Abstract
The past decades have witnessed the evolution of electronic and photonic integrated circuits, from application specific to programmable1,2. Although liquid-phase DNA circuitry holds the potential for massive parallelism in the encoding and execution of algorithms3,4, the development of general-purpose DNA integrated circuits (DICs) has yet to be explored. Here we demonstrate a DIC system by integration of multilayer DNA-based programmable gate arrays (DPGAs). We find that the use of generic single-stranded oligonucleotides as a uniform transmission signal can reliably integrate large-scale DICs with minimal leakage and high fidelity for general-purpose computing. Reconfiguration of a single DPGA with 24 addressable dual-rail gates can be programmed with wiring instructions to implement over 100 billion distinct circuits. Furthermore, to control the intrinsically random collision of molecules, we designed DNA origami registers to provide the directionality for asynchronous execution of cascaded DPGAs. We exemplify this by a quadratic equation-solving DIC assembled with three layers of cascade DPGAs comprising 30 logic gates with around 500 DNA strands. We further show that integration of a DPGA with an analog-to-digital converter can classify disease-related microRNAs. The ability to integrate large-scale DPGA networks without apparent signal attenuation marks a key step towards general-purpose DNA computing.
Similar content being viewed by others
Main
Liquid-phase biocomputing exploiting biomolecular interactions has been actively explored due to its massive parallelism and intrinsic compatibility with biological systems. For example, computational DNA reaction networks including automata5, logic circuits6,7,8, decision-making machines9,10 and neural networks11 have been realized, which have demonstrated potential in molecular information processing4,12, synthetic intelligent devices13 and biomedical applications14,15,16. Despite this progress, most of these computing systems are tailored in hardware to implement a specific algorithm or a limited number of computational tasks.
General-purpose electronic integrated circuits allow software programming rather than application-specific custom hardware fabrication to perform a certain function, providing a higher-level platform for prototyping computational machines without the requirement of previous knowledge of the underlying physics. Notably, classic silicon-based and emerging carbon nanotube-based computers and quantum computers have undergone similar evolution from application-specific (for example, application-specific integrated circuit) to general-purpose (for example, field-programmable gate array, FPGA)2,17,18,19,20. Programmability and scalability constitute two critical factors in achieving general-purpose computing. Programmability enables specification of the device to perform various algorithms whereas scalability allows the handling of a growing amount of work by the addition of resources to the system. Unlike electronic integrated circuits, in which gates are physically localized and universal electrical signals are transmitted in a directional manner, biomolecular components in DNA integrated circuits (DICs) diffuse and mix in solution21, which hinders the development of scalable and programmable biocomputing devices. In a typical DNA computing system, limitation in the orthogonality of DNA components and difficulty in controlling the intrinsically random collision of molecules make it practically challenging to realize general-purpose DNA computing.
Intense efforts have been made to explore the programmability of DNA circuits22,23. However, due to the general lack of directionality for the integration of these liquid-phase systems, scalability is yet to be explored for programmable DNA systems. Analogous to the subcomponent assembly in electronic24 or quantum25 circuits, spatial compartmentation has been introduced in cellular26 and synthetic27,28,29 molecular reaction systems to increase directionality, which nevertheless has shown limited scalability at the system level29. Inspired by silicon-based FPGAs in which electrons form a universal signal for intradevice programming and interdevice communication, here we developed highly scalable, DNA-based, programmable gate arrays (DPGAs; Extended Data Fig. 1) by employing generic single-stranded DNA oligonucleotides as a uniform transmission signal (DNA–UTS).
Scalable DPGA architecture
In a typical design for a scalable DPGA architecture enabled by DNA–UTS, a task circuit that could not be implemented with one DPGA was divided into subcircuits, each mapped to and executed by one DPGA (Fig. 1). As a mimic of electronic integrated circuit instructions, we established a molecular instruction set with approximately 1,000 instructions (over 2,000 oligonucleotides) that defines all legal wires on DPGAs. Molecular instructions for each subcircuit were mixed with involved computing units, forming a unique routing pattern that implements the function of the subcircuit. Twenty-four addressable logic gates of four types (AND, OR, NOT and XOR) were designed as computing units, the combination of which constituted a complete set of Boolean functions (Supplementary Fig. 2), offering high programming space on single DPGAs (Supplementary Text 6.2). The operation of DPGAs was based on receiving and sending DNA–UTS between gates and DPGAs along a programme-configured pathway. To avoid inter-DPGA crosstalk we further designed a DNA origami register to direct asynchronous computing processing of cascade DPGAs. Analogous to its electronic counterpart, intermediate values calculated from an upstream DPGA were written to the DNA origami register via DNA strand displacement, which were then transmitted to a downstream DPGA.
A complex task circuit beyond the capability of a single DPGA was divided into subcircuits, for which molecular instructions were generated. Involved computing units of each subcircuit, as shown by the logical arrangement in the scheme, were called by their logical address and the function was implemented via intra-DPGA routing with molecular instructions. The transmission of intermediate output between subcircuits was realized via DNA origami register-mediated inter-DPGA routing.
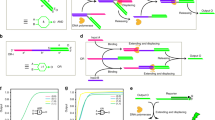
In this design, dual-rail logic gates characteristic of dual-rail input/output ports, which allow two DNA strands representing high and low signals to pass through simultaneously (Fig. 2a and Extended Data Fig. 3), were adopted to realize DPGAs. The uniformity of molecular design for input/output ports of all gates allowed arbitrary routing and integration of DPGAs, which was based on the given logic function followed by formulaic generation of corresponding DNA sequences (Supplementary Text 3.3 and 3.4). For example, a dual-rail AND gate was implemented by two hybridized DNA molecules with three specific domains that functioned as switches: one domain (blue) responded to low signal of either input whereas two-series ones (yellow) responded to high signal for each input port, respectively (Fig. 2b). During implementation of DPGAs, the input of DNA strands triggered strand displacement reactions (SDRs)30 to realize logic functions (Extended Data Fig. 4). Configuration of DPGAs was realized by addressing and connecting the required gates for a target circuit. Under these circumstances a configured DPGA could be abstracted as a higher-level computing unit, in which input ports for logic gates at the input layer and output ports for logic gates at the output layer function as DPGA-level input and output ports, respectively (Fig. 2a).
a, Computing units in DPGAs, shown by their logical arrangement, possess uniform input/output interfaces with packaged internal implementation for required logic functions. Of note, a configured DPGA functions as a higher-level integrable computing unit with input/output interfaces similar to a logic gate. b, Modular and automated molecular implementation of computing units. For a dual-rail AND gate, the switching scheme was generated according to the required logic function and then DNA molecules comprising the corresponding molecular switches were generated. c, SDRs for series switches and potential leak pathway. d, A gapped S3/S4/S5 structure with a 1 nt gap was employed for the realization of series switches to optimize both computing speed and signal leakage. e, Molecular dynamics simulations for the leak reaction pathway in c with 0, 1 or 2 nt gaps showing that a 1 nt gap could effectively suppress leak reaction similar to the 0 nt gap. f, Experimental measurement of kinetics for a dual-rail AND gate with all four possible input combinations. g, Relationship between dual-rail result and two output signals. Regions within green dashed lines represent correct outputs for both outH and outL, which lead to the correct dual-rail result (below 0.4 or above 0.6). Regions between red and green dashed lines represent one correct output and another undefined or wrong; dual-rail results can still show the correct value. Dual-rail results for the AND gate showing that all four input combinations fell within the correct regions. h, All four types of dual-rail computing unit generated correct high- and low-output signals. Edge colour indicates input combination, marker shape indicates gate type and face colour indicates the obtained dual-rail result.
Sequential SDRs were used to realize series switches, in which the first input (in2H) hybridized with ‘S3’, which displaced ‘S5’ to expose the toehold for in1H; in1H then hybridized and released ‘S4’, which acted as the output (Fig. 2c). We optimized the molecular design structurally and found that a gapped structure formed by S3, S4 and S5 with one unpaired base in S3 could suppress the leak reaction pathway and also allow a high computing speed (Fig. 2d,e, Extended Data Fig. 5 and Supplementary Text 4.1).
We first tested individual dual-rail gates, the function of which lays the foundation for DPGA programming. Both normalized high (outH)- and low (outL)-output signals generated the correct results, which were consistent with the truth tables within 20 min for all four types of gate (Fig. 2f and Supplementary Fig. 25), showing that the 1 nt gap design provided higher computing speed than, and comparable leakage to, previously reported SDR-based AND gates in large-scale DICs6,31. Importantly, even without any purification following strand annealing (Supplementary Figs. 21–23), we observed very low leakage (below 0.1) for all gates, which reflects the fault tolerance of DPGA. To interpret the outcome of one given execution, the dual-rail result was defined by the difference between outH and outL (dual-rail result, (outH – outL + 1)/2). The conversion map from outH and outL to dual-rail result is shown in Fig. 2g. In particular, the dual-rail results of the AND gate under all four input combinations fell within the correct ON and OFF state regions. Measurements of all types of dual-rail gate showed dual-rail results lower than 0.2 for all logical FALSE conditions and values higher than 0.8 for all TRUE conditions (Fig. 2h).
Having demonstrated the programmability within single gates, we next explored whether DNA–UTS could wire intergate and inter-DPGA transmission to implement computing circuits. For intergate transmission within single DPGAs, input values that enter a configured circuit via input ports are processed to generate output values transmitting to output ports for readout. Three types of wire—input port-to-gate, gate-to-gate and gate-to-output port—are thus required for programming of DPGAs to carry out a calculation (Fig. 3a). We established a molecular instruction set that defines all legal wires on DPGAs, containing three types of wiring instructions: type 1 instruction (WIR1) conducts an input to a gate; type 2 instruction (WIR2) conducts an output signal from an upstream gate to an input port of a downstream gate; and type 3 instruction (WIR3) conducts an output signal from a gate to an output port of DPGAs (Fig. 3b and Supplementary Text 2.1). All legal wires can be synthesized and assembled beforehand, allowing programming of the DPGA with the addition of the corresponding wires. WIR1 was implemented with an ssDNA input that entered an entry gate. Following execution of this gate, the resulting DNA–UTS as the output was transmitted to a downstream gate via WIR2, which was implemented with ten ssDNA strands (Fig. 3b). We designed noise thresholds and signal amplifiers for molecular implementations of wiring instructions (Supplementary Text 2.2 and Extended Data Fig. 6) to suppress signal decay during transmission along the reaction pathway that potentially limits the depth of DICs27,32.
a, A DPGA contains 24 dual-rail logic gates (six each of OR, AND, NOT and XOR, shown by the logical arrangement) as computing units, in which intra-DPGA programming is realized using three types of wiring instruction (WIR1, WIR2 and WIR3). Cascaded DPGAs asynchronously operate with signal transmission mediated by WIR4s. b, Schematic illustration and molecular implementation of WIR2. WIR2 contains two functional modules: a threshold that filters out noise and a converter and amplifier that convert and restore signal to a fixed level. Two DNA–UTSs were transmitted in parallel through high and low channels via SDRs. Orange-bordered box shows the DNA–UTS transmission process through the low channel. Dashed lines indicate the upstream binding region. c, A WIR4 contains a DNA origami register and a WIR2 to wire two configured DPGAs. Dashed box shows SDRs of the write and read processes for the DNA origami register. d, Transduction performance of a WIR2 showing nonlinear response in which output lower than 0.4 was suppressed by threshold and that higher than 0.6 was amplified toward 1. n.u., normalized units. e, Circuit diagram (left) and computing kinetics (right) of a four-layered circuit involving WIR1s, WIR2s and WIR3s. f, Atomic force microscopy (AFM) images showing temporal storage and readout of an output signal with a DNA origami register. Scale bar, 100 nm. g, Circuit diagram of a cascade circuit containing 11 layers of dual-rail gates, which was divided and implemented by three configured DPGAs. h, Computing kinetics of one- to six-layer subcircuits implemented by a single DPGA without WIR4, with input 0 (left) and 1 (right) for gate 12, respectively. i, Dual-rail results of the cascade circuit showing decayed performance when implemented by one DPGA and maintained performance when it was implemented by three DPGAs connected by WIR4s.
Signal transduction of WIR2 was realized via SDRs. The exhibition of a steep ramp suggested that we could effectively suppress leaked signal (below 0.4) to approach 0 and amplify genuine output (above 0.6) to approach 1 (Fig. 3d). Similarly, WIR3 was implemented with ten DNA strands to transduce signals via SDRs (Supplementary Fig. 4). Cascaded gates with increased layers were challenged to test the cooperation of type 1, 2 and 3 wiring instructions (Fig. 3e and Supplementary Figs. 28–31). Low- and high-output signals for all input combinations went to the correct states within 1 h. Dual-rail results were higher than 0.8 for those supposed to be 1 and lower than 0.2 for those supposed to be 0. Hence we established that programmable and reliable intra-DPGA DNA–UTS transmission could be realized with a subset of wiring instructions.
For inter-DPGA wiring we adopted a fourth type of wiring instructions (WIR4s) to realize inter-DPGA DNA–UTS transmission—that is, output signals from upstream DPGA1 were transmitted to downstream DPGA2. To suppress inter-DPGA crosstalk and signal attenuation, each WIR4 was implemented in a signal relay-like manner with a DNA origami register followed by a WIR2 (Fig. 3c). The spatial confinement and addressability of DNA origami33,34 controls the intrinsic stochasticity of molecular interactions in solution. In a typical WIR4 the DNA origami register temporally stores the outputs from DPGA1, then a downstream WIR2 transmits the output to the entry gate of DPGA2, providing the directionality for asynchronous execution of cascaded DPGAs. Output signal was written onto DNA origami via SDR that was mediated by the exposed toehold (5 nt), which was read out by a retrieval strand with a longer toehold (7 nt) at the opposite end and was confirmed both visually (Fig. 3f and Supplementary Text 5.8) and quantitively (Supplementary Figs. 44 and 45). Asynchrony was realized by isolation of DNA origami after capturing the output from the upstream DPGA and releasing the stored signal to the downstream DPGA (Supplementary Fig. 6). Importantly, we found that the use of WIR4s effectively improved circuit depth as illustrated by the performance of an 11-layer circuit consisting of 11 dual-rail gates with different addresses (Fig. 3g–i).
Thus, we reason that the use of DNA origami registers for physical separation of DPGAs restricts transient binding between unmatched DNA strands, which greatly attenuates signal decay at enlarged circuit sizes. In addition, WIR4 facilitates reaction kinetics by dividing a long reaction pathway into shorter ones, which shortens time delay in a signal relay-like manner. Given that signal decay is dependent on the length of the reaction pathway, the use of WIR4 improves the depth scalability of DICs.
Multitask reconfiguration
Having established DPGA wiring with DNA–UTS, we next explored the reconfiguration of DPGAs for multitask operations. We developed a compiler to convert a programme in natural language or Boolean expressions to a subset of wiring instructions (Extended Data Fig. 7 and Supplementary Text 7). By taking the two-bit multiplying function as an example, we compiled the programme to a group of wiring instructions that called eight dual-rail gates with eight WIR1s, six WIR2s and four WIR3s (Fig. 4a). The routing pattern of DPGA is shown in Fig. 4a. We successfully tested this multiplying circuit on all 16 possible inputs with correct computing results (Fig. 4b and Supplementary Fig. 37). Dynamic range, defined by the difference between the highest OFF state value and the lowest ON state value, was used to quantify the ON/OFF contrast of computing results (Supplementary Fig. 64c). Of note, all four outputs showed dynamic ranges over 0.5 (Fig. 4b), suggesting that the multiplying circuit operated correctly with high ON/OFF contrast.
a, Workflow for programming an exemplary function using the DPGA compiler. The function s = a × b (s represents the product of a and b that needs to be computed) was input into the DPGA compiler. The compiler then parsed the command and generated wiring instructions, as shown in the red box, to configure DPGA. The molecular components of each instruction were automatically generated with a custom-written software. Experimental implementation of DPGA programming was achieved by mixing DNA strands representing these instructions and the involved logic gates. b, Computing results of two-bit multiplication. c, Schematic diagram and equivalent logic gate-level layout in DPGA for a two-bit adder circuit that receives two two-bit inputs and calculates their sum to generate a three-bit result. d, Computing results of the adder circuit for all possible input combinations. e, Schematic diagram and equivalent logic gate-level layout in DPGA for a two-bit comparison circuit generates three possible outputs: s = 0 when a is lower than b; s = 1 when a = b; and s = 2 when a is greater than b. f, Computing results of the comparison circuit for all possible input combinations. g, Vector proximity (VP) angle for each circuit is plotted versus the corresponding dynamic range; n = 103 (circuit information provided in Supplementary Table 6). Inset, scheme illustration of dynamic range and vector proximity angle for quantifying the performance of an implemented circuit (see Supplementary Fig. 61 for details). h, Scatter plot of dynamic ranges and vector proximity angles based on circuit size. i, Scatter plot of 792 computing results showing that all dual-rail results fell within the correct region, with nine of them corrected from an undefined state for outH or outL under binarization rules.
To evaluate the generality and robustness of multitask reconfiguration of DPGAs with wiring instructions, we experimentally tested 103 circuits with 792 calculations by reconfiguring a single DPGA (Supplementary Table 6), which implemented various basic circuit structures including single gates, cascaded gates, fan-in and fan-out of gates and complex functional circuits. The size of these DNA circuits ranged from a single gate with 17 strands to 11 gates with more than 300 strands in one reaction. Addition is a basic operation for digital computers. We implemented a two-bit addition algorithm by calling seven gates on DPGA (Fig. 4c). Three output ports generated correct results for all 16 cases (Fig. 4d). We also demonstrated the use of DPGAs for implementation of comparison operations that are popularly used in logical statements to determine equality or difference between variables (Fig. 4e). A two-bit comparison function computing the numeric comparison between variables a and b was implemented with a circuit involving 11 dual-rail gates on a DPGA, which were correctly performed for every possible pairing of a and b (Fig. 4f). Importantly, we observed correct computing results for all 792 calculations within 2 h of reactions (Supplementary Fig. 64b). By examining the robustness of DPGA reconfiguration using dynamic range and vector proximity when executing diverse functions, we demonstrate the realization of reliable computing of DICs even with the involvement of 11 dual-rail gates (Fig. 4g–i and Supplementary Text 6.1). By taking the circuit size of 11 gates as a practical upper limit, we estimated that the programming space of single DPGAs would reach 100 billion unique patterns for tree-structured circuits, at least two orders of magnitude higher than existing approaches (under 100,000 for MAYA-III22 and roughly 1 billion for IBC23; Supplementary Text 6.2).
To evaluate the capability of error-free computing with single-DPGA programming, we further inspected the distribution of generated results from 792 experiments. Although 2% of outH and outL fell within undefined states (0.4 below outH, outL under 0.6), all dual-rail results led to correct Boolean values based on binarization rules (Fig. 4i). Therefore, the circuits exhibited a high level of noise tolerance with dual-rail gates. We demonstrated large-scale computing for circuits containing up to 300 DNA strands in one tube, with a high DNA concentration (1× = 100 nM), and with all hybridized strands directly used (after annealing) without further purification. We reason that the combination of sequential displacement reactions with the thresholding and amplification in configured DPGAs contributes to improved scalability of DICs in a single reaction (Supplementary Fig. 63).
Parallel and serial integration of DPGAs
Given the modular nature of DPGAs, we further tested whether reconfigurable DPGAs could be integrated into larger-scale circuits for practically accessible computing capability. In principle, algorithms that are divisible into independent parts could be implemented with parallel DPGA circuits. As a proof of concept, we employed three differently configured entities of DPGAs in parallel to operate the square-rooting function to one decimal digit (Fig. 5a,b). In this algorithm, one decimal digit was represented by four bits using binary-coded decimal code. The square root r was represented by fixed-point binary, the integer digit by two bits and the decimal digit by four bits. The circuit was divided and implemented with three parallel configured DPGAs (Fig. 5c). We found that the whole DPGA circuit computed the results in under 2 h for all experiments (Fig. 5d and Supplementary Fig. 41). Of note, 21 dual-rail gates with over 300 DNA strands were involved in this DPGA circuit, in which the largest subcircuit contained 130 strands.
a, Schematic for a fixed-point square-rooting circuit. A one-digit decimal input (0–9) is represented by four binary digits. The integer digit of output is represented by two bits and the decimal digit by four bits. b, DPGA-level circuit diagram of the square-rooting circuit. c, Logic gate-level layouts of the three paralleled DPGAs. d, Computing results for all ten valid inputs. e, DPGA-level circuit diagram of a conditional branching algorithm with two cascaded DPGAs. The upstream DPGA1 executes the condition circuit to determine whether input equals 7 by generating an intermediate output c. The downstream DPGA2 performs adding-one or setting-one operation based on the received signal c from DPGA1. f, The equivalent logic gate-level layouts. g, Final computing results for all valid input combinations. h, An equation-solving circuit that calculates the larger root of a quadratic equation. The root formula for this equation was divided into five parts with each assigned to one DPGA. i, Computing kinetics of DPGAs on the third layer when solving equation x2 – 3x = 0. j, Computing kinetics of DPGAs on the third layer when solving equation x2 + x – 2 = 0. k, Comparison of VP angles obtained from DPGAs in the network wired by WIR4s and that run individually.
Because DPGAs can be integrated with WIR4s, configured DPGAs could be serially assembled to implement high-depth circuits. To demonstrate this we implemented a finite-state machine via conditional branching (Fig. 5e). Days of the week from Monday to Sunday were assigned numbers 1–7. The state machine computing the number representing the next day was implemented by two cascaded DPGAs, with one as condition circuit to decide whether the input was 7 and the other as processing circuit (Fig. 5f). Signal transmission through WIR4 was performed using polyethylene glycol (PEG) precipitation-based origami purification and magnetic field-mediated isolation (Supplementary Figs. 51 and 52). Both approaches resulted in correct computing results with nearly identical performance under all seven input combinations (Fig. 5g and Supplementary Fig. 52), suggesting that the scalability of DPGA is intrinsic and also independent of the solution transfer methodology. Thus, the potential for parallelly and serially connecting DPGAs paves the way for the construction of DPGA networks to implement complex algorithms including arbitrary digital operations.
The modular design of DPGAs allows the integration of multiple DPGAs into a network. We next developed a DPGA network to solve quadratic equations with the expression x2 – bx + c = 0, where b and c are two-bit integers (Fig. 5h). The larger of the solutions x can be calculated by the root formula for quadratic equations: \(x=(b+\sqrt{({b}^{2}-4c)})/2\). This algorithm was implemented with an 11-layered DIC containing 30 dual-rail gates. In our design, the circuit was divided and mapped to a network with five cascaded DPGA, where intermediate values s = b2−4c and r = √s. The function was confirmed experimentally with two functions: x2 – 3x = 0 and x2 + x – 2 = 0 (Fig. 5i,j). Comparing WIR4-mediated DPGA networks and individual DPGAs with directly added ideal input we found that, in the presence of WIR4s, downstream DPGAs maintained little deviation from ideal values despite roughly threefold reduction in operating speed (Fig. 5k and Extended Data Fig. 9). Hence the development of DPGA networks, albeit at the cost of time consumption, holds the potential for implementation of large-scale integration circuits for high-complexity operation.
DPGA-based nonlinear classifier
Molecular circuits have been developed for identification of disease-associated biomarkers15,16, but are generally linear and thus limited when processing information from multiple biomarkers that may not always be linearly separable. We sought to address this problem by exploiting the high integrability of DPGA to construct a nonlinear classifier. We targeted a cancer classifier based on miRNA expression levels and used training on miRNA expression data for kidney renal clear cell carcinoma (KIRC) from The Cancer Genome Atlas (TCGA) to develop a decision tree circuit with three miRNA inputs of mir-200c, mir-204 and mir-887 (Fig. 6a and Supplementary Text 5.12). The classifier was implemented by integration of DPGA and analog-to-digital converters (ADCs) (Fig. 6b), the latter converting an analog single-rail miRNA signal to a dual-rail DNA–UTS that could then be further processed by DPGAs (Fig. 6c). The DNA signals generated by ADCs were transmitted to DPGA via WIR4s. Using mir-204 as a model, we verified the testing of ADC conversion (Fig. 6d,e).
a, An in silico-trained nonlinear classifier model using three miRNAs as input. b, Circuit diagram of the KIRC classifier. Single-rail RNA inputs were converted to dual-rail DNA outputs via an ADC module. c, The SDR network of ADC. When the miRNA level was above threshold, the excess was amplified and converted to outH that served as the outcome of ADC. When the miRNA level was below threshold, no outH was generated and outL was preserved to generate a low signal. d, Verification of testing of the ADC conversion using mir-204 as a model. Normalized outL and outH levels with two miRNA levels (0.12 and 0.86, respectively) in the test set entered the ADC module with a threshold of 0.55. e, Plot of input versus high- and low-output signals of ADC. OutL was generated for input levels under 0.55 whereas outH was generated for input levels above 0.60. Only when the input level was 0.60, which is only slightly higher than 0.55, did outL and outH fail to reach the correct range throughout the test duration. f, Visualization of the trained classifier model and experimental results of 23 randomly selected cases from the test set. Yellow-bordered region indicates the space for KIRC from in silico training. Coloured dots represent experimentally tested cases from a test set with 119 cases (grey dots). Output levels are represented by the corresponding colour in the colour bar to the right. g, Computing the kinetics of DPGA for samples 3, 13 and 21, which are marked by dashed circles in f. h, Testing of the KIRC classifier for all 23 randomized samples shows that molecular computing output is consistent with model prediction and actual disease states in the TCGA database. a.u., arbitrary units; th, the concentration of threshold.
In experimental tests of the nonlinear classifier using synthetic miRNA molecules (18 KIRC and 5 healthy samples, represented by coloured dots in Fig. 6f), after receiving DNA–UTS from ADCs, DPGA computed correct outH and outL in 2 h for 23 tested samples (Fig. 6g and Extended Data Fig. 10). Analysis of the dual-rail output of the classifier showed agreement between the molecular computation, model prediction and actual disease states labelled in TCGA (Fig. 6h). Although these classifier tests involved synthetic miRNA in Tris-EDTA (TE) buffer, experiments could also be run with DPGA in cell culture (Supplementary Fig. 61) and with ADC receiving miRNA signals from the serum-containing solution (Supplementary Fig. 62), suggesting that it should be possible to interface DPGAs directly with cells and body fluids. Because the programmability and scalability of DPGAs allow them to be coupled with diverse types of ADC, we anticipate that integrated DPGAs will be able to process information from biomarkers beyond miRNAs and might prove useful for clinical and diagnostic applications.
Discussion
Our generic DNA computing system achieves effective signal transmission in DPGAs through the use of DNA–UTS, which functions like electrons or photons in electronic/photonic integrated circuits. The resultant programmable DIC, rather than simply implementing a specific algorithm, can execute large-scale computation paths with an instruction set containing over 2,000 oligonucleotides and reach the complexity threshold for general-purpose computing. Various applications, such as diagnosis based on nonlinear classification, can be readily targeted by simply coupling DPGA with specific ADCs.
DPGA circuits can be programmably generated to execute various functions (Extended Data Fig. 2). The programmability of DPGAs is enabled by the use of dual-rail gates featuring uniform input/output interfaces, which allows each gate to be addressed, analogous to that in FPGA. Gates are connected via the address information in both DPGA and FPGA. The sequence of wiring instructions is dependent on the addresses of upstream/downstream gates, and also independent of their logic functions. Also of note, the dual-rail gates in DPGA can be called multiple times (for example, gate 09 was used five times in the equation-solving circuit), allowing the circuit to be reconfigured without the need for redesign of DNA sequences. The reusability of circuits is functionally equivalent to the physical reorganization in FPGA. As the computing units of DPGAs, the instructions of these dual-rail gates are compatible with conventional compiling syntax, offering great potential for software development of programmable general-purpose DNA computers. In particular, because the sequences for all logic gates are orthogonal, the DNA strands can be mass produced via high-throughput DNA synthesis35 (Supplementary Fig. 29).
DNA origami registers that mediate signal transmission between spatiotemporally separated circuits are the key to DPGA-level integration. These can direct signal transmission in liquid-phase circuits to integrate and mediate asynchronous execution of cascaded DPGAs, at the expense of manual transmission of intermediate data. The use of DNA origami registers greatly suppresses both the transient binding that limits circuit size and the signal decay that limits circuit depth, thereby facilitating higher-ordered integration of multilayer DPGAs and increasing the programme complexity that can be realized. Large-scale integration of DPGA networks with up to five cascade DPGAs, 30 logic gates and around 500 participating strands, for example, are equivalent to the achievable circuit depth of 11 layers of logic gates and 30 steps of cascaded SDRs (Extended Data Fig. 10) and break the current limit of programmable DNA circuits (Supplementary Fig. 57). Although the DNA-based register design requires manual intervention, it allows for the introduction of all-DNA circuits with high complexity and scalability without having the requirement for other biomolecular components (for example, enzymes22, light/electronic transducers36 or microfluidics29).
We believe that the programmability and integrability of DICs can be extended in several areas. First, our DPGA compiler provides a highly generic and user-friendly interface for the development of DNA computing, which can automatically compile target functions and generate corresponding DNA instructions without previous knowledge of their internal implementation. Second, the hierarchy of DPGAs combined with data register could be transplanted to other molecular reaction systems. For example, rather than using SDRs for molecular implementation, DNA-processing enzymes that can generate and degenerate DNA strands provide new tools for implementation of feedback digital circuits5. Third, the manual transfer currently required for inter-DPGA transmission might be further automated—for example, by encapsulation of each DPGA within a vesicular compartment29 or localization of each DPGA on a DNA origami template27. The magnetic field-assisted method is potentially integrable36 in a microfluidic system, and fully automated execution of DICs might be possible by coupling DPGAs with droplet-based microfluidics37.
Methods
Preparation of dual-rail gates and wiring instructions
DNA oligonucleotides were dissolved in 1× TE buffer (nuclease free, pH 8.0, Sigma-Aldrich), quantified with ultraviolet/visible spectrometry by monitoring their absorption at 260 nm and storage at −20 °C. Oligonucleotides labelled with fluorescent dyes or quenchers were dissolved in deionized water (Milli-Q) and stored in deionized water at −20 °C. Single strands were hybridized to prepare corresponding double-stranded DNA. To suppress potential leakage caused by synthetic and quantification errors, we used a lower concentration of strands for those destined for release. Hybridized structures were prepared by mixing the required strands to the corresponding final concentrations in TE buffer (1× TE: 40 mM Tris base, 20 mM acetic acid, 2 mM EDTA adjusted to pH 8.0) with 12.5 mM MgCl2. The final concentrations are shown in Supplementary Table 1.
Threshold and amplifier complexes of wiring instructions were prepared by annealing separately at a concentration ratio of 1:1.2, the final concentration being shown in Supplementary Table 2. Reporters were prepared by mixing strands with quenchers and fluorophores, in which the former was 50% excess of the latter to reach final concentrations of 15 and 10 μM, respectively. The buffer for all experiments and annealed complexes was TE with 12.5 mM Mg2+. Mixed strands were annealed by heating to 95 °C for 2 min, with slow cooling to room temperature at a speed of 0.1 °C every 6 s before holding at 4 °C. Hybridized molecules were stored at 4 °C for further use.
Fluorescence measurements for DNA reaction kinetics
A Synergy H1 Hybrid Multi-Mode Reader (BioTek) and a Corning 96-well black assay plate were employed to perform fluorescence measurements. All calculations for a circuit were run in parallel on the instrument. Fluorescence kinetics data were collected every 1 or 2 min, depending on the overall duration of the experiment. Excitation (emission) wavelengths were 510 nm (540 nm) for dye TET and 640 nm (670 nm) for dye Cy5. in general all circuit components, except input strand(s), were mixed in TE buffer with 12.5 mM MgCl2. Experiments were performed in the 96-well black assay plate (Corning) with a 98 μl reaction mixture per well for all experiments. The initial value was recorded as baseline. The experiment was then paused for the addition of 2 μl of input strand(s) and subsequent mixing by shaking. The plate was then replaced in the hybrid reader and the experiment resumed. Experiments were performed at a standard concentration of 100 nM (1×) for all gate and threshold complexes at a standard concentration of 40 nM (0.4×) for individual OR, AND, NOT and XOR gates, two- and three-layered circuits, fan-in and -out circuits, four-layered circuits and threshold complexes at a standard concentration of 60 nM (0.6×) for full-subtractor, two-bit multiplication, two-bit addition, two-bit comparison operation and squaring-rooting operation. For amplifier in wiring instructions, 2× hybridized molecules and 10× fuel were used. For a dual-rail logic circuit, the output trajectories were read using two distinct fluorophores—quencher pair TET-BHQ2 and Cy5-BHQ2—at a standard concentration of 500 nM (5×). A standard concentration of 200 nM (2×) for inputs was used. For each input combination we simultaneously recorded the two fluorescence channels representing high and low signal, respectively. Combining the output trajectories from each pair of experiments into a single plot allows simultaneous observation of outputs produced by different input combinations. The temperature was maintained at 18 °C throughout the reaction.
Preparation and operation of DNA origami registers
According to Rothemund’s method38, 90 × 60 nm2 rectangular-shaped DNA origami structures were assembled from the staple and output-binding strands and M13m18 ssDNA (NEB). The positions and sequences of the staple and output-binding strands are representd by different colours in Supplementary Figs. 49, 54 and 55. In a 100 µl system we used 50 nM staple strand, 100 nM output-binding strand and 10 nM M13 ssDNA to form origami. DNA origami was annealed and assembled in 1× Tris-acetate-DNA (TAE)-Mg2+ buffer (Tris 40 mM, acetic acid 20 mM, EDTA 2 mM and magnesium acetate 12.5 mM, pH 8.0) in a Thermocycler (Bio-Rad) with the following setup: incubation at 95 °C for 2 min, slow cooling to 60 °C at 12 s per 0.1 °C, incubation at 60 °C for 12 min, slow cooling to 25 °C at 12 s per 0.1 °C then retention at 4 °C for up to 24 h. The assembled rectangular DNA origami structures were separated from excess staple and output-binding strands using PEG precipitation39. Briefly, DNA origami structures at 20 mM MgCl2 were mixed 1:1 (v/v) with a precipitation buffer containing 15% PEG8000 w/v (no. MW:8000, Sigma), 5 mM Tris, 1 mM EDTA and 505 mM NaCl. The solution was mixed in a 1.5 ml centrifuge tube and spun at 12,000 rpm and 4 °C for 15 min in a centrifuge (Eppendorf). The supernatant was removed by pipette. The precipitate was dissolved in 20 µl of 1× TAE-Mg2+ buffer and incubated overnight at 40 °C and 400 rpm. The concentration of the purified rectangular DNA origami was quantified with a microvolume ultraviolet-visible spectrophotometer (NanoDrop). We added a fivefold excess of block strands to the origami with incubation for 2 h at room temperature, followed by storage at 4 °C until further use.
To test the writing and reading efficiency of the DNA origami register, the concentrations of free-state output were recorded with a TET-BHQ2 reporter. (1) The output strand (100 nM) was incubated with DNA origami (10 nM), with one binding area containing 21 sites and a 500 nM reporter for 2 h at room temperature. (2) The output strand was incubated with DNA origami for 2 h, then 1 µM each of retrieval and reporter were added with incubation for a further 2 h at room temperature. (3) The output strand was incubated with reporter for 2 h. Fluorescence intensity for each reaction was measured.
Execution of cascaded DPGAs
All circuit components of the upstream DPGA were mixed and incubated for 2 h to complete the reaction. For PEG precipitation-based transfer, the DNA origami register was added to a final concentration of 10 nM with incubation for a further 1 h. The origami was then purified using PEG precipitation then quantified by NanoDrop and added to the reaction system of the downstream DPGA to a final concentration of 10 nM. The corresponding WIR2s in WIR4 were also added and incubated for 2 h, and then its output was transferred to the following DPGA using the same signal transfer operations. For magnetic field-mediated transfer, biotin-modified DNA origami registers were incubated with Streptavidin-modified Magnetic Beads (New England Biolabs, 4 mg ml–1) to form MB-origami registers. MB-origami register was added to a final concentration of 10 nM following incubation of upstream DPGA. After incubation for 1 h to write DPGA output to DNA origami register, a magnetic field was applied and the upstream DPGA solution removed. The retrieval strand was then added to retrieve the stored output, which was transferred to the downstream DPGA reaction system. After receiving output signals, the downstream DPGA was incubated for 2 h and its output transferred to the following DPGA using the same signal transfer operations. Reaction kinetics were recorded with fluorescence reporters.
Execution and results of the 11-layered circuit
The 11-layer circuit, consisting of 11 dual-rail gates with different addresses, facilitated either implementation of the circuit by one DPGA or division into multiple DPGAs. First, we increased circuit depth from left to right and performed each calculation in a single reaction system. We found that, when circuit depth reached five layers, computing speed dramatically slowed and the signal dropped to leakage level (Fig. 3h). Also, the dual-rail results decayed along with the circuit layer. The computing results significantly deviated from the normal ON or OFF states at the five-layer depth (Fig. 3i). Next, we divided the circuit into three subcircuits and implemented each with one configured DPGA, among which information was transmitted through WIR4s. We found that, when circuit depth was increased to 11 layers, the outcome remained within the correct range (Fig. 3i and Supplementary Fig. 48). Further analysis showed that WIR4s, especially the internal DNA origami registers, improved implementable circuit size and depth primarily by limiting transient binding and time delay (Extended Data Fig. 8 and Supplementary Fig. 47).
Data normalization and dual-rail result calculation
All data were normalized from the raw fluorescence level to relative concentrations of output signals when plotted, faciltating quantitative analysis of the data despite differences in instrument performance, circuit functions and molecular implementations. The microplate reader supports up to 96 parallel kinetics experiments, and the difference in fluorescence readout caused by the instrument was negligible among these parallel experiments. Each set of parallel experiments performed was for the same circuit but with different inputs. The minimum level (output 0) was determined by the minimum of all tested data points at time t = 0. For a given fluorophore, parallel experiments had at least one output signal that increased (that is, a maximal ON completion level); maximum level (output 1) was determined by the average of the last five data points for the highest signal. The fluorescence data shown in Fig. 1e were normalized in this way. The fluorescence level that corresponds to standard concentration (1×) was obtained from the highest signal produced from the reporter on a plate. Negligible concentration (0×) corresponds to the background fluorescence of the reaction mixture when any input strand/strands have been joined at t = 0, which was obtained from the first measurement of the lowest signal produced from a reporter on a plate. All experiments on a single plate were normalized together, allowing direct comparison between the output of a circuit for different input patterns. We used two different reporters of the same circuit to read high-signal outH and low-signal outL, and thus the obtained signals for outH and outL were normalized independently. Following normalization, the dual-rail result was calculated using this equation:
Error flags were calculated using equation:
where |…| represents the absolute value of inner contents.
AFM Imaging
The DNA rectangle origami were imaged with PeakForce mode on a Multimode VIII AFM machine (Bruker, Inc.). Before scanning the sample using a peak-force fluid tip, approximately 30 µl of TAE-Mg2+ buffer was added to the liquid cell to infiltrate the tip. For the writing of output strand on the DNA origami register, that and the output strand were mixed at a 1:10 molar ratio and incubated for 1 h, followed by PEG8000 purification to remove unbound strands. The retrieval strand (100×) was added with incubatoin for 4 h to replace the stored output strand. Streptavidin (Sigma) was used to visualize the storage process of a biotin-modified output strand on the DNA origami register which, before writing, after writing and after data retrieval, was mixed with streptavadin at a 1:10 ratio and incubated at room temperature for 30 min. Finally, 5 µl of the mixture solution was deposited on a freshly cleaved mica surface and incubated for 3 min. AFM imaging was performed after washing the sample by TAE-Mg2+ buffer five times.
Molecular dynamics simulation
Data on the leak mechanism were acquired through molecular simulations using the coarse-grained model oxDNA40. oxDNA is top-down parametrized and describes each nucleotide as a site with six anisotropic interactions: excluded volume, stacking, cross-stacking, hydrogen bonding, backbone connectivity and electrostatic repulsion. Here we used the updated oxDNA2 force field with explicit electrostatics41.
The initial structures of the six simulated systems were originally obtained in PDB format and then exported to oxDNA format using the TacoxDNA webserver42. Structures were relaxed in two steps. A Monte Carlo simulation was performed using the DNA_relax force fields and a further relaxation using the max_backbone_force option in a molecular dynamics simulation with the DNA2 force field. During the two processes, mutual traps based on the intended design were applied to enforce relaxation to the intended design. The external forces and backbone force limitations were then released and a production simulation run performed using the same force field. To reduce simulation time, a mutual trap was applied in systems with gaps of 0, 1 and 2 nt. The distances between T(21) and A(51) (distance 1), C(10) and G(62) (distance 2) and C(0) and G(72) (distance 3) were monitored (Supplementary Fig. 16).
Simulations for all simulated systems were performed in the canonical NVT ensemble at 300 K using an Anderson-like thermostat in a periodic cubic box of length 40.87 nm. The simulation time step for integration was 15.15 fs (0.005 oxDNA time units) and molecular dynamics steps were set between 3 × 108 and 2 × 109, which was sufficient to study the leak mechanism for each system. The particle translational diffusion coefficient was set to 2.5, which is about two orders of magnitude faster than in the experiment43, to accelerate diffusive dynamics and improve sampling. The Newtonian step of an Anderson-like thermostat was 103 and configurations were saved for analysis every 1 × 104 steps. The salt condition for all simulations in this paper was set to monovalent NaCl concentration as 1 M NaCl44. The suite of oxDNA analysis scripts (distance.py, bond_analysis.py and contact_map.py)45 was used for a detailed exploration of the leak mechanism.
Decision tree training and validation
We used miRNA-seq data from the TCGA KIRC dataset for training the decision tree classifier. We randomly divided 521 KIRC and 71 healthy samples of this dataset into a training set and a test set with an 8:2 ratio in silico model. The decision tree classifier was trained with miRNA expression values in base –2 logarithm of reads per million, and threshold values for each miRNA were obtained (training set included 414 KIRC and 59 healthy samples) to classify cancer and healthy groups using Scikit-learn (Sklearn) by Python. This classifier was validated using a test set (including 107 KIRC and 12 healthy samples). The expression values of mi204, 200c and 887 from the database were scaled to 0–100 nM. DPGA performs proof-of-concept nonlinear classification with these miRNA inputs.
Data availability
The data that support the findings of this study are available in the manuscript or the Supplementary Information. Source data are provided with this paper. All other data is available on request.
Code availability
Source codes used in this study (Visual DSD, MATLAB, Python) are available from GitHub (https://github.com/FeiWANG-SJTU/DPGA). All other codes are available from the corresponding authors on reasonable request.
References
Burks, A. W. in A History of Computing in the Twentieth Century (ed. Metropolis, N.) 311–344 (Elsevier, 1980).
Bogaerts, W. et al. Programmable photonic circuits. Nature 586, 207–216 (2020).
Chen, X. & Ellington, A. D. Shaping up nucleic acid computation. Curr. Opin. Biotechnol. 21, 392–400 (2010).
Li, J., Green, A. A., Yan, H. & Fan, C. Engineering nucleic acid structures for programmable molecular circuitry and intracellular biocomputation. Nat. Chem. 9, 1056–1067 (2017).
Benenson, Y. et al. Programmable and autonomous computing machine made of biomolecules. Nature 414, 430–434 (2001).
Qian, L. & Winfree, E. Scaling up digital circuit computation with DNA strand displacement cascades. Science 332, 1196–1201 (2011).
Seelig, G., Soloveichik, D., Zhang, D. Y. & Winfree, E. Enzyme-free nucleic acid logic circuits. Science 314, 1585–1588 (2006).
Wang, F. et al. Implementing digital computing with DNA-based switching circuits. Nat. Commun. 11, 121 (2020).
Thubagere, A. J. et al. A cargo-sorting DNA robot. Science 357, eaan6558 (2017).
Chao, J. et al. Solving mazes with single-molecule DNA navigators. Nat. Mater. 18, 273–279 (2019).
Cherry, K. M. & Qian, L. L. Scaling up molecular pattern recognition with DNA-based winner-take-all neural networks. Nature 559, 370–376 (2018).
Kishi, J. Y., Schaus, T. E., Gopalkrishnan, N., Xuan, F. & Yin, P. Programmable autonomous synthesis of single-stranded DNA. Nat. Chem. 10, 155–164 (2018).
Zhang, Y. et al. Selective transformations between nanoparticle superlattices via the reprogramming of DNA-mediated interactions. Nat. Mater. 14, 840–847 (2015).
Douglas, S. M., Bachelet, I. & Church, G. M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 335, 831–834 (2012).
Lopez, R., Wang, R. & Seelig, G. A molecular multi-gene classifier for disease diagnostics. Nat. Chem. 10, 746–754 (2018).
Zhang, C. et al. Cancer diagnosis with DNA molecular computation. Nat. Nanotechnol. 15, 709–715 (2020).
Hills, G. et al. Modern microprocessor built from complementary carbon nanotube transistors. Nature 572, 595–602 (2019).
Debnath, S. et al. Demonstration of a small programmable quantum computer with atomic qubits. Nature 536, 63–66 (2016).
Athanas, P. M. & Silverman, H. F. Processor reconfiguration through instruction-set metamorphosis. Computer 26, 11–18 (1993).
Ruiz-Rosero, J., Ramirez-Gonzalez, G. & Khanna, R. Field programmable gate array applications—a scientometric review. Computation 7, 63 (2019).
Benenson, Y. Biomolecular computing systems: principles, progress and potential. Nat. Rev. Genet. 13, 455–468 (2012).
Pei, R., Matamoros, E., Liu, M., Stefanovic, D. & Stojanovic, M. N. Training a molecular automaton to play a game. Nat. Nanotechnol. 5, 773–777 (2010).
Woods, D. et al. Diverse and robust molecular algorithms using reprogrammable DNA self-assembly. Nature 567, 366–372 (2019).
Rabaey, J. M. Digital Integrated Circuits: A Design Perspective (Prentice Hall, 1995).
Wan, N. H. et al. Large-scale integration of artificial atoms in hybrid photonic circuits. Nature 583, 226–231 (2020).
Klosin, A. et al. Phase separation provides a mechanism to reduce noise in cells. Science 367, 464–468 (2020).
Chatterjee, G., Dalchau, N., Muscat, R. A., Phillips, A. & Seelig, G. A spatially localized architecture for fast and modular DNA computing. Nat. Nanotechnol. 12, 920–927 (2017).
Bian, X., Zhang, Z., Xiong, Q., De Camilli, P. & Lin, C. A programmable DNA-origami platform for studying lipid transfer between bilayers. Nat. Chem. Biol. 15, 830–837 (2019).
Joesaar, A. et al. DNA-based communication in populations of synthetic protocells. Nat. Nanotechnol. 14, 369–378 (2019).
Zhang, D. Y. & Winfree, E. Control of DNA strand displacement kinetics using toehold exchange. J. Am. Chem. Soc. 131, 17303–17314 (2009).
Li, W., Zhang, F., Yan, H. & Liu, Y. DNA based arithmetic function: a half adder based on DNA strand displacement. Nanoscale 8, 3775–3784 (2016).
Song, T. et al. Fast and compact DNA logic circuits based on single-stranded gates using strand-displacing polymerase. Nat. Nanotechnol. 14, 1075–1081 (2019).
Lund, K. et al. Molecular robots guided by prescriptive landscapes. Nature 465, 206–210 (2010).
Ke, Y. G., Lindsay, S., Chang, Y., Liu, Y. & Yan, H. Self-assembled water-soluble nucleic acid probe tiles for label-free RNA hybridization assays. Science 319, 180–183 (2008).
Praetorius, F. et al. Biotechnological mass production of DNA origami. Nature 552, 84–87 (2017).
Lauback, S. et al. Real-time magnetic actuation of DNA nanodevices via modular integration with stiff micro-levers. Nat. Commun. 9, 1446 (2018).
Genot, A. J. et al. High-resolution mapping of bifurcations in nonlinear biochemical circuits. Nat. Chem. 8, 760–767 (2016).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Wagenbauer, K. F. et al. How we make DNA origami. ChemBioChem 18, 1873–1885 (2017).
Ouldridge, T. E., Louis, A. A. & Doye, J. P. K. Structural, mechanical, and thermodynamic properties of a coarse-grained DNA model. J. Chem. Phys. 134, 085101 (2011).
Snodin, B. E. K. et al. Introducing improved structural properties and salt dependence into a coarse-grained model of DNA. J. Chem. Phys. 142, 234901 (2015).
Suma, A. et al. TacoxDNA: a user-friendly web server for simulations of complex DNA structures, from single strands to origami. J. Comput. Chem. 40, 2586–2595 (2019).
Sulc, P. Coarse-grained Modelling of Nucleic Acids (Univ. of Oxford, 2014).
Doye, J. P. et al. The oxDNA coarse-grained model as a tool to simulate DNA origami. Methods Mol. Biol. 2639, 93–112 (2023).
Poppleton, E. et al. Design, optimization and analysis of large DNA and RNA nanostructures through interactive visualization, editing and molecular simulation. Nucleic Acids Res. 48, e72 (2020).
Acknowledgements
We thank L. Qian (Caltech) and Y. Huang (Peking University) for helpful discussions. This work was supported by the National Key R&D Program of China (grant no. 2021YFF1200300), the National Natural Science Foundation of China (grant nos. T2188102, 21991134, 21904060, 22025404 and 22104088), the Science Foundation of Shanghai Municipal Science and Technology Commission (grant nos. 20dz1101000 and 21TQ1400222) and the New Cornerstone Investigator Program. Molecular dynamics simulations were run on the π2.0 cluster supported by the Center for High Performance Computing at Shanghai Jiao Tong University.
Author information
Authors and Affiliations
Contributions
C.F. and F.W. conceived the research. F.W. designed the circuits and wrote the sequence-generating and -compiling programmes. H.L. performed the majority of the experiments. N.X. performed AFM experiments. C.S. and Q.Z. performed magnetic field-related experiments. M.L. and F.W. performed simulations. L.Z. trained the nonlinear classification model. F.W., C.F. and H.L. analysed data and wrote the manuscript. M.D., J.L. and H.C. participated in data analysis and discussions. C.F., H.C., J.L., N.X., M.D. and X.Z. reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Outline of the design strategy for general-purpose DICs.
a, Architecture for electronic chips integration. b, Hierarchical illustration of scalable DPGA integration (shown by the logical arrangement). We referred architectural properties of programmable electronic integrated circuits to design general-purpose DICs. In electronic integrated circuits, general-purpose chips can be physically integrated, with the information exchange between chips and storage realized via electrons. Analogous to electronic signal, DNA-UTS is used to transmit information. Inter-gate and inter-DPGA information transmission are all enabled by DNA-UTS. With uniform transmitted signals, integrability is permitted both at the gate and the DPGA levels. Asynchronous execution of cascaded DPGAs interdicts molecular diffusion between DPGAs, allowing DPGA integration. Hence, the scalability is enabled with the use of DNA-UTS and DNA origami register. In addition, arbitrary gate connection is allowed in a DPGA, providing rich programming space. In all, the programmability and the scalability support general-purpose computing with DICs.
Extended Data Fig. 2 Representative DNA circuits experimentally implemented via multi-level programming of DPGAs.
The DIC is composed of three levels: DNA gates; DPGAs and DPGA networks.
Extended Data Fig. 3 Demonstrated advantages of dual-rail gates.
a, A single-rail gate receives one molecule as an input and generates one molecule as output. The concentration of output increases rapidly when the output is 1 while slowly when the output is 0. b, A dual-rail gate receives a molecule representing 1 or another molecule representing 1 as an input, and generates one molecule as output = 0 or another molecule as output = 1. The output signal is represented by the difference of two output signals. When output = 1, the output signal increases; when output = 0, the output signal decreases. c, Implemention of a dual-rail XOR gate with AND-OR gates requires six gates. d, Left, heatmap showing the result values with all possible combinations of high and low signals, when a result is supposed to be 0. Triangular region above the upper red line represents the obtained dual-rail results smaller than 0.4. Right, heatmap of error flag with all possible combinations of high and low signals. Regions inside the green box have error flag values lower than 0.4. e, Five possible computing states for a result supposed to be 0. f, Left, heatmap showing the result values with all possible combinations of high and low signals, when a result is supposed to be 1. Triangular region below the lower red line represents the obtained dual-rail result larger than 0.6. Right, heatmap of error flag with all possible combinations of high and low signals. Regions inside green boxed have error flag values lower than 0.4. g, Five possible computing states for a result supposed to be 1.
Extended Data Fig. 4 Internal structures and operating mechanisms of the four dual-rail gates.
a, Schematics showing internal structures of the dual-rail gates. AND gate has two series switches to respond to in1H and in2H respectively, generating outH in the presence of both in1H and in2H. Another switch responds to both L inputs, generating ouL in the presence of either in1L or in2L. OR gate has an opposite internal structure of AND gate. NOT gate has two switches, one for L input and one for H input. XOR contains four input-controlled switches. b–e, Signal transmission paths with all possible input combinations for AND (b), OR (c), NOT (d) and XOR (e) gates. High signals are shown in red and low signals in green. Single-stranded inputs bind to logic gates and release the corresponding output strands through SDRs. For AND gate, both in1L (representing in1 = 0) and in2L hybridize with s1 to displace s2 (=outL). in2H hybridizes with S3 to displace S5, exposing toehold γ for in1H to replace S4 (=outH).
Extended Data Fig. 5 The complete simulated time trajectories (a) for Fig. 2e and distance distributions during the simulation time (b).
With 1-nt gap, the toehold domain bound reversibly to S3 (middles panel of Distance 1 trajectory, inset), which did not lead to further branch migration (high fluctuation for Distance 2 and 3). Therefore, the presence of only in1H cannot generate a fault result, permitting neglectable leakage similar to that of 0-nt gap. However, 2-nt gap allowed stable binding of in1H (left panel of Distance 1 trajectory, indicated by H-bond arrow), and the output strand was replaced via branch migration (left panels of Distance 2 and 3 trajectories, indicated by H-bond arrows). Insets in (b): Distributions of Distance 1 ranging from 0.5 nm to 2.5 nm.
Extended Data Fig. 6 Experimental optimization and performance evaluation of WIR2 and WIR3.
a, Molecular reactions for wiring (for WIR2) or reading out (for WIR3) an output signal from a gate. Low (green) and high (red) signals are transmitted independently. The dashed lines indicate the upstream binding region. b, To introduce the threshold-over-amplifier binding priority, the duplex region was shortened by 2 bp, which minimized non-specific input-threshold binding and the leakage. c, Without Threshold, output from a gate could be amplified to close to 1. However, weak signal leakage could also be amplified, leading to false result. Thus, the threshold is essential to suppress leakage before amplification. d, We used a Threshold molecule (Th) that could interact with output quickly. With 0.4× leakage signal, we found Th with a concentration higher than 0.4× can effectively suppress leakage. With 1× output signal, we found signal transmission speed decreased with Th concentration. To balance leakage suppression and computing speed, we used 0.4× to 0.6× Threshold for experiments. e, Signal wiring for High signal (left) and Low signal (right) using 0.4× threshold.
Extended Data Fig. 7 A representative compiling process of the DNA Compiler.
Statements containing different type of operations and different priorities can be compiled into wiring instructions to configure DPGA.
Extended Data Fig. 8 The signal relay function of WIR4s.
a, Possible transient binding reactions between unmatched molecules that may affect the circuit performance. b, Numerical simulation of the computing process from upstream inputs to downstream inputs considering the transient binding. With the increase of the circuit size, the exposed toeholds have a higher chance of being occupied by unmatched strands via transient binding, leading to reduced computing speed. c, Experimental computing kinetics of an OR gate when 0, 500, 1000 and 1500 nM unmatched threshold molecules were added, respectively. d, Dual-rail results of the OR gate followed by a WIR3 at different unmatched threshold levels. e, The 11-layer cascade circuit in Fig. 3g when implemented with a single DPGA. f, Simulated computing kinetics of 1- to 11-layer subcircuits implemented with a single DPGA without WIR4. g, Simulated dual-rail results of the cascade circuit showing decayed performance with the increase of the circuit depth, which is generally consistent with the experimental results. h, Circuit diagram of a cascade circuit containing 11 layers of dual-rail gates, which was divided and implemented by three configured DPGAs. i, Simulated computing kinetics of the subcircuits in the cascaded DPGAs connected by WIR4s.
Extended Data Fig. 9 Signal decay during the cascade of DPGAs.
a–b, Illustration of the circuit depth. The largest circuit depth (a) and the corresponding SDR pathway that contains 30 steps of reactions (b), beyond what can be achieved with a single reaction. c, Schematic illustrations showing tested systems with direct ideal input and that transmitted by WIR4s. d, Paired comparison showing the increase of the half-competition time (t1/2). d–g, Output, leakage and error showing insignificant difference between DPGAs using direct input and transmitted input.
Extended Data Fig .10 Computing kinetics of 20 experimentally tested samples in addition to that in Fig. 6g in test set.
The values in bracket show the normalized concentration of mir-200c, mir-204 and mir-887, respectively.
Supplementary information
Supplementary Information
Methods, texts, Figs. 1–62, Tables 1–7 and references that extend and support the data and discussion presented in the main text.
Supplementary Data 1
DNA sequences used in each figure.
Supplementary Data 2
DNA sequences for dual-rails gates and wiring instructions.
Supplementary Data 3
Information and experimental results of 103 circuits for robustness evaluation.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lv, H., Xie, N., Li, M. et al. DNA-based programmable gate arrays for general-purpose DNA computing. Nature 622, 292–300 (2023). https://doi.org/10.1038/s41586-023-06484-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-023-06484-9
- Springer Nature Limited
This article is cited by
-
DNA as a universal chemical substrate for computing and data storage
Nature Reviews Chemistry (2024)
-
Chemical unclonable functions based on operable random DNA pools
Nature Communications (2024)
-
Chemical reservoir computation in a self-organizing reaction network
Nature (2024)
-
A spatially localized DNA linear classifier for cancer diagnosis
Nature Communications (2024)
-
Template-dependent DNA ligation for the synthesis of modified oligonucleotides
Nature Communications (2024)