Lin 2006), and then derive our proposed label-aligned multi-task feature selection model, as well as the corresponding optimization algorithm. Finally, we use the multi-kernel support vector machine for classification. Figure 1 gives the overview of the proposed classification method.
Multi-task feature selection
Denote X m = [x m1 , …, x m i , …, x m N ]T ∈ ℝN × d as the training data matrix on the m -th modality, where \( {\boldsymbol{x}}_i^m \) represents the corresponding (column) feature vector of the i -th subject, d is the dimension of features, and N is the number of subjects. Let Y = [y 1, …, y i , …, y N ]T ∈ ℝN be the label vector corresponding to N training samples, where the value of \( {y}_i \) is +1 or −1 (i.e., patient or normal control). Then, the objective function of multi-task feature selection (MTFS) model is as follows (Yuan and Lin 2006):
where w m ∈ ℝd is the regression coefficient vector for the m -th modality and the coefficient vectors for all \( M \) modalities form a coefficient matrix, W = [w 1, …, w m, …, w M] ∈ ℝd × M, and \( M \) is the total number of modalities. In (1), \( {\left\Vert \boldsymbol{W}\right\Vert}_{2,1} \) is the ℓ 2,1-norm of matrix W defined as ‖W‖2,1 = ∑ d i = 1 ‖w j ‖2, where w j is the j -th row of matrix W. Here, \( {\uplambda}_1 \) is a regularization parameter controlling the relative contributions of the two terms.
The \( {\mathrm{\ell}}_{2,1} \) -norm \( {\left\Vert \boldsymbol{W}\right\Vert}_{2,1} \) can be seen as the sum of the ℓ 2 -norms of the rows of matrix W (Yuan and Lin 2006), which encourages the weights corresponding to the same feature across different modalities to be grouped together and then a small number of common features will be jointly selected. So, the solution of MTFS results in a weight matrix W whose elements in many rows are all zeros for the characteristic of ‘group sparsity’. It is worth noting that when there is only one modality (i.e., \( M \) =1), the MTFS model will degenerate into the least absolute shrinkage and selection operator (LASSO) model (Tibshirani 1994).
Label-aligned multi-task feature selection
One limitation of the standard multi-task feature selection model is that only the relationship between modalities of the same subjects is considered, while ignoring the important relationship among labeled subjects. To address this issue, we introduce a new term called label-aligned regularization term, which minimizes the distance between within-class subjects in the feature-reduced space as follows:
where, \( {S}_{ij} \) is defined as:
The regularization term (2) can be explained as follows. ‖(w p)T x p i − (w q)T x q j ‖ 22 S ij measures the distance between x p i and x q j in the projected space. It implies that if x p i and x q j are from the same class, the distance between them should be as small as possible in the projected space. It is worth noting that 1) when \( p=q \) the local geometric structure of the same modality data is preserved in the feature-reduced space; 2) when \( p<q \) the complementary information provided from different modalities are used to guide the estimation of the feature-reduced space. Therefore, the Eq. (2) preserves the intrinsic label relatedness among multi-modality data and also explores the complementary information conveyed by different modalities. Generally speaking, the goal of (2) is to preserve label relatedness by aligning paired within-class subjects from multiple modalities.
By incorporating the regularizer (2) into (1), we can obtain the objective function of our label-aligned multi-task feature selection model as below:
where \( {\uplambda}_1 \) and \( {\uplambda}_2 \) are the two positive constants that control the sparseness and the degree of preserving the distance between subjects, respectively. From (4), we can not only jointly select a subset of common features from multi-modality data, but also preserve label relatedness by aligning paired within-class subjects. Figure 2 illustrates the used relationships among modalities and subjects in our proposed model as compared with the traditional multi-modality methods. In Fig. 2a, traditional multimodal methods only concern the relationships of different modalities (i.e., the single line connecting MRI and PET) from the same subject. As we can see from Fig. 2b, our proposed method can preserve not only the multi-modality relationship from the same subject, but also the correlation across modalities between different subjects.
Optimization algorithm
At present, there are several algorithms developed to solve the optimization problem in (4). Here, we choose the widely applied Accelerated Proximal Gradient (APG) method (Nesterov 2003; Chen et al. 2009) to get the solution of our proposed method. Specifically, we separate the objective function in (4) to the smooth part:
and non-smooth part:
Then, the following function is constructed for approximating the composite function f(W) + g(W):
where \( {\left\Vert \cdot \right\Vert}_F \) is the Frobenius norm, \( \nabla f\left({\boldsymbol{W}}_k\right) \) is the gradient of \( f\left(\boldsymbol{W}\right) \) at point W k of the k -th iteration, and l is the step size. Finally, the update step of AGP algorithm is defined as:
where l can be determined by line search, and \( {\boldsymbol{U}}_k={\boldsymbol{W}}_k-\frac{1}{l}\nabla f\left({\boldsymbol{W}}_k\right) \).
The key of AGP algorithm is how to solve the update step efficiently. The study in (Liu and Ye 2010) shows that this problem can be decomposed into d separate subproblems, and the analytical solutions of these sub-problems can be easily obtained.
In addition, according to the technique in (Chen et al. 2009), instead of computing (7) based on W k , we use Q k to calculate \( {\Omega}_l\left(\boldsymbol{W},{\boldsymbol{Q}}_k\right) \) and the search point Q k is defined as:
where \( {\eta}_k=\frac{\left(1-{\gamma}_{k-1}\right){\gamma}_k}{\gamma_{k-1}} \) and \( {\gamma}_k=\frac{2}{k+3} \). The algorithm for Eq. (4) can achieve a convergence rate of \( O\left(1/{K}^2\right) \), where \( K \) is the maximum iteration.
Multi-kernel support vector machine
Multi-kernel SVM can effectively integrate data from multiple modalities for classification of Alzheimer’s disease (Zhang et al. 2011). Given a set of training subjects, m = 1, … M, k m(z m i , z m j ) = ϕ m(z m i )T ϕ m(z m j ) is the kernel function for the subjects z m i and z m j of the m -th modality. Linear combined kernel, k(z i , z j ) = ∑ M m = 1 β m k m(z m i , z m j ) is adopted for fusing information from different modalities. Here \( {\beta}_m \) is the combining weight of the m -th kernel and ∑ M m = 1 β m = 1. In our experiments, the optimal \( {\beta}_m \) is determined via a coarse-grid search through cross-validation on the training set.
Experiments and results
We test the performance of the proposed method on 202 ADNI participants with corresponding baseline MRI and FDG-PET data. Classification performance is assessed between three clinically relevant pairs of diagnostic groups (AD vs. NC, MCI vs. NC, and MCI-C vs. MCI-NC). The proposed method is compared with three existing multi-kernel-based multimodal classification methods, including multi-kernel method (Zhang et al. 2011) without performing feature selection (denoted as Baseline), multi-kernel method with LASSO feature selection performed independently on single modalities (denoted as SMFS), and multi-kernel method using multi-modal feature selection method (denoted as MMFS) proposed in (Zhang and Shen 2012). We also directly concatenate 93 features from MRI and 93 features from FDG-PET into a 186 dimensional vector, and then perform t-test and LASSO as feature selection methods, followed by the standard SVM with linear kernel for classification (with the corresponding methods denoted as t-test and LASSO, respectively). It is worth noting that the same training and test subjects are used in all methods for fair comparison.
Validation
In our experiments, we use a 10-fold cross-validation strategy to evaluate the effectiveness of our proposed method. Specifically, the whole set of subject samples are equally partitioned into 10 subsets. For each cross-validation, the nine subsets are chosen for training and the remaining subjects are used for testing. The process is independently repeated 10 times to avoid any bias introduced by randomly partitioning the dataset in cross-validation. We evaluate the performance of different methods by computing the classification accuracy (ACC), as well as the sensitivity (SEN), the specificity (SPE) and the area under receiver operating characteristic (ROC) curve (AUC). Here, the accuracy measures the proportion of subjects correctly classified among the whole population, the sensitivity represents the proportion of AD or MCI patients correctly classified, and the specificity denotes the proportion of normal controls correctly classified. The SVM classifier is implemented using the LIBSVM toolbox (Chang and Lin 2007), with a linear kernel and a default value for the parameter C (i.e., \( C=1 \)). The optimal values of regularization parameters \( {\uplambda}_1 \), \( {\uplambda}_2 \) and the weights in the multi-kernel classification method are determined by another 10-fold cross-validation on the training subjects.
Results of AD/MCI vs. NC classification
The classification results of AD vs. NC and MCI vs. NC produced by different methods are listed in Table 2. As can be seen from Table 2, our proposed method consistently achieves better performance than other methods for the classification between AD/MCI patients and normal controls. Specifically, for classifying AD from NC, our proposed method achieves a classification accuracy of 95.95 %, while the best accuracy of other methods is only 92.25 % (obtained by SMFS). In addition, for classifying MCI from NC, our proposed method achieves a classification accuracy of 80.26 %, while the best accuracy of other methods is only 74.34 % (obtained by Baseline). Furthermore, we perform the significance test using paired t-test on the classification accuracies between our proposed method and other compared methods, with the corresponding results given in Table 2. From Table 2, we can see that our proposed method is significantly better than the compared methods (i.e., the corresponding p values are very small).
For further validation, in Fig. 3 we plot the ROC curves of four multi-modality based classification methods for AD/MCI vs. NC classification. Figure 3 shows that our proposed method consistently achieves better classification performances than other multi-modality based methods for both AD vs. NC and MCI vs. NC classifications. Specifically, as can be seen from Table 2, our method achieves the area under the ROC curve (AUC) of 0.97 and 0.81 for AD vs. NC and MCI vs. NC classifications, respectively, showing better classification ability compared with other methods.
Results of MCI conversion prediction
The classification results for MCI-C vs. MCI-NC are shown in Table 3. As can be seen from Table 3 and Fig. 4, our proposed method consistently outperforms other methods in MCI-converter classification. Specifically, our proposed method achieves a classification accuracy of 69.78 %, while the best one of other methods is only 61.67 %, which is obtained by SMFS. The classification accuracy of our proposed method is significantly (p < 0.001) higher than any compared methods.
Figure 4 plots the corresponding ROC curves of four multi-modality based methods for MCI-C vs. MCI-NC classification. We can see from Fig. 4 that the superior classification performance is obtained by our proposed method. Table 3 also lists the area under the ROC curve (AUC) of different classification methods. As can be seen from Table 3, AUC achieved by our proposed method is 0.69 for MCI-C vs. MCI-NC classification, while the best one of other methods is only 0.64, obtained by t-test, indicating the outstanding classification performance of our proposed method.
The most discriminative brain regions
The most discriminative regions are defined as those that are most frequently selected in cross-validation. For each selected discriminative feature, the standard paired t-test is performed to evaluate its discriminative power between patients and normal control groups. Top 10 ROIs detected from both MRI and FDG-PET data for MCI classification are listed in Table 4. Figure 5 plots these regions in the template space. As can be seen from Table 4 and Fig. 5, the most important regions for MCI classification include hippocampal, amygdale, etc., which are in agreement with other recent AD/MCI studies (Sole et al. 2008; Derflinger et al. 2011; Al 2008; Poulina et al. 2011; Wolf et al. 2003).
Discussion
In this paper, we proposed a novel label-aligned multi-task feature learning method for multimodal classification of Alzheimer’s disease and mild cognitive impairment. The experimental results on the ADNI database show that our proposed method achieves high classification accuracies of 95.95, 80.26, and 69.78 % for AD vs. NC, MCI vs. NC and MCI-C vs. MCI-NC classifications, in comparison with several state-of-the-art multimodal AD/MCI classification methods.
Multi-task learning
Multi-task learning is a recently developed technique in machine learning field, which can jointly learn multiple tasks via a shared representation. Because the domain information or some commonality is contained in the learning tasks, multi-task learning can usually improve the performances by learning classifiers for multiple tasks together.
Recently, multi-task learning has been introduced into medical imaging field. For example, Zhang et al. (Zhang and Shen 2012) applied multi-task learning for joint prediction of both regression variables (i.e., clinical scores) and classification variable (i.e., class labels) in Alzheimer’s disease. In their method, multi-task feature selection was first used to select the common subset features corresponding to different tasks, and then multi-kernel SVM was performed for final regression and classification. It is worth noting that the feature selection step in (Zhang and Shen 2012) was performed separately for each modality, while ignoring the potential relationship among different modalities. Afterwards, Liu et al. (2014) considered the inter-modality relationship within each subject to preserve the complementary information among modalities. However, in their method only information corresponding to individual subject is concerned. Suk et al. (2014) first assumed the data classes were multipeak distribution, and then formulated a multi-task learning problem in a l-2,1 framework with new label encodings obtained by clustering. However, the method in (Suk et al. 2014) still did not consider the potential information across different modalities. More recently, Jie et al. (2015) proposed a manifold regularized multi-task feature learning method, which only considered the manifold information in each modality separately and thus cannot reflect the information across different modalities. It is worth noting that our proposed method and Jie et al.’s method are developed based on different considerations. Jie et al.’s method only concerns preserving the manifolds existing in each modality of the data. Different from Jie et al.’s method, the proposed approach not only takes the structure information of each modality into account, but also regards the relationship across different modalities as extra information. Hence, Jie et al.’s method can be regarded as a special case of our proposed method. Although our proposed method has a more general feature selection framework compared with Jie et al.’s approach, the objective function of our method is still convex. Thus, the optimal solution can still be obtained, i.e., by using Accelerated Proximal Gradient (APG) method.
In contrast, our proposed label-aligned multi-task feature learning method can preserve the relationships not only across different modalities in the same subjects but also among different modalities in different subjects. Our proposed method is evaluated on the ADNI database using baseline MRI and FDG-PET data for three clinical groups classifications including AD vs. NC, MCI vs. NC and MCI-C vs. MCI-NC, and the experimental results demonstrate the effectiveness of our proposed method.
Comparison with existing methods
To compare our proposed method with existing methods, in this section we perform the comparisons between the results of our proposed method and those of existing state-of-the-art multi-modality methods, as shown in Table 5. As can be seen from Table 5, Hinrichs et al. (2011) used 48 AD subjects and 66 NC subjects, and obtained an accuracy of 87.6 % by using two modalities (MRI + PET). Huang et al. (2011) used 49 AD patients and 67 NC with MRI and PET modalities for AD classification, achieving an accuracy of 94.3 %. In (Gray et al. 2012), authors used 37 AD patients, 75 MCI patients and 35 NC and reported classification accuracies of 89.0, 74.6 and 58.0 % for AD, MCI and MCI-converter classification, respectively, using four different modalities (MRI + PET + CSF + genetic). Jie et al. (2015) achieved the accuracies of 95.03, 79.27 and 68.94 % for classification of AD/NC, MCI/NC and MCI-C/MCI-NC, respectively. Liu et al. (2014) obtained the accuracies of 94.37, 78.80 and 67.83 % for AD, MCI and MCI-converter classifications, respectively. It is worth noting that the dataset used in (Jie et al. 2015) and (Liu et al. 2014) are the same as that in the current study. Table 5 indicates that our proposed method consistently outperform other methods, which further validate the efficacy of our proposed method for AD diagnosis.
The effect of regularization parameters
In our method, there are two regularization items, i.e., the sparsity regularizer \( {\uplambda}_1 \) and label-aligned regularization term \( {\uplambda}_2 \). The two parameters control the relative contribution of those regularization terms. Here, the values of \( {\uplambda}_1 \) and \( {\uplambda}_2 \) are set from 0 to 50 at a step size of 10, respectively, to observe the effect of the regularization parameters on the classification performance of our proposed method. Figure 6 shows the classification results with respect to different values of \( {\uplambda}_1 \) and \( {\uplambda}_2 \). When \( {\uplambda}_1=0 \), all features extracted from MRI and FDG-PET data are used for classification, and thus our method will degenerate to multi-kernel method proposed in (Zhang et al. 2011). Also, when λ2 = 0, no label-aligned regularization item is introduced, and thus our method will degenerate to the MMFS method proposed in (Zhang and Shen 2012).
The classification accuracy with regularization parameters \( {\uplambda}_1 \) and \( {\uplambda}_2 \). a AD classification, b MCI classification, and c MCI conversion classification. Each curve denotes the performance for different selected value for \( {\uplambda}_1 \). X-axis represents diverse values for \( {\uplambda}_2 \)
As we can observe from Fig. 6, under all values of \( {\uplambda}_1 \) and \( {\uplambda}_2 \), our proposed method consistently outperforms the MMFS methods on three classification tasks (i.e., AD vs. NC, MCI vs. NC, and MCI-C vs. MCI-NC), which further indicates the advantage of using label-aligned regularization term. Also, Fig. 6 shows that when fixing the value of \( {\uplambda}_1 \), the curves corresponding to different values of \( {\uplambda}_2 \) are very smooth on three classification tasks, which shows our method is relatively robust to the regularization parameter \( {\uplambda}_2 \). Finally, as can be seen from Fig. 6, when fixing the value of \( {\uplambda}_2 \), the results on three classification tasks are largely affected with different values of \( {\uplambda}_1 \), which implies that the selection of \( {\uplambda}_1 \) is very important for final classification results. This is reasonable since \( {\uplambda}_1 \) controls the sparsity of model and thus determines the size of the optimal feature subset.
The effect of weights for multimodal classification
We investigate how the two combining kernel weights \( {\beta}_{\mathrm{MRI}} \) and \( {\beta}_{\mathrm{PET}} \) affect the classification performance of our proposed method. The combining kernel weights are set from 0 to 1 at a step size of 0.1, with the constraint of β MRI + β PET = 1. Figure 7 shows the classification accuracy and AUC value under different combination of kernel weights of MRI and PET. As we can observe from Fig. 7, the relative high classification performance is obtained in the middle part, which demonstrates the effectiveness of combining two modalities for classification. Moreover, the intervals with higher performance mainly lie in a larger interval of [0.2, 0.8], implying that each modality is indispensable for achieving good classification performances.
Limitations
There are several limitations that should be further considered in the future study. First, in the current study, we only investigated binary classification problem (i.e., AD vs. NC, MCI vs. NC, and MCI-C vs. MCI-NC), and did not test the ability of the classifier for the multi-class classification of AD, MCI and normal controls. Although multi-class classification is more challenging than binary-class classification, it is very important to diagnose different stage of dementia. Second, the proposed method requires the same number of features from different modalities. Other modalities in ADNI database, such as CSF and genetic data, which have different feature numbers, may also carry important pathological information that can help further improve the classification performance. Finally, longitudinal data may contain very important information for classification, while our proposed method can only deal with the baseline data.
Conclusion
This paper proposed a novel multi-task feature learning method for jointly selecting features from multi-modality neuroimaging data for AD/MCI classification. By introducing the label-aligned regularization term into the multi-task learning framework, the proposed method can utilize the relationships across both modalities and subjects to seek out the most discriminative features subset. Experimental results on the ADNI database demonstrate that our proposed method outperforms the state-of-the-art methods for multimodal classification of AD/MCI.
References
Al, N. F. E. (2008). Principal component analysis of FDG PET in amnestic MCI. European Journal of Nuclear Medicine and Molecular Imaging, 35(12), 2191–2202 (2112).
Apostolova, L. G., Hwang, K. S., Andrawis, J. P., Green, A. E., Babakchanian, S., Morra, J. H., et al. (2010). 3D PIB and CSF biomarker associations with hippocampal atrophy in ADNI subjects. Neurobiology of Aging, 31(8), 1284–1303.
Bouwman, F. H., van der Flier, W. M., Schoonenboom, N. S. M., van Elk, E. J., Kok, A., Rijmen, F., et al. (2007). Longitudinal changes of CSF biomarkers in memory clinic patients. Neurology, 69(10), 1006–1011.
Brookmeyer, R., Johnson, E., Ziegler-Grahamm, K., Arrighi, H. M., Brookmeyer, R., & Johnson, E. (2007). O1-02-01 forecasting the global burden of Alzheimer’s disease. Alzheimers & Dementia the Journal of the Alzheimers Association, 3(3), 186–191.
Chang, C. C., & Lin, C. J. (2007). LIBSVM: a library for support vector machines. ACM Transactions on Intelligent Systems and Technology, 2(3), 389–396.
Chen, X., Pan, W., Kwok, J. T., & Carbonell, J. G. (2009). Accelerated gradient method for multi-task sparse learning problem. Proceedings of the International Conference on Data Mining, 746–751.
Chételat, G., Desgranges, B., Sayette, V., La, D., Viader, F., Eustache, F., & J-C, B. (2003). Mild cognitive impairment: Can FDG-PET predict who is to rapidly convert to Alzheimer’s disease? Neurology, 60(8), 1374–1377.
Dai, Z., Yan, C., Wang, Z., Wang, J., Xia, M., Li, K., et al. (2012). Discriminative analysis of early Alzheimer’s disease using multi-modal imaging and multi-level characterization with multi-classifier (M3). NeuroImage, 59(3), 2187–2195.
De, S. S., de Leon, M. J., Rusinek, H., Convit, A., Tarshish, C. Y., Roche, A., et al. (2001). Hippocampal formation glucose metabolism and volume losses in MCI and AD. Neurobiology of Aging, 22(4), 529–539.
Derflinger, S., Sorg, C., Gaser, C., Myers, N., Arsic, M., Kurz, A., et al. (2011). Grey-matter atrophy in Alzheimer’s disease is asymmetric but not lateralized. Journal of Alzheimers Disease, 25(2), 347–357.
Desikan, R. S., Cabral, H. J., Hess, C. P., Dillon, W. P., Glastonbury, C. M., Weiner, M. W., et al. (2009). Automated MRI measures identify individuals with mild cognitive impairment and Alzheimer’s disease. Brain, 132(Part 8), 2048–2057.
Du, A. T., Schuff, N., Kramer, J. H., Rosen, H. J., Gorno-Tempini, M. L., Rankin, K., et al. (2007). Different regional patterns of cortical thinning in Alzheimer’s disease and frontotemporal dementia. Brain, 130(4), 1159–1166.
Evgeniou, T., & Pontil, M. (2004). Regularized multi—task learning. In Proceedings of the tenth ACM SIGKDD international conference on Knowledge discovery and data mining, (pp. 109–117).
Fan, Y., Shen, D., Gur, R. C., Gur, R. E., & Davatzikos, C. (2007). COMPARE: classification of morphological patterns using adaptive regional elements. IEEE Transactions on Medical Imaging, 26(1), 93–105.
Fjell, A. M., Walhovd, K. N. C., Mcevoy, L. K., Hagler, D. J., Holland, D., Brewer, J. B., et al. (2010). CSF biomarkers in prediction of cerebral and clinical change in mild cognitive impairment and Alzheimer’s disease. Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 30(6), 2088–2101.
Foster, N. L., Heidebrink, J. L., Clark, C. M., Jagust, W. J., Arnold, S. E., Barbas, N. R., et al. (2007). FDG-PET improves accuracy in distinguishing frontotemporal dementia and Alzheimer’s disease. Brain, 130(10), 2616–2635 (2620).
Gerardin, E., Chételat, G. l., Chupin, M., Cuingnet, R., Desgranges, B., Kim, H. S., et al. (2009). Multidimensional classification of hippocampal shape features discriminates Alzheimer’s disease and mild cognitive impairment from normal aging. NeuroImage, 47(4), 1476–1486.
Gray, K. R., Aljabar, P., Heckemann, R. A., Hammers, A., & Rueckert, D. (2012). Random forest-based similarity measures for multi-modal classification of Alzheimer’s disease. NeuroImage, 65, 167–175.
Higdon, R., Foster, N. L., Koeppe, R. A., DeCarli, C. S., Jagust, W. J., Clark, C. M., et al. (2004). A comparison of classification methods for differentiating fronto-temporal dementia from Alzheimer’s disease using FDG-PET imaging. Statistics in Medicine, 23(2), 315–326. doi:10.1002/sim.1719.
Hinrichs, C., Singh, V., Xu, G., & Johnson, S. C. (2011). Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population. NeuroImage, 55(2), 574–589.
Huang, S., Li, J., Ye, J., Wu, T., Chen, K., & Fleisher, A., et al. (2011). Identifying Alzheimer s disease-related brain regions from multi-modality neuroimaging data using sparse composite linear discrimination analysis. In J. Shawe-Taylor, R. S. Zemel, P. L. Bartlett, F. Pereira, & K. Q. Weinberger (Eds.), Advances in neural information processing systems 24. Curran Associates, Inc.
Jack, C. R., Jr., Knopman, D. S., Jagust, W. J., Shaw, L. M., Aisen, P. S., Weiner, M. W., et al. (2010). Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurology, 9(1), 119–128.
Jie, B., Zhang, D., Cheng, B., & Shen, D. (2015). Manifold regularized multitask feature learning for multimodality disease classification. Human Brain Mapping, 36(2), 489–507.
Kumar, A., & Daume Iii, H. (2012). Learning task grouping and overlap in multi-task learning. Computer Science - Learning.
Landau, S. M., Harvey DMadison, C. M., Reiman, E. M., Foster, N. L., Aisen, P. S., Petersen, R. C., et al. (2010). Comparing predictors of conversion and decline in mild cognitive impairment. Neurology, 75(3), 230–238.
Leon, M. J. D., Mosconi, L., Li, J., Santi, S. D., Yao, Y., Tsui, W. H., et al. (2007). Longitudinal CSF isoprostane and MRI atrophy in the progression to AD. Journal of Neurology, 254(12), 1666–1675.
Liu, J., & Ye, J. (2010). Efficient L1/Lq norm regularization. Cambridge University Pub.
Liu, F., Wee, C. Y., Chen, H., & Shen, D. (2014). Inter-modality relationship constrained multi-modality multi-task feature selection for Alzheimer’s disease and mild cognitive impairment identification. NeuroImage, 84, 466–475.
Magnin, B. t., Mesrob, L., Kinkingnéhun, S., Pélégrini-Issac, M., Colliot, O., Sarazin, M., et al. (2009). Support vector machine-based classification of Alzheimer’s disease from whole-brain anatomical MRI. Neuroradiology, 51(2), 73–83.
Mattsson, N., Zetterberg, H., Hansson, O., Andreasen, N., Parnetti, L., Jonsson, M., et al. (2009). CSF biomarkers and incipient Alzheimer disease in patients with mild cognitive impairment. JAMA: The Journal of the American Medical Association, 302(4), 385–393.
Mcevoy, L. K., Fennema-Notestine, C., Roddey, J. C., Hagler, D. J., Jr., Holland, D., Karow, D. S., et al. (2009). Alzheimer disease: quantitative structural neuroimaging for detection and prediction of clinical and structural changes in mild cognitive impairment1. Radiology, 251(1), 195–205.
MJ, W., Kawas, C. H., Stewart, W. F., Rudow, G. L., & Troncoso, J. C. (2004). Hippocampal neurons in pre-clinical Alzheimer’s disease. Neurobiology of Aging, 25(25), 1205–1212.
Morris, J., Storandt, M., Miller, J., McKeel, D., Price, J., Rubin, E., et al. (2001). Mild cognitive impairment represents early-stage Alzheimer disease. Archives of Neurology, 58(3), 397–405.
Nesterov, Y. (2003). Introductory lectures on convex optimization: a basic course. Computer Programming(Oct), 49–50.
Nestor, P. J., Scheltens, P., & Hodges, J. R. (2004). Advances in the early detection of Alzheimer’s disease. Nature Medicine, 10 suppl(7suppl), S34–S41.
Obozinski, G., Jordan, M., & Taskar, B. (2006). Multi-task feature selection. The Workshop of Structural Knowledge Transfer for Machine Learning in International Conference on Machine Learning, 7(2), 1693–1696.
Obozinski, G., Taskar, B., & Jordan, M. I. (2010). Joint covariate selection and joint subspace selection for multiple classification problems. Statistics and Computing, 20(2), 231–252.
Oliveira, P. P. D., Nitrini, R., Busatto, G., Buchpiguel, C., Sato, J. R., & Amaro, E. (2010). Use of SVM methods with surface-based cortical and volumetric subcortical measurements to detect Alzheimer’s disease. Journal of Alzheimers Disease, 19(4), 1263–1272. doi:10.3233/jad-2010-1322.
Petersen, R. C., Smith, G. E., Waring, S. C., Ivnik, R. J., Tangalos, E. G., & Kokmen, E. (1999). Mild cognitive impairment: clinical characterization and outcome. Archives of Neurology, 56(3), 303–308.
Poulina, S., Dautoffb, R., Morris, J., Barrett, L., & Dickersona, B. (2011). Amygdala atrophy is prominent in early Alzheimer’s disease and relates to symptom severity. Psychiatry Research: Neuroimaging, 194(1), 7–13.
Shattuck, D. W., Sandor-Leahy, S. R., Schaper, K. A., Rottenberg, D. A., & Leahy, R. M. (2001). Magnetic resonance image tissue classification using a partial volume model. In Neuroimage, pp. 856–876.
Shaw, L. M., Vanderstichele, H., Knapik‐Czajka, M., Clark, C. M., Aisen, P. S., Petersen, R. C., et al. (2009). Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Annals of Neurology, 65(4), 403–413.
Shen, D., & Davatzikos, C. (2002). HAMMER: hierarchical attribute matching mechanism for elastic registration. In IEEE Trans. on Medical Imaging pp. 1421–1439.
Sled, J. G., Zijdenbos, A. P., & Evans, A. C. (1997). A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Transactions on Medical Imaging, 17(1), 87–97.
Smith, & Stephen, M. (2002). Fast robust automated brain extraction. Human Brain Mapping, 17(3), 143–155.
Sole, A. D., Clerici, F., Chiti, A., Lecchi, M., Mariani, C., Maggiore, L., et al. (2008). Individual cerebral metabolic deficits in Alzheimer’s disease and amnestic mild cognitive impairment: an FDG PET study. European Journal of Nuclear Medicine and Molecular Imaging, 35(7), 1357–1366.
Suk, H. I., Lee, S. W., & Shen, D. (2014). Subclass-based multi-task learning for Alzheimer’s disease diagnosis. Frontiers in Aging Neuroscience, 6(6), 168.
Tibshirani, R. (1994). Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society, 58(1), 267–288.
Walhovd, K. B., Fjell, A. M., Dale, A. M., Mcevoy, L. K., Brewer, J., Karow, D. S., et al. (2010). Multi-modal imaging predicts memory performance in normal aging and cognitive decline. Neurobiology of Aging, 31(7), 1107–1121.
Westman, E., Muehlboeck, J. S., & Simmons, A. (2012). Combining MRI and CSF measures for classification of Alzheimer’s disease and prediction of mild cognitive impairment conversion. NeuroImage, 62(1), 229–238.
Wolf, H., Jelic, V., Gertz, H. J., Nordberg, A., Julin, P., & Wahlund, L. O. (2003). A critical discussion of the role of neuroimaging in mild cognitive impairment. Acta Neurologica Scandinavica, 179(Supplement s179), 52–76.
Yuan, M., & Lin, Y. (2006). Model selection and estimation in regression with grouped variables. Journal of the Royal Statistical Society, 68(1), 49–67. As the access to this document is restricted, you may want to look for a different version under “Related research” (further below) orfor a different version of it.
Yuan, L., Wang, Y., Thompson, P. M., Narayan, V. A., & Ye, J. (2012). Multi-source feature learning for joint analysis of incomplete multiple heterogeneous neuroimaging data ☆. NeuroImage, 61(3), 622–632.
Zhang, D., & Shen, D. (2012). Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer’s disease. NeuroImage, 59(2), 895–907.
Zhang, Y., Brady, M., & Smith, S. (2001). Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Transactions on Medical Imaging, 20(1), 45–57.
Zhang, D., Wang, Y., Zhou, L., Yuan, H., & Shen, D. (2011). Multimodal classification of Alzheimer’s disease and mild cognitive impairment. NeuroImage, 55, 856–867.
Acknowledgments
This work was supported in part by the National Natural Science Foundation of China (Nos. 61422204, 61473149, 61170151), the Jiangsu Natural Science Foundation for Distinguished Young Scholar (No. BK20130034), the Specialized Research Fund for the Doctoral Program of Higher Education (No. 20123218110009), and the NUAA Fundamental Research Funds (No. NE2013105), and by NIH grants EB006733, EB008374, EB009634, MH100217, AG041721, and AG042599.
Author information
Authors and Affiliations
Consortia
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Zu, C., Jie, B., Liu, M. et al. Label-aligned multi-task feature learning for multimodal classification of Alzheimer’s disease and mild cognitive impairment. Brain Imaging and Behavior 10, 1148–1159 (2016). https://doi.org/10.1007/s11682-015-9480-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11682-015-9480-7