Abstract
The programmable nature of DNA chemistry makes it an attractive framework for the implementation of unconventional computing systems. Our early work in this area was among the first to use oligonucleotide-based logic gates to perform computations in a bulk solution. In this chapter we chart the development of this technology over the course of almost 15 years. We review our work on the implementation of DNA-based logic gates and circuits, which we have used to demonstrate digital logic circuits, autonomous game-playing automata, trainable systems and, more recently, decision-making circuits with potential diagnostic applications.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1.1 Introduction
The development of electronic digital logic was one of the greatest technological achievements of the 20th century, and exponential increases in the computational power of commercially-available microprocessors meant that electronic computers are now ubiquitous and indispensable in the modern world. Contemporaneous advances in molecular biology made it clear that information processing is a fundamental capability of all biological systems. Subsequent rapid progress in that field progressed in parallel with the development of consumer electronics, and elucidated many of the mechanisms behind biological information processing [7]. Given that the information processing capabilities of biological systems were evolved over millions of years, it is fascinating to consider whether we can construct synthetic molecular computers that may be more compact and more robust than their natural or electronic counterparts.
When we say that molecules compute, what we usually mean is that an assembly of molecules detects certain inputs, typically the presence or absence of other molecules, and responds by producing one or more output signals, which may take the form of the release of an output molecule or the generation of a detectable fluorescent signal. The use of fluorescence is merely a convenient means of detecting the circuit response in an experimental setting, and plays no role in the actual computation. The goal of a molecular computer scientist is to engineer the intervening molecular system so that the pattern of output signals is related to the pattern of input signals by the desired logic function.
From an unconventional computing perspective, the development of molecular computers offers intriguing possibilities to implement extremely low-power computation [8, 88] and to implement autonomous computational systems that can survive and thrive in environments hostile or inaccessible to silicon microprocessors, such as within the bloodstream or within living cells . The compact nature of DNA has been previously exploited to demonstrate high-density information storage [37], but in our context the fact that billions of molecules exist in each experimental system may make it feasible to execute massively parallel computations in a very small volume, or to implement novel computational architectures that compute using the dynamics of interactions between molecular circuit components.
Our experimental work focuses on catalytic nucleic acid chemistry, in particular, DNAzymes (also known as deoxyribozymes), which are DNA-based enzymes that can cleave or combine other nucleic acid strands. DNAzymes are not known to occur in nature, and the known DNAzyme catalytic motifs have been isolated in in vitro evolution experiments [12, 77, 81]. We turned DNAzymes into logic gates by augmenting them with up to three input-binding modules that regulated the catalytic activity of the DNAzyme based on the pattern of input strands observed in the solution. The cleavage reaction catalyzed by the DNAzyme served as the reporting channel, and we exploited the combinatorial chemistry of DNA to enable us to build systems that processed multiple signals simultaneously in a single solution, with the different information streams identified by different DNA sequences. Thus, each DNAzyme unit implemented a logic gate with up to three inputs, and we constructed a set of such gates complete for Boolean logic [103].
In this chapter we review our designs for DNAzyme-based molecular computers , their integration in large-scale parallel gate arrays exhibiting sophisticated logical and temporal behaviors, and our recent attempts to diversify into sequential logic cascades . We begin by describing our early approach to molecular computing [20, 47], including the first reported complete set of nucleic acid -based logic gates [103]. We then describe how these gates were used to produce autonomous molecular computing systems that implement well-known logic circuits such as adders [58, 106] and large-scale game-playing automata [66, 82, 107]. This approach has been previously reviewed [108], including in the popular literature [67]. We then discuss how, in recent years, we have further developed this approach to achieve signal propagation in DNAzyme signaling cascades , and how we have begun to apply these new techniques to biodetection applications.
1.2 Developing DNAzyme-Based Logic Gates and Circuits
The historical context for our work was set by the publication of Adleman’s seminal paper [1], which demonstrated that combinatorial nucleic acid chemistry could be used to solve a small instance of the Hamiltonian path constraint satisfaction problem . By synthesizing a library of DNA strands to represent the vertices and directed edges of the graph, Adleman’s approach relied on combinatorial hybridization of these molecules to produce linear structures that encoded paths of various lengths through the graph. There followed an extensive sequence of purification and analysis of the resulting molecules that encoded the possible paths through the directed graph, to locate any paths of the correct length that visited every vertex. The fundamental insight behind this approach was to parallelize the “generate” phase of the “generate and test” paradigm for solving computationally intractable problems such as Hamiltonian path . However, the laborious nature of the “test” phase limited the practical applicability of this incarnation of molecular computing .
In our early work, we adopted an alternative approach to molecular computation. We were inspired by timely reviews [12, 77] on nucleic acid catalysts and aptamers , which got us thinking about using external inputs to control nucleic acid catalysis : an idea that was ripe for implementation [92, 110]. Thus, rather than using combinatorial chemical reactions to search for solutions to computationally hard problems, we instead used large populations of DNA logic gates to compute Boolean logic functions, using bulk fluorescence readouts to assay the result of the computations. This approach greatly simplified the experimental protocols and enabled us to execute relatively sophisticated computations, with human intervention required only to provide external data inputs.
DNAzyme reactions and sensors. a DNAzyme structure and DNAzyme-catalyzed cleavage of a substrate molecule . DNAzymes (here the “E6” catalytic motif [13]) consist of conserved catalytic core sequence (labeled \(\mathsf {core}\)) flanked by two substrate binding arms (labeled \(\mathsf {a}_{1}\) and \(\mathsf {a}_{2}\)). The corresponding substrate consists of sequences complementary to the substrate binding arms \(\mathsf {a}_{1}^{*}\) and \(\mathsf {a}_{2}^{*}\), with a cleavage site in the middle (denoted by a small disc). To produce a fluorescent readout of the cleavage reaction, the substrate is labeled with a fluorophore on one end (F) and a quencher on the other (Q). In its uncleaved state, when the fluorophore is excited (here by light of 530 nm wavelength), the energy is transferred to the quencher by Förster resonance energy transfer (FRET). Hence, no output fluorescence is observed. The first step of the cleavage reaction is for the DNAzyme to bind to the substrate (reaction 1). Then, the DNAzyme cleaves the substrate into two shorter product strands (reaction 2). Subsequent unbinding of the products (reaction 3) recycles the active DNAzyme into solution, whereupon it may proceed to interact with further substrates. In the cleaved state, the fluorophore is separated from the quencher, so when the fluorophore is excited, it re-emits the light at its output wavelength (here, 580 nm), which can be observed using standard optical techniques. b Molecular beacon reactions. The molecular beacon consists of a stem enclosing a loop whose sequence is complementary to that of the input strand. In the absence of the input, the beacon adopts the energetically favorable hairpin conformation. When the input is added, it binds to the loop and causes the stem to open. c A sensor (yes gate ) constructed by grafting a molecular beacon input detection module onto a DNAzyme such that the closed stem of the molecular beacon blocks one of the substrate binding arms (\(\mathsf {a}_{1}\)). Thus, in the absence of input \(\text {i}_{1}\), the blocked binding arm prevents the DNAzyme from binding to, and cleaving, its substrate, so no fluorescence is observed. However, in the presence of input \(\text {i}_{1}\), the stem of the molecular beacon is opened, exposing the substrate binding arm, so that the DNAzyme can bind and cleave its input, resulting in the generation of a fluorescent output signal. Hence, the yes gate computes the identity function of its input, as shown by the truth table
We constructed molecular logic gates using RNA-cleaving DNAzymes , which are single strands of DNA that can catalyze the cleavage of specific substrate molecules . The various parts of a DNAzyme strand are illustrated in Fig. 1.1a, using the “E6” catalytic motif [13] as the example. The central catalytic core sequence is largely fixed (with the exception of a small central loop in the E6 motif): this sequence is believed to coordinate the binding of metal ion cofactors (here \(\text {Mg}^{2+}\)) that are required for the cleavage reaction to occur. The catalytic core is flanked by two variable substrate binding arms , which recognize and bind to a complementary substrate molecule and position it correctly so that cleavage may take place. Figure 1.1a shows the means by which a DNAzyme binds to a substrate molecule, cleaves the substrate at the cleavage site (marked by a single RNA base in the DNA strand), and unbinds from the two shorter product molecules. We can monitor the progress of the cleavage reaction by labeling the substrate with a fluorescent tag on one end and a corresponding quencher molecule on the other end: when the substrate is cleaved the fluorophore and quencher are separated, which reduces the efficiency of the quenching reaction and causes an increase in observed fluorescence when the fluorophore is excited by a laser. It is important to note that the DNAzyme strand is unchanged by the cleavage reaction and may bind and cleave additional substrates in a “multiple turnover” reaction which provides an innate signal amplification capability. The efficiency of this process is determined by the lengths of the substrate binding arms : too long, and the post-cleavage unbinding reaction is slowed due to increased stability of the DNAzyme-product complex; too short, and the pre-cleavage binding reaction is slowed due to decreased stability of the DNAzyme-substrate complex. In addition to our work on using DNAzymes to construct molecular logic systems, the catalytic properties of DNAzymes have also been exploited to build systems of self-avoiding molecular walkers [65, 74, 83, 97].
There are three parts of an E6 DNAzyme strand that may be independently modified to control its behavior, the two substrate binding arms and the small loop in the catalytic core . We modified these parts of the strand by functionalizing them with molecular beacons , which are DNA structures that function as input recognition elements. The basic operation of a molecular beacon is illustrated in Fig. 1.1b. In its native state, it is energetically favorable for the mutually complementary ends of the beacon strand to bind to each other, forming a structure known as a hairpin . The single-stranded loop of the hairpin can then serve as an input recognition element: a strand that is complementary to the loop region can bind to the loop and thereby induce a conformational change (by converting the loop from a flexible single-stranded region to a rigid double-stranded region) that opens the hairpin stem. The classical use of molecular beacons in molecular biology is to generate a fluorescent response to the input binding event [115].
However, our interest in molecular beacons was as a means of regulating the catalytic activity of DNAzymes . Thus, our first published result [104] was a logic gate that sensed a single input oligonucleotide and activated a DNAzyme in response (Fig. 1.1c). This logic gate design incorporated a single molecular beacon module that, in the native state, blocks one of the substrate binding arms and thereby prevents the DNAzyme binding to the substrate . However, when the complementary input strand is present, it binds to the molecular beacon module and opens the stem, which exposes the substrate binding arm , enabling the DNAzyme to bind to the reporter substrate and cleave it, which we can detect via fluorescence. We call a gate that senses the input \(\text {i}_{1}\) in this way a yes \(\text {i}_{1}\) gate [104] (occasionally, a signal detector, sensor, or basic catalytic molecular beacon). Here and henceforth, we represent an input value of 1 by the presence of the corresponding input oligonucleotide and an input value of 0 by its absence. Similarly, we represent an output value of 1 by DNAzyme-catalyzed cleavage of the corresponding substrate molecule (observed via fluorescence) and an output value of 0 by no cleavage taking place. Thus, from a logical perspective, the yes gate simply computes the identity function of its input. Furthermore, by adding a different molecular beacon that blocks the other arm of the yes gate , we obtain a DNAzyme that is only activated when the complementary inputs for both molecular beacons are present, as both substrate binding arms must be available for the DNAzyme to bind to the substrate . Thus, such a DNAzyme will function as an and logic gate , as shown in Fig. 1.2b which implements \(\text {i}_{1}\mathbin {\wedge }\text {i}_{2}\). Thus, our DNAzyme-based molecular logic gates are switched by oligonucleotide input signals, just as electronic logic gates are switched by their respective electrical input signals.
DNAzyme logic gates . a and gate. By extending the yes gate motif with a molecular beacon module on both substrate binding arms, we obtain an and gate. This gate implements and logic because both substrate binding arms must be exposed for successful binding to the substrate, which is only possible when both inputs are present. Thus this gate computes \(\text {i}_{1}\mathbin {\wedge }\text {i}_{2}\), as shown in the truth table. b not gate . Extending the small loop in the catalytic core of the “E6” DNAzyme to a full-size input-binding loop allows us to implement a not gate. In the absence of input \(\text {i}_{3}\), the catalytic core structure folds as normal and the DNAzyme is functional. However, when \(\text {i}_{3}\) is added, the loop is opened, which distorts the catalytic core and prevents the DNAzyme from binding to the substrate . Thus, the catalytic activity of the DNAzyme is negatively regulated by the input, which computes \(\lnot {\text {i}_{3}}\), as shown in the truth table. c andandnot gate . By placing loops in all three possible positions, we combine the and gate with the not gate to produce an andandnot gate, which is active only when inputs \(\text {i}_{1}\) and \(\text {i}_{2}\) are present but \(\text {i}_{3}\) is not present. Thus, this gate computes \(\text {i}_{1}\mathbin {\wedge }\text {i}_{2}\mathbin {\wedge }\lnot \text {i}_{3}\), as shown in the truth table. d andand gate . By pre-binding the logic gate with strands complementary to the true input strands, we can invert the sense of control of any of the input-binding loops. In this example, the andandnot gate from c is converted into an andand gate by reversing the action of the \(\text {i}_{3}\) input. This is achieved by pre-binding the logic gate with the \(\text {c}_{3}\) strand, which binds to the input-binding loop in the catalytic core and deforms the catalytic core. Then, when input \(\text {i}_{3}\), which is complementary to \(\text {c}_{3}\), is added, it binds to \(\text {c}_{3}\) via the short, single-stranded, exposed “toehold” and removes \(\text {c}_{3}\) from the logic gate via toehold-mediated strand displacement [133, 139], which allows the catalytic core to refold. This reaction effectively converts the \(\text {i}_{1}\) and \(\text {i}_{2}\) and \(\text {i}_{3}\) gate into an \(\text {i}_{1}\) and \(\text {i}_{2}\) andnot \(\text {i}_{3}\), while simultaneously removing input \(\text {i}_{3}\) from solution. The inputs \(\text {i}_{1}\) and \(\text {i}_{2}\) then bind to the other input-binding loops as normal. Thus, the DNAzyme is active only when all three inputs are present, which yields an andand gate that computes \(\text {i}_{1}\mathbin {\wedge }\text {i}_{2}\mathbin {\wedge }\text {i}_{3}\), as shown in the truth table
Any set of complete Boolean logic gates must include some form of negation, and for this we turned to the third of the potential modification sites on the E6 DNAzyme strand , the small loop within the catalytic core . It turns out that (at least in the E6 catalytic motif) this loop can be enlarged to the same size as the loops from our other molecular beacon control modules without adversely affecting the catalytic activity of the DNAzyme . Thus, we were able to use this loop to negatively regulate DNAzyme catalysis, producing a not gate that computes \(\lnot \text {i}_{3}\), as shown in Fig. 1.2b. In this gate design, when an input strand binds to the molecular beacon loop in the catalytic core, the resulting conformational change distorts the catalytic core by pushing the two halves of the catalytic core (and the two substrate binding arms) apart, so that the DNAzyme cannot successfully bind and cleave its substrate . Conversely, the closed state of the molecular beacon forms the correct catalytic core structure so that substrate cleavage may occur, thus, the DNAzyme is active when the input is absent and inactive when the input is present, as required for a not gate [103].
As is well known, and and not gates are complete for Boolean logic , assuming that we can form circuits with arbitrary connections between the gates. We will return to the question of gate connectivity when we discuss our more recent work in Sect. 1.4, but here we observe that it is possible to implement more sophisticated information processing using unconnected DNAzyme units. In particular, we constructed three-input logic gates from a single DNAzyme by simultaneously modifying all three potential control sites (the two substrate binding arms and the catalytic core) with molecular beacons . Figure 1.2c shows the basic three-input logic gate, which is an andandnot gate [58, 103] that computes the logic function \(\text {i}_{1}\mathbin {\wedge }\text {i}_{2}\mathbin {\wedge }\lnot \text {i}_{3}\). In this case, the catalytic activity of the DNAzyme is positively regulated by the two input binding loops on the substrate binding arms and negatively regulated by the loop in the catalytic core .
The above examples of logic gate design show how direct application of molecular beacon input-binding modules to DNAzymes can be used to implement certain one-, two-, and three-input Boolean logic gates. To broaden our repertoire of logic gates, we employed a strategy of pre-binding gates with blocking strands complementary to certain input binding loops, so that they are initially held open as opposed to folding closed. Then, by adding complementary inputs to strip off the blocking strands via toehold-mediated strand displacement [133, 139], so that the effect of those inputs was negated compared to our previous designs. Strand displacement is an alternative technique for realizing molecular computation , which has been used previously to implement digital logic circuits [89, 96], catalytic cycles [140], artificial neural networks [91], chemical reaction networks [19, 101] and nanomachines [134]. As an example of this approach to DNAzyme logic gate construction, Fig. 1.2d illustrates an andand gate that computes the function \(\text {i}_{1}\mathbin {\wedge }\text {i}_{2}\mathbin {\wedge }\text {i}_{3}\), by using a pre-bound blocking strand to reverse the sense of action of the molecular beacon in the catalytic core . Thus, using this technique, a single DNAzyme can implement any Boolean formula that is a conjunction of one, two, or three literals .
It is important to note that we can vary the sequences of substrate binding arms (and of the corresponding substrate), which allows us to produce DNAzymes that cleave different substrates, so we can simultaneously monitor the outputs of different DNAzymes using different fluorophores . Similarly, we can vary the sequences of the inputs (and of the corresponding input-binding loops) without affecting their behavior: this allows us to replicate our logic gate motifs to compute the same logic function for different sets of inputs. The only proviso here is that the chosen sequences must form the correct structures when they are prepared in the test tube, and that there is no unintended cross-reactivity between the sequences. This is a general challenge in the field of DNA nanotechnology, and there are mature algorithms and software packages available to design nucleotide sequences that will fold to produce the desired structures [25–27, 125, 135, 136].
Furthermore, we can implement or gates implicitly by creating multiple gates with different input patterns that cleave the same substrate . Since the substrate will be cleaved when any of the corresponding gates are activated, this gives an implicit or connection between the gates. We can exploit these properties to construct systems of large numbers of DNAzyme-based logic gates that operate in parallel arrays connected by implicit or connections, and we can predict the behavior of the ensemble compositionally in terms of the behavior of the original logic gates.
Thus, we can implement any Boolean formula that can be converted into disjunctive normal form (DNF) such that each clause contains at most three literals . This abstraction of biochemical interactions as logic functions is a powerful organizing motif that has since been adopted by many other groups working on solution-phase molecular computation using other molecular computing frameworks [49, 71, 139].
Having developed a collection of elementary logic gates using DNAzymes , we used them to assemble some straightforward demonstration systems. An early circuit that we constructed was a half-adder [106], which took two bits as inputs and produced an output bit for the sum and an output bit for the carry. We used two oligonucleotide inputs \(\text {i}_{1}\) and \(\text {i}_{2}\) to represent the two input bits: the presence of \(\text {i}_{1}\) denoted a value of 1 for the first input bit and its absence denoted a value of 0 the first input bit. Similarly, presence or absence of \(\text {i}_{2}\) encoded values of 1 or 0 for the second input bit, respectively. The output bits were reported via different substrates, cleaved by DNAzymes with different substrate binding arm sequences, which were labeled with fluorescent molecules that emitted different colored light, which we could monitor simultaneously. The collection of logic gates that implement the half adder is presented in Fig. 1.3a: the value of the sum bit is the xor of the two input bits, which we implemented using two parallel andnot gates with opposite inputs (\({\textsc {sum}}=({\text {i}_{1}\mathbin {\wedge }\lnot \text {i}_{2}})\mathbin {\vee }({\text {i}_{2}\mathbin {\wedge }\lnot \text {i}_{1}})\)), and the carry bit was generated by a straightforward and gate (\({\textsc {carry}}=\text {i}_{1}\mathbin {\wedge }\text {i}_{2}\)).
DNAzyme binary adder circuits. a A half adder circuit can be constructed using three parallel DNAzyme logic gates . The sum bit is computed by an \(\text {i}_{1}\) andnot \(\text {i}_{2}\) gate and a \(\text {i}_{2}\) andnot \(\text {i}_{1}\) gate which, together, compute the xor of the two inputs. These gates cleave one output substrate, labeled with fluorophore \({{\mathbf {\mathsf{{{F}}}}}}_{{{\mathbf {\mathsf{{{1}}}}}}}\) and the corresponding quencher \({{\mathbf {\mathsf{{{Q}}}}}}_{{\mathbf {\mathsf{{{{1}}}}}}}\). The carry bit is computed by a single \(\text {i}_{1}\) and \(\text {i}_{2}\) gate , which cleaves another output substrate, labeled with a different fluorophore \({{\mathbf {\mathsf{{{F}}}}}}_{{{\mathbf {\mathsf{{{2}}}}}}}\) that emits in a different part of the electromagnetic spectrum from \({{\mathbf {\mathsf{{{F}}}}}}_{{{\mathbf {\mathsf{{{1}}}}}}}\), along with the corresponding quencher \({{\mathbf {\mathsf{{{Q}}}}}}_{{\mathbf {\mathsf{{{{2}}}}}}}\). b The full adder circuit extends the half adder design to additionally process the carry input \(\text {i}_{3}\)
We subsequently developed a larger system that implements a full adder [58], which extends the half adder with an extra carry input bit \(\text {i}_{3}\), such that
The logic gates that make up the full adder system are presented in Fig. 1.3b. Since the sum output bit depends on the two input bits as well as the carry-in bit, this system makes full use of the techniques from Fig. 1.2 for implementing arbitrary three-input logic gates. With further progress on circuit designs that enable information transmission in DNAzyme cascades (see Sect. 1.4), multiple full-adder units could be chained together with the carry output of one serving as the carry input of the next, producing a molecular system that can add larger binary numbers.
As we shall see below, there are alternatives to using molecular beacons for controlling DNAzyme catalysis. Particularly noteworthy is work on multi-component DNAzymes [10, 30–32, 46, 51, 52, 70, 76, 121] in which the DNAzyme strand is split within the catalytic core to produce two parts that, individually, possess no catalytic activity. Assembly of a catalytically active DNAzyme from these parts is controlled by inputs that can implement logic functions. This idea has also been generalized to self-assembling structures with more than two components [53, 54, 118].
1.3 Towards Wide Circuits Via Parallel Gate Arrays
To move towards implementing larger-scale systems, we decided initially to expand our circuits by adding additional gates with implicit or connections via shared substrate molecules. This can be viewed as increasing the “width” of the circuit, as opposed to the “depth”, i.e., there is no sequential information transfer (or cascading) between DNAzyme gates. Specifically, we worked on developing large-scale arrays of molecular logic gates that function as molecular automata, playing games of strategy against human opponents. In this context, a key aspect of circuit design is to render game strategies as Boolean formulae suitable for implementation using the available molecular logic gates [102]. As the resulting formulae tend to be large and complex, the challenge designing these automata was a good test of the extent to which molecular logic arrays may be engineered to implement large formulae.
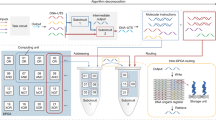
MAYA-I, an automaton that plays a symmetry-pruned game of tic-tac-toe. a Distribution of logic gates in wells. The center well (5) contains a DNAzyme without any logic gate attachments, while the other wells contain logic gates . Gates used in the example game from b are boxed. b Example gameplay for a game in which the human does not play perfectly and therefore loses
We have built three generations of game-playing automata , MAYA-I–III (originally standing for “Molecular Array of yes and and gates”). In all three, the human interacts with the automaton by adding input oligonucleotides corresponding to the next human move, and these stimuli cause the logic gates comprising the automaton to change state. These state changes record the history of moves and enable the automaton to signal its move by activating certain DNAzymes to produce a fluorescent response . The original MAYA-I automaton [107] played a symmetry-pruned game of tic-tac-toe (Fig. 1.4), and the subsequent MAYA-II automaton [66] played an unrestricted version of tic-tac-toe using a richer encoding of inputs (not shown). MAYA-III [82] could be trained to play specific strategies in a specially designed simple game, though we will not discuss MAYA-III in detail here.
In MAYA-I, the tic-tac-toe board is represented by 9 wells in a \(3 \times 3\) section of a well plate, which we number 1–9 (Fig. 1.4). We assume that the automaton moves first and always claims the center well (well 5), and that the human’s first move is either well 1 (if claiming a corner) or well 4 (if claiming a side). This symmetry pruning restricts play to just 19 legal games, which made it feasible to exhaustively test the automaton [107]. The automaton is programmed with an optimal strategy , so that the automaton will win 18 of the 19 possible games, with the human earning a draw only by playing perfectly.
The board is prepared by adding the requisite DNAzyme logic gates (Fig. 1.4a) to each of the 9 wells: there are 23 logic gates in total. The game is initiated by adding the required \(\text {Mg}^{2+}\) ions to all wells; since well 5 is the only well containing a non-logic-gated DNAzyme , that DNAzyme immediately activates and produces a fluorescent signal indicating that MAYA has claimed well 5. Human moves are represented by eight input oligonucleotides , corresponding to the 8 remaining wells, which are added to all wells to signal the next human move. Thus, each well records all moves made by the human, and after each human move each well independently computes whether it will be the next well claimed by the automaton. This is possible because the automaton’s strategy is fixed, and the automaton’s program is such that a single well will activate in response to each human move. An example of a gameplay sequence for the MAYA-I system is presented in Fig. 1.4b, and the Boolean logic functions computed by the logic gates in each of the nine wells are shown in Fig. 1.5.
Our follow-up work on the MAYA-II automaton removed the restrictions on the moves available to the human player, so that the initial human move could claim any of the 8 peripheral wells. This increased the number of legal games from 19 for MAYA-I to 76 for MAYA-II. We also increased the number of inputs from 8 to 32, encoding not just the well number but also the order of selection of that well in the game sequence. Thus, the number of logic gates implementing the game strategy increased from 23 for MAYA-I to 96 for MAYA-II. We introduced a second set of logic gates that respond to human moves by fluorescing in a second color, using 32 yes gates (4 in each of the 8 peripheral wells). Aside from these changes, the basic mechanism of game playing is the same as for MAYA-I, so we do not discuss it further here.
From an information processing perspective, MAYA-II is not significantly more capable than MAYA-I. However, the main advance in the development of MAYA-II was the engineering feat of scaling the system up from 23 gates in MAYA-I to a total of 128 gates in MAYA-II. In scaling up the system we learned that individual DNAzymes that work perfectly in isolation may fail due to unwanted interference when placed in a parallel or-gate array with other DNAzymes. Predicting such cross-reactivity and designing to avoid it is a major challenge in the implementation of molecular computing systems, as discussed in Sect. 1.2. In the context of a molecular automaton such as MAYA-I or MAYA-II, however, each gate must be designed not just against the other gates present in the same well, but also against the constraints imposed by shared sequences that may appear in other wells, such as input binding loops and substrate binding arms . Recent work on computational optimization of nucleic acid structures [25, 125, 135, 136] may aid future work in this direction, although it seems likely that designing the large number of gates present in the MAYA-II system would remain challenging even with the assistance of such computational tools.
The MAYA-I and MAYA-II automata were hardwired to play a fixed strategy in the game of tic-tac-toe . In contrast, for the MAYA-III automaton [82] we started with a blank slate and invented a simple retributive game, which we called tit-for-tat , which is played on a \(2 \times 2\) board, and for which there are 81 different winning strategies. Our goal was to demonstrate a molecular computing system that could be “trained by example” to play a particular strategy in the tit-for-tat game . We will not go into the details of the tit-for-tat game and of the MAYA-III implementation that plays it, except to say that the system consisted of 4 yes gates and 12 and gates implementing the game logic, that were each augmented with an additional input binding loop that detects the training inputs. This allows a non-expert user to select the automaton’s strategy in a training phase that mimicks the real gameplay but using the training inputs instead of the gameplay inputs, which “teaches” the system by only activating the required gates in individual wells of the automaton to enable the correct responses for the intended strategy. From a computational standpoint, the training inputs are simply an instance of “staging” the inputs to the system, although in the MAYA-III design the training inputs were designed with an additional overhanging toehold, making it possible to retrain a trained system (before it has been used for gameplay) by removing the training inputs via strand displacement . This capability relies on the fact that the input-binding loops will refold into their “closed” hairpin conformation once the training input has been stripped away.
1.4 Towards Deep Circuits Via Signaling Cascades
Our experience developing the MAYA series of molecular automata was valuable in that we learned that engineering large-scale assemblies of parallel DNAzyme logic gates is possible, although sometimes challenging. A particular limitation of the MAYA approach is that parallel arrays of the available molecular logic gates cannot implement all possible Boolean formulae: any formula whose DNF representation contains a clause with four or more literals cannot be implemented using our available logic gates using a parallel or-gate array. This is a limitation of our DNAzyme-based framework, as we only had three available locations for functionalization of the DNAzyme with input detection modules. For formulae that are not in DNF, functional completeness [124] can only be achieved by multi-layer circuits, which require arbitrarily many layers to implement arbitrary formulae, in general. The natural remedy for this limitation is to develop methods to connect DNAzyme logic gates via signal propagation cascades , which would enable us to connect DNAzymes into “deep circuits” so that we could, at least in principle, construct a circuit to implement any Boolean logic formula .
Contemporaneously with the work described in Sect. 1.3, we carried out some initial investigations into connecting DNAzymes into signaling cascades and multi-layer logic circuits. We initially developed a two-layer logic cascade in which an upstream ligase DNAzyme (which joins two short substrates into a longer product) activated a downstream phosphodiesterase (substrate cleaving) DNAzyme [105]. This demonstrated signal transmission but precluded building circuits deeper than two levels, as a different kind of DNAzyme was used in the two levels. We also explored the construction of networked DNAzymes attached to microspheres [132], whereby substrate molecules were cleaved from a microsphere once the nearby DNAzyme logic gates were activated, allowing the substrates to diffuse to another microsphere and serve as inputs for the DNAzyme logic gates attached to the second microsphere , thereby achieving signal transmission. This provided a more promising framework for the implementation of deep circuits , as well as offering the opportunity for sequence reuse on different microspheres ; however, signal attenuation was significant because the DNAzymes and their substrates were both attached to fixed points on the microsphere , reducing the amount of turnover by activated DNAzymes and hence limiting the potential for scaling up the system.
Thus, we looked for an alternative approach to implement robust, scalable mechanisms for signal transmission in deep circuits of DNAzyme logic gates . We began from the observation that, if we are to implement signal transmission from one substrate-cleaving DNAzyme to another, then the substrate cleavage reaction must enable some downstream reaction involving the cleavage products that was not possible with the uncleaved substrate molecule. Since the cleavage products will both be sub-sequences of the longer uncleaved substrate, this is a non-trivial engineering problem. We addressed this problem by developing substrate molecules that were structured , as opposed to the linear, unstructured substrates used in our prior work on DNAzyme logic gates (Sect. 1.2). Thus, the full sequence of the downstream activator strand is present in the uncleaved substrate structure but is folded up in the structure and is therefore prevented from reacting with any other DNAzymes in the system.
Design of structured chimeric substrates for deep DNAzyme circuits . a The design of our structured chimeric substrate (SCS) molecule consists of an inner stem and loop and an outer stem and loop, and an external toehold . The inner stem and loop sequester the downstream activator sequence, and the outer stem and loop comprise the upstream DNAzyme binding and cleavage sites. The outer loop also contains the toehold that will enable the downstream activator to bind to the downstream DNAzyme . We write \(\mathsf {core}'\) for a sequence consisting of part of the catalytic core sequence of the 8-17 DNAzyme , rather than the full core sequence. b The mechanism of the SCS cleavage reaction . Binding of the upstream 8-17 DNAzyme to the SCS initially displaces the outer stem (reaction 1), opening the outer loop so that the upstream DNAzyme can bind to that part of the SCS (reaction 2). This positions the catalytic core of the DNAzyme correctly with respect to the SCS cleavage site, so that the DNAzyme can cleave the SCS (reaction 3). Unbinding of the DNAzyme from the cleaved SCS recycles the DNAzyme and produces a short waste strand and a downstream activator (reaction 4). This activator is structurally weaker than the SCS because it only contains a single stem, and can therefore interconvert into a linearized form. c The linearized activator strand interacts with a downstream DNAzyme that has been inhibited by hybridization with a partially complementary inhibitor strand. The activator binds to a complementary toehold on the downstream inhibitor strand and initiates a strand displacement reaction that displaces an active downstream DNAzyme and produces an inert waste complex. The displaced DNAzyme can then cleave its own substrate, which may be a fluorescently-labeled linear readout substrate (as shown here) or another SCS molecule that enables further signal propagation
The design of structured DNAzyme substrates is technically challenging because the structure must balance pre-cleavage stability (to prevent undesired activation without cleavage of the substrate) with post-cleavage instability (to promote rapid activation when the substrate has been cleaved). We worked on a number of potential designs for these structured substrates , which we summarized previously [55], before settling on a design for a substrate that provided a workable compromise between these concerns [15]. Our structured chimeric substrate (SCS) design is summarized in Fig. 1.6a, and consists of a dual stem-loop design. The inner stem and loop sequester the sequence that will activate the downstream DNAzyme , and the outer stem and loop contain the binding and cleavage sites for the upstream DNAzyme . We found that the enhanced stability conferred by the combination of the two stems was vital to maintain stability of the SCS structure prior to cleavage.
The mechanism by which the upstream DNAzyme binds and cleaves the structured substrate is illustrated in Fig. 1.6b. The upstream DNAzyme binds to the SCS via the external toehold and initiates a strand displacement reaction with one of its substrate binding arms , thereby opening the outer stem of the SCS (reaction 1). The second arm of the upstream DNAzyme can then bind to the outer loop and position the SCS cleavage site correctly with respect to the catalytic core of the upstream DNAzyme (reaction 2). Following cleavage of the SCS strand (reaction 3), the upstream DNAzyme unbinds (reaction 4), leaving a short waste strand along with the remainder of the SCS strand, minus the outer stem, which we call the activator strand. The structure of the activator is significantly weaker than that of the uncleaved SCS , therefore, the downstream effector sequence contained within the activator structure is made available to interact with downstream DNAzyme gates. Thus the catalytic activity of the upstream DNAzyme causes a covalent modification to the SCS molecule which alters its structure, causing signal propagation to a downstream logic gate by release of the effector sequence.
We used the SCS design from Fig. 1.6 to implement several “deep” circuits that were beyond the capabilities of our previous framework of parallel logic gates (Sect. 1.3). In this context we used a different design for DNAzyme logic gates that were inhibited via direct hybridization with an inhibitor strand and activated via toehold-mediated strand displacement [16], as shown in Fig. 1.6c. The practical reason for this was that using toehold-mediated strand displacement allows us to control the binding pathway in the activation reaction: this information was helpful in determining which parts of the activator sequence to protect, and how, when designing the SCS molecules [55]. We also based these logic gates on the \(\text {Zn}^{2+}\)-dependent “8–17” DNAzyme motif [93], which is more compact than E6 and has a higher catalytic rate.
Multi-layer DNAzyme signaling cascades . a Signaling cascade reactions. In each layer of the cascade, an active DNAzyme cleaves the corresponding SCS , producing an activator that activates the downstream DNAzyme via a strand displacement reaction, thereby propagating the activating signal to the next layer of the cascade. In the final layer (layer 1), the activated DNAzyme cleaves a linear fluorescent reporter substrate to generate an output signal. b Experimental data from DNAzyme signaling cascades. The mean fluorescence signal (solid lines) from multi-layer DNAzyme signaling cascades with equal concentrations (100 nM) of each DNAzyme from each layer. The dashed lines represent the same reaction without the top-layer active DNAzyme, which measures the non-specific activation (leakage) of the cascade. c Further experimental data from multi-layer DNAzyme signaling cascades with increasing DNAzyme concentrations in each layer (25 nM in layer 4, 50 nM in layer 3, 75 nM in layer 2, and 100 nM in layer 1), to demonstrate signal amplification. In both b and c, the dotted linesrepresent the 95 % confidence interval from three replicate experiments
As a proof-of-concept for depthwise scaling of DNAzyme circuits using our SCS design, we implemented multi-stage linear signaling cascades up to five layers deep, as outlined in Fig. 1.7a. The cascade consists of a series of inactive DNAzymes , which are activated in turn by a cleaved SCS in a strand displacement reaction, and proceed to cleave their own SCS molecule to activate the DNAzyme in the next layer. This system was inspired by protein signaling cascades such as the MAP kinase phosphorylation cascades [85, 95]. Experimental results from this system for two-, three-, four-, and five-layer variants of the cascade are shown in Fig. 1.7b, c. This cascade comprised only strand-displacement “yes” gates : we have also used the SCS approach to connect DNAzymes controlled via molecular beacons , such as those from Sect. 1.2—see the Supporting Information from [15] for details. This work demonstrates that multi-layer DNAzyme networks may be implemented provided that the information transmission interfaces between DNAzymes are designed carefully, and opens the possibility of scaling up the parallel or-gate arrays discussed above by connecting the logic gates in multi-layer circuits.
Multi-layer diagnostic logic circuits for the detection of sequences from the genomes of all four dengue serotypes . a Circuit design template. The diagnostic circuit for serotype DEN-k (\(k\in \{{1,2,3,4}\}\)) requires the presence of two conserved sequences from the dengue genomes (which we call “DengueA” and “DengueB”) and one sequence specific to the serotype of interest (which we call “DEN-k”). The circuit consists of two DNAzyme-based AND gates , whose inhibitors contain mismatched bases to promote rapid activation [16]. When both upstream inputs are present, the upstream DNAzyme is displaced by a cooperative strand displacement reaction involving both input strands simultaneously [138]. The active upstream DNAzyme cleaves an SCS molecule, producing an activator that serves as one input to the downstream logic gate. If the second conserved dengue sequence is also present, it serves as the second input to the downstream logic gate. When the downstream DNAzyme is activated, it generates a fluorescent output signal as before. Thus, all three inputs must be present to produce an output, which would increase confidence in the diagnosis in a practical application. We derived detection circuits for all four dengue serotypes (DEN-\(1-4\)) by modifying part of the upstream logic gate and the SCS molecule. b Experimental data for all four dengue serotyping circuits, showing correct operation of all four instantiations of circuit template using all eight combinations of the two conserved sequences and the correct serotype-specific sequence. Error bars represent the 95 % confidence interval from three replicate experiments
Furthermore, to demonstrate the inclusion of logical processing into SCS circuits, we designed a two-layer, three-input and circuit using two strand displacement-based and gates that require two inputs to activate each DNAzyme [16] via a cooperative strand displacement reaction in which the two input strands simultaneously displace part of the sequestered DNAzyme strand [138], as shown in Fig. 1.8a. As a demonstration of our systems’ potential applicability for virus detection , we used the circuit template from Fig. 1.8a to implement four logic circuits to detect and distinguish all four serotypes of dengue virus [9, 100]. We chose four target sequences unique to the four serotypes and two generic sequences conserved across the four serotypes , and designed four three-input and systems that require both generic inputs and a particular serotype-specific input to be present before the fluorescent output is triggered. Experimental results for these circuits are summarized in Fig. 1.8b, showing that the and logic functions correctly. Furthermore, each version of the circuit is only sensitive to one serotype —see the Supporting Information from [15] for details.
Our approach to depthwise scaling of DNAzyme circuits using the SCS approach was successful because of the use of the SCS as an intermediary between the communicating DNAzymes . This removed the need for direct interaction between the DNAzymes , which allowed us to standardize the SCS design and enabled simpler scaling of the circuit. We believe that this approach could be deployed to implement a range of interesting dynamical behaviors such as DNAzyme feedback systems. An interesting direction for future work would be to engineer inhibitory connections between DNAzymes, which would permit the implementation of more kinds of circuit derived from gene networks explored in systems biology [7], such as DNAzyme oscillators [33, 38].
We are gratified that other groups have also begun to explore DNAzyme-based computation cascades . For instance, the Kolpashchikov group has published [35] a design similar to our structured substrate molecule. In that work, cleavage of the structured substrate directly released a downstream DNAzyme that generates an output signal via a color change in the solution, which limits the possibilities for scaling up to larger systems. Additionally, the Willner group has developed logic cascades based on DNAzymes which used a two-strand substrate structure [30]: see [121] and citations therein. Finally, other workers have explored DNAzyme cascades in which each DNAzyme directly activates the next by cleaving away an inhibitor strand that was holding the DNAzyme in an inactive, quasi-circular conformation [10], and cross-catalytic amplification cycles in which circularized DNAzymes are linearized by the cleavage reaction [61].
1.5 Towards Applications in Biodetection
In the previous section, we described a two-layer, three-input and circuit that detected synthetic oligonucleotide targets with sequences corresponding to segments of the four dengue virus genomes. In this section, we briefly review recent work, by ourselves and others, on moving DNAzyme-based logic systems toward practical biodetection applications.
Modular, DNAzyme-based biosensor design and experimental data. a Our biosensor design consists of orthogonal “detection” and “reporter” modules. DNA detection targets, such as single-stranded oligonucleotides or denatured double-stranded DNA , bind to the detection module by toehold-mediated strand displacement and, in doing so, expose the secondary toehold in the reporter module. In both cases, this allows the fuel strand to bind and complete displacement of the DNAzyme strand from the inhibitor , which can then fold into a catalytically active conformation and generate an amplified fluorescent output by cleaving fluorescently-labeled substrates . b and c Detection of genes on denatured plasmid DNA extracted from bacteria . As a demonstration, we designed five biosensors using a common reporter module but varying the detection module, to detect five genetic sequences from plasmids encoding GFP-fusion protein variants: a commercially available Emerald GFP plasmid (“emGFP”) and a Pinpoint Xa plasmid containing a SNAP25-GFP fusion protein [94] (“SNAP25”). Three biosensors targeted genetic sequences common to both plasmids (which we called C1–3), one targeted a sequence specific to emGFP (which we called E), and the final biosensor targeted a sequence specific to SNAP25 (which we called S). Results from detecting the emGFP and SNAP25 plasmids using the five biosensors individually are shown in b and c, respectively. In b we observe a strong response from C1–3 and E, and a weak response from S, as expected. Similarly, in c we observe a strong response from C1–3 and S, and a weak response from E. Thus, our biosensors are specific to their detection target sequences and can be used to detect bacterial DNA in realistic assay scenarios
We used as a basis for our biodetection work the strand displacement-based DNAzyme yes gates described in Sect. 1.4. We extended this gate design by incorporating a separate “input binding” module that, when activated, releases a secondary toehold from a loop so that a secondary “fuel” strand can complete the displacement of an active DNAzyme from the complex, as shown in Fig. 1.9a. This two-step process allowed us to retain the strand displacement mechanism of activation while separating the input sequence from that of the DNAzyme itself, so that each may be changed independently of the other. We used this gate design to demonstrate detection of oligonucleotides and of small molecules (such as ATP) via aptamer binding, whereby the small molecule target binds to a particular DNA subsequence that is known to have high binding affinity for the target molecule. We also used this sensor platform to detect genes on DNA extracted from bacteria [14]. Some results from the detection of genetic elements on bacterial DNA are shown in Fig. 1.9b, c, and demonstrate sequence specificity as well as the possibility of detecting a technically challenging double-stranded target . This is a realistic model biodetection assay: most viable protocols for pathogen detection will include similar sample preparation steps.
The use of DNAzymes for biodetection and other analytical chemistry applications has also been explored extensively by other groups, as the innate catalytic activities of DNAzymes make them useful for implementing both detection and output functionalities. The Willner group has published a range of papers on analytical applications of DNAzymes and DNAzyme logic [31, 64, 112, 118–120], as have the Lu group [44, 57, 60, 63, 123, 126–128, 130, 141], the Li group [2–6, 40, 68, 113, 114], the Kolpashchikov group [35, 36, 51, 52], and the Liu group [41–43]. A good example of the output capabilities of DNAzymes is the peroxidase-mimicking DNAzyme which, when activated, turns the solution from clear to colorless. This DNAzyme has been used to produce a visual output from several molecular logic systems [28, 35]. In addition, one of us (D.S.) has worked on applying the molecular beacon-based DNAzyme motifs from Sect. 1.2 to the problem of virus detection [86], demonstrating the use of well-plates as fluorescent seven-segment displays for multiplexed detection of oligonucleotide inputs corresponding to representative viral sequences.
1.6 Conclusions
In conclusion, we have reviewed almost fifteen years of work in our laboratories on the development of DNAzyme-based logic systems. Our early research on solution-phase molecular logic systems was among the first work in this direction, and this approach to molecular logic has now become broadly accepted in the community. That said, recent work [73, 90, 111] hints at a move towards implementing entire molecular computing systems on a single surface . This line of research aims to combine the advantages of autonomous molecular logic, as described here, with the additional benefits conferred by confinement to a surface, such as the possibility for direct sequence reuse in different parts of the circuit. Our work on large-scale molecular automata , which we characterize here as a move towards “wide” circuits of gates operating in parallel, demonstrated that molecular systems can implement non-trivial interactive systems, and we are hopeful that there will be future incarnations of the MAYA series of automata, subject to the difficulty of rendering game strategies as Boolean formulae [102].
Our subsequent work on DNAzyme cascades and multi-layer logic circuits, which we characterize here as a move towards “deep” circuits, illustrates the challenges involved in connecting substrate-cleaving DNAzymes into sequential logic circuits. Inspired by protein cascades , our chosen mechanism relied on the secondary structure of the uncleaved substrate molecule to sequester the downstream activator sequence prior to cleavage. However, there may be other potential approaches: in particular, our previous work on DNAzyme ligase logic gates [105] suggests a possible way forward, in which DNAzyme processing units assemble activators (or inhibitors ) from short, inactive strands to regulate other DNAzymes. The difficulty with this approach is that turnover in ligase systems is severely restricted by the stability of the bond between the DNAzyme and the ligated product strand. These issues notwithstanding, the sheer variety of chemical reactions that can be catalyzed by DNAzymes [11, 17, 18, 34, 39, 59, 72, 78, 87, 98, 99, 122, 131] offers intriguing possibilities for DNAzyme information processing units as components of a “synthetic metabolism” in artificial living systems [109, 129]; here we may draw inspiration from fascinating work on self-replicating ribozymes [45, 48, 56, 62, 75, 79, 80, 116, 117, 137]. Ribozymes are RNA enzymes, which are similar to DNAzymes but occur naturally, and which may have formed a key component of prebiotic “life”.
Our work, and the work of others, on DNAzyme biosensors highlights a key practical advantage of DNAzyme technology: despite the wide array of reactions that DNAzymes catalyze, they are still just short, single strands of DNA and are therefore relatively simple to design, very cheap to synthesize, and robust to degradation in the laboratory. In particular, the efficient RNA-cleaving activity of many DNAzymes makes them attractive for use in gene silencing applications, as they can cleave the RNA molecules that serve as an intermediary in the gene expression process, thereby rendering them inoperable. Preliminary work has demonstrated that DNAzymes can function in the intracellular chemical environment [46]: potential therapeutic applications of DNAzymes include cancer therapy [21–24, 29, 50, 69] and antiviral applications [84]. This work offers a direct route to the development of logic-based molecular therapeutics that perform non-trivial information processing based on the observed intracellular chemical environment before making an informed decision about whether to activate its therapeutic gene-silencing DNAzyme payload. This offers the potential for highly targeted therapy, so that well-defined cell types (such as tumor cells) may be eliminated with minimal side effects. Thus, there is great potential for future research in DNAzyme-based computing systems in a wide range of fields, from logic and artificial life to biomedical diagnostics and therapeutics .
References
Adleman, L.M.: Molecular computation of solutions to combinatorial problems. Science 266, 1021–1024 (1994)
Aguirre, S.D., Ali, M.M., Kanda, P., Li, Y.: Detection of bacteria using fluorogenic DNAzymes. J. Vis. Exp. 63, e3961 (2012). doi:10.3791/3961
Aguirre, S.D., Ali, M.M., Salena, B.J., Li, Y.: A sensitive DNA enzyme-based fluorescent assay for bacterial detection. Biomolecules 3, 563–577 (2013). doi:10.3390/biom3030563
Ali, M.M., Aguirre, S.D., Lazim, H., Li, Y.: Fluorogenic DNAzyme probes as bacterial indicators. Angew. Chem. Int. Ed. 50, 3751–3754 (2011)
Ali, M.M., Aguirre, S.D., Mok, W.W.K., Li, Y.: Developing fluorogenic RNA-cleaving DNAzymes for biosensing applications. In: Hartig, J.S., (ed.) Ribozymes, Methods in Molecular Biology, vol. 848, pp. 395–418. Springer, New York (2012)
Ali, M.M., Li, Y.: Colorimetric sensing using allosteric DNAzyme-coupled rolling circle amplification and a peptide nucleic acid-organic dye probe. Angew. Chem. Int. Ed. 48, 3512–3515 (2009)
Alon, U.: An Introduction to Systems Biology: Design Principles of Biological Circuits. Chapman & Hall/CRC, Boca Raton (2007)
Bennett, C.H.: The thermodynamics of computation–a review. Int. J. Theor. Phys. 21(12), 905–940 (1982)
Bhatt, S., Gething, P.W., Brady, O.J., Messina, J.P., Farlow, A.W., Moyes, C.L., Drake, J.M., Brownstein, J.S., Hoen, A.G., Sankoh, O., Myers, M.F., George, D.B., Jaenisch, T., Wint, G.R.W., Simmons, C.P., Scott, T.W., Farrar, J.J., Hay, S.I.: The global distribution and burden of dengue. Nature 496, 504–507 (2013). doi:10.1038/nature12060
Bone, S.M., Hasick, N.J., Lima, N.E., Erskine, S.M., Mokany, E., Todd, A.V.: DNA-only cascade: A universal tool for signal amplification, enhancing the detection of target analytes. Anal. Chem. 86(18), 9106–9113 (2014). doi:10.1021/ac501811r
Brandsen, B.M., Velez, T.E., Sachdeva, A., Ibrahim, N.A., Silverman, S.K.: DNA-catalyzed lysine side chain modification. Angew. Chem. Int. Ed. 53(34), 9045–9050 (2014). doi:10.1002/anie.201404622
Breaker, R.R.: In vitro selection of catalytic polynucleotides. Chem. Rev. 97(2), 371–390 (1997)
Breaker, R.R., Joyce, G.F.: A DNA enzyme with Mg\({}^{2+}\)-dependent RNA phosphoesterase activity. Chem. Biol. 2(10), 655–656 (1995)
Brown III, C.W., Lakin, M.R., Fabry-Wood, A., Horwitz, E.K., Baker, N.A., Stefanovic, D., Graves, S.W.: A unified sensor architecture for isothermal detection of double-stranded DNA, oligonucleotides, and small molecules. ChemBioChem 16, 725–730 (2015). doi:10.1002/cbic.201402615
Brown III, C.W., Lakin, M.R., Horwitz, E.K., Fanning, M.L., West, H.E., Stefanovic, D., Graves, S.W.: Signal propagation in multi-layer DNAzyme cascades using structured chimeric substrates. Angew. Chem. Int. Ed. 53(28), 7183–7187 (2014). doi:10.1002/anie.201402691
Brown III, C.W., Lakin, M.R., Stefanovic, D., Graves, S.W.: Catalytic molecular logic devices by DNAzyme displacement. ChemBioChem 15, 950–954 (2014). doi:10.1002/cbic.201400047
Chandra, M., Sachdeva, A., Silverman, S.K.: DNA-catalyzed sequence-specific hydrolysis of DNA. Nat. Chem. Biol. 5(10), 718–720 (2009)
Chandrasekar, J., Silverman, S.K.: Catalytic DNA with phosphatase activity. Proc. Natl. Acad. Sci. U.S. Am. 110(14), 5315–5320 (2013). doi:10.1073/pnas.1221946110
Chen, Y.J., Dalchau, N., Srinivas, N., Phillips, A., Cardelli, L., Soloveichik, D., Seelig, G.: Programmable chemical controllers made from DNA. Nat. Nanotechnol. 8, 755–762 (2013). doi:10.1038/nnano.2013.189
Credi, A.: Molecules that make decisions. Angew. Chem. Int. Ed. 46(29), 5472–5475 (2007)
Dass, C.R.: Deoxyribozymes: cleaving a path to clinical trials. Trend. Pharmacol. Sci. 25(8), 395–397 (2004)
Dass, C.R., Choong, P.F., Khachigian, L.M.: DNAzyme technology and cancer therapy: cleave and let die. Mol. Cancer Ther. 7(2), 243–251 (2008). doi:10.1158/1535-7163.MCT-07-0510
Dass, C.R., Galloway, S.J., Choong, P.F.: Dz13, a c-jun DNAzyme, is a potent inducer of caspase-2 activation. Oligonucleotides 20(3), 137–146 (2010). doi:10.1089/oli.2009.0226
Dass, C.R., Saravolac, E.G., Li, Y., Sun, L.Q.: Cellular uptake, distribution, and stability of 10–23 deoxyribozymes. Antisense Nucl. Acid Drug Dev. 12, 289–299 (2002)
Dirks, R.M., Bois, J.S., Schaeffer, J.M., Winfree, E., Pierce, N.A.: Thermodynamic analysis of interacting nucleic acid strands. SIAM Rev. 49, 65–88 (2007)
Dirks, R.M., Pierce, N.A.: A partition function algorithm for nucleic acid secondary structure including pseudoknots. J. Comput. Chem. 24, 1664–1677 (2003)
Dirks, R.M., Pierce, N.A.: An algorithm for computing nucleic acid base-pairing probabilities including pseudoknots. J. Comput. Chem. 25, 1295–1304 (2004)
Eckhoff, G., Codrea, V., Ellington, A.D., Chen, X.: Beyond allostery: catalytic regulation of a deoxyribozyme through an entropy-driven DNA amplifier. J. Syst. Chem. 1, 13 (2010). doi:10.1186/1759-2208-1-13
Elahy, M., Dass, C.R.: Dz13: c-jun downregulation and tumour cell death. Chem. Biol. Drug Des. 78, 909–912 (2011). doi:10.1111/j.1747-0285.2011.01166.x
Elbaz, J., Lioubashevski, O., Wang, F., Remacle, F., Levine, R.D., Willner, I.: DNA computing circuits using libraries of DNAzyme subunits. Nat. Nanotechnol. 5(6), 417–422 (2010)
Elbaz, J., Moshe, M., Shlyahovsky, B., Willner, I.: Cooperative multicomponent self-assembly of nucleic acid structures for the activation of DNAzyme cascades: A paradigm for DNA sensors and aptasensors. Chemistry–A. Eur. J. 15, 3411–3418 (2009)
Elbaz, J., Wang, F., Remacle, F., Willner, I.: PH-programmable DNA logic arrays powered by modular DNAzyme libraries. Nano Lett. 12, 6049–6054 (2012). doi:10.1021/nl300051g
Farfel, J., Stefanovic, D.: Towards practical biomolecular computers using microfluidic deoxyribozyme logic gate networks. In: Carbone, A., Pierce, N.A. (eds.) Proceedings of the 11th International Meeting on DNA Computing, Lecture Notes in Computer Science, vol. 3892, pp. 38–54. Springer, Heidelberg (2006)
Flynn-Charlebois, A., Wang, Y., Prior, T.K., Rashid, I., Hoadley, K.A., Coppins, R.L., Wolf, A.C., Silverman, S.K.: Deoxyribozymes with 2’-5’ RNA ligase activity. J. Am. Chem. Soc. 125, 2444–2454 (2003). doi:10.1021/ja028774y
Gerasimova, Y.V., Cornett, E.M., Edwards, E., Su, X., Rohde, K.H., Kolpashchikov, D.M.: Deoxyribozyme cascade for visual detection of bacterial RNA. ChemBioChem 14, 2087–2090 (2013). doi:10.1002/cbic.201300471
Gerasimova, Y.V., Kolpashchikov, D.M.: Folding of 16S rRNA in a signal-producing structure for the detection of bacteria. Angew. Chem. Int. Ed. 52, 10586–10588 (2013). doi:10.1002/anie.201303919
Goldman, N., Bertone, P., Chen, S., Dessimoz, C., LeProust, E.M., Sipos, B., Birney, E.: Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. Nature 494, 77–80 (2013). doi:10.1038/nature11875
Goudarzi, A., Lakin, M.R., Stefanovic, D.: DNA reservoir computing: a novel molecular computing approach. In: Soloveichik, D., Yurke, B. (eds.) Proceedings of the 19th International Conference on DNA Computing and Molecular Programming, Lecture Notes in Computer Science, vol. 8141, pp. 76–89. Springer, Heidelberg (2013). doi:10.1007/978-3-319-01928-4_6
Gu, H., Furukawa, K., Weinberg, Z., Berenson, D.F., Breaker, R.R.: Small, highly active DNAs that hydrolyze DNA. J. Am. Chem. Soc. 135, 9121–9129 (2013). doi:10.1021/ja403585e
He, S., Qu, L., Shen, Z., Tan, Y., Zeng, M., Liu, F., Jiang, Y., Li, Y.: Highly specific recognition of breast tumors by an RNA-cleaving fluorogenic DNAzyme probe. Anal. Chem. 87(1), 569–577 (2014). doi:10.1021/ac5031557
Huang, P.J.J., Liu, M., Liu, J.: Functional nucleic acids for detecting bacteria. Rev. Anal. Chem. 32(1), 77–89 (2013). doi:10.1515/revac-2012-0027
Huang, P.J.J., Vazin, M., Liu, J.: In vitro selection of a new lanthanide-dependent DNAzyme for ratiometric sensing lanthanides. Anal. Chem. 86(19), 9993–9999 (2014). doi:10.1021/ac5029962
Huang, P.J.J., Vazin, M., Matuszek, Z., Liu, J.: A new heavy lanthanide-dependent DNAzyme displaying strong metal cooperativity and unrescuable phosphorothioate effect. Nucleic Acid. Res. 43(1), 461–469 (2015). doi:10.1093/nar/gku1296
Hwang, K., Wu, P., Kim, T., Lei, L., Tian, S., Wang, Y., Lu, Y.: Photocaged DNAzymes as a general method for sensing metal ions in living cells. Angew. Chem. Int. Ed. 53, 13798–13802 (2014)
Hayden, J.E., Riley, C.A., Burton, A.S., Lehman, N.: RNA-directed construction of structurally complex and active ligase ribozymes through recombination. RNA 11(11), 1678–1687 (2005). doi:10.1261/rna.2125305
Kahan-Hanum, M., Douek, Y., Adar, R., Shapiro, E.: A library of programmable DNAzymes that operate in a cellular environment. Sci. Rep. 3, 1535 (2013)
Katz, E., Privman, V.: Enzyme-based logic systems for information processing. Chem. Soc. Rev. 39(5), 1835–1857 (2010)
Kim, D.E., Joyce, G.F.: Cross-catalytic replication of an RNA ligase ribozyme. Chem. Biol. 11, 1505–1512 (2004). doi:10.1016/j.chembiol.2004.08.021
Kim, J., Winfree, E.: Synthetic in vitro transcriptional oscillators. Mol. Syst. Biol. 7, 465 (2011). doi:10.1038/msb.2010.119
Kim, S.H., Dass, C.R.: Induction of caspase-2 activation by a DNA enzyme evokes tumor cell apoptosis. DNA Cell Biol. 31(1) (2012). doi:10.1089/dna.2011.1323
Kolpashchikov, D.M.: A binary deoxyribozyme for nucleic acid analysis. ChemBioChem 8, 2039–2042 (2007)
Kolpashchikov, D.M.: Binary probes for nucleic acid analysis. Chem. Rev. 110, 4709–4723 (2010)
Kolpashchikov, D.M., Gerasimova, Y.V., Khan, M.S.: DNA nanotechnology for nucleic acid analysis: DX motif-based sensor. ChemBioChem 12, 2564–2567 (2011)
Lake, A., Shang, S., Kolpashchikov, D.M.: Molecular logic gates connected through DNA four-way junctions. Angew. Chem. Int. Ed. 49, 4459–4462 (2010). doi:10.1002/anie.200907135
Lakin, M.R., Brown III, C.W., Horwitz, E.K., Fanning, M.L., West, H.E., Stefanovic, D., Graves, S.W.: Biophysically inspired rational design of structured chimeric substrates for DNAzyme cascade engineering. PLoS ONE 9(10), e110986 (2014). doi:10.1371/journal.pone.0110986
Lam, B.J., Joyce, G.F.: Autocatalytic aptazymes enable ligand-dependent exponential amplification of RNA. Nat. Biotechnol. 27(3), 288–292 (2009). doi:10.1038/nbt.1528
Lan, T., Furuya, K., Lu, Y.: A highly selective lead sensor based on a classic lead DNAzyme. Chem. Commun. 46, 3896–3898 (2010)
Lederman, H., Macdonald, J., Stefanovic, D., Stojanovic, M.N.: Deoxyribozyme-based three-input logic gates and construction of a molecular full adder. Biochemistry 45(4), 1194–1199 (2006)
Lee, C.S., Mui, T.P., Silverman, S.K.: Improved deoxyribozymes for synthesis of covalently branched DNA and RNA. Nucleic Acid. Res. 39(1), 269–279 (2011). doi:10.1093/nar/gkq753
Lee, J.H., Wang, Z., Liu, J., Lu, Y.: Highly sensitive and selective colorimetric sensors for uranyl (\({\rm {UO}}_{2}^{2+}\)): development and comparison of labeled and label-free DNAzyme-gold nanoparticle systems. J. Am. Chem. Soc. 130, 14217–14226 (2008)
Levy, M., Ellington, A.D.: Exponential growth by cross-catalytic cleavage of deoxyribozymogens. Proc. Natl. Acad. Sci. U.S. Am. 100(11), 6416–6421 (2003). doi:10.1073/pnas.1130145100
Lincoln, T.A., Joyce, G.F.: Self-sustained replication of an RNA enzyme. Science 323, 1229–1232 (2009)
Liu, J., Cao, Z., Lu, Y.: Functional nucleic acid sensors. Chem. Rev. 109, 1948–1998 (2009). doi:10.1021/cr030183i
Lu, C.H., Wang, F., Willner, I.: Zn\(^{2+}\)-ligation DNAzyme-driven enzymatic and nonenzymatic cascades for the amplified detection of DNA. J. Am. Chem. Soc. 134(25), 10651–10658 (2012)
Lund, K., Manzo, A.J., Dabby, N., Michelotti, N., Johnson-Buck, A., Nangreave, J., Taylor, S., Pei, R., Stojanovic, M.N., Walter, N.G., Winfree, E., Yan, H.: Molecular robots guided by prescriptive landscapes. Nature 465, 206–209 (2010)
Macdonald, J., Li, Y., Sutovic, M., Lederman, H., Pendri, K., Lu, W., Andrews, B.L., Stefanovic, D., Stojanovic, M.N.: Medium scale integration of molecular logic gates in an automaton. Nano Lett. 6(11), 2598–2603 (2006)
Macdonald, J., Stefanovic, D., Stojanovic, M.N.: DNA computers for work and play. Sci. Am. 299(5), 84–91 (2008)
McManus, S.A., Li, Y.: Turning a kinase deoxyribozyme into a sensor. J. Am. Chem. Soc. 135(19), 7181–7186 (2013)
Mitchell, A., Dass, C.R., Sun, L.Q., Khachigian, L.M.: Inhibition of human breast carcinoma proliferation, migration, chemoinvasion and solid tumour growth by DNAzymes targeting the zinc finger transcription factor EGR-1. Nucleic Acid. Res. 32(10), 3065–3069 (2004). doi:10.1093/nar/gkh626
Mokany, E., Bone, S.M., Young, P.E., Doan, T.B., Todd, A.V.: MNAzymes, a versatile new class of nucleic acid enzymes that can function as biosensors and molecular switches. J. Am. Chem. Soc. 132, 1051–1059 (2010). doi:10.1021/ja9076777
Montagne, K., Plasson, R., Sakai, Y., Fujii, T., Rondelez, Y.: Programming an in vitro DNA oscillator using a molecular networking strategy. Mol. Syst. Biol. 7, 466 (2011). doi:10.1038/msb.2011.12
Mui, T.P., Silverman, S.K.: Convergent and general one-step DNA-catalyzed synthesis of multiply branched DNA. Organ. Lett. 10(20), 4417–4420 (2008)
Muscat, R.A., Strauss, K., Ceze, L., Seelig, G.: DNA-based molecular architecture with spatially localized components. In: ISCA ’13: Proceedings of the 40th Annual International Symposium on Computer Architecture, pp. 177–188. ACM, New York (2013)
Olah, M.J., Stefanovic, D.: Superdiffusive transport by multivalent molecular walkers moving under load. Phys. Rev. E 87, 062713 (2013). doi:10.1103/PhysRevE.87.062713
Olea, Jr. C., Horning, D.P., Joyce, G.F.: Ligand-dependent exponential amplification of a self-replicating lRNA enzyme. J. Am. Chem. Soc. 134, 8050–8053 (2012). doi:10.1021/ja302197x
Orbach, R., Remacle, F., Levine, R.D., Willner, I.: DNAzyme-based 2:1 and 4:1 multiplexers and 1:2 demultiplexer. Chem. Sci. 5, 1074–1081 (2014)
Osborne, S.E., Ellington, A.D.: Nucleic acid selection and the challenge of combinatorial chemistry. Chem. Rev. 97(2), 349–370 (1997)
Parker, D.J., Xiao, Y., Aguilar, J.M., Silverman, S.K.: DNA catalysis of a normally disfavored RNA hydrolysis reaction. J. Am. Chem. Soc. 135(23), 8472–8475 (2013). doi:10.1021/ja4032488
Paul, N., Joyce, G.F.: A self-replicating ligase ribozyme. Proc. Natl. Acad. Sci. U.S. Am. 99(20), 12733–12740 (2002). doi:10.1073/pnas.202471099
Paul, N., Joyce, G.F.: Minimal self-replicating systems. Curr. Opin. Chem. Biol. 8, 634–639 (2004). doi:10.1016/j.cbpa.2004.09.005
Paul, N., Springsteen, G., Joyce, G.F.: Conversion of a ribozyme to a deoxyribozyme through in vitro evolution. Chem. Biol. 13, 329–338 (2006). doi:10.1016/j.chembiol.2006.01.007
Pei, R., Matamoros, E., Liu, M., Stefanovic, D., Stojanovic, M.N.: Training a molecular automaton to play a game. Nat. Nanotechnol. 5, 773–777 (2010)
Pei, R., Taylor, S.K., Stefanovic, D., Rudchenko, S., Mitchell, T.E., Stojanovic, M.N.: Behavior of polycatalytic assemblies in a substrate-displaying matrix. J. Am. Chem. Soc. 128(39), 12693–12699 (2006)
Peracchi, A.: Prospects for antiviral ribozymes and deoxyribozymes. Rev. Med. Virol. 14, 47–64 (2004). doi:10.1002/rmv.415
Plotnikov, A., Zehorai, E., Procaccia, S., Seger, R.: The MAPK cascades: Signaling components, nuclear roles and mechanisms of nuclear translocation. Biochimica et Biophysica Acta 1813, 1619–1633 (2011)
Poje, J.E., Kastratovic, T., Macdonald, A.R., Guillermo, A.C., Troetti, S.E., Jabado, O.J., Fanning, M.L., Stefanovic, D., Macdonald, J.: Visual displays that directly interface and provide read-outs of molecular states via molecular graphics processing units. Angew. Chem. Int. Ed. 53(35), 9222–9225 (2014)
Purtha, W.E., Coppins, R.L., Smalley, M.K., Silverman, S.K.: General deoxyribozyme-catalyzed synthesis of native 3’-5’ RNA linkages. J. Am. Chem. Soc. 127, 13124–13125 (2005). doi:10.1021/ja0533702
Qian, L., Soloveichik, D., Winfree, E.: Efficient Turing-universal computation with DNA polymers. In: Sakakibara, Y., Mi, Y. (eds.) Proceedings of the 16th International Conference on DNA Computing and Molecular Programming, Lecture Notes in Computer Science, vol. 6518, pp. 123–140. Springer, New York (2011)
Qian, L., Winfree, E.: Scaling up digital circuit computation with DNA strand displacement cascades. Science 332, 1196–1201 (2011). doi:10.1126/science.1200520
Qian, L., Winfree, E.: Parallel and scalable computation and spatial dynamics with DNA-based chemical reaction networks on a surface. In: Murata, S., Kobayashi, S. (eds.) Proceedings of the 20th International Conference on DNA Computing and Molecular Programming, Lecture Notes in Computer Science, vol. 8727, pp. 114–131. Springer, New York (2014)
Qian, L., Winfree, E., Bruck, J.: Neural network computation with DNA strand displacement cascades. Nature 475, 368–372 (2011). doi:10.1038/nature10262
Robertson, M.P., Ellington, A.: In vitro selection of an allosteric ribozyme that transduces analytes to amplicons. Nat. Biotechol. 17(1), 62–66 (1999)
Santoro, S.W., Joyce, G.F.: A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. U.S. Am. 94, 4262–4266 (1997)
Saunders, M.J., Edwards, B.S., Zhu, J., Sklar, L.A., Graves, S.W.: Microsphere-based flow cytometry protease assays for use in protease activity detection and high-throughput screening. In: Robinson, J.P. (ed.) Current Protocols in Cytometry Unit 13.12. Wiley, Hoboken (2010). doi:10.1002/0471142956.cy1312s54
Schaeffer, H.J., Weber, M.J.: Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol. Cell. Biol. 19(4), 2435–2444 (1999)
Seelig, G., Soloveichik, D., Zhang, D.Y., Winfree, E.: Enzyme-free nucleic acid logic circuits. Science 314, 1585–1588 (2006). doi:10.1126/science.1132493
Semenov, O., Olah, M.J., Stefanovic, D.: Cooperative linear cargo transport with molecular spiders. Nat. Comput. 12(2), 259–276 (2013). doi:10.1007/s11047-012-9357-2
Silverman, S.K.: Deoxyribozymes: DNA catalysts for bioorganic chemistry. Organ. Biomol. Chem. 2, 2701–2706 (2004)
Silverman, S.K.: Catalytic DNA (deoxyribozymes) for synthetic applications—current abilities and future prospects. Chem. Commun. pp. 3467–3485 (2008). doi:10.1039/b807292m
Simmons, C.P., Farrar, J.J., van Vin Chau, N., Wills, B.: Dengue. New Engl. J. Med. 366, 1423–1432 (2012)
Soloveichik, D., Seelig, G., Winfree, E.: DNA as a universal substrate for chemical kinetics. Proc. Natl. Acad. Sci. U.S. Am. 107(12), 5393–5398 (2010). doi:10.1073/pnas.0909380107
Stefanovic, D., Stojanovic, M.N.: Computing game strategies. In: Computability in Europe: The Nature of Computation, pp. 383–392. Milano (2013)
Stojanovic, M.N., Mitchell, T.E., Stefanovic, D.: Deoxyribozyme-based logic gates. J. Am. Chem. Soc. 124, 3555–3561 (2002)
Stojanovic, M.N., de Prada, P., Landry, D.W.: Catalytic molecular beacons. ChemBioChem 2, 411–415 (2001)
Stojanovic, M.N., Semova, S., Kolpashchikov, D., Macdonald, J., Morgan, C., Stefanovic, D.: Deoxyribozyme-based ligase logic gates and their initial circuits. J. Am. Chem. Soc. 127, 6914–6915 (2005). doi:10.1021/ja043003a
Stojanovic, M.N., Stefanovic, D.: Deoxyribozyme-based half adder. J. Am. Chem. Soc. 125(22), 6673–6676 (2003)
Stojanovic, M.N., Stefanovic, D.: A deoxyribozyme-based molecular automaton. Nat. Biotechnol. 21(9), 1069–1074 (2003)
Stojanovic, M.N., Stefanovic, D., Rudchenko, S.: Exercises in molecular computing. Acc. Chem. Res. 47, 1845–1852 (2014)
Tabor, J.J., Levy, M., Ellington, A.D.: Deoxyribozymes that recode sequence information. Nucleic Acid. Res. 34, 2166–2172 (2006)
Tang, J., Breaker, R.R.: Rational design of allosteric ribozymes. Chem. Biol. 4(6), 453–459 (1997)
Teichmann, M., Kopperger, E., Simmel, F.C.: Robustness of localized DNA strand displacement cascades. ACS Nano 8(8), 8487–8496 (2014)
Teller, C., Shimron, S., Willner, I.: Aptamer-DNAzyme hairpins for amplified biosensing. Anal. Chem. 81, 9114–9119 (2009)
Tram, K., Kanda, P., Li, Y.: Lighting up RNA-cleaving DNAzymes for biosensing. J. Nucleic Acid. p. 958683 (2012). doi:10.1155/2012/958683
Tram, K., Kanda, P., Salena, B.J., Huan, S., Li, Y.: Translating bacterial detection by DNAzymes into a litmus test. Angew. Chem. Int. Ed. 53(47), 12799–12802 (2014). doi:10.1002/anie.201407021
Tyagi, S., Kramer, F.R.: Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14(3), 303–309 (1996)
Vaidya, N., Manapat, M.L., Chen, I.A., Xulvi-Brunet, R., Hayden, E.J., Lehman, N.: Spontaneous network formation among cooperative RNA replicators. Nature 491, 72–77 (2012). doi:10.1038/nature11549
Vaidya, N., Walker, S.I., Lehman, N.: Recycling of informational units leads to selection of replicators in a prebiotic soup. Chem. Biol. 20(2), 241–252 (2013). doi:10.1016/j.chembiol.2013.01.007
Wang, F., Elbaz, J., Orbach, R., Magen, N., Willner, I.: Amplified analysis of DNA by the autonomous assembly of polymers consisting of DNAzyme wires. J. Am. Chem. Soc. 133, 17149–17151 (2011)
Wang, F., Elbaz, J., Teller, C., Willner, I.: Amplified detection of DNA through an autocatalytic and catabolic DNAzyme-mediated process. Angew. Chem. Int. Ed. 50, 295–299 (2011)
Wang, F., Elbaz, J., Willner, I.: Enzyme-free amplified detection of DNA by an autonomous ligation DNAzyme machinery. J. Am. Chem. Soc. 134, 5504–5507 (2012)
Wang, F., Lu, C.H., Willner, I.: From cascaded catalytic nucleic acids to enzyme-DNA nanostructures: controlling reactivity, sensing, logic operations, and assembly of complex structures. Chem. Rev. 114, 2881–2941 (2014). doi:10.1021/cr400354z
Wang, Y., Silverman, S.K.: Deoxyribozymes that synthesize branched and lariat RNA. J. Am. Chem. Soc. 125, 6880–6881 (2003). doi:10.1021/ja035150z
Wang, Z., Lee, J.H., Lu, Y.: Label-free colorimetric detection of lead ions with a nanomolar detection limit and tunable dynamic range by using gold nanoparticles and DNAzyme. Adv. Mater. 20(17), 3263–3267 (2008)
Wernick, W.: Complete sets of logical functions. Trans. Am. Math. Soc. 51, 117–132 (1942)
Wolfe, B.R., Pierce, N.A.: Sequence design for a test tube of interacting nucleic acid strands. ACS Synth. Biol. 4(10), 1086–1100 (2015). doi:10.1021/sb5002196
Wu, P., Hwang, K., Lan, T., Lu, Y.: A DNAzyme-gold nanoparticle probe for uranyl ion in living cells. J. Am. Chem. Soc. 135, 5254–5257 (2013). doi:10.1021/ja400150v
Xiang, Y., Lu, Y.: Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nat. Chem. 3, 697–703 (2011). doi:10.1038/nchem.1092
Xiang, Y., Lu, Y.: Expanding targets of DNAzyme-based sensors through deactivation and activation of DNAzymes by single uracil removal: Sensitive fluorescent assay of uracil-DNA glycosylase. Anal. Chem. 84, 9981–9987 (2012). doi:10.1021/ac302424f
Xiang, Y., Wang, Z., Xing, H., Lu, Y.: Expanding DNAzyme functionality through enzyme cascades with applications in single nucleotide repair and tunable DNA-directed assembly of nanomaterials. Chem. Sci. 4, 398–404 (2013). doi:10.1039/c2sc20763j
Xiang, Y., Wu, P., Tan, L.H., Lu, Y.: DNAzyme-functionalized gold nanoparticles for biosensing. In: Gu, M.B., Kim, H.S. (eds.) Biosensors Based on Aptamers and Enzymes, Advances in Biochemical Engineering/Biotechnology, vol. 140, pp. 93–120. Springer, New York (2014)
Xiao, Y., Wehrmann, R.J., Ibrahim, N.A., Silverman, S.K.: Establishing broad generality of DNA catalysts for site-specific hydrolysis of single-stranded DNA. Nucleic Acid. Res. 40(4), 1778–1786 (2012). doi:10.1093/nar/gkr860
Yashin, R., Rudchenko, S., Stojanovic, M.N.: Networking particles over distance using oligonucleotide-based devices. J. Am. Chem. Soc. 129(50), 15581–15584 (2007)
Yurke, B., Mills Jr., A.P.: Using DNA to power nanostructures. Genet. Program. Evol. Mach. 4, 111–122 (2003). doi:10.1023/A:1023928811651
Yurke, B., Turberfield, A.J., Mills Jr., A.P., Simmel, F.C., Neumann, J.L.: A DNA-fuelled molecular machine made of DNA. Nature 406, 605–608 (2000). doi:10.1038/35020524
Zadeh, J.N., Steenberg, C.D., Bois, J.S., Wolfe, B.R., Pierce, M.B., Khan, A.R., Dirks, R.M., Pierce, N.A.: NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 32, 170–173 (2011)
Zadeh, J.N., Wolfe, B.R., Pierce, N.A.: Nucleic acid sequence design via efficient ensemble defect optimization. J. Comput. Chem. 32, 439–452 (2011)
Zenisek, S.F.M., Hayden, E.J., Lehman, N.: Genetic exchange leading to self-assembling RNA species upon encapsulation in artificial protocells. Artif. Life 13(3), 279–289 (2007). doi:10.1162/artl.2007.13.3.279
Zhang, D.Y.: Cooperative hybridization of oligonucleotides. J. Am. Chem. Soc. 133, 1077–1086 (2011). doi:10.1021/ja109089q
Zhang, D.Y., Seelig, G.: Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 3, 103–113 (2011). doi:10.1038/nchem.957
Zhang, D.Y., Turberfield, A.J., Yurke, B., Winfree, E.: Engineering entropy-driven reactions and networks catalyzed by DNA. Science 318, 1121–1125 (2007)
Zhang, X.B., Kong, R.M., Lu, Y.: Metal ion sensors based on DNAzymes and related DNA molecules. Annu. Rev. Anal. Chem. 4, 105–128 (2011)
Acknowledgments
We acknowledge our other experimental collaborators, in particular, Joanne Macdonald, Sergei Rudchenko, Steven Graves, and Carl Brown, III. This material is based upon work supported by the National Science Foundation under grants 1027877, 1028238, and 1318833. M.R.L. gratefully acknowledges support from the New Mexico Cancer Nanoscience and Microsystems Training Center.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Lakin, M.R., Stojanovic, M.N., Stefanovic, D. (2017). Implementing Molecular Logic Gates, Circuits, and Cascades Using DNAzymes. In: Adamatzky, A. (eds) Advances in Unconventional Computing. Emergence, Complexity and Computation, vol 23. Springer, Cham. https://doi.org/10.1007/978-3-319-33921-4_1
Download citation
DOI: https://doi.org/10.1007/978-3-319-33921-4_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-33920-7
Online ISBN: 978-3-319-33921-4
eBook Packages: EngineeringEngineering (R0)