Abstract
The application of microarray technology to the diagnosis of cancer has been a challenge for computational techniques because the datasets obtained have high dimension and a few examples. In this paper two computational techniques are applied to tumor datasets in order to carry out the task of diagnosis of cancer (classification task) and identifying the most promising candidates among large list of genes (gene prioritization). Both techniques obtain good classification results but only one provides a ranking of genes as additional information and thus, more interpretable models, being more suitable for jointly addressing both tasks.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Tumor Classification from Gene Expression Data
The challenge of cancer treatment has been to target specific therapies to pathogenetically distinct tumor types, to maximize efficacy and minimize toxicity. Improvements in cancer classification have thus been central to advances in cancer treatment. Cancer classification is divided into two challenges: class discovery and class prediction. Class discovery refers to defining previously unrecognized tumor subtypes. Class prediction refers to the assignment of particular tumor examples to already-defined classes. In the early days, cancer classification has been relying on subjective judgment from experienced pathologists. When microarray technology was discovered began to be applied to cancer diagnosis. The most important application of the microarray technique is to discriminate the normal and cancerous tissue samples according to their expression levels, identify a small subset of genes that are responsible for the disease and to discover potential drugs [15].
Experimental techniques based on oligonucleotide or cDNA arrays now allow the expression level of thousands of genes to be monitored in parallel [1]. To use the full potential of such experiments, it is important to develop the ability to process and extract useful information from large gene expression datasets.
Constantly improving gene expression profiling technologies are expected to provide understanding and insight into cancer related cellular processes. Gene expression data is also expected to significantly aid in the development of efficient cancer diagnosis and classification platforms. Gene expression data can help in better understanding of cancer. Normal cells can evolve into malignant cancer cells through a series of mutations in genes that control the cell cycle, apoptosis, and genome integrity, to name only a few. As determination of cancer type and stage is often crucial to the assignment of appropriate treatment [16], a central goal of the analysis of gene expression data is the identification of sets of genes that can serve, via expression profiling assays, as classification or diagnosis platforms.
Another important purpose of gene expression studies is to improve understanding of cellular responses to drug treatment. Expression profiling assays performed before, during and after treatment, are aimed at identifying drug responsive genes, indications of treatment outcomes, and at identifying potential drug targets [9]. More generally, complete profiles can be considered as a potential basis for classification of treatment progression or other trends in the evolution of the treated cells.
Data obtained from cancer related gene expression studies typically consists of expression level measurements of thousands of genes. This complexity calls for data analysis methodologies that will efficiently aid in extracting relevant biological information. Previous gene expression analysis work emphasizes clustering techniques (nonsupervised classification), which aim at partitioning the set of genes into subsets that are expressed similarly across different conditions. On the other hand, supervised classification techniques (also called class prediction or class discrimination) with the aim to assign examples to predefined categories [12, 16, 19].
The objectives of supervised classification techniques are: (1) to build accurate classifiers that enable the reliable discrimination between different cancer classes, (2) to identify biomarkers of diseases, i.e. a small set of genes that leads to the correct discrimination between different cancer states. This second purpose of supervised classification can be achieved by classifiers that provide understandable results and indicate which genes contribute to the discrimination.
Following this line, in this paper the goal is to apply two techniques with embedded capacity to discard input features and thus propose a subset of discriminative genes (embedded methods [20]). We apply them to classify and select features to tumor datasets in order to carry out an analysis of these datasets and to obtain the information that provide understandable results. These techniques are the Fuzzy Random Forest method (FRF) proposed in [3, 7] and the Feature Selection Fuzzy Random Forest method (FRF-fs) proposed in [6].
This paper is organized as follows. First, in Sect. 2 some techniques applied to gene expression data reported in literature are briefly described. Next, in Sect. 3, the applied methods are described. Then, in Sect. 4 we perform an analysis of two tumor datasets using these methods. Finally, in Sect. 5 remarks and conclusions are presented.
2 Machine Learning and Gene Expression Data
In this section, we describe some of the machine learning techniques used for the management of gene expression data.
2.1 Cluster Analysis Based Techniques
Clustering is one of the primary approaches to analyze such large amount of data to discover the groups of co-expressed genes. In [18] an attempt to improve a fuzzy clustering solution by using SVM classifier is presented. In this regard, two fuzzy clustering algorithm, VGA and IFCM have been used.
In [1] a clustering algorithm to organize the data in a binary tree is used. The algorithm was applied to both the genes and the tissues, revealing broad coherent patterns that suggest a high degree of organization underlying gene expression in these tissues. Coregulated families of genes clustered together. Clustering also separated cancerous from noncancerous tissue.
In [16] a SOM to divide the leukemia examples into cluster is used. First, they applied a two-cluster SOM to automatically discovering the two types of leukemia. Next, they applied a four-cluster SOM. They subsequently obtained immunophenotype data on the examples and found that the four classes largely corresponded to AML, T-lineage ALL, B-lineage ALL, and B-lineage ALL, respectively. The four-cluster SOM thus divided the examples along another key biological distinction.
In [2] a clustering based classifier is built. The clustering algorithm on which the classifier is constructed is the CAST algorithm that takes as input a threshold parameter t, which controls the granularity of the resulting cluster structure, and a similarity measure between the tissues. To classify a example they cluster the training data and example, maximizing compatibility to the labeling of the training data. Then they examine the labels of all elements of the cluster the example belongs to and use a simple majority rule to determine the unknown label.
2.2 Techniques for Feature Selection and Supervised Classification
Discovering novel disease genes is still challenging for constitutional genetic diseases (a disease involving the entire body or having a widespread array of symptoms) for which no prior knowledge is available. Performing genetic studies frequently result in large lists of candidate genes of which only few can be followed up for further investigation. Gene prioritization establishes the ranking of candidate genes based on their relevance with respect to a biological process of interest, from which the most promising genes can be selected for further analysis [19]. This is a special case of feature selection, a well-known problem in machine learning.
In [16] a procedure that uses a fixed subset of “informative genes” is developed. These “informative genes” are chosen based on their correlation with the class distinction.
In [12], a Random Forest ensemble is used to carry out the feature selection process for classification from gene expression data. The technique calculates a measure of importance for each feature based on how the permutation of the values of that feature in the dataset affects to the classification of the out-of-bag (OOB) dataset of each decision tree of ensemble [5]. Following this study, in [14], a Random forest ensemble which solves the problems existing in [12] is proposed.
In [13] a study of classification of gene expression data using metaheuristics is presented. The authors show that gene selection can be casted as a combinatorial search problem, and consequently be handled by these optimization techniques.
In [19], four different strategies to prioritize candidate genes are proposed. These strategies are based on network analysis of differential expression using distinct machine learning approaches to determine whether a gene is surrounded by highly differentially expressed genes in a functional association or protein-protein interaction network.
Another work to select genes is proposed in [10]. This paper shows that a systematic and efficient algorithm, mixed integer linear programming based hyper-box enclosure (HBE) approach, can be applied to classification of different cancer types efficiently.
3 Classification and Feature Selection by Fuzzy Random Forest
In this section, we describe the methods that we will use in this paper.
3.1 Fuzzy Random Forest for Classification
We briefly describe the Fuzzy Random Forest (FRF) ensemble proposed in [3, 7]. FRF ensemble was originally presented in [3], and then extended in [7], to handle imprecise and uncertain data. We describe the basic elements of the FRF ensemble and the types of data that are supported by this ensemble in both learning and classification phases.
Fuzzy Random Forest Learning
Let E be a dataset. FRF learning phase uses Algorithm 1 to generate the FRF ensemble whose base classifier is a Fuzzy Decision Tree (FDT). Algorithm 2 shows the FDT learning algorithm [8].

Algorithm 2 has been designed so that the FDTs can be constructed without considering all the features to split the nodes. Algorithm 2 is an algorithm to construct FDTs where the numerical features have been discretized by a fuzzy partition. The domain of each numerical feature is represented by trapezoidal fuzzy sets, \(F_1,\ldots ,F_f\) so each internal node of the FDTs, whose division is based on a numerical feature, generates a child node for each fuzzy set of the partition. Moreover, Algorithm 2 uses a function, denoted by \(\chi _{t,N}(e)\), that indicates the degree with which the example e satisfies the conditions that lead to node N of FDT t. Each example e is composed of features which can be crisp, missing, interval, fuzzy values belonging (or not) to the fuzzy partition of the feature. Furthermore, we allow the class feature to be set-valued. These examples (according to the value of their features) have the following treatment:

-
Each example e used in the training of the FDT t has assigned an initial value \(\chi _{t,root}(e)\). If an example has a single class this value is 1. If an example has a set-valued class, it is replicated with a weight according to the available knowledge about the classes.
-
According to the membership degree of the example e to different fuzzy sets of partition of a split based on a numerical feature:
-
If the value of e is crisp, the example e may belong to one or two children nodes, i.e., \(\mu _{fuzzy\_set\_partition}(e)>0\). In this case \(\chi _{t,child node}(e)=\chi _{t,node}(e)\cdot \mu _{fuzzy\_set\_partition}(e)\).
-
If the value of e is a fuzzy value matching with one of the sets of the fuzzy partition of the feature, e will descend to the child node associated. In this case, \(\chi _{t,child node}(e)=\chi _{t,node}(e)\).
-
If the value of e is a fuzzy value different from the sets of the fuzzy partition of the feature, or the value of e is an interval value, we use a similarity measure, \(\mu _{simil}(\cdot )\), that, given the feature “Attr” to be used to split a node, measures the similarity between the values of the fuzzy partition of the feature and fuzzy values or intervals of the example in that feature. In this case, \(\chi _{t,childnode}(e)=\chi _{t,node}\cdot \mu _{simil}(e)\).
-
When the example e has a missing value, the example descends to each child node \(node_h\), \(h=1,\ldots ,H_i\) with a modified value proportionately to the weight of each child node. The modified value for each \(node_h\) is calculate as \(\chi _{node_h}(e)=\chi _{node}(e) \cdot \frac{T\chi _{node_{h}}}{T\chi _{node}}\) where \(T\chi _{node}\) is the sum of the weights of the examples with known value in the feature i at node and \(T\chi _{node_{h}}\) is the sum of the weights of the examples with known value in the feature i that descend to the node \(node_h\).
-
Fuzzy Random Forest Classification
The fuzzy classifier module operates on FDTs of the FRF ensemble using one of these two possible strategies: Strategy 1—Combining the information from the different leaves reached in each FDT to obtain the decision of each individual FDT and then applying the same or another combination method to generate the global decision of the FRF ensemble; and Strategy 2—Combining the information from all leaves reached from all FDTs to generate the global decision of the FRF ensemble.
3.2 Fuzzy Random Forest for Feature Selection
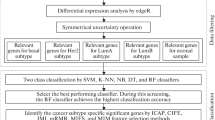
The FRF-fs method [6] is classified as a hybrid method that combines the filter and wrapper methods. The framework (Fig. 1) consists of main steps: (1) Scaling and discretization process of the feature set; and feature pre-selection using the discretization process; (2) The feature pre-selection ranking process using information given by Fuzzy Random Forest ensemble; and (3) Wrapper feature selection using a classification technique. Starting from the ordered features, this wrapper method constructs an ascending sequence of sets of candidate features, by invoking and testing the features stepwise. The different feature subsets obtained by this process are evaluated by a machine learning method. In each step, the method obtains information useful to the user: pre-selected feature subset, feature subsets ranking and optimal feature subset.
In the filter method, we use the method proposed in [8]. From the feature subset and the dataset obtained with the filter method, we apply FRF method. Once FRF ensemble has been obtained, we have all the information about each FDT. Algorithm 3 describes how information provided for each FDT of the ensemble is compiled and used to measure the importance of each feature.
More specifically, the information we get from each FDT t for each feature a is the following:
-
Information gain of node N for the feature a (\(IG_{Na}\)) where the feature a has been selected as the best candidate to split it.
-
Depth level of node N (\(P_{Na}\)) where feature a has been selected as the best candidate to split it.
-
Classification accuracy \(Acc_t\) of FDT t when classifying the dataset \(OOB_t\).

Algorithm 4 details how the information INF obtained from the FRF ensemble is combined to obtain an importance measure of the features where \(p_i\) is the weight we assign to feature a depending on the place where it appears in the FDT t. After the information is combined, the output of this algorithm is a matrix (IMP) where for each FDT t and for each feature a, the importance value obtained in the FDT t for the feature a is stored.

The idea behind the measure of importance of each feature is that it uses the features of the FDTs obtained and the decision nodes built with them in the following way. The importance of a feature is determined by its depth in a FDT. Therefore a feature that appears on the top of a FDT is more important in that FDT than another feature that appears in the lower nodes. And, a FDT that has a classification accuracy greater than another to classify the corresponding OOB (dataset independent of the training dataset) is a better FDT. The final decision is agreed by the information obtained for all FDTs.
As a result of Algorithm 4, we obtain for each FDT of FRF ensemble an importance ranking of features. Specifically, we will have TN importance rankings for each feature a. Applying an operator OWA, we add them into one ranking. This final ranking indicates the definitive importance of the features.
OWA operators (Ordered Weighted Averaging) were introduced by Yager in 1988 [22]. OWA operators are known as compensation operators. They are aggregation operators of numerical information that consider the order of the assessments that will be added. In our case, we have TN ordered sets. Given a weight vector W, the vector RANK represents the ranking of the pre-selected feature subset and is obtained as follows (the vector RANK is ordered in descending order: \(RANK_{\sigma }\)):
3.3 Wrapper for Feature Final Selection
Once the ranking of the pre-selected feature subset, \(RANK_{\sigma }\), is obtained, we have to find an optimal subset of features. One option to search the optimal subset is by adding a single feature at a time following a process that uses \(RANK_{\sigma }\). The several feature subsets obtained by this process are evaluated by a machine learning method that supports low quality data (called \(Classifier_{LQD}\)) with a process of cross-validation. The detailed process of the proposed wrapper method is shown in Algorithm 5.
Starting from the ordered feature pre-selected, construct an ascending sequence of FRF models, by invoking and testing the features stepwise. We perform a sequential feature introduction in two phases:
-
In the first phase two feature subsets are built: the feature subsets \(CF_{base}\) and \(CF_{comp}\). A feature \(f_i\) is added to the \(CF_{base}\) subset only if the decrease of the error rate using the features of \(CF_{base}\cup \{f_i\}\) subset exceeds a threshold \(\delta _1\). The idea is that the error decrease by adding \(f_i\) must be significant for that feature to belong to the \(CF_{base}\) subset. If when we classify using the subset \(CF_{base}\cup \{f_i\}\), an error decrease smaller than a threshold \(\delta _1\) or an error increase smaller than a threshold \(\delta _2\) is obtained, \(f_i\) becomes part of the subset \(CF_{comp}\).
-
The second phase starts with both \(CF_{base}\) and \(CF_{comp}\) sets. We fix \(CF_{base}\) and add feature subgroups from \(CF_{comp}\) to build several FRF models. This phase determines the final feature set with minimum error according to the conditions reflected on line 22 of Algorithm 5. These conditions are interpreted as “select the subset that decrements the error in an amount over threshold \(\delta _3\) or decrements the error in an amount below \(\delta _3\) but using a smaller number of features.”
4 FRF and Tumor Classification
In this section we examine the performance of the FRF ensemble for classification and feature selection from gene expression data.
4.1 Gene Expression Data
In this section, we describe the three datasets that we will analyze. The first dataset involves comparing tumor and normal examples of the same tissue, the second one involves examples from two variants of the same disease and the third one contains measurements of the gene expression of cancer patients and healty men.

Colon Cancer, Leukemia and Prostate Datasets. Colon tumor is a disease in which cancerous growths are found in the tissues of the colon epithelial cells. The Colon dataset contains 62 examples. Among them, 40 tumor biopsies are from tumors (labeled as “negative”) and 22 normal (labeled as “positive”) biopsies are from healthy parts of the colons of the same patients. The final assignments of the status of biopsy examples were made by pathological examination. The total number of genes to be tested is 2000 [1].
In the 1960s was provided the first basis for classification of acute leukemias into those arising from lymphoid precursors (acute lymphoblastic leukemia, ALL) or from myeloid precursors (acute myeloid leukemia, AML). The Leukemia dataset is a collection of expression measurements reported by [16]. The dataset contains 72 examples. These examples are divided to two variants of leukemia: 25 examples of acute myeloid leukemia (AML) and 47 examples of acute lymphoblastic leukemia (ALL). The source of the gene expression measurements was taken from 63 bone marrow examples and 9 peripheral blood examples. Gene expression levels in these 72 examples were measured using high density oligonucleotide microarrays. The expression levels of 7129 genes are reported.
Prostate dataset contains gene expression data (6033 genes for 102 examples) from the microarray study reported by [21]. The obtained results support the notion that the clinical behavior of prostate cancer is linked to underlying gene expression differences that are detectable at the time of diagnosis. This dataset contains measurements of gene expression of 52 prostate patients and 50 healty men.
4.2 Estimating Prediction Errors
We apply the cross-validation method to evaluate the prediction accuracy of the classification method. To apply this method, we partition the dataset E into k sets of examples, \(C_1,\ldots ,C_k\). Then, we construct a data set \(D_i=E-C_i\), and test the accuracy of a model obtained from \(D_i\) on the examples in \(C_i\). We estimate the accuracy of the method by averaging the accuracy over the k cross-validation trials.
There are several possible choices of k. A common approach is to set \(k=\)number of examples. This method is known as leave one out cross validation (LOOCV). We will use the LOOCV method.
Although our purpose is not to compare the results with other methods, as a sample, in Table 1 we show the accuracy estimates for the different methods applied to the three datasets. The results obtained in [12, 14] are calculated with the 0.632+bootstrap method, and the Leukemia dataset has 38 examples and 3051 features.
Estimates of classification accuracy give only a partial insight on the performance of a method. Also, we treat all errors as having equal penalty. In the problems we handle, however, errors have asymmetric weights. We distinguish false positive error- normal tissues classified as tumor, and false negative errors - tumor tissues classified as normal. In diagnostic applications, false negative errors can be detrimental, while false positives may be tolerated.
ROC curves are used to evaluate the “power” of a classification method for different asymmetric weights [4, 17]. Since the area under the ROC curve (denoted by AUC) is a portion of the area of the unit square, its value will always be between 0.0 and 1.0. A realistic classifier should not have an AUC less than 0.5 (area under the diagonal line between (0,0) and (1,1)). The AUC has an important statistical property: the AUC of a classifier is equivalent to the probability that the classifier will rank a randomly chosen positive instance higher than a randomly chosen negative instance. This is equivalent to the Wilcoxon test of ranks [17].
The confusion matrixes obtained by applying FRF to the three datasets are shown in Table 2.
Confusion matrix of Colon dataset shows five errors, and a Specificity of 0.9091 and Sensibility of 0.9250. Confusion matrix of Leukemia dataset shows one error, and a Specificity of 1.0 and Sensibility of 0.9787. Confusion matrix of Prostate dataset shows four errors, and a Specificity of 0.98 and Sensibility of 0.9423.
ROC curves with all features are shown in Fig. 2 and AUC values for (a) Colon, (b) Leukemia and (c) Prostate datasets are 0.9761, 0.9991 and 0.9983 respectively.
4.3 Gene Selection
It is clear that the expression levels of many of the genes in our datasets are irrelevant to the distinction between tumors. Taking such genes into account during classification increases the dimensionality of the classification problem, presents computational difficulties, and introduces noise to the process. Another issue with a large number of genes is the interpretability of the results. If our methods to distinguish tumor from normal tissues is encoded in the expression levels of few genes, then we might be able to understand the biological significance of these genes.
Thus, it is crucial to recognize whether a small number of genes can suffice for good classification. The gene expression datasets are problematic in that they contain a large number of genes (features) and thus methods that search over subsets of features can be expensive. Moreover, these datasets contain only a small number of examples, so the detection of irrelevant genes can suffer from statistical instabilities.
Significance of a Gene and Ranking. The FRF-fs method [6] to feature selection obtains a feature ranking based on an importance measurement of each feature, and from that ranking, an optimal feature subset. The vector RANK (see Sect. 3.2) contains the importance measure of the features. In Table 5 a portion of that ranking of features and their importance values is shown.
Gene Prioritization in Cancer Data. In the final phase of the FRF-fs method [6] an optimal feature subset is obtained.
In the Colon dataset the optimal feature subset is \(\{\)419, 765, 824, 1168, 513, 1772, 571, 1546, 1423, 1761, 1939, 1990, 377, 1668, 1346, 1586, 548, 474, 802, 1867\(\}\). In addition, to give more interpretability, FRF-fs method obtains a feature partition. In Table 6 the partition obtained for this optimal features subset is shown. The first column shows the gene number while the second one shows the different partitions for this gene.
In the Leukemia dataset the optimal feature subset is \(\{\)3252, 4847, 2288, 2354, 6041, 6376, 4644\(\}\). In Table 7 the partition obtained for this optimal features subset is shown.
In the Prostate dataset the optimal feature subset is \(\{\)2619, 5016, 1881, 1359, 4335, 4183, 4087, 4287, 3616, 4136, 3946, 3606\(\}\). In Table 8 the partition obtained for this optimal features subset is shown.
Classifying with Selected Subsets. Now, the classification procedure is applied using the training data restricted to the subset of selected genes.
In Table 3 we show the accuracy estimates for FRF method applied to the three datasets with/without the selected features.
The confusion matrixes obtained by applying FRF to the three datasets with the selected features are shown in Table 4.
Confusion matrix of Colon dataset shows four errors, and a Specificity of 0.9091 and Sensibility of 0.9500. Confusion matrix of Leukemia dataset shows one error, and a Specificity of 0.9600 and Sensibility of 1.0. Confusion matrix of Prostate dataset shows three errors, and a Specificity of 0.98 and Sensibility of 0.9615. ROC curves are shown in Fig. 2. AUC values for Colon, Leukemia and Prostate are 0.9710, 0.9987 and 0.9954 respectively.
Following the methods proposed in [11, 17], we conclude that there are no significant differences between the results obtained when using all features or the selected ones.
We can therefore conclude that the selection of features does not cause loss of accuracy but significantly decreases the number of features.
5 Conclusions
In this paper we have applied a fuzzy decision tree ensemble to tumor datasets with gene expression data.
On the one hand, we have applied the ensemble to the classification of examples described by the set of all features. On the other hand, we have applied the ensemble to select a gene subset and to classify examples only described with the selected genes. The classification accuracies, in both cases, are high. These results are validated statistically by the ROC curve and AUC area.
When we work with a fuzzy decision tree ensemble, in addition to achieve good results, these one are provided in a highly interpretable way.
As part of the solution, the method provides a partition of numerical features of the problem and a ranking of importance of these features which permits the identification of sets of genes that can serve as classification or diagnosis platforms.
References
Alon, U., Barkai, N., Notterman, D.A., Gish, K., Ybarra, S., Mack, D., Levine, A.J.: Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc. Natl. Acad. Sci. U.S.A. 96, 6745–6750 (1999)
Ben-Dor, A., Bruhn, L., Friedman, N., Nachman, I., Schummer, M., Yakhini, Z.: Tissue classification with gene expression profiles. J. Comput. Biol. 7(3–4), 559–583 (2004)
Bonissone, P.P., Cadenas, J.M., Garrido, M.C., Díaz-Valladares, R.A.: A fuzzy random forest. Int. J. Approximate Reasoning 51(7), 729–747 (2010)
Brandley, A.P.: The use of the area under the roc curve in the evaluation of machine learning algorithms. Pattern Recogn. 30(7), 1145–1159 (1997)
Breiman, L.: Random forests. Mach. Learn. 45, 5–32 (2001)
Cadenas, J.M., Garrido, M.C., Martínez, R.: Feature subset selection filter-wrapper based on low quality data. Expert Syst. Appl. 40, 1–10 (2013)
Cadenas, J.M., Garrido, M.C., Martínez, R., Bonissone, P.P.: Extending information processing in a fuzzy random forest ensemble. Soft Comput. 16(5), 845–861 (2012)
J.M. Cadenas, M.C. Garrido, R. Martínez, P.P. Bonissone, Ofp\(\_\)class: a hybrid method to generate optimized fuzzy partitions for classification. Soft Comput. 16(4), 667–682 (2012)
Clarke, P.A., George, M., Cunningham, D., Swift, I., Workman, P.: Analysis of tumor gene expression following chemotherapeutic treatment of patients with bowel cancer. Nat. Genet. 23(3), 39–39 (1999)
Dagliyan, O., Uney-Yuksektepe, F., Kavakli, I.H., Turkay, M.: Optimization based tumor classification from microarray gene expression data. PLoS ONE 6(2), e14579 (2011)
DeLong, E.R., DeLong, D.M., Clarke-Pearson, D.L.: Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44(3), 837–845 (1988)
Diaz-Uriarte, R., Alvarez de Andrés, S.: Gene selection and classification of microarray data using random forest. BMC Bioinform. 7(3), (2006)
Duval, B., Hao, J.K.: Advances in metaheuristics for gene selection and classification of microarray data. Briefings Bioinform. 11(1), 127–141 (2010)
Genuer, R., Poggi, J.M., Tuleau-Malot, C.: Variable selecting using random forest. Pattern Recogn. Lett. 31(14), 2225–2236 (2010)
Ghoraia, S., Mukherjeeb, A., Duttab, P.K.: Gene expression data classification by VVRKFA. Procedia Technol. 4, 330–335 (2012)
Golub, T.R., Slonim, D.K., Tamayo, P., Huard, C., Gaasenbeek, M., Mesirov, J.: P, Coller H., Loh M., Downing J. R., Caligiuri M. A., Bloomfield C. D., Lander E.S.: Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286(5439), 531–537 (1999)
Hanley, J.A., McNeil, B.J.: The meaning and use of the area under a receiver operating characteristic (roc) curve. Radiology 143(1), 29–36 (1982)
Mukhopadhyaya, A., Maulikb, U.: Towards improving fuzzy clustering using support vector machine: Application to gene expression data. Pattern Recogn. 42(11), 2744–2763 (2009)
Nitsch D., Gonzalves J. P., Ojeda F., De Moor B., Moreau Y.: Candidate gene prioritization by network analysis of differential expression using machine learning approaches. BMC Bioinform. 11(460), (2010)
Saeys, Y., Inza, I., Larraaga, P.: A review of feature selection techniques in bioinformatics. Bioinformatics 23(19), 2507–2517 (2007)
Singh D., Febbo P. G., Ross K., Jackson D. G. et all: Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 1(2), 203–209 (2002)
Yager, R.R.: On ordered weighted averaging aggregation operators in multicriteria decision making. IEEE Trans. Syst. Man Cybern. 18(1), 183–190 (1988)
Acknowledgments
Supported by the projects TIN2011-27696-C02-01 and TIN2011-27696-C02-02 of the Ministry of Economy and Competitiveness of Spain. Thanks also to “Agencia de Ciencia y Tecnología de la Región de Murcia” (Spain) for the support given to Raquel Martínez by the scholarship program FPI.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this paper
Cite this paper
Cadenas, J.M., Garrido, M.C., Martínez, R., Pelta, D., Bonissone, P.P. (2016). Gene Priorization for Tumor Classification Using an Embedded Method. In: Madani, K., Dourado, A., Rosa, A., Filipe, J., Kacprzyk, J. (eds) Computational Intelligence. IJCCI 2013. Studies in Computational Intelligence, vol 613. Springer, Cham. https://doi.org/10.1007/978-3-319-23392-5_20
Download citation
DOI: https://doi.org/10.1007/978-3-319-23392-5_20
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-23391-8
Online ISBN: 978-3-319-23392-5
eBook Packages: EngineeringEngineering (R0)