Abstract
In food industries, due to improper cleaning or control practices, along with certain other factors, pathogenic microbes and contaminants such as (e.g., Bacillus cereus) can form biofilms on surfaces. These biofilm matrixes can potentially cause food spoilage and disease transmission, especially during their detachment stage. It is best to use efficient sterilizing techniques when biofilms are still young since they are harder to destroy as they age. Moreover, biofilm microbes show increased resistance to disinfectants and many antimicrobial compounds, rendering them far more challenging to remove. By synthesizing antimicrobial substances with modern approaches to action, the biotechnology industry is attempting to address the issue of biofilms. Various biotechnological gadgets can aid in the identification of different stages of biofilm development on relevant surfaces. Under the umbrella of “green nanotechnology,” antioxidants, essential oils, dyes, and other ingredients are being utilized for creating anti-biofilm technologies. The use of essential oils as bactericides and preservatives is expanding in the food business. Essential oil metabolites can be loaded with various nanoparticles of embedded polymers and lipids to breakdown biofilm layers. It can be concluded that novel biotechnological advances for preventing biofilm formation in the food industry hold out hope for the future. This chapter discusses the present understanding of the significant microbes responsible for the colonization and maturing of biofilms in the food industry, their related health challenges in ready-to-eat convenience meals, dairies, and other food arenas along with biotechnological applications to control and prevent these biofilms.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Food Biofilms
What Is a Biofilm?
Many microorganisms have a proclivity to attach to surfaces and form a complex made up of extracellular matrix, in various environmental settings. These complex ecosystems with embedded bacteria, are known as biofilms. Furthermore, microbes in biofilm state often times are quite dissimilar in their phenotypic traits and exhibit high antimicrobial tolerability, In contrast to planktonic organisms (Abdallah et al., 2014; Galié et al., 2018) It has been demonstrated that mixed biofilms are resistant to biocides and disinfectants, such as quaternary ammonium compounds.
Impact of Biofilms in the Food Sector
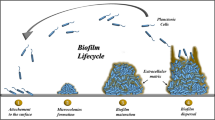
In the food industry setup, biofilms can prove to be a notorious issue to tackle. The type of surface material and the reversible attachment of microbial cells to it are the initial processes involved. After that, the attachment becomes irreversible, with embedded microcolonies. Finally, the biofilm presents as a tridimensional assembly (Fig. 9.1), creating a sophisticated ecosystem that is prepared for dispersal.
Attachment Mechanisms
A variety of biological, chemical, as well as physical mechanisms control the formation of biofilms. The terms adhesion and cohesion are used to describe different types of cell attachment in a biofilm. The adhesive and cohesive qualities that a biofilm will display are ultimately determined by the mechanisms underlying these types of attachment. The different steps involved in biofilm formation are (Marshall, 1986):
(i) adsorption, or the buildup of an organism on a substrate or other collecting surface (deposition); (ii) attachment, which is the stabilization of bacterium-collector contact and frequently entails the building of polymeric bridging between the two; (iii) colonization, i.e., the growth and division of microbes on the collector surfaces.
-
(a)
Conditioning layers: The conditioning layer, which may be made up of several inorganic or organic particulates, serves as the substrate onto which a biofilm can develop (Characklis & Cooksey, 1983). Through gravitational force or flow movement, anything that might be present in the main fluid may settle down on a substrate and eventually becoming a component of the conditioning layer. This layer changes the substrata to make it readily accessible to microorganisms. Surface charge, potential, and tension can be positively affected by the interaction between the conditioning layer and substrate. The substrate gives the bacterial community support and nutrients to help it grow.
-
(b)
Reversible adhesion: Initially, physical traits, including flagella and other bacterial appendages are used to move planktonic microbial cells from the main medium/fluid, to the conditioned surface. Reversible adsorption occurs in a portion of the available bacterial cells that reach the surface. Microbial adherence is locally influenced by characterizing factors, like the amount of energy available, the functionality of the surface, the orientation of the bacteria, and the temperature and pressure levels. The bacteria will separate from the surface if the attracting forces are stronger than the repulsive ones. This is more likely to happen prior to substrate conditioning.
Bacterial desorption has a low activation energy, making it likely to happen and illuminating the bonds’ frailty. DVLO (Derjaguin, Verwey, Landau and Overbeek) forces, also known as van der Waals forces, along with electrostatic (double layer) interactions and steric interactions, are involved in defining the extent of individual bacterial adhesions (Rutter & Vincent, 1984). Van der Waals interactions, whether they be associated with attraction, or repulsion, form an overlap amid the substratum and the cell’s electrical double layer. These interactions defined by the DVLO theory, describe cell to flat surface net interactions, as a balance between dual influential factors (Hermansson, 1999). Also, these physical interactions are classified as long-distance forces, or physisorption. Thermodynamic interaction has also been discussed in terms of a modified DVLO theory, which further considers hydrophobic/hydrophilic and osmotic interactions (Chang & Chang, 2002).
-
(c)
Irreversible adhesion: Some reversibly absorbed cells undergo real-time immobilization and develop irreversible adsorptive bonds. Fimbriae, flagella, pili and other such bacterial appendages have been said to outweigh the electrical double layer’s physical repulsive forces (de Weger et al., 1987). They come into contact with the conditioning layer’s overall framework, promoting oxidation, hydration and other such chemical reactions to strengthen the bacterial cell-to-surface interaction. According to some research, the hydrophilic-hydrophobic characteristics of involved surfaces have a significant impact on microbial adherence.
-
(d)
Population growth: Daughter cells move out and up from their initial attachment sites to cluster up as even more stationary cells continue to undergo binary division (Hall-Stoodley & Stoodley, 2002). These proliferative interactions typically result in the formation of a mushroom-like structure within the growing biofilm. It is thought that the mushroom-shaped structure enables the delivery of nutrients to microorganisms deep within a biofilm.
A rapid population expansion, also known as ‘exponential growth phase’, proceeds an initial ‘lag phase’. This is dependent on the environment’s physical and chemical makeup. The substrate’s and bulk fluid’s nearby nutrients are used up in the process of the rapid growth. Majority of the chemical or physical activities of the initial attachment stage reach a steady halt, and are replaced with other biological communications. Stronger cell bonds are created as a result of the interaction between polysaccharide intercellular adhesion (PIA) polymer excretions and bivalent cations (Dunne Jr., 2002).
The planktonic and sessile state of a bacterial cell differ in terms of gene expression. For instance, because mobility is limited and no longer required in sessile species, the development of surface appendages is hindered. A number of genes that produce excretion products and proteins on cell surfaces are simultaneously expressed more frequently. Opr C and Opr E are examples of surface proteins (porins) that facilitate the outward passage of excretory products (e.g., some polysaccharides), as well as inward passage of necessary extracellular products into individual bacterial cells of biofilms (Hancock et al., 1990).
Final Stages of Biofilm Formation
When cell division and cell death rates are identical, a phase of growth is said to be stationary. Biofilms employ various cellular signaling pathways known as quorum sensing, which become active at significant cell densities (Bassler, 1999). The process of quorum sensing stimulates genetic expressions of mechanical, as well as enzymatic processors of alginates. These play vital roles within the extracellular matrix and consist of diverse auto-inducers, including chemical and peptide signals such as homoserine lactones, which are present in elevated concentrations.
The biofilm is destroyed during the dying phase. By breaking down the polysaccharidal bindings, the biofilm, together with the help of available enzymes nearby, is capable of causing the surface bacteria to be actively released for colonizing onto new substrates. Enzymes involved in the breakdown of the biofilm matrix include alginate lyase produced by Pseudomonas (Ps.) aeruginosa and Ps. fluorescens, hyaluronidase derived from Streptococcus equi, and N-acetyl-heparosan lyase sourced from Escherichia coli (E. coli) (Sutherland, 1999). Various porin-related genes are down-regulated; they play a role in the genetic cycle for adherence and cohesive properties in different biofilms. On the other hand, flagellin-related operons are up-regulated; they provide the bacteria with the machinery for motion.
Resistance Mechanisms
Increased antimicrobial resistance is a problematic property of biofilm-bacteria. Microbes exhibit strong resistance when attached in form of biofilm, making bacterial cells 10–1000 times more resistant to different antibiotics, as compared to the same bacteria produced in free-floating, planktonic forms (e.g., in culture) (Mah & O’Toole, 2001). Major mechanisms involved in this resistance are briefly shown in Fig. 9.2.
Drug Resistance in biofilms: The yellow area stands for the extracellular polysaccharide, whereas blue polygons represent biofilm-bacteria. Nutrient gradients, oxygen and waste products can be found in biofilms, which characterize their heterogeneity (shown by colored crosses). The cellularly dense biofilm, along with its physical expulsion of many antimicrobial classes encompass resistance mechanisms. Additionally, individual biofilm-microorganisms might adapt physiologically to increase their resistance to biocides
-
(a)
Difficult antibiotic penetration of biofilm: The exopolysaccharide matrix (EPM) is a vital feature of any microbial biofilm. A proposition suggests that the EPM serves multiple functions, including acting as a barrier to prevent antibiotics from reaching the bacterial cells. The distribution of antimicrobial compounds, towards the biofilm’s cells, can be constrained by either the compound’s interaction with or sorption to various biofilm constituents. Although past mathematical models initially predicted that there shouldn’t be any obstacles to the diffusion of many antibiotics through a biofilm, modern trials have revealed the apparent inability of some antibacterials to reach their targets in/on the biofilm cells. In the case of a mixed biofilm associated with Klebsiella pneumoniae and Ps. aeruginosa, ordinary disinfectants like chlorine were observed to reach less than 20% of the concentration present in the surrounding bulk media. This evaluation was conducted using a microelectrode specifically designed to detect chlorine levels (De Beer et al., 1994). Theoretically, penetration profile indicated that a substrate may have been consumed inside the matrix. Also, infrared spectroscopy was utilized by Suci, et al. to demonstrate that ciprofloxacin was transported to a colonized surface at a slower pace than it was to a sterile surface (Suci et al., 1994). These scientists hypothesized that the ciprofloxacin was attaching to the elements of the biofilm.
According to Larsen (2002), when Porphyromonas gingivalis’s planktonic populations were evaluated in terms of cell density levels, comparable to those of biofilm populaces (107–108 cells mL−1), the minimum inhibitory concentrations of antimicrobials (metronidazole, amoxicillin and doxycycline) were noticeably heightened. This finding suggested that an inoculum impact contributes to the enhanced bacterial resistance, but it does not fully account for the changes in susceptibilities between planktonic and biofilm organisms. Larsen’s experiments also discovered the tested biofilm populaces possessed two to eight times more resistance to antimicrobials such as doxycycline and amoxicillin, as opposed to equal amounts of planktonic bacteria.
-
(b)
Stress responses and slow growth: A bacterial cell culture’s growth is slowed when it lacks any necessitated nutrient. Antibiotic resistance typically increases as growth goes from exponential to sluggish or no growth (Tuomanen et al., 1986a, b). In established biofilms, the bacteria have been seen to proliferate at slowed rates. It’s been anticipated that this biological alteration is one of the explanations to why biofilms have resistance to many antibiotics; cells growing in biofilms are expectantly encounter varying grades of nutritional constraint.
Modern researches have explicitly explored the significance of survival of biofilm cells, with slowed growth rates, even in the presence of antibiotics. They’ve closely observed growth phases of biofilm-cells as well as planktonic-cells. Gilbert and colleagues (Duguid et al., 1992; Evans et al., 1991) recorded the differing growth rates of biofilms versus planktonic cultures of E. coli, Ps. aeruginosa and Staphylococcus epidermidis. They discovered that susceptibilities to drugs (i.e., ciprofloxacin, tobramycin) increased in parallel to the growth rate, for both biofilm and planktonic-cells. This confirmed a fundamental concept: ‘biofilm-cells’ slowed growth rates act as shields against active antimicrobial action’. With biofilm and planktonic forms of Ps. aeruginosa cells, sub-optimal growth rates yielded similar ciprofloxacin resistance. The cells in planktonic phase, however, stood more vulnerable to ciprofloxacin than the biofilm cells as the development rate was accelerated. This finding lends support to the hypothesis that the biofilm’s documented resistance to antimicrobial treatment can be attributed to factors beyond its gradual formation, indicating the presence of additional contributing characteristics (Desai et al., 1998). Desai et al. conducted a comparative analysis between the resilience of biofilm and planktonic cells at multiple stages during an exponential growth phase, extending until the initiation of a stationary phase. Their investigation revealed an increasing resistance in both biofilm cells and planktonic cultures as they approached the stationary phase. The bacterial cells in biofilms had antimicrobial resistance which was 15 times more than the planktonic cells during the stationary phase of both cultures, which is when resistance was at its highest. These findings indicate that the level of resistance to certain factors is influenced by variables beyond just the growth rate of cells. It appears that delayed growth also plays a role in providing extra protection. One possible explanation for this phenomenon is the observed increase in cellular densities during later stages of exponential growth, which could be linked to this additional protective factor (Brooun et al., 2000).
-
(c)
Quorum sensing (QS): Deeper comprehension pertaining to the concept of quorum-sensing (QS) in bacteria, has paved the way for advancements in knowledge on regulatory mechanisms governing drug resistance processes. The fight against drug resistance have been made possible by this discovery. Study findings demonstrate that the quorum-sensing system controls a number of cellular processes in microorganisms, including extracellular polysaccharide (EPS) synthesis, toxigenic protein production and pathogenic gene expression (Bäuerle et al., 2018; Turan et al., 2017). It also plays significant roles in the functioning of drug efflux pumps and in the development of bacterial biofilms.
Previous research of Davies and colleagues demonstrated a Ps. aeruginosa strain’s mutation in its lasR-lasI QS system, was incapable of biofilm production with a typical structure (Davies et al., 1998). Furthermore, these authors provided evidence that lasI-mutant biofilms were abnormally susceptible to SDS treatment, albeit they did not address the issue of whether these mutant-laden biofilms had changed drug resistance (Davies et al., 1998). Bacterial efflux pump regulation by the QS system has been verified by different researches (Subhadra et al., 2019; Wang et al., 2019).
-
(d)
Physiologically different biofilm cells: A developing theory in the field states that a subset of the community is given a biofilm-specific phenotype, which ignites causes active mechanisms to counter the negative effects of bactericides. Many researches of the present century are aimed at reporting activated or suppressed genes associated with biofilms and comparing planktonic-cell genes upon cellular attachment onto vulnerable surfaces (Kuchma & O’Toole, 2000). Additionally, it’s feasible to say that almost all of these biofilm cells will exhibit heightened antibiotic resistance. High cell density, specific types of stress, nutritional restriction, or a combination of these factors may all contribute to the development of this resistant phenotype. Antimicrobial drugs that are unrelated chemically can be expelled from the cell through multidrug efflux pumps.
The mar operon is upregulated in E. coli, which leads to a multidrug-resistant phenotype. AcrAB is assumed to be the efflux pump in charge of this resistance. By using lacZ fusion (Maira-Litrán et al., 2000), mar expression was tracked in batch, chemostat and biofilm growths to look for any linkage between this well-known mechanism of multi-drug resistance and biofilm resistance to antimicrobials. Mar detected was lower in biofilms than it was in comparable stationary-phase culture batches, contradicting the impression that mar operon is elevated in biofilms.
Ps. aeruginosa has three essentially known multi-drug efflux pumps, and the Ps. aeruginosa genome project has also discovered numerous more putative pumps. One study made a case for the significance of one of these pumps in the development of ofloxacin resistance (Brooun et al., 2000). It demonstrated that, at lesser ofloxacin concentrations, biofilms without the pump were more responsive to this medication, than biofilms overexpressing it; Ps. aeruginosa strains that either lacked or overexpressed the MexAB-OprM pump were used. There was no change, though, for ciprofloxacin, a different quinolone. Therefore, just as with the E. coli investigations, more research is needed to determine whether stimulation of pumps is factually a crucial modification giving resistance to biofilms, or not.
The modification of the composition of membrane proteins due to antibiotics is another mechanism for resistance inducible in bacterial cells of biofilms. This alteration might make cells less permeable to certain substances. E. coli strains with mutated genes of ompB (incharge of regulation for outer membrane porin genes: OmpF, OmpC), as well as ompF, experience greater β-lactam resistance. OmpF-deficient mutants have been found to have higher levels of tetracycline and chloramphenicol resistance. Additionally, the relative ratios of the two main E. coli porins, (OmpC and OmpF) (Jaffe et al., 1982) were changed in starved cells, favoring the production of OmpC, a smaller porin (Liu & Ferenci, 1998). The results mentioned above lend credence to the idea that changing porin expression alters how resistant bacteria are by nature to antimicrobials. In the past, it was also demonstrated that biofilm bacteria expressed more of the omp C gene than planktonic cells did for three other osmotically regulated genes (Pugsley & Schnaitman, 1978). These findings suggest that biofilm-microbes indeed do survive in osmotically stressful microenvironments. Therefore, a biofilm’s external factors may cause changes in cell membranes, shielding the cell from the negative effects of antibacterial drugs.
Effect of Food Biofilms on Health
Many studies have shown the significance and effects of biofilms on the food industry. Biofilms showcase higher antibiotic resistance patterns than their planktonic counterparts, by up to 1000-fold, accounting for almost 20% of cases of food poisoning (Lebeaux et al., 2014). Cross-contamination between relevant food products with food pathogens, such as Bacillus cereus, Campylobacter jejuni, E. coli O157:H7, Listeria monocytogenes, Salmonellae, Staphylococci and Yersinia enterocolitica, are common (Anand et al., 2014). Apart from infections, biofilm may be associated with intoxications. For instance, in food processing units, biofilms can emit toxins, contaminating food matrices, with resultant intoxications in single or multiple persons (outbreaks). In both situations, the presence of biofilms in a food manufacturing facility poses a significant risk to public health. The level of risk associated with these persistent, complex living structures, known as biofilms, is contingent upon the specific bacterial species involved. The formation of biofilms in different types of factories is influenced by factors specific to each facility. In particular, the main areas prone to biofilm development is in proximity include: animal carcasses, assembly-line tops, contact surfaces, dispensing tubing, packing materials, pasteurizer plates, reverse osmosis membranes, storage silos for raw materials, pipelines carrying milk, water or other liquids, etc. (Colagiorgi et al., 2017).
Biofilm Microorganisms
A variety of microorganisms can flourish on foods and throughout the network of the food industry. Varying microbe species may have different capabilities for adhering to and/or forming biofilm on various surfaces. Biofilms have vastly intricate microstructures and are made up of a variety of these symbiotic microbes, including some that are pathogenic to humans, as revealed by various detection technologies and microscopic techniques. The primary microorganisms involved in the early development, colonization and spread of biofilms in the food business are discussed in this part, along with the health problems they may cause (for instance, in association with pre-prepped foods, dairies and other food matrixes) (Galié et al., 2018).
Major bacteria including, Bacillus cereus, Campylobacter jejuni, Clostridium perfringens, Enterococci, Escherichia coli, Listeria monocytogenes, Salmonella, Staphylococcus aureus, Vibrio and others, have been found to establish biofilms on foods and food-contact surfaces, raising serious questions about food safety (Sharan et al., 2022). Biofilms may be essential in ecophysiology, because they promote colonization of a variety of environmental microhabitats, such as farm-food surfaces. They may be linked to symptomless, direct colonization among some hosts, or even transmission through food systems with subsequent infection (Ahmed et al., 2013). Throughout the food business, biofilm-producing microbes may be potentially harmful to humans as they create tough complexes in manufacturing settings. These microorganisms use the processing parameters used in the food sectors, such as glass, polyethylene, stainless-steel, wood and rubber as artificial substrates (Abdallah et al., 2015; Colagiorgi et al., 2017). When disinfecting and sterilizing procedures are taken into consideration, the parameters of different microbial growth forms on foodstuffs in a processing facility involve several propensities. Choosing the best method might be challenging when attempting to prevent biofilm development in the food sector (Carrascosa et al., 2021). Table 9.1 gives brief descriptions of several of these pertinent biofilm-producing microorganisms in the food sector.
Bacillus cereus
The Gram-positive, spore-producing, facultatively anaerobic, Bacillus cereus (B. cereus), can grow in a variety of settings at temperatures ranging from 40 to 120 °F. B. cereus is immune to chemicals, irradiation and thermal treatments (Bottone, 2010). It is a soil-dwelling organism that is typically isolated from food and food-related goods like dairies, meats, rice and vegetables. B. cereus releases toxin elements which are capable of causing diarrheal food poisoning (gastroenteritis) in people (Carrascosa et al., 2021).
B. cereus-laden biofilm matrices are found on materials that come into touch with food, like storage tanks, industrial belts, machinery and metal pipes. These intricate biofilm communities are probably crucial to B. cereus’ capacity to colonize many habitats. In conjunction with their spores, they give the microbe a high level of tolerance and adhesiveness on a number of substrates, namely stainless-steel, a substance that is of frequent use in industrial food lines. B. cereus can survive for extended periods of time and can endure sanitization techniques in these settings (Majed et al., 2016). They may also build up as submerged or as floating biofilms that secrete bacteriocins, proteases, lipases, metabolites and surfactants capable of altering the sensorial properties of food (Dincer et al., 2020). Their flagellar motility provide access to surfaces that are favorable for biofilm formation, and promote propagation onto uncolonized surfaces. B. cereus flagellar mechanisms, however, haven’t been discovered to be necessarily related with adhesion to glass, but their motile activity can play a significant role in biofilm production (Houry et al., 2010).
Bacillus cereus is among the most common causes of gastroenteritis outbreaks, manifesting predominantly as diarrheal or emetic in nature. B. cereus variants associated with emetic predominance are capable of secreting non-ribosomal cyclic, heat-stable toxin peptides into food, that when consumed, causes vomiting. Simply speaking, they tolerate cooking temperatures and contribute to vomiting episodes when consumed (Ehling-Schulz et al., 2015). In accordance with the current paradigm of diarrheal B. cereus food poisoning, foods contaminated with spores are consumed and germination inside the gut is possible, where bacterial spores proliferate alongside the release of enterotoxins. This is particularly true for diarrheal B. cereus strains which secrete cytotoxin K, hemolysin BL and non-hemolytic enterotoxin (Stenfors Arnesen et al., 2008).
Campylobacter jejuni
Campylobacter species are Gram-negative, thermophilic, motile, curved bacilli, with the most common strain being Campylobacter jejuni (C. jejuni) (Klančnik et al., 2021). Aerobic as well as microaerophilic environments allow C. jejuni to form biofilms (Téllez, 2010). C. jejuni is of fastidious nature, but it can remain viable beyond the avian gut before it infects a human host. Biofilm production is triggered by a variety of external conditions, which are then influenced by a number of intrinsic variables (Tram et al., 2020).
According to the European Union One Health 2019 Zoonoses Report, C. jejuni is a frequent commensal in cattle, hens and turkeys. It is regarded as an opportunistic infectious agent and is considered to be a common cause of food-borne illness in humans (Authority & European Centre for Disease Prevention and Control, 2021). C. jejuni enters the host body via infecting and colonizing the gut, subsequently inducing food-borne illness, as a result of contaminated food processing units, water or raw milk (Chlebicz & Śliżewska, 2018).
Escherichia coli
The majority of these Gram-negative, Escherichia coli (E. coli) strains are found as non-pathogenic bacilli in the human gut microflora. The pathogenic/virulent strains consist of: enterohemorrhagic, enteroinvasive, enteropathogenic, enterotoxigenic and vero-cytotoxigenic E. coli (EHEC, EIEC, EPEC, ETEC, VTEC, respectively) (Gould et al., 2013). It is vital to keep in mind that a variety of E. coli strains have the potential to infect people, with EHEC strains being important in the food processing industry. The most prevalent from the EHEC serotype, is the O157:H7 strain, responsible for hemolytic uremic syndrome (HUS) and epidemics of bloody diarrhea around the world. They are transmissible via consumption of contaminated dark green vegetables, drinks, fruits such as melons, meats and unpasteurized milk (Galié et al., 2018; Gould et al., 2013).
E. coli’s propensity to form biofilms is, in large part, what causes it to flourish widely in natural habitats. With their flagella, membrane proteins and pili, E. coli rods are able to attach onto non-living surfaces, creating an extracellular polymeric substance (EPS) which promotes antimicrobial and disinfectant resistance (Lim et al., 2019). Despite the fact that EHEC strains can create biofilms on a variety of surfaces involved in the industrial food sector, there isn’t currently a credible way to stop E.coli biofilms from forming, or a better way to treat EHEC infections because using the limited available medications tends to raise the risk of renal damage and hemolytic uremic syndrome (Lee et al., 2007).
Listeria monocytogenes
Listeria monocytogenes (Gram-positive rod) is a frequently identified bacterium found in animals, decaying vegetation, food, soil, water and responsible for food-borne infection. When consumed, it can cause major difficulties in the old and young, as well as abortions in pregnant females. The infection can spread to many food forms, including cheeses, chicken, dairy goods, fish, fruits, meats, pre-packaged meals, raw milk, frozen goods including ice-creams and vegetables (Rothrock et al., 2017). The primary method of Listeria monocytogenes transmission to people, is through the consumption of contaminated food items (ready-to-eat food products, dairy, meats, poultry and vegetables) (Andreoletti et al., 2007).
L. monocytogenes can produce biofilm on a variety of surfaces utilized in the food business, which poses a major risk to consumer health as this could act as a source of infection. Numerous processed foodstuffs have been found to contain L. monocytogenes, and cooked foods can potentially become compromised as a consequence of subsequent contamination (D’Ostuni et al., 2016; Jofré et al., 2016; Vitas & Garcia-Jalon, 2004; Vongkamjan et al., 2016). Listeria monocytogenes can adhere to a variety of surfaces that come into contact with food, including glass, polystyrene and stainless-steel (Di Bonaventura et al., 2008). It’s been discovered to linger in food sectors for decades, where it may frequently cross-contaminate food stuffs (Ferreira et al., 2014).
The pathogen can exist in complex microbial biofilms or simply as monomicrobial biofilms, and readily thrives at low temperatures (Chmielewski & Frank, 2003). L. monocytogenes may persist in biofilms devoid of oxygen and also can endure low-pH environments for extended periods of time. Depending on the other contending microbial population in biofilms, its population may increase or decrease (Raheem & Raheem, 2016). Most strains of L. monocytogenes in food industrial environments have strong adhesive abilities due to their flagellar, membrane protein and pili properties (Lemon et al., 2007).
Pseudomonas Species
Pseudomonas are heterotrophic, motile, Gram-negative bacilli which may be found as common spoiling microbes in high-pH dairy goods, on fruits, meats, vegetables, as well as in the drainage and flooring of food production facilities (Chmielewski & Frank, 2003; González-Rivas et al., 2018). These bacteria, with their extracellular filamentous extensions, have distinct effects on surface interaction and the adhesion process. Research on their flagellar and pili properties is extensive, relevant with context to biofilms, food spoilage and infections (Amina & Bensoltane, 2015). Pseudomonas aeruginosa (1–5 mm in length; 0.5–1 mm in width), can serve as a model bacterium for exploring the development of biofilms and how quorum sensing pathways guide them. With nitrate serving as a final electron acceptor, they are able to grow as facultative aerobes, through both anaerobic and aerobic means (Golovlev, 2002). Massive volumes of EPS are synthesized by Pseudomonas, which are also known to create biofilms which adhere onto stainless steels. As part of mixed-species biofilms, they may interact with certain other bacteria, increasing their own stability and giving rise to resistance patterns (Chmielewski & Frank, 2003). On soft cheeses, Pseudomonas fluorescens-containing biofilms are often accompanied by pyocyanin production, a unique bluish pigment (Carrascosa et al., 2015).
Salmonella enterica
Salmonella enterica (S. enterica) are facultatively aerobic, flagellated, Gram-negative bacilli, which are often present as colonizers, but are also known to be associated with gastroenteritis, as well as some cases of septicemia (Lamas et al., 2018). Salmonellae produce curli fibers, that are proteinaceous extracellular substances associated with cell to cell and cell to surface communications (Ćwiek et al., 2019). Other than curli, many fimbriae adhesin proteins have been reported, having different biofilm-promoting properties depending on various serotypes (Grigore-Gurgu et al., 2019). In 1966, the first occurrence of complex multicellular biofilms on edible surfaces was discovered and the importance of these bacteria as a food pathogen was brought to light (Duguid et al., 1966). Food-borne salmonellosis presents as one of many common causes of food-related illness. In environments where food is handled or processed, contaminated surfaces may create biofilms that increase the danger of Salmonella poisoning (Corcoran et al., 2014).
From all the serotypes of S. enterica, the Enteritidis serovar is the one most commonly associated with febrile symptoms, abdominal pain, diarrhea, nausea and vomiting (Nguyen et al., 2014). According to a study by Russo et al., despite thorough decontamination and sterilization practices, a strain of Salmonella enterica subspecies enterica’s serotype, Agona was accountable for continuing outbreaks of food-borne illness in the microenvironments of a food manufacturing plant (Russo et al., 2013). Additionally, Salmonella’s Agona serovar has been responsible in the past, for recurrent outbreaks of salmonellosis (Brouard et al., 2007; Killalea et al., 1996; Russo et al., 2013; Shohat et al., 1996; Threlfall et al., 1996). The potential of different disinfection methods to limit the persistence of Salmonella on food surfaces was of interest in light of a significant global outbreak of S. Agona in 2008 (Nicolay et al., 2011). S. enterica strains have the potential to contaminate food streamlines, giving rise to large-scale outbreaks linked with morbidities in immunocompromised populations. It readily develops as multi-dimensional layers on stainless-steel substrates, with varying morphologies, such as reticular colonies produced when cultivated on tryptic soy broth (Wang et al., 2013). On food surfaces in industrial contexts, food poisoning-linked Salmonella strains speedily form biofilms, conferring Salmonella’s persistence in the long-run (Cogan et al., 1999; Møretrø et al., 2012; Reij & Den Aantrekker, 2004; Rodrigues et al., 2011). These biofilms may serve as reservoirs for persistent microbial contamination in food production plants, with potential to give rise to outbreaks of food poisoning (Corcoran et al., 2014).
Staphylococcus aureus
The Gram-positive coccus, Staphylococcus aureus (S. aureus), is a facultative anaerobe that can produce enterotoxins between 10 and 46 °C. S. aureus, when examined under the microscope, are usually visible in a cluster or grape-like arrangement. Their colonies possess a carotenoid pigment which gives them a golden color on nutrient agar, and hence the name, ‘aureus,’ which is Latin for ‘gold’ (Masalha et al., 2001).
Food handlers’ mucous membranes and skin may get colonized by S. aureus, which can cause serious problems in food manufacturing facilities (Giaouris et al., 2015). Heat-stable enterotoxins can indeed be released alongside the bacterium, in meals accidentally contaminated by food handlers. S. aureus thrives well in foods containing heightened sugars or salt levels with limited water activity (Kadariya et al., 2014). Dairy, meat and poultry products provide favorable microenvironments for S. aureus’ virulent toxin-producing strains (Adams et al., 2000). Enteric toxins produced by S. aureus are well recognized as having class II major histocompatibility complex-binding properties (in T-cells), increasing the risk of acute toxic shock syndrome with diarrheal illness (Schelin et al., 2017). Animals, dust, food handlers, unprocessed foods, water, etc., are known contamination sources in the food industry (Todd et al., 2009). Furthermore, it is well-established that biofilms linger onto equipment and surfaces that come into touch with food, operating as constant sources of contamination. Moreover, it was shown that factors like nutrition availability, pH, surface characteristics and temperature have an impact on subsequent microbial growth, virulence and biofilm development in this industry (Abdallah et al., 2014). According to various studies, S. aureus biofilms were shown to colonize food-contact surfaces in meat, poultry, dairy, pasteurization belts and seafood sectors (Gutiérrez et al., 2012; Latorre et al., 2010; Sharma & Anand, 2002).
Polysaccharide intercellular adhesins are well expressed in these cocci, and controlled by intercellular adhesion genes (ica) associated with the formation of biofilms. The fact that S. aureus strains lacking ica gene may still produce biofilms, nevertheless, suggests the existence of a different route, possibly the Bap (biofilm-associated protein) linked one (Toledo-Arana et al., 2005). S. aureus’s quorum regulator factor, SarA, is involved in positively controlling the Bap-linked pathway and increasing the transcription of ica operon (Trotonda et al., 2005).
Other Menaces and Synergisms
The non-pathogenic, Gram-positive bacillus, Anoxybacillus flavithermus, is a spore-forming, thermophilic, facultative anaerobe, occasionally reported as a contaminant in dairies (Strejc et al., 2020). It often presents as a challenge for sectors involved with processing of milk powders, since large concentrations will make milk powder products unfit when under food quality standards for marketing (Murphy et al., 1999). Vegetative cells of A. flavithermus may thrive at temperatures as high as 65 °C, and with skimmed milk there is a rise in adherent microbial cells attached onto stainless-steel surfaces, implying that milk has a favorable impact on the development of A. flavithermus biofilm (Sadiq et al., 2017).
The Gram-positive rods of Geobacillus stearothermophilus (G. Stearothermophilus; also known as Bacillus stearothermophilus), are heat-resistant, spore-producing, facultative anaerobes (Flint et al., 2001). They develop biofilms by attaching to stainless-steel substrates on manufacturing lines, with eventual release of thermophilic colonies into manufactured products (Wu et al., 2019). Most of the biofilms found in milk or milk-product sectors worldwide, have the prevalence of thermophilic A. flavithermus and/or G. stearothermophilus strains (Burgess et al., 2009; Sadiq et al., 2017).
Gram-negative, anaerobic, Pectinatus bacilli have been encountered in biofilms, and linked to poor sanitation conditions in many breweries (Paradh et al., 2011). These spoilage-associated microbes were initially found in unpasteurized beer at 30 °C in a beer producing facility in Colorado (Lee et al., 1978). Since then, Pectinatus cerevisiiphilus species have been habitually isolated in many European breweries (Paradh et al., 2011).
Within food sectors, biofilms can be formed as a result of microbial synergisms. Certain microorganisms are capable of coexisting in food manufacturing settings, as complex microbial biofilms, from which infectious as well as spoilage microbes may contaminate edibles (Sterniša et al., 2019). For example, heterogeneous pathogens (i.e., Aeromonas hydrophila, S. enterica, L. monocytogenes, Vibrio species, etc.) can develop biofilms on fresh seafoods in fisheries, that may cause serious adverse consequences on both, the economy and health (Mizan et al., 2015). Burkholderia caryophylli, E. coli and Ralstonia insidiosa, have been found to have synergistic interactions, giving rise to tough poly-microbial biofilms in fresh-cut production facilities (Lee et al., 2007). Acylases and acyl-homoserine lactones in microorganisms, help regulate the development of polymicrobial biofilms (Lee et al., 2007).
Microorganisms adhere onto surfaces (abiotic or biotic) and utilize quorum sensing mechanisms. These mechanisms promote better cellular integration of biofilms and their dispersion via cell-to-cell communication signals (Toushik et al., 2020). Quorum sensing-regulated exopolysaccharide production as is with biofilm-forming strains of Vibrio cholera, has further supported the notion that cell signaling is essential for the development of bacterial biofilms (González-Rivas et al., 2018).
Numerous scientists have extensively analyzed and contrasted biofilm developmental stages of multiple microbe communities, with those of each microbe under single-microbial biofilm settings. They have discovered evidence of synergistic properties amongst certain pathogens. By examining poly-microbial biofilms, such as those containing Candida albicans strains, different researchers have discovered beneficial synergies in various investigations (Pammi et al., 2013; Zupančič et al., 2018). Researchers have observed that Acinetobacter junii and Pseudomonas aeruginosa found on various materials, serve as superior biofilm builders, including their attachment-deficient strains, which are capable of increasing biofilm development in commonly tested microbe communities.
Consequences of biofilms, in terms of corrosion of metals, lipase or protease-directed modification of organoleptic qualities and pathogenicity are extremely significant throughout the industrial food sector. Butter homogenizers, cheese tanks, packaging machines, pasteurizers, food belts, pipelines and raw milk stores, for instance, can serve as contact substrates for bacterial biofilm matrixes at varying temperatures and include a variety of heterogeneous colonizers and pathogens. As a result, it is crucial to establish precise techniques for detecting biofilms in situ in order to safeguard against contamination and guarantee the quality of foods.
Detection and Monitoring Techniques
Standard Methods
Traditional platforms use macro, as well as micro-scale reactors (with flow cells or static cells), to measure the development and growth of biofilms. Multi-well plates and other static instruments are frequently used in microbiology labs (Melo et al., 2012). However, the inability to consistently refill the culture media over time may cause planktonic cells along with unwanted remnants to build up, hindering any ongoing, active measurement. Alternatively, flow cells, in the form of Robbin’s device (Kharazmi et al., 1999), the Calgary device (Ceri et al., 1999) and the Centers for Disease Control biofilm reactor (Goeres et al., 2005), enable only end-point disruptive analysis while providing more repeatable and controlled biofilm formation. These traditional systems can be scaled down to reduce some of their drawbacks and can be combined with brand-new sensor technologies. These innovations include portability, high-output analysis, probability, smaller test volumes and the capacity to conduct nondestructive, real-time biofilm descriptions. These traits can be used to obtain new understanding of biofilm development, interactions amongst biofilm cells and identify any likely antibiotic resistance processes taking place in tested biofilms (Meyer et al., 2011; Paredes et al., 2014a).
Additionally, with great repeatability and throughput, microfluidic devices have been used to investigate how factors like pH, flow rate, and temperature affect the production of biofilms (Gashti et al., 2016; Pousti et al., 2018). They give researchers the chance to examine biofilms in carefully regulated micro-settings, so as to replicate natural settings or in-vivo circumstances (Shumi et al., 2010). Although these systems can occasionally function independently, they frequently require integration with established biofilm tests that often use microscopy, semi-quantification (colony forming units), crystal violet staining, or other such experimentations. Microfluidic designs for nondisruptive analysis in real-time framework for biofilm testing are very limited, since these approaches demand labelling of agents, sample treatment and intrusive procedures with optical corrections (Subramanian et al., 2020). For instance, microscopy-based approaches require ongoing lens focus correction in order to record changes in biofilm depth.
Sensing Devices
Advancements in sensing technologies, such as optical, electrochemical, and mechanical transducers, have facilitated the non-invasive evaluation of microbial biofilms. The high detection sensitivity of optical detection systems, for instance, comes at the expense of lengthy data collecting and analysis (Pu et al., 2021). Confocal reflection microscopy (CRM) (Yawata et al., 2010), SR-FTIR—synchrotron radiation-based Fourier transform infrared (Holman et al., 2009; Keirsse et al., 2003), white-light interferometry, Brillouin spectroscopy, SPR—surface plasmon resonance, LSPR—localized surface plasmon resonance (LSPR), fiber optics and spectro-microscopy are a few examples (Zhong et al., 2016, 2019).
Electrochemical sensors also partake biofilm testing (Clark & Lyons, 1962). Some detection methods come under the umbrella of ‘impedance microbiology’, built on the impedimetric sensing principle, i.e., the use of impedance variations amongst exposed electrodes to determine any involved bacteria (Firstenberg-Eden & Eden, 1984). Using these methods, researchers have been able to monitor changes in the capacitance of bacterial inoculants, conductance or impedance in order to track bacterial development in real time (Blanco-Cabra et al., 2021; Bruchmann et al., 2015; Jain et al., 2021; Liu et al., 2018; Paredes et al., 2013, 2014a, b; Pavanello et al., 2011; Poma et al., 2021; Robb et al., 2018; Ward et al., 2014; Zheng et al., 2013). In a nutshell, the biofilm-bacteria along with its extracellular elements are of dielectric nature, influential to the micro-system’s overall impedance (Paredes et al., 2014a). Therefore, the development of biofilms, including any subsequent metabolic or compositional alterations within, can be inferred by tracking the impedance in the bacterial solution over time. Amperometry and potentiometry based electrochemical devices are based on measuring in terms of faradaic current generated by biofilms’ redox species when the tested biofilms are laden onto electrode surfaces. These methods possess the beneficial capacity to record not only the early cell adhesion phases (Bayoudh et al., 2008; Palmer et al., 2007), but also the electroactive metabolites (Bellin et al., 2016), henceforth, monitoring specific bacterial biochemical activities on a real-time basis (Saccomano et al., 2021).
Interfacial rheometric devices, QCM—quartz crystal microbalances, quartz tuning fork oscillators, quartz tuning fork sensors, SAW—surface acoustic wave sensors and tensiometer-based devices, are also amongst the extensive list of biofilm detecting devices (de Wouters et al., 2015; Hollenbeck et al., 2014; Rühs et al., 2013, 2014). When organic or inorganic speciated cells (biofilm components) are adsorbed, surface-bound, and/or deposited onto exclusive device surfaces containing piezoelectric factors, electrical responses are generated, which are recorded accordingly. These devices are employed to real-time, with high temporal resolution and very inexpensively capture the growth of biofilms. Additionally, certain QCM-D systems are available, which are QCM-systems with added permittance of dissipation monitoring (Ripa et al., 2020). These offers additional details on mechanical characteristics of biofilm adherence, and are essential when improving eradication processes or creating surfaces that are bacterial repellant in food processing facilities or other settings. Bacterial adhesion properties and adsorption kinetics pertaining to specific surfaces may be inferred by recording mechanical responses. These responses may be measured in terms of ‘interfacial tension’ and ‘interfacial elasticity’ of adsorbed biofilm-cells on a test surface, by means of interfacial rheometric devices, or by tensiometer-based devices (de Wouters et al., 2015; Hollenbeck et al., 2014; Rühs et al., 2013, 2014).
Emerging Methods
For fully comprehending geographic and temporal dynamics of certain metabolites in biofilms, newer detecting methods are also being explored (Saccomano et al., 2021). High variabilities in phenotypic properties of involved biofilm bacteria within the biofilm, EPS matrix structure as well as biochemical heterogeneities and the generally heterogeneousness of different microbial biofilms, all contribute to inherent challenges. Additionally, biofilm activities can be quickly impacted by intrusive procedures used to evaluate the features of the EPS matrix, microbial cells and their densities (Gloag et al., 2020). As an illustration, microelectrode probes have occasionally been used to pierce the biofilm, disrupting its structural integrity and changing the permeability of the cells (McLean et al., 2008; Peter Revsbech, 2005).
As part of some detection procedures, test biofilms are often pretreated with chemicals. This can be hindering, because, for example, when a biofilm is subjected to particular dyes, its microbial cell components may undergo biophysical and/or biochemical modifications. The challenges mentioned above encourage scientists to create less invasive methods, such as those based on pH or oxygen level trackers throughout biofilm depths. Electrical and microelectrode sensory apparatuses partake in monitoring biofilm pH, oxygen levels, ions (such as ammonium, nitrite, and nitrate), glucose, temperature changes, Ca2+ concentrations and other variables. Although these sensors are reliable and adaptable, interference or cross-sensitivity can have an impact (e.g., pH and temperature) (Wei et al., 2019). Additionally, as electrical sensors frequently have to impale through biofilms, this can decrease the measuring repeatability and alter its accuracy.
Planar optodes, or polymeric films implanted with oxygen-sensitive, luminous probes (Wolfbeis, 2015), as well as labelled micro- and nanoparticles, have been employed in optical imaging systems centered on fluorescence and confocal microscopies to monitor changes in pH, oxygen and other parameters in biofilms. Planar optodes, which allow for biofilms’ cellular attachments onto two-dimensioned polymeric films, are unable, however, to reveal detailed information on inside mechanisms of test biofilms (Glud et al., 1998; Khosravi et al., 2020; Kühl et al., 2007; Staal et al., 2011). In contrast, biofilms can be subjected to nanoparticles and microparticles laced with oxygen or pH-sensitive luminescing or fluorescing dyes. These can be distributed into EPS matrices to provide in-depth three-dimensional mapping of tested parameters (e.g., oxygen concentration, pH levels) inside the entire biofilm. In-situ biofilm studies can be conducted with less stress thanks to their nondestructive nature (Acosta et al., 2009; Jewell et al., 2019; Sønderholm et al., 2018). The detection of heavy metal adsorption (Fe3+, Cu2+, Zn2+, and Hg2+) by biofilm microbes is pertinent to bioremediation strategies, which may be carried out with specialized fluorescent probes. C-di-GMP—cyclic di-GMP, glucose and HSL—N-acyl homoserine lactone, have drawn the most interest among the biofilm metabolites. Since glucose is the main carbon and energy source for microorganisms, its consumption directly equates with evidence of any active metabolism taking place in a biofilm. As a result, glucose has been regularly identified using conventional electrochemical sensors. C-di-GMP and HLS are significant signaling proteins (Cronenberg & van den Heuvel, 1991; Horn & Hempel, 1997). In some studies, c-di-GMPn in biofilms has been detected with electrochemical, fluorescence or luminescence-based sensors (Wang et al., 2016; Xie et al., 2015; Dippel et al., 2018). Das et al. has quantified test biofilm HLS levels using fluorescence-based sensors and Struss et al. has used colorimetric sensors for the same (Das et al., 2018; Struss et al., 2010). Dynamics of certain parameters (such as ions, metabolite products, oxygen levels, pH, etc.) within microbial biofilms have been tracked using a variety of sensing techniques.
The choice of the most suitable method for a specific application relies on a multitude of variables, including target preference and the resistance levels to intrusive procedures. For example, when aiming for high sensitivity and reliability, electrochemical sensors utilizing microelectrodes prove to be highly effective. Obtaining measurements from biofilms typically involves direct contact and penetration of the biofilm, which may pose a risk to its structural integrity. On the other hand, optical techniques that employ planar optodes and minute particles offer a less intrusive alternative for conducting such measurements. However, due to the toxic potential of the applied chemicals/dyes on microorganisms, caution is advised. Interfacial tension and elasticity-based apparatuses are accurate predictors of microbe adsorption mechanisms and biofilm development in terms of mechanical systems (Rühs et al., 2014). To be more precise, microbe adsorption at a hydrophobic interface brings about reduced interfacial tension along with increased interfacial elasticity. Rheological and tensiometric techniques can be used to measure these material properties, which can then be connected with bacterial adhesion traits.
Methods for Detecting Biofilms on Food Surfaces
There are two types of biofilm evaluation approaches in use (Fig. 9.3): direct and indirect. Examples of direct methods are: enzymatic reaction tests, contact plates, laser scanning confocal, epifluorescence microscopy, scanning electron microscopy (SEM), atomic force microscopy, environmental-SEM, cryo-SEM, focused ion beam-SEM. These allow direct observance of associated biofilm microcolonies and their involved bacteria (Van Houdt & Michiels, 2010). The foundation of indirect approaches is the separation of involved bacteria from their contact interfaces, prior to quantifying them. They consist of methods for determining the viability of microorganisms, namely: conventional plate counting tests like TEMPOR (bioM’erieux, Marcy l’Etoile, France), methods for measuring impedance, staining biofilm biomass using safranin or crystal violet and performing metabolite tests (XTT assay, Alamar Blue assay) (Verran, 2002). Though each method has benefits as well as drawbacks, merging several techniques ensures improved detection (Van Houdt & Michiels, 2010).
Biofilm Disrupting Technologies and Techniques
Bacteriocins
Bacteriocins are proteinaceous or peptide toxins normally produced by bacteria to prevent any related bacterial strains around them. Around a 100 years ago (1925), Andre Gratia discovered bacteriocins (Gratia, 2000). Bacteriocins are commonly synthesized as non-biological active precursor peptides possessing N-terminal leader sequence; however, in some cases, precursors undergo post-translational modification (Soltani et al., 2021) . The ensuing molecular event may lead to the cleavage of the leader region and the export outside the cell (Mokoena, 2017). The generated antimicrobial peptides exhibit bactericidal or bacteriostatic properties directed against the closely related strain (Hatakka et al., 2008). The beauty of bacteriocin-producing cells is that they set up a mechanism to prevent them from being harmed or destroyed by their own toxins. For self-defense, they may produce immunomodulatory proteins, utilizing efflux pumps or a combination of both systems to stay safe (Bastos et al., 2015).
Classification of Bacteriocins
Bacteriocins are classified based on strain, resistance mechanism, and killing mechanism.
Recently, Cotter et al. categorized the bacteriocins of both Gram-positive and Gram-negative bacteria (Cotter et al., 2013).
Bacteriophages
Bacteriophages, commonly known as phages, typically infect and replicate in bacterial cells. Bacteriophages are viruses that exist widely throughout the environment and are recognized as the most abundant biological entities on Earth (Schmaljohn & McClain, 1996). Genome-wise, bacteriophages are exceptionally diverse, containing proteins that encapsulate DNA/RNA. They may encode a few genes (MS2) or several hundred genes based on genome size. Bacteriophages display a species-specific nature concerning their hosts, primarily infecting single bacterial strains or particular species. Replication of phages occurs within the bacterium following the introduction of their genome into the cytoplasm.
The resistance of biofilms to bacteriophages is attributed to the impermeability of the biofilm matrix, which hinders the phages’ ability to penetrate and effectively target the bacterial cells within the biofilm. It is a well-known fact that the genome of bacteriophages possesses genes of enzymes capable of breaking down the matrix of biofilm (Leiman et al., 2009; Sillankorva et al., 2011). Sometimes soluble enzymes usually target the host bacterial cell wall and release the host cell. These enzymes have the potential to degrade the biofilm extracellular polymeric substance while releasing it from the lysed host cell. During the process, cells also liberate DNA, and DNAses may contribute to the biofilm matrix. Bacteriophages like T4 and HK260 (bacteriophages of E. coli) encompass an enzyme on the tail of the virus particle, through which they penetrate the bacterial cell wall. The enzyme may play an important role in degrading the biofilm matrix and is often concealed until the tail reconfigures during and after infection and impart localized action (Leiman et al., 2004).
These proteins are specific as they fit within the virus structure and function accordingly. Yan et al. presented a ‘common model bacteriophage (tailed) infection’. Here, constituents of the tail first recognize and then digest the capsular polysaccharide. The tail penetrates the cell membrane and injects the bacterial genome inside. The tail structure of bacteriophage usually possesses the polysaccharide depolymerase protein, and tail spike protein has endoglycosidase activity which hydrolyses the polysaccharide receptors.
Bacteriophages possess the ability to induce gene expression of depolymerase enzyme in the host bacteria (Topka-Bielecka et al., 2021).
Phage-based management is currently being exploited to combat biofilm via numerous mechanisms. Previously it has been reported that phages are strictly species-specific viruses where they infect bacteria and are totally dependent upon the host during the replication process. Recently, with the emergence of antimicrobial resistance and subsequent discoveries of new antibiotics, phage and phage therapy have been brought into mainstream research to combat the new species of bacteria. Several thousand phages have been discovered recently (Ackermann & Prangishvili, 2012). Based on basic structural forms, phages are classified into four basic categories, namely (i) tailed phages, (ii) polyhedral phages, (iii) filamentous phages, and (iv) pleomorphic phages (Basic Phage Electron Microscopy | Springer Nature Experiments, n.d.). The interaction between phage and host cells relies entirely upon the receptor-binding protein present in the tail fiber of phages (Zinke et al., 2022).
Two enzymes mainly responsible for the antibacterial activity of phages are depolymerases and lysins. Depolymerases degrade capsular polysaccharides, while lysins destroy peptidoglycan present in the cell wall of bacteria (Schmelcher et al., 2012). At the tip of phage (tail fibers), the domain of depolymerase is frequently exhibited; however, the lysins are encoded either on the tail or inside of virion particles and can cut the peptidoglycan of cell wall from outside or inside respectively (Sharma et al., 2017).
Lysins
Phage lysins, depending on their target bacteria, are categorized into two types: Gram-positive or Gram-negative lysins. These lysins are hydrolytic enzymes that are produced towards the end of the phage’s lytic replication cycle. Their main function is to cleave the bacterial cell wall from within the cell, leading to the liberation of new phage particles. Moreover, lysins can exhibit an external action by assisting in the permeation of the bacterial cell by the parental phage. In addition to their phage-related functions, lysins possess the ability to dismantle the extracellular polymeric matrix of biofilms and effectively target the bacteria residing at the periphery of the matrix. Due to the absence of an outer membrane (OM) in Gram-positive bacteria, lysins work efficiently, while in Gram-negative bacteria, OM hampers lysin penetration to reach the peptidoglycan.
However, recent research has presented that four Gram-negative bacteria target endolysins (LysAm24, LysAp22, LysECD7, and LysSi3) exhibiting antibacterial activity in vitro as well as in vivo.
In the case of Gram-positive lysins, the cell binding domain (CBD) harboring at C-terminus is accountable for interacting with the cell wall, while the enzymatically active domain (EAD) present on the N-terminus responsible for the hydrolysis of peptidoglycan. Gram-negative lysin does not interfere with CBD and typically employs a globular conformation having a single EAD to hinder bacterial cell walls (Becker et al., 2008). Currently, lysins are manipulated as free enzymes as an alternative to antibacterial drugs in treating biofilms. Lysins have also been used extensively in the multi-drug resistant S. aureus in clinical settings. Chimeric lysins such as ClyH and ClyF have shown a large percentage of biofilm mass reduction (Yang et al., 2014, 2017).
Depolymerases
Depolymerases are the class of enzymes that interfere with and degrade the capsular polysaccharide of Gram-negative bacteria. Depolymerase is generally encoded as a portion of phage structure. Various known depolymerases work against a range of bacterial species and have been recently employed for biofilm destruction. Phage depolymerases have further subdivided into two groups based on different mechanisms: hydrolases and lyases (Knecht et al., 2019). Hydrolases function in cleaving the substrates, which utilizes hydrolysis with the involvement of water (Knecht et al., 2020). In the previous section, we discussed the main component of biofilm, i.e., EPS, which forms 50–90% of the total biofilm organic component and thereby can interfere with biofilm formation (Flemming & Wingender, 2010).
Depolymerases derived from phage may demonstrate two approaches against antibiofilm management, namely (i) free enzyme and (ii) tail spike protein (TSP). Free depolymerase shows a certain degree of advantage over TSP as it provides extended molecular stability, efficiently delivers via diffusion and diminishes chances of resistance development (Chen et al., 2022). Depolymerase (Dpo42) extracted from the ORF42 of the vB_EcoM_ECOO78 E. coli phage. After subsequent purification and expression via E. coli BL21 as a free protein, it was determined that Dpo42 successfully degraded the capsular polysaccharide encompassing the E. coli and prevented biofilm formation (Guo et al., 2017). The specificity of depolymerases is that it degrades bacterial capsules and the glycocalyx, the main constituent of biofilm (Chan & Abedon, 2015).
TSP depolymerase opened the vista for medical device application. Recent studies on A. baumannii-adhered catheters have shown effective inhibition of bacteria within a few hours (4 h) of treatment (Shahed-Al-Mahmud et al., 2021). It was also found that TSP derived from φAB6 may postulate potential management against MDR A. baumannii infection in the next decade.
Phages possess the inherent capability to destroy bacterial hosts and thereby inhibit the formation of biofilm (Domingo-Calap & Delgado-Martínez, 2018). The existing biofilm can also be penetrated by phages and destroy the biofilm structure with or without killing the resident bacteria.
Broadly, biofilm disruption with the application of phages has been cand be divided into types, namely (Chan & Abedon, 2015):
-
extra- to intra- cellular degradation of bacterial structure
-
intra- to extra- cellular degradation of bacterial structure
-
chemical dispersion of biofilm matrix—particularly of EPS
Biosurfactants
Biosurfactants prevent biofilm formation by various mechanisms like (i) changing the cell adhesion capability, (ii) membrane disruption, and (iii) inhibiting the electron transport chain (Satpute et al., 2016). Biosurfactants are microorganism specific and exhibit antifungal, antibacterial, and antibiofilm activities depending upon the species (Paraszkiewicz et al., 2021). Biosurfactants decrease the growth of biofilm produced by S. aureus by controlling the expression of genes like dltB, cidA, and icaA (Yan et al., 2018). A significant reduction of gene expression of cidA gene was shown from the biosurfactants obtained from Lactobacillus plantarum at a concentration of 12.5 mg/mL (Yan et al., 2018). Similarly, biosurfactants obtained from Pediococcus acidilactiti at a concentration of 50 mg/mL affect gene expression by downregulating autoinducer-2 signaling molecules, accessory gene regulator, and staphylococcal accessory regulator (Yan et al., 2018). Liposome-derived Lactobacillus-based biosurfactants exhibit more inhibition of S. aureus biofilm formation and elimination as compared to free biosurfactants (Giordani et al., 2019).
Recently identified lipopeptides from Acinetobacter junii capable of self-aggregate to form sheet rich biosurfactant vesicle having thermostable properties and less toxic are utilized as promising antibiofilm agents (Ohadi et al., 2020). Another lipopeptide biosurfactant isolated from Bauveria bassiana significantly removes biofilm in ex vivo surroundings for M. canis (Abdul-Aziz et al., 2020). Here, biosurfactants disturb the integrity of the cell membrane and affect cell membrane permeability. Inexpensive biosurfactants of B. bassiana are normally produced from steep corn liquor and are widely used against recalcitrant dermatophytosis. Surfactin (cyclic lipopeptide) was reported to have promising results against C. albicans biofilm-associated infections. Surfactin controls the expression of several genes required for hyphae production and acts by reducing the surface hydrophobicity of cells (Janek et al., 2020).
Rhamnolipids obtained from Pseudomonas aeruginosa MN1 possess higher antibiofilm and antiadhesive properties compared to Surfactin (Abdollahi et al., 2020). Glycolipid fabricated from Burkhoderia sp. WYAT7, an endophyte of Artemisia nilagirica (Clarke) Pamp, shows antibiofilm activities versus S. aureus (Ashitha et al., 2020). Glycolipoprotein, rich in LeuHis- Trp amino acids isolated from Acinetobacter indicus M6, may remove more than 80% of biofilm at a concentration of 500 μg/mL (Karlapudi et al., 2020).
Biosurfactants are exploited as a coating agent for medical devices such as urinal catheters and bone implants to prevent biofilm formation from the pathogenic organism. Rhamnolipids and sorphorolipids hinder biofilm formed by Gram-positive and Gram-negative bacteria (Sharma et al., 2021). Biosurfactants isolated from Lactobacillus acidophilus restrict biofilm generation of S. aureus and Proteus vulgaris on polydimethylsiloxane-based implants (Satpute et al., 2016).
Blockage of Quorum Sensing
Quorum sensing (QS) is the phenomenon to detect, respond, and communicate within the bacterial community by regulating gene expression. QS plays a significant role in the regulation of diverse cellular properties in bacteria, such as bioluminescence, antibiotic resistance, virulence gene expression, and biofilm formation (Li et al., 2012). QS is an efficient strategy to restrict biofilm formation where cell-cell communication stops (Chen et al., 2022; Sharma et al., 2021).
QS system is broadly divided into three main categories (Brackman & Coenye, 2015):
-
Acyl homoserine lactone—i.e., AHL (Gram-negative organisms)
-
Autoinducing peptide—i.e., AIP (Gram-positive organisms)
-
Autoinducer-2—i.e., AI-2 (Gram-staining bacteria)
Homoserine lactones are an important class of cellular signaling molecules implicated in QS and AHL-dependent QS primarily exhibited by Gram-negative bacteria (Li & Tian, 2012). Interestingly, AHLs are produced by particular cognate AHL synthetase, and increasing concentration of AHLs are correlated with substantial growth of bacteria. AIPs are also signaling molecules synthesized by Gram-positive bacteria and secreted by membrane transporters. Once the concentration of AIPs increases in the bacteria, they interact with histidine kinase sensors and phosphorylates. Due to phosphorylation, gene expression takes place and is strictly regulated by an accessory gene regulator (agr), which is associated with the secretion of AIPs.
Potential Anti-biofilm Nanotechnologies
Chemical Processes
There are various chemicals that can disrupt biofilms, and they can be broadly categorized into enzymes, surfactants, quorum sensing inhibitors, and antimicrobial agents. Enzymes such as DNase, protease, and dispersin B can degrade the extracellular matrix components of biofilms and weaken their structure. Surfactants such as sodium dodecyl sulfate (SDS) can penetrate the biofilm and disrupt the cell membrane, leading to cell death and biofilm disruption (Flemming & Wingender, 2010; Kaplan, 2010; Vasilev et al., 2009). Quorum sensing inhibitors (QSIs) are chemicals that can interfere with the cell-to-cell communication mechanism of bacteria, which is essential for biofilm formation (Wu et al., 2015). Examples of QSIs include furanones, halogenated furanones, and azithromycin. These compounds can disrupt biofilms by preventing the production of extracellular polymeric substances and inhibiting cell adhesion (Tateda et al., 2003). Antimicrobial agents such as antibiotics and biocides can also disrupt biofilms. However, their efficacy is often limited by the biofilm’s ability to create a protective barrier that reduces their penetration and neutralizes their effect. Therefore, higher concentrations of antimicrobial agents are required to disrupt biofilms compared to planktonic bacteria (Mah & O’Toole, 2001).
Overall, the disruption of biofilms by chemicals is a complex and challenging task, and the choice of the appropriate chemical will depend on the type of biofilm and the specific microorganisms involved.
Enzymatic Interference
Enzymes are a type of protein that can interact with non-protein molecules called cofactors, and they have the ability to accelerate the speed of chemical reactions in biological systems. In other words, they act as catalysts, facilitating reactions without being consumed in the process.
The eradication of biofilms typically necessitates rigorous mechanical, physical, or chemical interventions, which may not be viable for delicate medical equipment, such as endoscopes, rendering them vulnerable to bacterial colonization (Stiefel et al., 2016). Moreover, the application of harsh methods is not always feasible for eliminating biofilms caused by pathogens within the human body, thereby contributing to the persistence of infections, chronic wounds, and malfunctioning medical devices (Del Pozo, 2018; Metcalf & Bowler, 2013).
Therefore, enzymes can be utilized as an alternative to chemical and mechanical means for the dispersion of biofilms under mild conditions, such as physiological temperatures. Through enzymatic treatment, biofilms on tank and pipe surfaces can be effectively removed by breaking down the essential components of the biofilm matrix (Lequette et al., 2010; Simões et al., 2010). These enzymes are designed to specifically target the primary constituents of biofilms, which include exopolysaccharides, proteins, and nucleic acids. Enzymes work by breaking down the various structures that make up the extracellular polymeric substance (EPS) of a biofilm, which ultimately results in a reduction in the biofilm’s physical integrity. To achieve an effective removal of the biofilm, it is crucial to first identify the specific structural components of the EPS before applying the enzymes (Molobela et al., 2010).
There exist four distinct classes of enzymes that are commonly employed for the purpose of eliminating biofilms. These enzyme types include proteolytic enzymes, which target proteins, polysaccharide-degrading enzymes, which break down complex carbohydrates, oxidative enzymes, which trigger oxidation reactions, and anti-quorum sensing enzymes, which inhibit the signalling mechanisms utilized by biofilm-forming bacteria (Bzdrenga et al., 2017; Johansen et al., 1997; Thallinger et al., 2013).
Essential Oils
Essential oils are volatile and aromatic compounds that are derived from various parts of plants such as petals, seeds, leaves, stems, and roots through natural processes, and are considered to be their essence or fundamental nature.
Essential oils are endowed with properties that enable them to inhibit the growth of plasmodium, fungi, and bacteria (Utchariyakiat et al., 2016). It possess properties that enable them to inhibit the growth of food spoilage and foodborne pathogenic bacteria, thus making them effective as food preservatives also (Bai & Vittal, 2014). There are a variety of naturally occurring substances and medicinal plants that can be found in fruits, spices, and phytochemicals. These substances have the ability to inhibit quorum sensing, which is a process used by certain bacteria to communicate with one another and coordinate their behavior. Essentially, these natural products contain compounds that can disrupt the ability of bacteria to communicate with one another, potentially leading to a reduction in harmful bacterial activity (Adonizio et al., 2008; Sybiya Vasantha Packiavathy et al., 2012; Vandeputte et al., 2010; Vattem et al., 2007).
EOs exhibit strong antibacterial effects against both Gram-positive and Gram-negative bacteria, whether they are in a stationary or mobile state (Essential oils against bacterial isolates from cystic fibrosis patients by means of antimicrobial and unsupervised machine learning approaches | Scientific Reports, n.d.; Millezi et al., 2016). Essential oils are known for their volatile nature, which gives them the ability to produce vapor that exhibits potential antimicrobial properties. Various studies have shown that the vapor phase of essential oils such as cassia, cinnamon, cherry laurel, origanum, and thyme, possess inhibitory effects against a diverse range of bacteria. Additionally, these oils have been found to be effective in preventing the growth of molds in food products and combating bacteria that form biofilms (Benzaid et al., 2019; Ji et al., 2019; Maruzzella & Sicurella, 1960).
Measures to Block Quorum Sensing
Quorum sensing is a sophisticated mechanism used by microorganisms to communicate with each other and coordinate their behavior. This communication system enables microorganisms to sense when their population reaches a certain density or “quorum”, and respond accordingly by regulating gene expression and producing specific molecules that can affect the behavior of neighbouring cells. These signaling molecules act as chemical messengers, much like hormones in higher organisms, and allow microorganisms to act in a synchronized and cooperative manner. By using quorum sensing, microorganisms are able to coordinate their activities, such as forming biofilms or carrying out group behaviours, in a way that maximizes their chances of survival and success (Bandara et al., 2012; Hawver et al., 2016; Hense et al., 2007; Redfield, 2002).
Biofilms can be formed as a result of this communication system, and these infections can be difficult to treat with antibiotics. Bacterial infections can potentially be prevented or treated by blocking quorum sensing. Numerous studies have demonstrated with convincing evidence that QS inhibitors are capable of effectively impeding the formation of biofilms (Chen et al., 2018b; Ouyang et al., 2016).
Quorum Sensing Inhibitors
These are molecules that interfere with the quorum sensing signalling pathways and prevent bacteria from communicating with each other. There are many different types of quorum sensing inhibitors, such as natural products, synthetic compounds, and peptides.
Plants Based QS Inhibitors
There exists a plethora of natural substances that can effectively inhibit biofilm formation by interfering with the process of QS. These compounds are predominantly sourced from plants. As an illustration, curcumin, a compound found in the Curcuma longa plant, has been observed to impede the development of biofilms in various uropathogens, including Pseudomonas aeruginosa PAO1, Escherichia coli, Serratia marcescens, and Proteus mirabilis. This is achieved by curcumin’s ability to decrease the production of exopolysaccharide through the inhibition of quorum sensing. Moreover, curcumin also shown the ability impede bacterial motility, which further slows down the formation of biofilms (Packiavathy et al., 2014).
Likewise, the natural compound resveratrol has been found to interfere with QS signaling in P. aeruginosa PAO1 by binding to the protein receptor LasR, thereby hindering the formation of biofilms (Vasavi et al., 2017). Similarly, carvacrol was shown to inhibit biofilm formation and pyocyanin production in P. aeruginosa (Tapia-Rodriguez et al., 2019). Additionally, naturally occurring furocoumarins sourced from grapefruit have demonstrated inhibitory effects on the biofilm formation of E. coli O157:H7, P. aeruginosa, and Salmonella enterica serovar Typhimurium (Girennavar et al., 2008).
Since these nontoxic, natural, biofilm inhibitors pose no harm to environment and the host, they offer great capability for application in diverse fields.
Synthetic QS Inhibitors
Apart from natural compounds, synthetic compounds have also been identified to have the ability to inhibit QS signalling pathways.
For instance, synthetic compounds such as furanone C-30 have been studied. Furanone C-30 has been identified as an effective biofilm inhibitor in S. mutans (He et al., 2012). Similarly, 2(5H) Furanone has been found to reduce microbial motilities and biofilms of C. jejuni strains by disrupting QS activities (Castillo et al., 2015). Another synthetic compound, Meta-bromo-thiolactone, has been demonstrated to inhibit the production of pyocyanin and biofilm formation in P. aeruginosa. This inhibition is achieved through the compound’s ability to bind to two QS signal receptors, namely LasR and RhlR (O’Loughlin et al., 2013).
Biofilm inhibitors that target QS pathways have been extensively utilized for inhibiting a wide range of biofilms. However, there are still numerous QS inhibitors currently under development that hold potential for treating infections caused by biofilms or eliminating biofilms that form on tissue implants (Luo et al., 2017; Yu et al., 2018).
In addition to the use of QS-based biofilm inhibitors, alternative strategies exist for controlling biofilm formation. One such approach involves combining QS inhibitors with antibiotics to achieve superior biofilm control (Thomann et al., 2016).
Quorum Quenching
As a result of the increasing prevalence of antibiotic-resistant bacteria resulting from overuse of antibiotics, it has become crucial to explore alternative methods of fighting microbial infections. One such approach is quorum quenching (QQ), which involves interfering with the process of microbial communication. By using QQ-driving molecules, it is possible to reduce or completely inhibit the production of virulence factors, such as biofilm formation. This can prevent bacterial populations from coordinating their activities and limit their ability to cause infections. Thus, QQ has emerged as a promising strategy for developing new antimicrobial therapies that can effectively combat antibiotic-resistant bacteria by targeting their communication mechanisms.
The enzymatic breakdown of AHL molecules is the most well-known mechanism of quorum quenching, and this process is facilitated by four different groups of enzymes: lactonases and acylases, which break down the HSL ring and amide bond of AHL, respectively, and reductases and oxidases, which modify the activity of AHL without fully breaking it down (Rehman & Leiknes, 2018). Inducer antagonists represent an additional mechanism for disrupting bacterial communication. These molecules can inhibit the transmission of signals between cells by either binding to the receptor in competition with inductors or by non-competitively blocking the inductor-mediated signal transmission into the cell (Bodede et al., 2018). Various approaches to quorum quenching have been identified, including inhibition of signal molecule synthesis, such as AHL, through the use of C8-HSL to impede LuxI enzymatic activity (Hirakawa & Tomita, 2013); blocking of signal transduction cascades using small molecule inhibitors like savrin, which interferes with AgrA and inhibits the production of RNAIII and virulence factors (Sully et al., 2014) and inhibition of QS signal molecules in Gram-positive bacteria through kinase inhibitors like closantel, RWJ-49815, and LY266500 (Brackman & Coenye, 2015).
Use of Anti-quorum Sensing Antibodies
AHL and AI-2 signaling activation can trigger programmed cell death by affecting the host’s immune system (Gupta et al., 2011; Khajanchi et al., 2011), but researchers have discovered ways to interfere with this process using monoclonal antibodies. For instance, the RS2-1G9 antibody can bind to 3-oxo-C12-HSL in the extracellular environment of Pseudomonas aeruginosa to reduce the host’s inflammatory response (Park et al., 2007), while the XYD-11G2 antibody catalyzes the hydrolysis of 3-oxo-C12-HSL signaling, inhibiting pyocyanin production by Gram-negative bacteria (Koul et al., 2016; Praneenararat et al., 2012). Additionally, the AP4-24H11 monoclonal antibody can block the QS signal of Gram-positive Staphylococcus aureus by interfering with AIP IV, which has been shown to attenuate tissue necrosis in infected models (Grandclément et al., 2016; Park et al., 2007). Although promising, the use of these monoclonal antibodies for treating bacterial diseases is still in the early stages.
Non-thermal Plasma
In settings where nosocomial biofilms need to be removed, traditional methods such as high heat and chemical exposure may not be ideal due to the potential for surface damage and environmental contamination with toxic chemicals. However, a promising alternative technique, called non-thermal plasma (NTP), has the potential to effectively decontaminate or sterilize nosocomial biofilms (Thapa & Ayan, 2019).
NTP is an emerging tool for improved biofilm sterilization (Koban et al., 2011; Thapa & Ayan, 2019). Plasma, the fourth fundamental state of matter, contains free radicals, reactive oxygen and nitrogen species (Jha et al., 2017), and positive and negative ions (Gaunt et al., 2006; Graves, 2012), which act as potential antimicrobial agents. Two distinct types of plasma, namely thermal and non-thermal, can be distinguished based on the relative energy levels of electrons and heavy particles they contain (Moreau et al., 2008), Thermal plasma is characterized by having both electrons and heavy particles at the same temperature, which is achieved through high pressure and power conditions. On the other hand, NTP consists of electrons at higher temperatures while heavy particles remain at room temperature. This state is produced under low-pressure and low-power conditions (Hoffmann et al., 2013; Moreau et al., 2008).
Thermal plasma has been utilized for purposes such as tissue removal, sterilization, and cauterization. However, the high heat production associated with thermal plasma can result in tissue and surface damage. On the other hand, NTP such as DBD and jet plasmas can carry out the same functions without causing harm or side effects (Keidar et al., 2013), making it suitable for biological and medical applications. Recent studies have revealed encouraging outcomes regarding the sterilization and decontamination of biofilms formed by various bacterial species using NTP (Ayan, 2009).
There are non-thermal jet plasma devices that use atmospheric pressure plasma, which are available for commercial use (Weltmann et al., 2009). One such device is the kINPen®, designed for biomedical applications, that allows for precise and arbitrary movements in three dimensions (Bekeschus et al., 2016). Applying a high-frequency voltage to the pin-type electrode generates the plasma, which is considered electrically safe as it is certified and compliant with EU standards (Weltmann et al., 2009). kINPen® plasma, which primarily uses argon gas but can also use other gases in smaller amount (Reuter et al., 2015), is a safe and effective medical device for antimicrobial purposes and wound healing, as demonstrated by clinical studies on both animals and humans. Its predecessor, kINPen®MED, was the first atmospheric pressure plasma jet device to receive accreditation as a medical device for patient use (Bekeschus et al., 2016).
Only few other plasma sources, including SteriPlas (AdTec Ltd., Japan), PlasmaDerm (Cinogy GmbH, Duderstadt, Germany), and Plasma One (Medical Systems GmbH, Bad Ems, Germany) (Bekeschus et al., 2016), have been certified as medical devices, and they have been used for various biomedical applications such as wound healing, chronic leg ulcers (Brehmer et al., 2015; Heinlin et al., 2013), reducing bacterial populations in wounds (Isbary et al., 2010), and biofilm decontamination or sterilization (Thapa & Ayan, 2019).
Coating Surfaces
Applying a coating to the surfaces where microbial attachment occurs can serve as an effective strategy to prevent the adherence of microorganisms and the subsequent formation of biofilms.
The wettability of a surface, which is determined by its surface free energy (SFE), can have a significant impact on the attachment of microorganisms. Microbes can attach to surfaces by forming biofilms, and surfaces with high SFE are more hydrophilic and thus more attractive to microbes for attachment (Nakamura et al., 2016).
However, other factors such as surface roughness, charge, and chemistry can also affect microbial attachment. Therefore, considering the SFE and wettability of a surface is important when designing materials to prevent microbial attachment, especially in fields like medical devices, food processing equipment, and water treatment systems.
A research scenario involves modifying the SFE of denture materials by applying salivary and/or blood plasma proteins. This alteration can effectively hinder the attachment of Candida albicans and prevent the formation of biofilms (da Silva et al., 2015). Microorganisms face challenges in establishing colonies on surfaces that exhibit superhydrophilic properties (Almaguer-Flores et al., 2012).
The application of a coating consisting of small molecules has the potential to modify the adhesive properties of the underlying surface materials. One way to modify the properties of silicone rubber surfaces, which are commonly utilized for creating tissue implants, is by applying a thin layer of a chemical mixture called monomeric trimethylsilane (TMS)/O2. This alteration has been observed to greatly impact the way in which certain microbial surface proteins adhere to the surface, ultimately inhibiting the formation of biofilms by the bacteria S. aureus (Xu et al., 2015). The TMS/O2 coating technique is a highly effective and eco-friendly method, with significant potential for use in various clinical applications.
The use of an antimicrobial peptide coating represents a valuable approach to prevent the formation of biofilms. It has been discovered that titanium discs, which are chemically bonded with GL13K, demonstrate remarkable antimicrobial properties against Streptococcus gordonii. Additionally, the coating effectively prevents the attachment of S. gordonii to the treated surface (Chen et al., 2018a) GL13K, a cationic peptide with bactericidal properties derived from BPIFA2, a secretory protein produced by the parotid gland in humans (Hirt & Gorr, 2013), as a coating on titanium surfaces, has been found to be highly effective in reducing the growth of two types of bacteria—Fusobacterium nucleatum and Porphyromonas gingivalis. Moreover, this coating also prevents the formation of biofilms by these organisms (Li et al., 2017).
Therefore, coating the surface of tissue implants and medical devices with certain materials is an effective method of preventing bacterial infections. A reduction in microbe attachment to materials, prevention of biofilm formation, and reduced risk of bacterial infections can all be achieved this way.
Potential Anti-biofilm Nanotechnologies
Due to the intricate nature of biofilm, traditional methods are unable to completely eliminate them (Chaudhary et al., 2020). Moreover, due to their resistance to antibiotics, higher therapeutic doses may be necessary, which increases the risk of systemic toxicity. Hence, researchers aim to overcome these constraints by utilizing various methods, such as nanoparticle-based drug delivery systems and interference with bacterial communication pathways using small molecules that regulate biofilm formation (Diab et al., 2015; Eleraky et al., 2020; Lopez-Leban et al., 2010; Tamilvanan et al., 2008).
Nanoparticles exhibit two primary mechanisms for better anti-biofilm properties: (1) direct interaction with single cells and (2) interaction with or denaturation of the EPS matrix. The unique properties of nanoparticles make them suitable for controlling biofilm infections. The size and shape, as well as the surface and interior properties of nanoparticles, are essential factors in the control of biofilm infections (Diab et al., 2015; Moghadas-Sharif et al., 2015; Ramos et al., 2018; Tan et al., 2020).
Nanoparticles possess unique physical and chemical properties due to their small size, which makes them an attractive research area for many fields including photochemistry, electrochemistry, and biomedicine (Haruna et al., 2016; Liu et al., 2019). Their high surface area and distinct electronic properties make them stand out from their bulk counterparts. Experts have determined that the optimal size range for Nps, which are used to control biofilm infections, is between 5 and 200 nm. It is important to note that Nps should not exceed 500 nm in size to ensure maximum effectiveness (Liu et al., 2019).
Nanoparticles have diverse therapeutic applications and can be synthesized from various inorganic and organic compounds. Inorganic materials are essential for simultaneous therapy and diagnosis due to their easy modification, high drug loading capacity, and stability (Saleh, 2014). The utilization of nanoparticles in the pharmaceutical sector, including its use in drug delivery systems, is well known (Huang et al., 2008).
Zinc Oxide Nanoparticles (ZnO-NPs)
ZnO NPs are highly effective in preventing biofilm infections due to their potent antibacterial properties (Padmavathy & Vijayaraghavan, 2008). Numerous studies have confirmed their ability to inhibit the growth of various bacteria, including P. aeruginosa (Dwivedi et al., 2014; Lee et al., 2014), S. pneumonia (Bhattacharyya et al., 2018), B. subtilis, E. coli, and P. vulgaris (Abinaya et al., 2018; Hsueh et al., 2015; Ishwarya et al., 2018). Moreover, research has also shown that ZnO-NPs can significantly reduce biofilm growth of certain fungi such as Alternaria alternate, Penicillium chrysogenum, and Penicillium pinophilum. However, their impact on Aspergillus niger was not as significant (Gambino et al., 2017).
Magnesium Oxide Nanoparticles (MgO-NPs)
MgO-NPs are a promising option for combating bacterial infections because they are non-toxic and readily available. Various studies have investigated their antimicrobial and inhibitory effects against both Gram-negative and Gram-positive bacteria, as well as yeasts (Cai et al., 2018; Hayat et al., 2018). In addition to MgO-NPs, magnesium fluoride NPs (MgF2-NPs) have also been explored as a means of inhibiting biofilm formation through surface modification and have shown potential as antibacterial agents in several studies (Lellouche et al., 2009, 2012; Tamilvanan et al., 2008).
Iron Oxide Nanoparticles (IO-NPs)
Iron oxide nanoparticles (IO-NPs) have unique magnetic properties, high biocompatibility, and a large surface-to-volume ratio that make them well-suited for various bioprocess applications (Ebrahimi et al., n.d.; Ebrahiminezhad et al., 2016). Studies have shown that IO-NPs are effective in reducing biofilm growth on implant surfaces, which is a common cause of implant failure (Thukkaram et al., 2014). Furthermore, research has demonstrated that IO-NPs coated with 3-aminopropyltriethoxy silane (IO-NPs3-APTES), in contrast to naked IO-nanoparticles (i.e., superparamagnetic iron oxide (SIONPs)) can effectively disrupt stubborn biofilms.
Additionally, both, SIONPs, as well as IO-NPs, have been found to exhibit anti-microbial/biofilm properties against bacteria such as P. aeruginosa and S. aureus (Akbari & Ali, 2017; Sathyanarayanan et al., 2013).
In additional, Numerous studies have highlighted the potential of various metal and metal oxide nanoparticles for disrupting or inhibiting microbial biofilms. For instance, Shakibaie et al. synthesized selenium nanoparticles (Se-NPs) and tested their effectiveness against biofilms caused by multiple strains of P. aeruginosa, S. aureus, and P. mirabilis (Shakibaie et al., 2015). Copper nanoparticles (Cu-NPs), nanofibers containing copper, and tungsten (W) and molybdenum (Mo) nanoparticles dispersed in alkyl alkoxysilane polymer have also demonstrated antibiofilm effects (Ahire et al., 2016; Chari et al., 2017; Ghasemian et al., 2015; Ogawa et al., 2017). Additionally, Chrzanowska et al. have identified the relevant nanoparticles, namely, zirconium oxide, as well as aluminum oxide to have activities against biofilm (Chrzanowska & Załęska-Radziwiłł, 2014). Moreover, some NPs (e.g., titanium dioxide and calcium fluoride) have shown capability to reduce the formation of biofilms and hence, may be useful in different industrial set-ups (Kulshrestha et al., 2016; Maurer-Jones et al., 2013).
References
Ashitha, A., Radhakrishnan, E. K., & Mathew, J. (2020). Characterization of biosurfactant produced by the endophyte Burkholderia sp. WYAT7 and evaluation of its antibacterial and antibiofilm potentials. Journal of Biotechnology, 313, 1–10. https://doi.org/10.1016/j.jbiotec.2020.03.005. Epub 2020 Mar 6. PMID: 32151643
Abdallah, M., Benoliel, C., Drider, D., Dhulster, P., & Chihib, N.-E. (2014). Biofilm formation and persistence on abiotic surfaces in the context of food and medical environments. Archives of Microbiology, 196(7), 453–472. https://doi.org/10.1007/s00203-014-0983-1
Abdallah, M., Khelissa, O., Ibrahim, A., Benoliel, C., Heliot, L., Dhulster, P., & Chihib, N.-E. (2015). Impact of growth temperature and surface type on the resistance of Pseudomonas aeruginosa and Staphylococcus aureus biofilms to disinfectants. International Journal of Food Microbiology, 214, 38–47. https://doi.org/10.1016/j.ijfoodmicro.2015.07.022
Abdollahi, T., Pedram Razi, S., Pahlevan, D., Yekaninejad, M. S., Amaniyan, S., Leibold Sieloff, C., & Vaismoradi, M. (2020). Effect of an ergonomics educational program on musculoskeletal disorders in nursing staff working in the operating room: A quasi-randomized controlled clinical trial. International Journal of Environmental Research and Public Health, 17, 7333. https://doi.org/10.3390/ijerph17197333
Abdul-Aziz, M. H., Alffenaar, J.-W. C., Bassetti, M., Bracht, H., Dimopoulos, G., Marriott, D., Neely, M. N., Paiva, J.-A., Pea, F., Sjovall, F., Timsit, J. F., Udy, A. A., Wicha, S. G., Zeitlinger, M., De Waele, J. J., Roberts, J. A., & Infection Section of European Society of Intensive Care Medicine (ESICM), Pharmacokinetic/Pharmacodynamic and Critically Ill Patient Study Groups of European Society of Clinical Microbiology and Infectious Diseases (ESCMID), Infectious Diseases Group of International Association of Therapeutic Drug Monitoring and Clinical Toxicology (IATDMCT), Infections in the ICU and Sepsis Working Group of International Society of Antimicrobial Chemotherapy (ISAC). (2020). Antimicrobial therapeutic drug monitoring in critically ill adult patients: a position paper. Intensive Care Medicine, 46, 1127–1153. https://doi.org/10.1007/s00134-020-06050-1
Abinaya, M., Vaseeharan, B., Divya, M., Sharmili, A., Govindarajan, M., Alharbi, N. S., Kadaikunnan, S., Khaled, J. M., & Benelli, G. (2018). Bacterial exopolysaccharide (EPS)-coated ZnO nanoparticles showed high antibiofilm activity and larvicidal toxicity against malaria and Zika virus vectors. Journal of Trace Elements in Medicine and Biology, 45, 93–103. https://doi.org/10.1016/j.jtemb.2017.10.002
Ackermann, H.-W., & Prangishvili, D. (2012). Prokaryote viruses studied by electron microscopy. Archives of Virology, 157, 1843–1849. https://doi.org/10.1007/s00705-012-1383-y
Acosta, M. A., Ymele-Leki, P., Kostov, Y. V., & Leach, J. B. (2009). Fluorescent microparticles for sensing cell microenvironment oxygen levels within 3D scaffolds. Biomaterials, 30(17), 3068–3074. https://doi.org/10.1016/j.biomaterials.2009.02.021
Adams, M. R., Moss, M. O., & Moss, M. O. (2000). Food microbiology. Royal Society of Chemistry.
Adonizio, A., Kong, K.-F., & Mathee, K. (2008). Inhibition of quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa by South Florida plant extracts. Antimicrobial Agents and Chemotherapy, 52, 198–203. https://doi.org/10.1128/AAC.00612-07
Ahire, J. J., Hattingh, M., Neveling, D. P., & Dicks, L. M. T. (2016). Copper-containing anti-biofilm nanofiber scaffolds as a wound dressing material. PLoS One, 11, e0152755. https://doi.org/10.1371/journal.pone.0152755
Ahmed, D., Islam, M. S., Begum, Y. A., Janzon, A., Qadri, F., & Sjöling, Å. (2013). Presence of enterotoxigenic Escherichia coli in biofilms formed in water containers in poor households coincides with epidemic seasons in Dhaka. Journal of Applied Microbiology, 114(4), 1223–1229. https://doi.org/10.1111/jam.12109
Akbari, K. R. A., & Ali, A. A. (2017). Study of antimicrobial effects of several antibiotics and iron oxide nanoparticles on biofilm producing pseudomonas aeruginosa. Nanomedicine Journal, 4, 37–43. https://doi.org/10.22038/nmj.2017.8051
Almaguer-Flores, A., Olivares-Navarrete, R., Wieland, M., Ximénez-Fyvie, L. A., Schwartz, Z., & Boyan, B. D. (2012). Influence of topography and hydrophilicity on initial oral biofilm formation on microstructured titanium surfaces in vitro: Selective bacterial colonization on microstructured titanium. Clinical Oral Implants Research, 23, 301–307. https://doi.org/10.1111/j.1600-0501.2011.02184.x
Amina, M., & Bensoltane, A. (2015). Review of pseudomonas attachment and biofilm formation in food industry. Poultry, Fisheries & Wildlife Sciences, 3. https://doi.org/10.4172/2375-446X.1000126
Anand, S., Singh, D., Avadhanula, M., & Marka, S. (2014). Development and control of bacterial biofilms on dairy processing membranes. Comprehensive Reviews in Food Science and Food Safety, 13(1), 18–33. https://doi.org/10.1111/1541-4337.12048
Andreoletti, O., Budka, H., Buncic, S., Colin, P., Collins, J., & De, A. (2007). Request for updating the former SCVPH opinion on Listeria monocytogenes risk related to ready-to-eat foods and scientific advice on different levels of Listeria monocytogenes in ready-to-eat foods and the related risk for human. EFSA Journal, 599, 1–42.
Authority, E. F. S., & European Centre for Disease Prevention and Control. (2021). The European Union One Health 2019 Zoonoses report. EFSA Journal, 19(2), e06406. https://doi.org/10.2903/j.efsa.2021.6406
Ayan, H., 2009. Uniform dielectric barrier discharge with nanosecond pulse excitation for biomedical applications. https://doi.org/10.17918/etd-3078
Bai, A. J., & Vittal, R. R. (2014). Quorum sensing inhibitory and anti-biofilm activity of essential oils and their in vivo efficacy in food systems. Food Biotechnology, 28, 269–292. https://doi.org/10.1080/08905436.2014.932287
Bandara, H. M. H. N., Lam, O. L. T., Jin, L. J., & Samaranayake, L. (2012). Microbial chemical signaling: A current perspective. Critical Reviews in Microbiology, 38, 217–249. https://doi.org/10.3109/1040841X.2011.652065
Basic Phage Electron Microscopy | Springer Nature Experiments. (n.d.). [WWW Document] URL https://experiments.springernature.com/articles/10.1007/978-1-60327-164-6_12. Accessed 2 May 2023.
Bassler, B. L. (1999). How bacteria talk to each other: Regulation of gene expression by quorum sensing. Current Opinion in Microbiology, 2(6), 582–587. Scopus. https://doi.org/10.1016/S1369-5274(99)00025-9.
Bastos, M. d. C. d. F., Coelho, M. L. V., & Santos, O. C. d. S. (2015). Resistance to bacteriocins produced by Gram-positive bacteria. Microbiology (Reading), 161, 683–700. https://doi.org/10.1099/mic.0.082289-0
Bäuerle, T., Fischer, A., Speck, T., & Bechinger, C. (2018). Self-organization of active particles by quorum sensing rules. Nature Communications, 9(1), 3232. https://doi.org/10.1038/s41467-018-05675-7
Bayoudh, S., Othmane, A., Ponsonnet, L., & Ben Ouada, H. (2008). Electrical detection and characterization of bacterial adhesion using electrochemical impedance spectroscopy-based flow chamber. Colloids and Surfaces A: Physicochemical and Engineering Aspects, 318(1), 291–300. https://doi.org/10.1016/j.colsurfa.2008.01.005
Becker, P., Egea, E., & Eeckhaut, I. (2008). Characterization of the bacterial communities associated with the bald sea urchin disease of the echinoid Paracentrotus lividus. Journal of Invertebrate Pathology, 98, 136–147. https://doi.org/10.1016/j.jip.2007.12.002
Bekeschus, S., Schmidt, A., Weltmann, K.-D., & Von Woedtke, T. (2016). The plasma jet kINPen – A powerful tool for wound healing. Clinical Plasma Medicine, 4, 19–28. https://doi.org/10.1016/j.cpme.2016.01.001
Bellin, D. L., Sakhtah, H., Zhang, Y., Price-Whelan, A., Dietrich, L. E. P., & Shepard, K. L. (2016). Electrochemical camera chip for simultaneous imaging of multiple metabolites in biofilms. Nature Communications, 7, 10535. https://doi.org/10.1038/ncomms10535
Benzaid, C., Belmadani, A., Djeribi, R., & Rouabhia, M. (2019). The effects of Mentha × piperita essential oil on C. albicans growth, transition, biofilm formation, and the expression of secreted aspartyl proteinases genes. Antibiotics (Basel), 8, 10. https://doi.org/10.3390/antibiotics8010010
Bhattacharyya, P., Agarwal, B., Goswami, M., Maiti, D., Baruah, S., & Tribedi, P. (2018). Zinc oxide nanoparticle inhibits the biofilm formation of Streptococcus pneumoniae. Antonie Van Leeuwenhoek, 111, 89–99. https://doi.org/10.1007/s10482-017-0930-7
Blanco-Cabra, N., López-Martínez, M. J., Arévalo-Jaimes, B. V., Martin-Gómez, M. T., Samitier, J., & Torrents, E. (2021). A new biofilm chip device for testing biofilm formation and antibiotic susceptibility. npj Biofilms and Microbiomes, 7(1), Article 1. https://doi.org/10.1038/s41522-021-00236-1
Bodede, O., Shaik, S., Chenia, H., Singh, P., & Moodley, R. (2018). Quorum sensing inhibitory potential and in silico molecular docking of flavonoids and novel terpenoids from Senegalia nigrescens. Journal of Ethnopharmacology, 216, 134–146. https://doi.org/10.1016/j.jep.2018.01.031
Bottone, E. J. (2010). Bacillus cereus, a volatile human pathogen. Clinical Microbiology Reviews, 23(2), 382–398. https://doi.org/10.1128/CMR.00073-09
Brackman, G., & Coenye, T. (2015). Quorum sensing inhibitors as anti-biofilm agents. Current Pharmaceutical Design, 21, 5–11. https://doi.org/10.2174/1381612820666140905114627
Brehmer, F., Haenssle, H. A., Daeschlein, G., Ahmed, R., Pfeiffer, S., Görlitz, A., Simon, D., Schön, M. P., Wandke, D., & Emmert, S. (2015). Alleviation of chronic venous leg ulcers with a hand-held dielectric barrier discharge plasma generator (PlasmaDerm® VU-2010): Results of a monocentric, two-armed, open, prospective, randomized and controlled trial (NCT01415622). Journal of the European Academy of Dermatology and Venereology, 29, 148–155. https://doi.org/10.1111/jdv.12490
Brooun, A., Liu, S., & Lewis, K. (2000). A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrobial Agents and Chemotherapy, 44(3), 640–646. https://doi.org/10.1128/AAC.44.3.640-646.2000
Brouard, C., Espié, E., Weill, F.-X., Kérouanton, A., Brisabois, A., Forgue, A.-M., Vaillant, V., & de Valk, H. (2007). Two consecutive large outbreaks of Salmonella enterica serotype Agona infections in infants linked to the consumption of powdered infant formula. The Pediatric Infectious Disease Journal, 26(2), 148–152. https://doi.org/10.1097/01.inf.0000253219.06258.23
Bruchmann, J., Sachsenheimer, K., Rapp, B. E., & Schwartz, T. (2015). Multi-channel microfluidic biosensor platform applied for online monitoring and screening of biofilm formation and activity. PLoS One, 10(2), e0117300. https://doi.org/10.1371/journal.pone.0117300
Burgess, S. A., Brooks, J. D., Rakonjac, J., Walker, K. M., & Flint, S. H. (2009). The formation of spores in biofilms of Anoxybacillus flavithermus. Journal of Applied Microbiology, 107(3), 1012–1018. https://doi.org/10.1111/j.1365-2672.2009.04282.x
Bzdrenga, J., Daudé, D., Rémy, B., Jacquet, P., Plener, L., Elias, M., & Chabrière, E. (2017). Biotechnological applications of quorum quenching enzymes. Chemico-Biological Interactions, 267, 104–115. https://doi.org/10.1016/j.cbi.2016.05.028
Cai, L., Chen, J., Liu, Z., Wang, H., Yang, H., & Ding, W. (2018). Magnesium oxide nanoparticles: Effective agricultural antibacterial agent against Ralstonia solanacearum. Frontiers in Microbiology, 9, 790.
Carrascosa, C., Millán, R., Jaber, J. R., Lupiola, P., del Rosario-Quintana, C., Mauricio, C., & Sanjuán, E. (2015). Blue pigment in fresh cheese produced by Pseudomonas fluorescens. Food Control, 54, 95–102. https://doi.org/10.1016/j.foodcont.2014.12.039
Carrascosa, C., Raheem, D., Ramos, F., Saraiva, A., & Raposo, A. (2021). Microbial biofilms in the food industry—A comprehensive review. International Journal of Environmental Research and Public Health, 18(4), Article 4. https://doi.org/10.3390/ijerph18042014
Castillo, S., Heredia, N., & García, S. (2015). 2(5H)-Furanone, epigallocatechin gallate, and a citric-based disinfectant disturb quorum-sensing activity and reduce motility and biofilm formation of Campylobacter jejuni. Folia Microbiologica, 60, 89–95. https://doi.org/10.1007/s12223-014-0344-0
Ceri, H., Olson, M. E., Stremick, C., Read, R. R., Morck, D., & Buret, A. (1999). The Calgary biofilm device: New technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. Journal of Clinical Microbiology, 37(6), 1771–1776. https://doi.org/10.1128/JCM.37.6.1771-1776.1999
Chan, B. K., & Abedon, S. T. (2015). Bacteriophages and their enzymes in biofilm control. Current Pharmaceutical Design, 21, 85–99. https://doi.org/10.2174/1381612820666140905112311
Chang, Y.-I., & Chang, P.-K. (2002). The role of hydration force on the stability of the suspension of Saccharomyces cerevisiae—Application of the extended DLVO theory. Colloids and Surfaces A: Physicochemical and Engineering Aspects, 211(1), 67–77. Scopus. https://doi.org/10.1016/S0927-7757(02)00238-8.
Characklis, W. G., & Cooksey, K. E. (1983). Biofilms and microbial fouling. In A. I. Laskin (Ed.), Advances in applied microbiology (Vol. 29, pp. 93–138). Academic Press. https://doi.org/10.1016/S0065-2164(08)70355-1
Chari, N., Felix, L., Davoodbasha, M., Sulaiman Ali, A., & Nooruddin, T. (2017). In vitro and in vivo antibiofilm effect of copper nanoparticles against aquaculture pathogens. Biocatalysis and Agricultural Biotechnology, 10, 336–341. https://doi.org/10.1016/j.bcab.2017.04.013
Chaudhary, S., Jyoti, A., Shrivastava, V., & Tomar, R. S. (2020). Role of nanoparticles as Antibiofilm agents: A comprehensive review. Current Trends in Biotechnology and Pharmacy, 14, 97–110. https://doi.org/10.5530/ctbp.2020.1.10
Chen, L., Li, X., Zhou, X., Zeng, J., Ren, Z., Lei, L., Kang, D., Zhang, K., Zou, J., & Li, Y. (2018a). Inhibition of Enterococcus faecalis growth and biofilm formation by molecule targeting cyclic di-AMP synthetase activity. Journal of Endodontia, 44, 1381–1388.e2. https://doi.org/10.1016/j.joen.2018.05.008
Chen, X., Zhang, L., Zhang, M., Liu, H., Lu, P., & Lin, K. (2018b). Quorum sensing inhibitors: a patent review (2014–2018). Expert Opinion on Therapeutic Patents, 28, 849–865. https://doi.org/10.1080/13543776.2018.1541174
Chen, X., Liu, M., Zhang, P., Xu, M., Yuan, W., Bian, L., Liu, Y., Xia, J., & Leung, S. S. Y. (2022). Phage-derived Depolymerase as an antibiotic adjuvant against multidrug-resistant Acinetobacter baumannii. Frontiers in Microbiology, 13, 845500.
Chlebicz, A., & Śliżewska, K. (2018). Campylobacteriosis, salmonellosis, yersiniosis, and listeriosis as zoonotic foodborne diseases: A review. International Journal of Environmental Research and Public Health, 15(5), Article 5. https://doi.org/10.3390/ijerph15050863
Chmielewski, R. A. N., & Frank, J. F. (2003). Biofilm formation and control in food processing facilities. Comprehensive Reviews in Food Science and Food Safety, 2(1), 22–32. https://doi.org/10.1111/j.1541-4337.2003.tb00012.x
Chrzanowska, N., & Załęska-Radziwiłł, M. (2014). The impacts of aluminum and zirconium nano-oxides on planktonic and biofilm bacteria. Desalination and Water Treatment, 52, 3680–3689. https://doi.org/10.1080/19443994.2014.884528
Clark, L. C., & Lyons, C. (1962). Electrode systems for continuous monitoring in cardiovascular surgery. Annals of the New York Academy of Sciences, 102, 29–45. https://doi.org/10.1111/j.1749-6632.1962.tb13623.x
Cogan, T. A., Bloomfield, S. F., & Humphrey, T. J. (1999). The effectiveness of hygiene procedures for prevention of cross-contamination from chicken carcases in the domestic kitchen. Letters in Applied Microbiology, 29(5), 354–358. https://doi.org/10.1046/j.1472-765X.1999.00656.x
Colagiorgi, A., Bruini, I., Di Ciccio, P. A., Zanardi, E., Ghidini, S., & Ianieri, A. (2017). Listeria monocytogenes biofilms in the wonderland of food industry. Pathogens, 6(3), Article 3. https://doi.org/10.3390/pathogens6030041
Corcoran, M., Morris, D., De Lappe, N., O’Connor, J., Lalor, P., Dockery, P., & Cormican, M. (2014). Commonly used disinfectants fail to eradicate Salmonella enterica biofilms from food contact surface materials. Applied and Environmental Microbiology, 80(4), 1507–1514. https://doi.org/10.1128/AEM.03109-13
Cotter, P. D., Ross, R. P., & Hill, C. (2013). Bacteriocins – A viable alternative to antibiotics? Nature Reviews. Microbiology, 11, 95–105. https://doi.org/10.1038/nrmicro2937
Cronenberg, C. C., & van den Heuvel, J. C. (1991). Determination of glucose diffusion coefficients in biofilms with micro-electrodes. Biosensors & Bioelectronics, 6(3), 255–262. https://doi.org/10.1016/0956-5663(91)80011-l
Ćwiek, K., Bugla-Płoskońska, G., & Wieliczko, A. (2019). Salmonella biofilm development: Structure and significance. Postępy Higieny i Medycyny Doświadczalnej, 73, 937–943. https://doi.org/10.5604/01.3001.0013.7866
D’Ostuni, V., Tristezza, M., De Giorgi, M. G., Rampino, P., Grieco, F., & Perrotta, C. (2016). Occurrence of listeria monocytogenes and Salmonella spp. in meat processed products from industrial plants in southern Italy. Food Control, 62, 104–109. https://doi.org/10.1016/j.foodcont.2015.10.025
da Silva, W. J., Leal, C. M. B., Viu, F. C., Gonçalves, L. M., Barbosa, C. M. R., & Del Bel Cury, A. A. (2015). Influence of surface free energy of denture base and liner materials on Candida albicans biofilms. Journal of Investigative and Clinical Dentistry, 6, 141–146. https://doi.org/10.1111/jicd.12079
Das, S., Sarkar, H. S., Uddin, M. R., Mandal, S., & Sahoo, P. (2018). A chemosensor to recognize N-acyl homoserine lactone in bacterial biofilm. Sensors and Actuators B: Chemical, 259, 332–338. https://doi.org/10.1016/j.snb.2017.12.040
Davies, D. G., Parsek, M. R., Pearson, J. P., Iglewski, B. H., Costerton, J. W., & Greenberg, E. P. (1998). The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science, 280(5361), 295–298. https://doi.org/10.1126/science.280.5361.295
De Beer, D., Srinivasan, R., & Stewart, P. S. (1994). Direct measurement of chlorine penetration into biofilms during disinfection. Applied and Environmental Microbiology, 60(12), 4339–4344. https://doi.org/10.1128/aem.60.12.4339-4344.1994
de Weger, L. A., van Der Vlugt, C. I. M., Wijfjes, A. H. M., Bakker, P. A., Schippers, B., & Lugtenberg, B. (1987). Flagella of a plant-growth-stimulating Pseudomonas fluorescens strain are required for colonization of potato roots. Journal of Bacteriology, 169(6), 2769–2773. Scopus. https://doi.org/10.1128/jb.169.6.2769-2773.1987.
de Wouters, T., Jans, C., Niederberger, T., Fischer, P., & Rühs, P. A. (2015). Adhesion potential of intestinal microbes predicted by physico-chemical characterization methods. PLoS One, 10(8), e0136437. https://doi.org/10.1371/journal.pone.0136437
Del Pozo, J. L. (2018). Biofilm-related disease. Expert Review of Anti-Infective Therapy, 16, 51–65. https://doi.org/10.1080/14787210.2018.1417036
Desai, M., Bühler, T., Weller, P. H., & Brown, M. R. (1998). Increasing resistance of planktonic and biofilm cultures of Burkholderia cepacia to ciprofloxacin and ceftazidime during exponential growth. Journal of Antimicrobial Chemotherapy, 42(2), 153–160. https://doi.org/10.1093/jac/42.2.153
Di Bonaventura, G., Piccolomini, R., Paludi, D., D’Orio, V., Vergara, A., Conter, M., & Ianieri, A. (2008). Influence of temperature on biofilm formation by Listeria monocytogenes on various food-contact surfaces: Relationship with motility and cell surface hydrophobicity. Journal of Applied Microbiology, 104(6), 1552–1561. https://doi.org/10.1111/j.1365-2672.2007.03688.x
Diab, R., Khameneh, B., Joubert, O., & Duval, R. (2015). Insights in nanoparticle-bacterium interactions: new frontiers to bypass bacterial resistance to antibiotics. Current Pharmaceutical Design, 21, 4095–4105. https://doi.org/10.2174/138161282128150922175445
Dincer, S., Özdenefe, M. S., & Arkut, A. (2020). Bacterial biofilms. BoD – Books on Demand.
Dippel, A. B., Anderson, W. A., Evans, R. S., Deutsch, S., & Hammond, M. C. (2018). Chemiluminescent biosensors for detection of second messenger cyclic di-GMP. ACS Chemical Biology, 13(7), 1872–1879. https://doi.org/10.1021/acschembio.7b01019
Domingo-Calap, P., & Delgado-Martínez, J. (2018). Bacteriophages: Protagonists of a post-antibiotic era. Antibiotics (Basel), 7, 66. https://doi.org/10.3390/antibiotics7030066
Duguid, J. P., Anderson, E. S., & Campbell, I. (1966). Fimbriae and adhesive properties in salmonellae. The Journal of Pathology and Bacteriology, 92(1), 107–137. https://doi.org/10.1002/path.1700920113
Duguid, I. G., Evans, E., Brown, M. R. W., & Gilbert, P. (1992). Growth-rate-independent killing by ciprofloxacin of biofilm-derived Staphylococcus epidermidis evidence for cell-cycle dependency. Journal of Antimicrobial Chemotherapy, 30(6), 791–802. https://doi.org/10.1093/jac/30.6.791
Dunne, W. M., Jr. (2002). Bacterial adhesion: Seen any good biofilms lately? Clinical Microbiology Reviews, 15(2), 155–166. Scopus. https://doi.org/10.1128/CMR.15.2.155-166.2002.
Dwivedi, S., Wahab, R., Khan, F., Mishra, Y. K., Musarrat, J., & Al-Khedhairy, A. A. (2014). Reactive oxygen species mediated bacterial biofilm inhibition via zinc oxide nanoparticles and their statistical determination. PLoS One, 9, e111289. https://doi.org/10.1371/journal.pone.0111289
Ebrahimi, N., Rasoul-Amini, S., Niazi, A., Erfani, N., Moghadam, A., Ebrahiminezhad, A., & Ghasemi, Y. (n.d.). Cytotoxic and apoptotic effects of three types of silver-iron oxide binary hybrid nanoparticles. Current Pharmaceutical Biotechnology, 17, 1049–1057.
Ebrahiminezhad, A., Varma, V., Yang, S., & Berenjian, A. (2016). Magnetic immobilization of Bacillus subtilis natto cells for menaquinone-7 fermentation. Applied Microbiology and Biotechnology, 100, 173–180. https://doi.org/10.1007/s00253-015-6977-3
Ehling-Schulz, M., Frenzel, E., & Gohar, M. (2015). Food–bacteria interplay: Pathometabolism of emetic Bacillus cereus. Frontiers in Microbiology, 6. https://www.frontiersin.org/articles/10.3389/fmicb.2015.00704
Eleraky, N. E., Allam, A., Hassan, S. B., & Omar, M. M. (2020). Nanomedicine fight against antibacterial resistance: An overview of the recent pharmaceutical innovations. Pharmaceutics, 12, 142. https://doi.org/10.3390/pharmaceutics12020142
Essential oils against bacterial isolates from cystic fibrosis patients by means of antimicrobial and unsupervised machine learning approaches | Scientific Reports. (n.d.). [WWW Document]. URL https://www.nature.com/articles/s41598-020-59553-8. Accessed 27 Apr 2023.
Evans, D. J., Allison, D. G., Brown, M. R. W., & Gilbert, P. (1991). Susceptibility of Pseudomonas aeruginosa and Escherichia coli biofilms towards ciprofloxacin: Effect of specific growth rate. Journal of Antimicrobial Chemotherapy, 27(2), 177–184. https://doi.org/10.1093/jac/27.2.177
Ferreira, V., Wiedmann, M., Teixeira, P., & Stasiewicz, M. J. (2014). Listeria monocytogenes persistence in food-associated environments: Epidemiology, strain characteristics, and implications for public health. Journal of Food Protection, 77(1), 150–170. https://doi.org/10.4315/0362-028X.JFP-13-150
Firstenberg-Eden, R., & Eden, G. (1984). Impedance microbiology. https://www.cabdirect.org/cabdirect/abstract/19850496519
Flemming, H.-C., & Wingender, J. (2010). The biofilm matrix. Nature Reviews. Microbiology, 8, 623–633. https://doi.org/10.1038/nrmicro2415
Flint, S., Palmer, J., Bloemen, K., Brooks, J., & Crawford, R. (2001). The growth of Bacillus stearothermophilus on stainless steel. Journal of Applied Microbiology, 90(2), 151–157. https://doi.org/10.1046/j.1365-2672.2001.01215.x
Franz, C. M. A. P., Stiles, M. E., Schleifer, K. H., & Holzapfel, W. H. (2003). Enterococci in foods—A conundrum for food safety. International Journal of Food Microbiology, 88(2–3), 105–122. https://doi.org/10.1016/s0168-1605(03)00174-0
Fruin, J. T. (1977). Significance of Clostridium perfringens in processed foods. Journal of Food Protection, 40(5), 330–332. https://doi.org/10.4315/0362-028X-40.5.330
Galié, S., García-Gutiérrez, C., Miguélez, E. M., Villar, C. J., & Lombó, F. (2018). Biofilms in the food industry: Health aspects and control methods. Frontiers in Microbiology, 9. https://www.frontiersin.org/articles/10.3389/fmicb.2018.00898
Gambino, M., Ahmed, M. A. A., Villa, F., & Cappitelli, F. (2017). Zinc oxide nanoparticles hinder fungal biofilm development in an ancient Egyptian tomb. International Biodeterioration & Biodegradation, 122, 92–99. https://doi.org/10.1016/j.ibiod.2017.05.011
Gashti, M. P., Asselin, J., Barbeau, J., Boudreau, D., & Greener, J. (2016). A microfluidic platform with pH imaging for chemical and hydrodynamic stimulation of intact oral biofilms. Lab on a Chip, 16(8), 1412–1419. https://doi.org/10.1039/c5lc01540e
Gaunt, L. F., Beggs, C. B., & Georghiou, G. E. (2006). Bactericidal action of the reactive species produced by gas-discharge nonthermal plasma at atmospheric pressure: A review. IEEE Transactions on Plasma Science, 34, 1257–1269. https://doi.org/10.1109/TPS.2006.878381
Ghasemian, E., Naghoni, A., Rahvar, H., Kialha, M., & Tabaraie, B. (2015). Evaluating the effect of copper nanoparticles in inhibiting Pseudomonas aeruginosa and listeria monocytogenes biofilm formation. Jundishapur Journal of Microbiology, 8. https://doi.org/10.5812/jjm.17430
Giaouris, E., Heir, E., Desvaux, M., Hébraud, M., Møretrø, T., Langsrud, S., Doulgeraki, A., Nychas, G.-J., Kačániová, M., Czaczyk, K., Ölmez, H., & Simões, M. (2015). Intra- and inter-species interactions within biofilms of important foodborne bacterial pathogens. Frontiers in Microbiology, 6. https://www.frontiersin.org/articles/10.3389/fmicb.2015.00841
Giordani, L., He, G. J., Negroni, E., Sakai, H., Law, J. Y. C., Siu, M. M., Wan, R., Corneau, A., Tajbakhsh, S., Cheung, T. H., & Le Grand, F. (2019). High-dimensional single-cell cartography reveals novel skeletal muscle-resident cell populations. Molecular Cell, 74, 609–621.e6. https://doi.org/10.1016/j.molcel.2019.02.026
Girennavar, B., Cepeda, M. L., Soni, K. A., Vikram, A., Jesudhasan, P., Jayaprakasha, G. K., Pillai, S. D., & Patil, B. S. (2008). Grapefruit juice and its furocoumarins inhibits autoinducer signaling and biofilm formation in bacteria. International Journal of Food Microbiology, 125, 204–208. https://doi.org/10.1016/j.ijfoodmicro.2008.03.028
Gloag, E. S., Fabbri, S., Wozniak, D. J., & Stoodley, P. (2020). Biofilm mechanics: Implications in infection and survival. Biofilms, 2, 100017. https://doi.org/10.1016/j.bioflm.2019.100017
Glud, R. N., Santegoeds, C. M., Beer, D. D., Kohls, O., & Ramsing, N. B. (1998). Oxygen dynamics at the base of a biofilm studied with planar optodes. Aquatic Microbial Ecology, 14(3), 223–233. https://doi.org/10.3354/ame014223
Goeres, D. M., Loetterle, L. R., Hamilton, M. A., Murga, R., Kirby, D. W., & Donlan, R. M. (2005). Statistical assessment of a laboratory method for growing biofilms. Microbiology (Reading, England), 151(Pt 3), 757–762. https://doi.org/10.1099/mic.0.27709-0
Golovlev, E. L. (2002). The mechanism of formation of Pseudomonas aeruginosa biofilm, a type of structured population. Microbiology, 71(3), 249–254. https://doi.org/10.1023/A:1015866123848
González-Rivas, F., Ripolles-Avila, C., Fontecha-Umaña, F., Ríos-Castillo, A. G., & Rodríguez-Jerez, J. J. (2018). Biofilms in the spotlight: Detection, quantification, and removal methods. Comprehensive Reviews in Food Science and Food Safety, 17(5), 1261–1276. https://doi.org/10.1111/1541-4337.12378
Gould, L. H., Mody, R. K., Ong, K. L., Clogher, P., Cronquist, A. B., Garman, K. N., Lathrop, S., Medus, C., Spina, N. L., Webb, T. H., White, P. L., Wymore, K., Gierke, R. E., Mahon, B. E., Griffin, P. M., & Emerging Infections Program Foodnet Working Group. (2013). Increased recognition of non-O157 Shiga toxin–producing Escherichia coli infections in the United States during 2000–2010: epidemiologic features and comparison with E. coli O157 infections. Foodborne Pathogens and Disease, 10(5), 453–460. https://doi.org/10.1089/fpd.2012.1401
Grandclément, C., Tannières, M., Moréra, S., Dessaux, Y., & Faure, D. (2016). Quorum quenching: Role in nature and applied developments. FEMS Microbiology Reviews, 40, 86–116. https://doi.org/10.1093/femsre/fuv038
Gratia, J.-P. (2000). André Gratia: A forerunner in microbial and viral genetics. Genetics, 156, 471–476. https://doi.org/10.1093/genetics/156.2.471
Graves, D. (2012). The emerging role of reactive oxygen and nitrogen species in redox biology and some implications for plasma applications to medicine and biology. Journal of Physics D: Applied Physics, 45, 263001. https://doi.org/10.1088/0022-3727/45/26/263001
Grigore-Gurgu, L., Bucur, F. I., Borda, D., Alexa, E.-A., Neagu, C., Nicolau, A. I., Grigore-Gurgu, L., Bucur, F. I., Borda, D., Alexa, E.-A., Neagu, C., & Nicolau, A. I. (2019). Biofilms formed by pathogens in food and food processing environments. In Bacterial biofilms. IntechOpen. https://doi.org/10.5772/intechopen.90176
Guo, J., Li, J., Chen, H., Bond, P. L., & Yuan, Z. (2017). Metagenomic analysis reveals wastewater treatment plants as hotspots of antibiotic resistance genes and mobile genetic elements. Water Research, 123, 468–478. https://doi.org/10.1016/j.watres.2017.07.002
Gupta, R. K., Chhibber, S., & Harjai, K. (2011). Acyl homoserine lactones from culture supernatants of Pseudomonas aeruginosa accelerate host immunomodulation. PLoS One, 6, e20860. https://doi.org/10.1371/journal.pone.0020860
Gutiérrez, D., Delgado, S., Vázquez-Sánchez, D., Martínez, B., Cabo, M. L., Rodríguez, A., Herrera, J. J., & García, P. (2012). Incidence of Staphylococcus aureus and analysis of associated bacterial communities on food industry surfaces. Applied and Environmental Microbiology, 78(24), 8547–8554. https://doi.org/10.1128/AEM.02045-12
Hall-Stoodley, L., & Stoodley, P. (2002). Developmental regulation of microbial biofilms. Current Opinion in Biotechnology, 13(3), 228–233. Scopus. https://doi.org/10.1016/S0958-1669(02)00318-X.
Hancock, R. E. W., Siehnel, R., & Martin, N. (1990). Outer membrane proteins of pseudomonas. Molecular Microbiology, 4(7), 1069–1075. Scopus. https://doi.org/10.1111/j.1365-2958.1990.tb00680.x.
Haruna, K., Saleh, T. A., Al Thagfi, J., & Al-Saadi, A. A. (2016). Structural properties, vibrational spectra and surface-enhanced Raman scattering of 2,4,6-trichloro- and tribromoanilines: A comparative study. Journal of Molecular Structure, 1121, 7–15. https://doi.org/10.1016/j.molstruc.2016.05.021
Hatakka, K., Holma, R., El-Nezami, H., Suomalainen, T., Kuisma, M., Saxelin, M., Poussa, T., Mykkänen, H., & Korpela, R. (2008). The influence of Lactobacillus rhamnosus LC705 together with Propionibacterium freudenreichii ssp. shermanii JS on potentially carcinogenic bacterial activity in human colon. International Journal of Food Microbiology, 128, 406–410. https://doi.org/10.1016/j.ijfoodmicro.2008.09.010
Hawver, L. A., Jung, S. A., & Ng, W.-L. (2016). Specificity and complexity in bacterial quorum-sensing systems. FEMS Microbiology Reviews, 40, 738–752. https://doi.org/10.1093/femsre/fuw014
Hayat, S., Muzammil, S., Rasool, M. H., Nisar, Z., Hussain, S. Z., Sabri, A. N., & Jamil, S. (2018). In vitro antibiofilm and anti-adhesion effects of magnesium oxide nanoparticles against antibiotic resistant bacteria. Microbiology and Immunology, 62, 211–220. https://doi.org/10.1111/1348-0421.12580
He, Z., Wang, Q., Hu, Y., Liang, J., Jiang, Y., Ma, R., Tang, Z., & Huang, Z. (2012). Use of the quorum sensing inhibitor furanone C-30 to interfere with biofilm formation by Streptococcus mutans and its luxS mutant strain. International Journal of Antimicrobial Agents, 40, 30–35. https://doi.org/10.1016/j.ijantimicag.2012.03.016
Heinlin, J., Zimmermann, J. L., Zeman, F., Bunk, W., Isbary, G., Landthaler, M., Maisch, T., Monetti, R., Morfill, G., Shimizu, T., Steinbauer, J., Stolz, W., & Karrer, S. (2013). Randomized placebo-controlled human pilot study of cold atmospheric argon plasma on skin graft donor sites. Wound Repair and Regeneration, 21, 800–807. https://doi.org/10.1111/wrr.12078
Hense, B. A., Kuttler, C., Müller, J., Rothballer, M., Hartmann, A., & Kreft, J.-U. (2007). Does efficiency sensing unify diffusion and quorum sensing? Nature Reviews. Microbiology, 5, 230–239. https://doi.org/10.1038/nrmicro1600
Hermansson, M. (1999). The DLVO theory in microbial adhesion. Colloids and Surfaces B: Biointerfaces, 14(1–4), 105–119. Scopus. https://doi.org/10.1016/S0927-7765(99)00029-6.
Hirakawa, H., & Tomita, H. (2013). Interference of bacterial cell-to-cell communication: a new concept of antimicrobial chemotherapy breaks antibiotic resistance. Frontiers in Microbiology, 4, 114. https://doi.org/10.3389/fmicb.2013.00114
Hirt, H., & Gorr, S.-U. (2013). Antimicrobial peptide GL13K is effective in reducing biofilms of Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy, 57, 4903–4910. https://doi.org/10.1128/AAC.00311-13
Hoffmann, C., Berganza, C., & Zhang, J. (2013). Cold atmospheric plasma: Methods of production and application in dentistry and oncology. Medical Gas Research, 3, 21. https://doi.org/10.1186/2045-9912-3-21
Hollenbeck, E. C., Fong, J. C. N., Lim, J. Y., Yildiz, F. H., Fuller, G. G., & Cegelski, L. (2014). Molecular determinants of mechanical properties of V. cholerae biofilms at the air-liquid interface. Biophysical Journal, 107(10), 2245–2252. https://doi.org/10.1016/j.bpj.2014.10.015
Holman, H.-Y. N., Miles, R., Hao, Z., Wozei, E., Anderson, L. M., & Yang, H. (2009). Real-time chemical imaging of bacterial activity in biofilms using open-channel microfluidics and synchrotron FTIR spectromicroscopy. Analytical Chemistry, 81(20), 8564–8570. https://doi.org/10.1021/ac9015424
Horn, H., & Hempel, D. C. (1997). Growth and decay in an auto-/heterotrophic biofilm. Water Research, 31(9), 2243–2252. https://doi.org/10.1016/S0043-1354(97)00081-X
Houry, A., Briandet, R., Aymerich, S., & Gohar, M. Y. (2010). Involvement of motility and flagella in Bacillus cereus biofilm formation. Microbiology, 156(4), 1009–1018. https://doi.org/10.1099/mic.0.034827-0
Hsueh, Y.-H., Ke, W.-J., Hsieh, C.-T., Lin, K.-S., Tzou, D.-Y., & Chiang, C.-L. (2015). ZnO nanoparticles affect Bacillus subtilis cell growth and biofilm formation. PLoS One, 10, e0128457. https://doi.org/10.1371/journal.pone.0128457
Huang, Z., Zheng, X., Yan, D., Yin, G., Liao, X., Kang, Y., Yao, Y., Huang, D., & Hao, B. (2008). Toxicological effect of ZnO nanoparticles based on bacteria. Langmuir, 24, 4140–4144. https://doi.org/10.1021/la7035949
Isbary, G., Morfill, G., Schmidt, H. U., Georgi, M., Ramrath, K., Heinlin, J., Karrer, S., Landthaler, M., Shimizu, T., Steffes, B., Bunk, W., Monetti, R., Zimmermann, J. L., Pompl, R., & Stolz, W. (2010). A first prospective randomized controlled trial to decrease bacterial load using cold atmospheric argon plasma on chronic wounds in patients. The British Journal of Dermatology, 163, 78–82. https://doi.org/10.1111/j.1365-2133.2010.09744.x
Ishwarya, R., Vaseeharan, B., Kalyani, S., Banumathi, B., Govindarajan, M., Alharbi, N. S., Kadaikunnan, S., Al-anbr, M. N., Khaled, J. M., & Benelli, G. (2018). Facile green synthesis of zinc oxide nanoparticles using Ulva lactuca seaweed extract and evaluation of their photocatalytic, antibiofilm and insecticidal activity. Journal of Photochemistry and Photobiology B: Biology, 178, 249–258. https://doi.org/10.1016/j.jphotobiol.2017.11.006
Jaffe, A., Chabbert, Y. A., & Semonin, O. (1982). Role of porin proteins OmpF and OmpC in the permeation of beta-lactams. Antimicrobial Agents and Chemotherapy, 22(6), 942–948. https://doi.org/10.1128/AAC.22.6.942
Jain, M. C., Nadaraja, A. V., Mohammadi, S., Vizcaino, B. M., & Zarifi, M. H. (2021). Passive microwave biosensor for real-time monitoring of subsurface bacterial growth. IEEE Transactions on Biomedical Circuits and Systems, 15(1), 122–132. https://doi.org/10.1109/TBCAS.2021.3055227
Janek, T., Drzymała, K., & Dobrowolski, A. (2020). In vitro efficacy of the lipopeptide biosurfactant surfactin-C15 and its complexes with divalent counterions to inhibit Candida albicans biofilm and hyphal formation. Biofouling, 36, 210–221. https://doi.org/10.1080/08927014.2020.1752370
Jewell, M. P., Galyean, A. A., Kirk Harris, J., Zemanick, E. T., & Cash, K. J. (2019). Luminescent nanosensors for ratiometric monitoring of three-dimensional oxygen gradients in laboratory and clinical Pseudomonas aeruginosa biofilms. Applied and Environmental Microbiology, 85(20), e01116–e01119. https://doi.org/10.1128/AEM.01116-19
Jha, N., Ryu, J. J., Choi, E. H., & Kaushik, N. K. (2017). Generation and role of reactive oxygen and nitrogen species induced by plasma, lasers, chemical agents, and other systems in dentistry. Oxidative Medicine and Cellular Longevity, 2017. https://doi.org/10.1155/2017/7542540
Ji, H., Kim, H., Beuchat, L. R., & Ryu, J. H. (2019). Synergistic antimicrobial activities of essential oil vapours against Penicillium corylophilum on a laboratory medium and beef jerky. International Journal of Food Microbiology, 291, 104–110. https://doi.org/10.1016/j.ijfoodmicro.2018.11.023
Jofré, A., Garriga, M., Aymerich, T., Pérez-Rodríguez, F., Valero, A., Carrasco, E., & Bover-Cid, S. (2016). Closing gaps for performing a risk assessment on listeria monocytogenes in ready-to-eat (RTE) foods: Activity 1, an extensive literature search and study selection with data extraction on L. monocytogenes in a wide range of RTE food. EFSA Supporting Publications, 13(12), 1141E. https://doi.org/10.2903/sp.efsa.2016.EN-1141
Johansen, C., Falholt, P., & Gram, L. (1997). Enzymatic removal and disinfection of bacterial biofilms. Applied and Environmental Microbiology, 63, 3724–3728. https://doi.org/10.1128/aem.63.9.3724-3728.1997
Kadariya, J., Smith, T. C., & Thapaliya, D. (2014). Staphylococcus aureus and staphylococcal food-borne disease: An ongoing challenge in public health. BioMed Research International, 2014, e827965. https://doi.org/10.1155/2014/827965
Kaplan, J. B. (2010). Biofilm dispersal: Mechanisms, clinical implications, and potential therapeutic uses. Journal of Dental Research, 89, 205–218. https://doi.org/10.1177/0022034509359403
Karlapudi, A. P., Venkateswarulu, T. C., Srirama, K., Kota, R. K., Mikkili, I., & Kodali, V. P. (2020). Evaluation of anti-cancer, anti-microbial and anti-biofilm potential of biosurfactant extracted from an Acinetobacter M6 strain. Journal of King Saud University – Science, 32, 223–227. https://doi.org/10.1016/j.jksus.2018.04.007
Keidar, M., Shashurin, A., Volotskova, O., Stepp, M. A., Srinivasan, P., Sandler, A., & Trink, B. (2013). Cold atmospheric plasma in cancer therapy. Physics of Plasmas, 20, 057101. https://doi.org/10.1063/1.4801516
Keirsse, J., Boussard-Plédel, C., Loréal, O., Sire, O., Bureau, B., Leroyer, P., Turlin, B., & Lucas, J. (2003). IR optical fiber sensor for biomedical applications. Vibrational Spectroscopy, 32(1), 23–32. https://doi.org/10.1016/S0924-2031(03)00044-4
Khajanchi, B. K., Kirtley, M. L., Brackman, S. M., & Chopra, A. K. (2011). Immunomodulatory and protective roles of quorum-sensing signaling molecules N-Acyl homoserine lactones during infection of mice with Aeromonas hydrophila. Infection and Immunity, 79, 2646–2657. https://doi.org/10.1128/IAI.00096-11
Kharazmi, A., Giwercman, B., & Høiby, N. (1999). Robbins device in biofilm research. Methods in Enzymology, 310, 207–215. https://doi.org/10.1016/s0076-6879(99)10018-1
Khosravi, Y., Kandukuri, R. D. P., Palmer, S. R., Gloag, E. S., Borisov, S. M., Starke, E. M., Ward, M. T., Kumar, P., de Beer, D., Chennu, A., & Stoodley, P. (2020). Use of an oxygen planar optode to assess the effect of high velocity microsprays on oxygen penetration in a human dental biofilms in-vitro. BMC Oral Health, 20(1), 230. https://doi.org/10.1186/s12903-020-01217-0
Killalea, D., Ward, L. R., Roberts, D., de Louvois, J., Sufi, F., Stuart, J. M., Wall, P. G., Susman, M., Schwieger, M., Sanderson, P. J., Fisher, I. S. T., Mead, P. S., Gill, O. N., Bartlett, C. L. R., & Rowe, B. (1996). International epidemiological and microbiological study of outbreak of Salmonella agona infection from a ready to eat savoury snack—I: England and Wales and the United States. BMJ, 313(7065), 1105–1107. https://doi.org/10.1136/bmj.313.7065.1105
Klančnik, A., Šimunović, K., Sterniša, M., Ramić, D., Smole Možina, S., & Bucar, F. (2021). Anti-adhesion activity of phytochemicals to prevent Campylobacter jejuni biofilm formation on abiotic surfaces. Phytochemistry Reviews, 20(1), 55–84. https://doi.org/10.1007/s11101-020-09669-6
Knecht, V. R., McGinniss, J. E., Shankar, H. M., Clarke, E. L., Kelly, B. J., Imai, I., Fitzgerald, A. S., Bittinger, K., Bushman, F. D., & Collman, R. G. (2019). Molecular analysis of bacterial contamination on stethoscopes in an intensive care unit. Infection Control & Hospital Epidemiology, 40, 171–177. https://doi.org/10.1017/ice.2018.319
Knecht, L., Veljkovic, M., & Fieseler, L. (2020). Diversity and function of phage encoded depolymerases. Frontiers in Microbiology, 10. https://doi.org/10.3389/fmicb.2019.02949
Koban, I., Holtfreter, B., Hübner, N.-O., Matthes, R., Sietmann, R., Kindel, E., Weltmann, K.-D., Welk, A., Kramer, A., & Kocher, T. (2011). Antimicrobial efficacy of non-thermal plasma in comparison to chlorhexidine against dental biofilms on titanium discs in vitro – Proof of principle experiment. Journal of Clinical Periodontology, 38, 956–965. https://doi.org/10.1111/j.1600-051X.2011.01740.x
Koul, S., Prakash, J., Mishra, A., & Kalia, V. C. (2016). Potential emergence of multi-quorum sensing inhibitor resistant (MQSIR) bacteria. Indian Journal of Microbiology, 56, 1–18. https://doi.org/10.1007/s12088-015-0558-0
Kuchma, S. L., & O’Toole, G. A. (2000). Surface-induced and biofilm-induced changes in gene expression. Current Opinion in Biotechnology, 11(5), 429–433. https://doi.org/10.1016/S0958-1669(00)00123-3
Kühl, M., Rickelt, L. F., & Thar, R. (2007). Combined imaging of bacteria and oxygen in biofilms. Applied and Environmental Microbiology, 73(19), 6289–6295. https://doi.org/10.1128/AEM.01574-07
Kulshrestha, S., Khan, S., Hasan, S., Khan, M. E., Misba, L., & Khan, A. U. (2016). Calcium fluoride nanoparticles induced suppression of Streptococcus mutans biofilm: An in vitro and in vivo approach. Applied Microbiology and Biotechnology, 100, 1901–1914. https://doi.org/10.1007/s00253-015-7154-4
Lamas, A., Miranda, J. M., Regal, P., Vázquez, B., Franco, C. M., & Cepeda, A. (2018). A comprehensive review of non-enterica subspecies of Salmonella enterica. Microbiological Research, 206, 60–73. https://doi.org/10.1016/j.micres.2017.09.010
Larsen, T. (2002). Susceptibility of Porphyromonas gingivalis in biofilms to amoxicillin, doxycycline and metronidazole. Oral Microbiology and Immunology, 17(5), 267–271. https://doi.org/10.1034/j.1399-302x.2002.170501.x
Latorre, A. A., Van Kessel, J. S., Karns, J. S., Zurakowski, M. J., Pradhan, A. K., Boor, K. J., Jayarao, B. M., Houser, B. A., Daugherty, C. S., & Schukken, Y. H. (2010). Biofilm in milking equipment on a dairy farm as a potential source of bulk tank milk contamination with Listeria monocytogenes. Journal of Dairy Science, 93(6), 2792–2802. https://doi.org/10.3168/jds.2009-2717
Lebeaux, D., Ghigo, J.-M., & Beloin, C. (2014). Biofilm-related infections: Bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiology and Molecular Biology Reviews: MMBR, 78(3), 510–543. https://doi.org/10.1128/MMBR.00013-14
Lee, S. Y., Mabee, M. S., & Jangaard, N. O. Y. (1978). Pectinatus, a new genus of the family Bacteroidaceae. International Journal of Systematic and Evolutionary Microbiology, 28(4), 582–594. https://doi.org/10.1099/00207713-28-4-582
Lee, J., Bansal, T., Jayaraman, A., Bentley, W. E., & Wood, T. K. (2007). Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-Hydroxyindole and stimulated by Isatin. Applied and Environmental Microbiology, 73(13), 4100–4109. https://doi.org/10.1128/AEM.00360-07
Lee, J.-H., Kim, Y.-G., Cho, M. H., & Lee, J. (2014). ZnO nanoparticles inhibit Pseudomonas aeruginosa biofilm formation and virulence factor production. Microbiological Research, 169, 888–896. https://doi.org/10.1016/j.micres.2014.05.005
Leiman, P. G., Chipman, P. R., Kostyuchenko, V. A., Mesyanzhinov, V. V., & Rossmann, M. G. (2004). Three-dimensional rearrangement of proteins in the tail of bacteriophage T4 on infection of its host. Cell, 118, 419–429. https://doi.org/10.1016/j.cell.2004.07.022
Leiman, P., Basler, M., Ramagopal, U., Bonanno, J., Sauder, J., Pukatzki, S., Burley, S., Almo, S., & Mekalanos, J. (2009). Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proceedings of the National Academy of Sciences, 106, 4154–4159. https://doi.org/10.1073/pnas.0813360106
Lellouche, J., Kahana, E., Elias, S., Gedanken, A., & Banin, E. (2009). Antibiofilm activity of nanosized magnesium fluoride. Biomaterials, 30, 5969–5978. https://doi.org/10.1016/j.biomaterials.2009.07.037
Lellouche, J., Friedman, A., Lahmi, R., Gedanken, A., & Banin, E. (2012). Antibiofilm surface functionalization of catheters by magnesium fluoride nanoparticles. International Journal of Nanomedicine, 7, 1175–1188. https://doi.org/10.2147/IJN.S26770
Lemon, K. P., Higgins, D. E., & Kolter, R. (2007). Flagellar motility is critical for listeria monocytogenes biofilm formation. Journal of Bacteriology, 189(12), 4418–4424. https://doi.org/10.1128/JB.01967-06
Lequette, Y., Boels, G., Clarisse, M., & Faille, C. (2010). Using enzymes to remove biofilms of bacterial isolates sampled in the food-industry. Biofouling, 26, 421–431. https://doi.org/10.1080/08927011003699535
Li, Y.-H., & Tian, X. (2012). Quorum sensing and bacterial social interactions in biofilms. Sensors (Basel), 12, 2519–2538. https://doi.org/10.3390/s120302519
Li, Y., Du, X.-F., Liu, C.-S., Wen, Z.-L., & Du, J.-L. (2012). Reciprocal regulation between resting microglial dynamics and neuronal activity in vivo. Developmental Cell, 23, 1189–1202. https://doi.org/10.1016/j.devcel.2012.10.027
Li, T., Wang, N., Chen, S., Lu, R., Li, H., & Zhang, Z. (2017). Antibacterial activity and cytocompatibility of an implant coating consisting of TiO2 nanotubes combined with a GL13K antimicrobial peptide. International Journal of Nanomedicine, 12, 2995–3007. https://doi.org/10.2147/IJN.S128775
Lim, E. S., Koo, O. K., Kim, M.-J., & Kim, J.-S. (2019). Bio-enzymes for inhibition and elimination of Escherichia coli O157:H7 biofilm and their synergistic effect with sodium hypochlorite. Scientific Reports, 9(1), Article 1. https://doi.org/10.1038/s41598-019-46363-w
Liu, X., & Ferenci, T. (1998). Regulation of porin-mediated outer membrane permeability by nutrient limitation in Escherichia coli. Journal of Bacteriology, 180(15), 3917–3922. https://doi.org/10.1128/JB.180.15.3917-3922.1998
Liu, L., Xu, Y., Cui, F., Xia, Y., Chen, L., Mou, X., & Lv, J. (2018). Monitoring of bacteria biofilms forming process by in-situ impedimetric biosensor chip. Biosensors & Bioelectronics, 112, 86–92. https://doi.org/10.1016/j.bios.2018.04.019
Liu, Y., Shi, L., Su, L., van der Mei, H. C., Jutte, P. C., Ren, Y., & Busscher, H. J. (2019). Nanotechnology-based antimicrobials and delivery systems for biofilm-infection control. Chemical Society Reviews, 48, 428–446. https://doi.org/10.1039/c7cs00807d
Lopez-Leban, F., Kiran, M. D., Wolcott, R., & Balaban, N. (2010). Molecular mechanisms of RIP, an effective inhibitor of chronic infections. The International Journal of Artificial Organs, 33, 582–589. https://doi.org/10.1177/039139881003300904
Luo, J., Dong, B., Wang, K., Cai, S., Liu, T., Cheng, X., Lei, D., Chen, Y., Li, Y., Kong, J., & Chen, Y. (2017). Baicalin inhibits biofilm formation, attenuates the quorum sensing-controlled virulence and enhances Pseudomonas aeruginosa clearance in a mouse peritoneal implant infection model. PLoS One, 12, e0176883. https://doi.org/10.1371/journal.pone.0176883
Mah, T.-F. C., & O’Toole, G. A. (2001). Mechanisms of biofilm resistance to antimicrobial agents. Trends in Microbiology, 9(1), 34–39. https://doi.org/10.1016/S0966-842X(00)01913-2
Maira-Litrán, T., Allison, D. G., & Gilbert, P. (2000). Expression of the multiple antibiotic resistance operon (mar) during growth of Escherichia coli as a biofilm. Journal of Applied Microbiology, 88(2), 243–247. https://doi.org/10.1046/j.1365-2672.2000.00963.x
Majed, R., Faille, C., Kallassy, M., & Gohar, M. (2016). Bacillus cereus biofilms—Same, only different. Frontiers in Microbiology, 7. https://www.frontiersin.org/articles/10.3389/fmicb.2016.01054
Marshall, K. C. (1986). Adsorption and adhesion processes in microbial growth at interfaces. Advances in Colloid and Interface Science, 25, 59–86. https://doi.org/10.1016/0001-8686(86)80002-1
Maruzzella, J. C., & Sicurella, N. A. (1960). Antibacterial activity of essential oil vapors. Journal of the American Pharmaceutical Association (Scientific ed.), 49, 692–694. https://doi.org/10.1002/jps.3030491103
Masalha, M., Borovok, I., Schreiber, R., Aharonowitz, Y., & Cohen, G. (2001). Analysis of transcription of the Staphylococcus aureus aerobic class Ib and anaerobic class III ribonucleotide reductase genes in response to oxygen. Journal of Bacteriology, 183(24), 7260–7272. https://doi.org/10.1128/JB.183.24.7260-7272.2001
Maurer-Jones, M. A., Gunsolus, I. L., Meyer, B. M., Christenson, C. J., & Haynes, C. L. (2013). Impact of TiO2 nanoparticles on growth, biofilm formation, and flavin secretion in Shewanella oneidensis. Analytical Chemistry, 85, 5810–5818. https://doi.org/10.1021/ac400486u
McLean, J. S., Ona, O. N., & Majors, P. D. (2008). Correlated biofilm imaging, transport and metabolism measurements via combined nuclear magnetic resonance and confocal microscopy. The ISME Journal, 2(2), 121–131. https://doi.org/10.1038/ismej.2007.107
Melo, L., Bott, T. R., Fletcher, M., & Capdeville, B. (2012). Biofilms—Science and technology. Springer.
Metcalf, D., & Bowler, P. (2013). Biofilm delays wound healing: A review of the evidence. Burns & Trauma, 1, 5–12. https://doi.org/10.4103/2321-3868.113329
Meyer, M. T., Roy, V., Bentley, W. E., & Ghodssi, R. (2011). Development and validation of a microfluidic reactor for biofilm monitoring via optical methods. Journal of Micromechanics and Microengineering, 21(5), 054023. https://doi.org/10.1088/0960-1317/21/5/054023
Millezi, A. F., Piccoli, R. H., Oliveira, J. M., & Pereira, M. O. (2016). Anti-biofim and antibacterial effect of essential oils and their major compounds. Journal of Essential Oil Bearing Plants, 19, 624–631. https://doi.org/10.1080/0972060X.2014.960262
Mizan, M. F. R., Jahid, I. K., & Ha, S.-D. (2015). Microbial biofilms in seafood: A food-hygiene challenge. Food Microbiology, 49, 41–55. https://doi.org/10.1016/j.fm.2015.01.009
Moghadas-Sharif, N., Fazly Bazzaz, B. S., Khameneh, B., & Malaekeh-Nikouei, B. (2015). The effect of nanoliposomal formulations on Staphylococcus epidermidis biofilm. Drug Development and Industrial Pharmacy, 41, 445–450. https://doi.org/10.3109/03639045.2013.877483
Mokoena, M. P. (2017). Lactic acid bacteria and their bacteriocins: Classification, biosynthesis and applications against uropathogens: A mini-review. Molecules, 22, 1255. https://doi.org/10.3390/molecules22081255
Molobela, I. P., Cloete, T. E., & Beukes, M. (2010). Protease and amylase enzymes for biofilm removal and degradation of extracellular polymeric substances (EPS) produced by Pseudomonas fluorescens bacteria. African Journal of Microbiology Research, 4(14), 1515–1524.
Moreau, M., Orange, N., & Feuilloley, M. G. J. (2008). Non-thermal plasma technologies: New tools for bio-decontamination. Biotechnology Advances, 26, 610–617. https://doi.org/10.1016/j.biotechadv.2008.08.001
Møretrø, T., Heir, E., Nesse, L. L., Vestby, L. K., & Langsrud, S. (2012). Control of Salmonella in food related environments by chemical disinfection. Food Research International, 45(2), 532–544. https://doi.org/10.1016/j.foodres.2011.02.002
Murphy, P. M., Lynch, D., & Kelly, P. M. (1999). Growth of thermophilic spore forming bacilli in milk during the manufacture of low heat powders. International Journal of Dairy Technology, 52(2), 45–50. https://doi.org/10.1111/j.1471-0307.1999.tb02069.x
Nakamura, M., Hori, N., Ando, H., Namba, S., Toyama, T., Nishimiya, N., & Yamashita, K. (2016). Surface free energy predominates in cell adhesion to hydroxyapatite through wettability. Materials Science & Engineering. C, Materials for Biological Applications, 62, 283–292. https://doi.org/10.1016/j.msec.2016.01.037
Nguyen, H. D. N., Yang, Y. S., & Yuk, H. G. (2014). Biofilm formation of Salmonella Typhimurium on stainless steel and acrylic surfaces as affected by temperature and pH level. LWT – Food Science and Technology, 55(1), 383–388. https://doi.org/10.1016/j.lwt.2013.09.022
Nicolay, N., Thornton, L., Cotter, S., Garvey, P., Bannon, O., McKeown, P., Cormican, M., Fisher, I., Little, C., Boxall, N., De Pinna, E., Peters, T. M., Cowden, J., Salmon, R., Mason, B., Irvine, N., Rooney, P., O’Flanagan, D. (2011). Salmonella enterica serovar Agona European outbreak associated with a food company. Epidemiology and Infection, 139(8), 1272–1280. https://doi.org/10.1017/S0950268810002360. Epub 2010 Oct 18. PMID: 20950515.
O’Loughlin, C. T., Miller, L. C., Siryaporn, A., Drescher, K., Semmelhack, M. F., & Bassler, B. L. (2013). A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proceedings of the National Academy of Sciences of the United States of America, 110, 17981–17986. https://doi.org/10.1073/pnas.1316981110
Ogawa, A., Kiyohara, T., Kobayashi, Y., Sano, K., & Kanematsu, H. (2017). Nickel, molybdenum, and tungsten nanoparticle-dispersed alkylalkoxysilane polymer for biomaterial coating: Evaluation of effects on bacterial biofilm formation and biosafety. Biomedical Research and Clinical Practice, 2. https://doi.org/10.15761/BRCP.1000138
Ohadi, M., Shahravan, A., Dehghannoudeh, N., Eslaminejad, T., Banat, I. M., & Dehghannoudeh, G. (2020). Potential use of microbial surfactant in microemulsion drug delivery system: A systematic review. Drug Design, Development and Therapy, 14, 541–550. https://doi.org/10.2147/DDDT.S232325
Ouyang, J., Sun, F., Feng, W., Sun, Y., Qiu, X., Xiong, L., Liu, Y., & Chen, Y. (2016). Quercetin is an effective inhibitor of quorum sensing, biofilm formation and virulence factors in Pseudomonas aeruginosa. Journal of Applied Microbiology, 120, 966–974. https://doi.org/10.1111/jam.13073
Packiavathy, S., Selvam, P., Pandian, S., & Arumugam, V. R. (2014). Inhibition of biofilm development of uropathogens by curcumin – An anti-quorum sensing agent from Curcuma longa. Food Chemistry, 148C, 453–460. https://doi.org/10.1016/j.foodchem.2012.08.002
Padmavathy, N., & Vijayaraghavan, R. (2008). Enhanced bioactivity of ZnO nanoparticles—An antimicrobial study. Science and Technology of Advanced Materials, 9, 035004. https://doi.org/10.1088/1468-6996/9/3/035004
Palmer, J., Flint, S., & Brooks, J. (2007). Bacterial cell attachment, the beginning of a biofilm. Journal of Industrial Microbiology & Biotechnology, 34(9), 577–588. https://doi.org/10.1007/s10295-007-0234-4
Pammi, M., Liang, R., Hicks, J., Mistretta, T.-A., & Versalovic, J. (2013). Biofilm extracellular DNA enhances mixed species biofilms of Staphylococcus epidermidis and Candida albicans. BMC Microbiology, 13(1), 257. https://doi.org/10.1186/1471-2180-13-257
Paradh, A. D., Mitchell, W. J., & Hill, A. E. (2011). Occurrence of pectinatus and megasphaera in the major UK breweries. Journal of the Institute of Brewing, 117(4), 498–506. https://doi.org/10.1002/j.2050-0416.2011.tb00497.x
Paraszkiewicz, K., Moryl, M., Płaza, G., Bhagat, D., Satpute, S. K., & Bernat, P. (2021). Surfactants of microbial origin as antibiofilm agents. International Journal of Environmental Health Research, 31, 401–420. https://doi.org/10.1080/09603123.2019.1664729
Paredes, J., Becerro, S., Arizti, F., Aguinaga, A., Del Pozo, J. L., & Arana, S. (2013). Interdigitated microelectrode biosensor for bacterial biofilm growth monitoring by impedance spectroscopy technique in 96-well microtiter plates. Sensors and Actuators B: Chemical, 178, 663–670. https://doi.org/10.1016/j.snb.2013.01.027
Paredes, J., Becerro, S., & Arana, S. (2014a). Label-free interdigitated microelectrode based biosensors for bacterial biofilm growth monitoring using petri dishes. Journal of Microbiological Methods, 100, 77–83. https://doi.org/10.1016/j.mimet.2014.02.022
Paredes, J., Becerro, S., & Arana, S. (2014b). Comparison of real time impedance monitoring of bacterial biofilm cultures in different experimental setups mimicking real field environments. Sensors and Actuators B: Chemical, 195, 667–676. https://doi.org/10.1016/j.snb.2014.01.098
Park, J., Jagasia, R., Kaufmann, G. F., Mathison, J. C., Ruiz, D. I., Moss, J. A., Meijler, M. M., Ulevitch, R. J., & Janda, K. D. (2007). Infection control by antibody disruption of bacterial quorum sensing signaling. Chemistry & Biology, 14, 1119–1127. https://doi.org/10.1016/j.chembiol.2007.08.013
Pavanello, G., Faimali, M., Pittore, M., Mollica, A., Mollica, A., & Mollica, A. (2011). Exploiting a new electrochemical sensor for biofilm monitoring and water treatment optimization. Water Research, 45(4), 1651–1658. https://doi.org/10.1016/j.watres.2010.12.003
Peter Revsbech, N. (2005). Analysis of microbial communities with electrochemical microsensors and microscale biosensors. In Methods in enzymology (Vol. 397, pp. 147–166). Academic Press. https://doi.org/10.1016/S0076-6879(05)97009-2
Poma, N., Vivaldi, F., Bonini, A., Salvo, P., Kirchhain, A., Ates, Z., Melai, B., Bottai, D., Tavanti, A., & Di Francesco, F. (2021). Microbial biofilm monitoring by electrochemical transduction methods. TrAC Trends in Analytical Chemistry, 134, 116134. https://doi.org/10.1016/j.trac.2020.116134
Pousti, M., Zarabadi, M. P., Amirdehi, M. A., Paquet-Mercier, F., & Greener, J. (2018). Microfluidic bioanalytical flow cells for biofilm studies: A review. Analyst, 144(1), 68–86. https://doi.org/10.1039/C8AN01526K
Praneenararat, T., Palmer, A. G., & Blackwell, H. E. (2012). Chemical methods to interrogate bacterial quorum sensing pathways. Organic & Biomolecular Chemistry, 10, 8189–8199. https://doi.org/10.1039/C2OB26353J
Pu, H., Xu, Y., Sun, D.-W., Wei, Q., & Li, X. (2021). Optical nanosensors for biofilm detection in the food industry: Principles, applications and challenges. Critical Reviews in Food Science and Nutrition, 61(13), 2107–2124. https://doi.org/10.1080/10408398.2020.1808877
Pugsley, A. P., & Schnaitman, C. A. (1978). Outer membrane proteins of Escherichia coli. VII. Evidence that bacteriophage-directed protein 2 functions as a pore. Journal of Bacteriology, 133(3), 1181–1189. https://doi.org/10.1128/jb.133.3.1181-1189.1978
Raheem, D., & Raheem, D. (2016). Outbreaks of listeriosis associated with deli meats and cheese: An overview. AIMS Microbiology, 2(3), 230–250. https://doi.org/10.3934/microbiol.2016.3.230
Ramos, M. A. D. S., Silva, P. B. D., Spósito, L., Toledo, L. G. D., Bonifácio, B. V., Rodero, C. F., Santos, K. C. D., Chorilli, M., & Bauab, T. M. (2018). Nanotechnology-based drug delivery systems for control of microbial biofilms: a review. IJN, 13, 1179–1213. https://doi.org/10.2147/IJN.S146195
Redfield, R. J. (2002). Is quorum sensing a side effect of diffusion sensing? Trends in Microbiology, 10, 365–370. https://doi.org/10.1016/s0966-842x(02)02400-9
Rehman, Z. U., & Leiknes, T. (2018). Quorum-quenching bacteria isolated from Red Sea sediments reduce biofilm formation by Pseudomonas aeruginosa. Frontiers in Microbiology, 9, 1354. https://doi.org/10.3389/fmicb.2018.01354
Reij, M. W., & Den Aantrekker, E. D. (2004). Recontamination as a source of pathogens in processed foods. International Journal of Food Microbiology, 91(1), 1–11. https://doi.org/10.1016/S0168-1605(03)00295-2
Reuter, S., Winter, J., Iséni, S., Schmidt-Bleker, A., Dünnbier, M., Masur, K., Wende, K., & Weltmann, K. (2015). The influence of feed gas humidity versus ambient humidity on atmospheric pressure plasma jet-effluent chemistry and skin cell viability. IEEE Transactions on Plasma Science, 43, 3185–3192. https://doi.org/10.1109/TPS.2014.2361921
Ripa, R., Shen, A. Q., & Funari, R. (2020). Detecting Escherichia coli biofilm development stages on gold and titanium by quartz crystal microbalance. ACS Omega, 5(5), 2295–2302. https://doi.org/10.1021/acsomega.9b03540
Robb, A. J., Vinogradov, S., Danell, A. S., Anderson, E., Blackledge, M. S., Melander, C., & Hvastkovs, E. G. (2018). Electrochemical detection of small molecule induced Pseudomonas aeruginosa biofilm dispersion. Electrochimica Acta, 268, 276–282. https://doi.org/10.1016/j.electacta.2018.02.113
Rodrigues, D., Teixeira, P., Oliveira, R., & Azeredo, J. (2011). Salmonella enterica Enteritidis biofilm formation and viability on regular and triclosan-impregnated bench cover materials. Journal of Food Protection, 74(1), 32–37. https://doi.org/10.4315/0362-028X.JFP-10-167
Rothrock, M. J., Davis, M. L., Locatelli, A., Bodie, A., McIntosh, T. G., Donaldson, J. R., & Ricke, S. C. (2017). Listeria occurrence in poultry flocks: Detection and potential implications. Frontiers in Veterinary Science, 4. https://www.frontiersin.org/articles/10.3389/fvets.2017.00125
Rühs, P. A., Böni, L., Fuller, G. G., Inglis, R. F., & Fischer, P. (2013). In-situ quantification of the interfacial rheological response of bacterial biofilms to environmental stimuli. PLoS One, 8(11), e78524. https://doi.org/10.1371/journal.pone.0078524
Rühs, P. A., Böcker, L., Inglis, R. F., & Fischer, P. (2014). Studying bacterial hydrophobicity and biofilm formation at liquid-liquid interfaces through interfacial rheology and pendant drop tensiometry. Colloids and Surfaces B: Biointerfaces, 117, 174–184. https://doi.org/10.1016/j.colsurfb.2014.02.023
Russo, E. T., Biggerstaff, G., Hoekstra, R. M., Meyer, S., Patel, N., Miller, B., Quick, R., & For the Salmonella Agona Outbreak Investigation Team. (2013). A recurrent, multistate outbreak of Salmonella serotype Agona infections associated with dry, unsweetened cereal consumption, United States, 2008. Journal of Food Protection, 76(2), 227–230. https://doi.org/10.4315/0362-028X.JFP-12-209
Rutter, P. R., & Vincent, B. (1984). Physicochemical interactions of the substratum, microorganisms, and the fluid phase. In K. C. Marshall (Ed.), Microbial adhesion and aggregation (pp. 21–38). Springer. https://doi.org/10.1007/978-3-642-70137-5_3
Saccomano, S. C., Jewell, M. P., & Cash, K. J. (2021). A review of chemosensors and biosensors for monitoring biofilm dynamics. Sensors and Actuators Reports, 3, 100043. https://doi.org/10.1016/j.snr.2021.100043
Sadiq, F. A., Flint, S., Yuan, L., Li, Y., Liu, T., & He, G. (2017). Propensity for biofilm formation by aerobic mesophilic and thermophilic spore forming bacteria isolated from Chinese milk powders. International Journal of Food Microbiology, 262, 89–98. https://doi.org/10.1016/j.ijfoodmicro.2017.09.015
Saleh, T. (2014). Detection: From electrochemistry to spectroscopy with chromatographic techniques, recent trends with nanotechnology. Detection, 02, 27–32. https://doi.org/10.4236/detection.2014.24005
Sathyanarayanan, M. B., Balachandranath, R., Genji Srinivasulu, Y., Kannaiyan, S. K., & Subbiahdoss, G. (2013). The effect of gold and iron-oxide nanoparticles on biofilm-forming pathogens. ISRN Microbiology, 2013, 272086. https://doi.org/10.1155/2013/272086
Satpute, S. K., Kulkarni, G. R., Banpurkar, A. G., Banat, I. M., Mone, N. S., Patil, R. H., & Cameotra, S. S. (2016). Biosurfactant/s from Lactobacilli species: Properties, challenges and potential biomedical applications: Biosurfactant/s from Lactobacilli species. Journal of Basic Microbiology, 56, 1140–1158. https://doi.org/10.1002/jobm.201600143
Schelin, J., Susilo, Y. B., & Johler, S. (2017). Expression of staphylococcal enterotoxins under stress encountered during food production and preservation. Toxins, 9(12), Article 12. https://doi.org/10.3390/toxins9120401
Schmaljohn, A. L., & McClain, D. (1996). Alphaviruses (Togaviridae) and Flaviviruses (Flaviviridae). In S. Baron (Ed.), Medical microbiology. University of Texas Medical Branch at Galveston.
Schmelcher, M., Donovan, D. M., & Loessner, M. J. (2012). Bacteriophage endolysins as novel antimicrobials. Future Microbiology, 7, 1147–1171. https://doi.org/10.2217/fmb.12.97
Shahed-Al-Mahmud, M., Roy, R., Sugiokto, F. G., Islam, M. N., Lin, M.-D., Lin, L.-C., & Lin, N.-T. (2021). Phage φAB6-borne depolymerase combats Acinetobacter baumannii biofilm formation and infection. Antibiotics (Basel), 10, 279. https://doi.org/10.3390/antibiotics10030279
Shakibaie, M., Forootanfar, H., Golkari, Y., Mohammadi-Khorsand, T., & Shakibaie, M. R. (2015). Anti-biofilm activity of biogenic selenium nanoparticles and selenium dioxide against clinical isolates of Staphylococcus aureus, Pseudomonas aeruginosa, and Proteus mirabilis. Journal of Trace Elements in Medicine and Biology, 29, 235–241. https://doi.org/10.1016/j.jtemb.2014.07.020
Sharan, M., Vijay, D., Dhaka, P., Bedi, J. S., & Gill, J. P. S. (2022). Biofilms as a microbial hazard in the food industry: A scoping review. Journal of Applied Microbiology, 133(4), 2210–2234. https://doi.org/10.1111/jam.15766
Sharma, M., & Anand, S. K. (2002). Characterization of constitutive microflora of biofilms in dairy processing lines. Food Microbiology, 19(6), 627–636. https://doi.org/10.1006/fmic.2002.0472
Sharma, S., Sharma, R., Sardesai, M. M., & Mishra, V. (2017). Anticancer potential of leafless mistletoe (Viscum angulatum) from Western Ghats of India. International Journal of Pharmaceutical Sciences and Research, 9. https://doi.org/10.13040/IJPSR.0975-8232.9(5).1000-05
Sharma, J., Sundar, D., & Srivastava, P. (2021). Biosurfactants: Potential agents for controlling cellular communication, motility, and antagonism. Frontiers in Molecular Biosciences, 8, 727070.
Shohat, T., Green, M. S., Merom, D., Gill, O. N., Reisfeld, A., Matas, A., Blau, D., Gal, N., & Slater, P. E. (1996). International epidemiological and microbiological study of outbreak of Salmonella agona infection from a ready to eat savoury snack—II: Israel. BMJ, 313(7065), 1107–1109. https://doi.org/10.1136/bmj.313.7065.1107
Shumi, W., Lim, J., Nam, S.-W., Lee, K., Kim, S. H., Kim, M.-H., Cho, K.-S., & Park, S. (2010). Environmental factors that affect Streptococcus mutans biofilm formation in a microfluidic device mimicking teeth. BioChip Journal, 4(4), 257–263. https://doi.org/10.1007/s13206-010-4401-8
Sillankorva, S., Kluskens, L. D., Lingohr, E. J., Kropinski, A. M., Neubauer, P., & Azeredo, J. (2011). Complete genome sequence of the lytic Pseudomonas fluorescens phage ϕIBB-PF7A. Virology Journal, 8, 142. https://doi.org/10.1186/1743-422X-8-142
Simões, M., Simões, L. C., & Vieira, M. J. (2010). A review of current and emergent biofilm control strategies. LWT – Food Science and Technology, 43, 573–583. https://doi.org/10.1016/j.lwt.2009.12.008
Soltani, S., Hammami, R., Cotter, P. D., Rebuffat, S., Said, L. B., Gaudreau, H., Bédard, F., Biron, E., Drider, D., & Fliss, I. (2021). Bacteriocins as a new generation of antimicrobials: Toxicity aspects and regulations. FEMS Microbiology Reviews, 45, fuaa039. https://doi.org/10.1093/femsre/fuaa039
Somerton, B., Flint, S., Palmer, J., Brooks, J., & Lindsay, D. (2013). Preconditioning with cations increases the attachment of Anoxybacillus flavithermus and Geobacillus species to stainless steel. Applied and Environmental Microbiology, 79(13), 4186–4190. https://doi.org/10.1128/AEM.00462-13
Sønderholm, M., Koren, K., Wangpraseurt, D., Jensen, P. Ø., Kolpen, M., Kragh, K. N., Bjarnsholt, T., & Kühl, M. (2018). Tools for studying growth patterns and chemical dynamics of aggregated Pseudomonas aeruginosa exposed to different electron acceptors in an alginate bead model. npj Biofilms and Microbiomes, 4, 3. https://doi.org/10.1038/s41522-018-0047-4
Staal, M., Borisov, S. M., Rickelt, L. F., Klimant, I., & Kühl, M. (2011). Ultrabright planar optodes for luminescence life-time based microscopic imaging of O2 dynamics in biofilms. Journal of Microbiological Methods, 85(1), 67–74. https://doi.org/10.1016/j.mimet.2011.01.021
Stenfors Arnesen, L. P., Fagerlund, A., & Granum, P. E. (2008). From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiology Reviews, 32(4), 579–606. https://doi.org/10.1111/j.1574-6976.2008.00112.x
Sterniša, M., Klančnik, A., & Smole Možina, S. (2019). Spoilage Pseudomonas biofilm with Escherichia coli protection in fish meat at 5 °C. Journal of the Science of Food and Agriculture, 99(10), 4635–4641. https://doi.org/10.1002/jsfa.9703
Stiefel, P., Rosenberg, U., Schneider, J., Mauerhofer, S., Maniura-Weber, K., & Ren, Q. (2016). Is biofilm removal properly assessed? Comparison of different quantification methods in a 96-well plate system. Applied Microbiology and Biotechnology, 100, 4135–4145. https://doi.org/10.1007/s00253-016-7396-9
Strejc, J., Kyselova, L., Cadkova, A., Matoulkova, D., Potocar, T., & Branyik, T. (2020). Experimental adhesion of Geobacillus stearothermophilus and Anoxybacillus flavithermus to stainless steel compared with predictions from interaction models. Chemical Papers, 74(1), 297–304. https://doi.org/10.1007/s11696-019-00880-0
Struss, A., Pasini, P., Ensor, C. M., Raut, N., & Daunert, S. (2010). Paper strip whole cell biosensors: A portable test for the semiquantitative detection of bacterial quorum signaling molecules. Analytical Chemistry, 82(11), 4457–4463. https://doi.org/10.1021/ac100231a
Subhadra, B., Oh, M. H., & Choi, C. H. (2019). RND efflux pump systems in Acinetobacter, with special emphasis on their role in quorum sensing. Journal of Bacteriology and Virology, 49(1), 1–11. https://doi.org/10.4167/jbv.2019.49.1.1
Subramanian, S., Huiszoon, R. C., Chu, S., Bentley, W. E., & Ghodssi, R. (2020). Microsystems for biofilm characterization and sensing—A review. Biofilms, 2, 100015. https://doi.org/10.1016/j.bioflm.2019.100015
Suci, P. A., Mittelman, M. W., Yu, F. P., & Geesey, G. G. (1994). Investigation of ciprofloxacin penetration into Pseudomonas aeruginosa biofilms. Antimicrobial Agents and Chemotherapy, 38(9), 2125–2133. https://doi.org/10.1128/AAC.38.9.2125
Sully, E. K., Malachowa, N., Elmore, B. O., Alexander, S. M., Femling, J. K., Gray, B. M., DeLeo, F. R., Otto, M., Cheung, A. L., Edwards, B. S., Sklar, L. A., Horswill, A. R., Hall, P. R., & Gresham, H. D. (2014). Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathogens, 10, e1004174. https://doi.org/10.1371/journal.ppat.1004174
Sutherland, I. W. (1999). Polysaccharases for microbial exopolysaccharides. Carbohydrate Polymers, 38(4), 319–328. Scopus. https://doi.org/10.1016/S0144-8617(98)00114-3
Sybiya Vasantha Packiavathy, I. A., Agilandeswari, P., Musthafa, K. S., Karutha Pandian, S., & Veera Ravi, A. (2012). Antibiofilm and quorum sensing inhibitory potential of Cuminum cyminum and its secondary metabolite methyl eugenol against Gram negative bacterial pathogens. Food Research International, 45, 85–92. https://doi.org/10.1016/j.foodres.2011.10.022
Tamilvanan, S., Venkateshan, N., & Ludwig, A. (2008). The potential of lipid- and polymer-based drug delivery carriers for eradicating biofilm consortia on device-related nosocomial infections. Journal of Controlled Release, 128, 2–22. https://doi.org/10.1016/j.jconrel.2008.01.006
Tan, Y., Ma, S., Leonhard, M., Moser, D., Ludwig, R., & Schneider-Stickler, B. (2020). Co-immobilization of cellobiose dehydrogenase and deoxyribonuclease I on chitosan nanoparticles against fungal/bacterial polymicrobial biofilms targeting both biofilm matrix and microorganisms. Materials Science and Engineering: C, 108, 110499. https://doi.org/10.1016/j.msec.2019.110499
Tapia-Rodriguez, M. R., Bernal-Mercado, A. T., Gutierrez-Pacheco, M. M., Vazquez-Armenta, F. J., Hernandez-Mendoza, A., Gonzalez-Aguilar, G. A., Martinez-Tellez, M. A., Nazzaro, F., & Ayala-Zavala, J. F. (2019). Virulence of Pseudomonas aeruginosa exposed to carvacrol: Alterations of the quorum sensing at enzymatic and gene levels. Journal of Cell Communication and Signaling, 13, 531–537. https://doi.org/10.1007/s12079-019-00516-8
Tateda, K., Ishii, Y., Horikawa, M., Matsumoto, T., Miyairi, S., Pechere, J. C., Standiford, T. J., Ishiguro, M., & Yamaguchi, K. (2003). The Pseudomonas aeruginosa autoinducer N-3-oxododecanoyl homoserine lactone accelerates apoptosis in macrophages and neutrophils. Infection and Immunity, 71, 5785–5793. https://doi.org/10.1128/IAI.71.10.5785-5793.2003
Téllez, S. (2010). Biofilms and their impact on food industry. VISAVET Outreach Journal. https://www.visavet.es/en/articles/biofilms-impact-food-industry.php
Thallinger, B., Prasetyo, E. N., Nyanhongo, G. S., & Guebitz, G. M. (2013). Antimicrobial enzymes: An emerging strategy to fight microbes and microbial biofilms. Biotechnology Journal, 8, 97–109. https://doi.org/10.1002/biot.201200313
Thapa, T., & Ayan, H. (2019). Application of non-thermal plasma on biofilm: A review. Applied Sciences, 9, 3548. https://doi.org/10.3390/app9173548
Thomann, A., de Mello Martins, A. G. G., Brengel, C., Empting, M., & Hartmann, R. W. (2016). Application of dual inhibition concept within looped autoregulatory systems toward antivirulence agents against Pseudomonas aeruginosa infections. ACS Chemical Biology, 11, 1279–1286. https://doi.org/10.1021/acschembio.6b00117
Threlfall, E. J., Hampton, M. D., Ward, L. R., & Rowe, B. (1996). Application of pulsed-field gel electrophoresis to an international outbreak of Salmonella agona. Emerging Infectious Diseases, 2(2), 130–132.
Thukkaram, M., Sitaram, S., Kannaiyan, S. K., & Subbiahdoss, G. (2014). Antibacterial efficacy of iron-oxide nanoparticles against biofilms on different biomaterial surfaces. International Journal of Biomaterials. https://doi.org/10.1155/2014/716080
Todd, E. C. D., Greig, J. D., Bartleson, C. A., & Michaels, B. S. (2009). Outbreaks where food workers have been implicated in the spread of foodborne disease. Part 6. Transmission and survival of pathogens in the food processing and preparation environment. Journal of Food Protection, 72(1), 202–219. https://doi.org/10.4315/0362-028x-72.1.202
Toledo-Arana, A., Merino, N., Vergara-Irigaray, M., Débarbouillé, M., Penadés, J. R., & Lasa, I. (2005). Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. Journal of Bacteriology, 187(15), 5318–5329. https://doi.org/10.1128/JB.187.15.5318-5329.2005
Topka-Bielecka, G., Nejman-Faleńczyk, B., Bloch, S., Dydecka, A., Necel, A., Węgrzyn, A., & Węgrzyn, G. (2021). Phage-bacteria interactions in potential applications of bacteriophage vB_EfaS-271 against enterococcus faecalis. Viruses, 13, 318. https://doi.org/10.3390/v13020318
Toushik, S. H., Mizan, M. F. R., Hossain, M. I., & Ha, S.-D. (2020). Fighting with old foes: The pledge of microbe-derived biological agents to defeat mono- and mixed-bacterial biofilms concerning food industries. Trends in Food Science & Technology, 99, 413–425. https://doi.org/10.1016/j.tifs.2020.03.019
Tram, G., Day, C. J., & Korolik, V. (2020). Bridging the gap: A role for Campylobacter jejuni biofilms. Microorganisms, 8(3), Article 3. https://doi.org/10.3390/microorganisms8030452
Trotonda, M. P., Manna, A. C., Cheung, A. L., Lasa, I., & Penadés, J. R. (2005). SarA positively controls bap-dependent biofilm formation in Staphylococcus aureus. Journal of Bacteriology, 187(16), 5790–5798. https://doi.org/10.1128/JB.187.16.5790-5798.2005
Tuomanen, E., Cozens, R., Tosch, W., Zak, O., & Tomasz, A. (1986a). The rate of killing of Escherichia coli by beta-lactam antibiotics is strictly proportional to the rate of bacterial growth. Journal of General Microbiology, 132(5), 1297–1304. https://doi.org/10.1099/00221287-132-5-1297
Tuomanen, E., Durack, D. T., & Tomasz, A. (1986b). Antibiotic tolerance among clinical isolates of bacteria. Antimicrobial Agents and Chemotherapy, 30(4), 521–527. https://doi.org/10.1128/AAC.30.4.521
Turan, N. B., Chormey, D. S., Büyükpınar, Ç., Engin, G. O., & Bakirdere, S. (2017). Quorum sensing: Little talks for an effective bacterial coordination. TrAC Trends in Analytical Chemistry, 91, 1–11. https://doi.org/10.1016/j.trac.2017.03.007
Utchariyakiat, I., Surassmo, S., Jaturanpinyo, M., Khuntayaporn, P., & Chomnawang, M. T. (2016). Efficacy of cinnamon bark oil and cinnamaldehyde on anti-multidrug resistant Pseudomonas aeruginosa and the synergistic effects in combination with other antimicrobial agents. BMC Complementary and Alternative Medicine, 16, 158. https://doi.org/10.1186/s12906-016-1134-9
Van Houdt, R., & Michiels, C. W. (2010). Biofilm formation and the food industry, a focus on the bacterial outer surface. Journal of Applied Microbiology, 109(4), 1117–1131. https://doi.org/10.1111/j.1365-2672.2010.04756.x
Vandeputte, O. M., Kiendrebeogo, M., Rajaonson, S., Diallo, B., Mol, A., El Jaziri, M., & Baucher, M. (2010). Identification of catechin as one of the flavonoids from Combretum albiflorum bark extract that reduces the production of quorum-sensing-controlled virulence factors in Pseudomonas aeruginosa PAO1. Applied and Environmental Microbiology, 76, 243–253. https://doi.org/10.1128/AEM.01059-09
Vasavi, H. S., Sudeep, H. V., Lingaraju, H. B., & Shyam Prasad, K. (2017). Bioavailability-enhanced Resveramax™ modulates quorum sensing and inhibits biofilm formation in Pseudomonas aeruginosa PAO1. Microbial Pathogenesis, 104, 64–71. https://doi.org/10.1016/j.micpath.2017.01.015
Vasilev, K., Cook, J., & Griesser, H. J. (2009). Antibacterial surfaces for biomedical devices. Expert Review of Medical Devices, 6, 553–567. https://doi.org/10.1586/erd.09.36
Vattem, D. A., Mihalik, K., Crixell, S. H., & McLean, R. J. C. (2007). Dietary phytochemicals as quorum sensing inhibitors. Fitoterapia, 78, 302–310. https://doi.org/10.1016/j.fitote.2007.03.009
Verran, J. (2002). Biofouling in food processing: Biofilm or biotransfer potential? Food and Bioproducts Processing, 80(4), 292–298. https://doi.org/10.1205/096030802321154808
Vitas, A. I., & Garcia-Jalon, V. A. e. I. (2004). Occurrence of Listeria monocytogenes in fresh and processed foods in Navarra (Spain). International Journal of Food Microbiology, 90(3), 349–356. https://doi.org/10.1016/S0168-1605(03)00314-3
Vongkamjan, K., Fuangpaiboon, J., Turner, M. P., & Vuddhakul, V. (2016). Various ready-to-eat products from retail stores linked to occurrence of diverse listeria monocytogenes and listeria spp. isolates. Journal of Food Protection, 79(2), 239–245. https://doi.org/10.4315/0362-028X.JFP-15-361
Wang, H., Ding, S., Wang, G., Xu, X., & Zhou, G. (2013). In situ characterization and analysis of Salmonella biofilm formation under meat processing environments using a combined microscopic and spectroscopic approach. International Journal of Food Microbiology, 167(3), 293–302. https://doi.org/10.1016/j.ijfoodmicro.2013.10.005
Wang, X. C., Wilson, S. C., & Hammond, M. C. (2016). Next-generation RNA-based fluorescent biosensors enable anaerobic detection of cyclic di-GMP. Nucleic Acids Research, 44(17), e139. https://doi.org/10.1093/nar/gkw580
Wang, Y., Liu, B., Grenier, D., & Yi, L. (2019). Regulatory mechanisms of the LuxS/AI-2 system and bacterial resistance. Antimicrobial Agents and Chemotherapy, 63(10), e01186–e01119. https://doi.org/10.1128/AAC.01186-19
Ward, A. C., Connolly, P., & Tucker, N. P. (2014). Pseudomonas aeruginosa can be detected in a polymicrobial competition model using impedance spectroscopy with a novel biosensor. PLoS One, 9(3), e91732. https://doi.org/10.1371/journal.pone.0091732
Wei, Y., Jiao, Y., An, D., Li, D., Li, W., & Wei, Q. (2019). Review of dissolved oxygen detection technology: From laboratory analysis to online intelligent detection. Sensors, 19(18), Article 18. https://doi.org/10.3390/s19183995
Weltmann, K.-D., Kindel, E., Brandenburg, R., Meyer, C., Bussiahn, R., Wilke, C., & von Woedtke, T. (2009). Atmospheric pressure plasma jet for medical therapy: Plasma parameters and risk estimation. Contributions to Plasma Physics, 49, 631–640. https://doi.org/10.1002/ctpp.200910067
Wolfbeis, O. S. (2015). Luminescent sensing and imaging of oxygen: Fierce competition to the Clark electrode. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology, 37(8), 921–928. https://doi.org/10.1002/bies.201500002
Wu, H., Moser, C., Wang, H.-Z., Høiby, N., & Song, Z.-J. (2015). Strategies for combating bacterial biofilm infections. International Journal of Oral Science, 7, 1–7. https://doi.org/10.1038/ijos.2014.65
Wu, P., Guo, Y., Golly, M. K., Ma, H., He, R., Luo, S., Zhang, C., Zhang, L., & Zhu, J. (2019). Feasibility study on direct fermentation of soybean meal by Bacillus stearothermophilus under non-sterile conditions. Journal of the Science of Food and Agriculture, 99(7), 3291–3298. https://doi.org/10.1002/jsfa.9542
Xie, Q., Zhao, F., Liu, H., Shan, Y., & Liu, F. (2015). A label-free and self-assembled electrochemical biosensor for highly sensitive detection of cyclic diguanylate monophosphate (c-di-GMP) based on RNA riboswitch. Analytica Chimica Acta, 882, 22–26. https://doi.org/10.1016/j.aca.2015.04.061
Xu, Y., Jones, J. E., Yu, H., Yu, Q., Christensen, G. D., Chen, M., & Sun, H. (2015). Nanoscale plasma coating inhibits formation of Staphylococcus aureus biofilm. Antimicrobial Agents and Chemotherapy, 59, 7308–7315. https://doi.org/10.1128/aac.01944-15
Yan, X., Gu, S., Cui, X., Shi, Y., Wen, S., Chen, H., & Ge, J. (2018). Antimicrobial, anti-adhesive and anti-biofilm potential of biosurfactants isolated from Pediococcus acidilactici and Lactobacillus plantarum against Staphylococcus aureus CMCC26003. Microbial Pathogenesis, 127. https://doi.org/10.1016/j.micpath.2018.11.039
Yang, G., Feng, L., Yang, Q., Zhu, L., Xu, J., & Xu, X. (2014). Startup pattern and performance enhancement of pilot-scale biofilm process for raw water pretreatment. Bioresource Technology, 172, 22–31. https://doi.org/10.1016/j.biortech.2014.08.116
Yang, G.-F., Feng, L.-J., Guo, C.-R., Xia, T., Xu, X.-Y., & Zhu, L. (2017). Performance improvement of raw water pretreatment process with pre-inoculation biofilm: Feasibility and limiting factors. Biodegradation, 28, 111–123. https://doi.org/10.1007/s10532-016-9781-6
Yawata, Y., Toda, K., Setoyama, E., Fukuda, J., Suzuki, H., Uchiyama, H., & Nomura, N. (2010). Monitoring biofilm development in a microfluidic device using modified confocal reflection microscopy. Journal of Bioscience and Bioengineering, 110(3), 377–380. https://doi.org/10.1016/j.jbiosc.2010.04.002
Yu, S., Zhu, X., Zhou, J., & Cai, Z. (2018). Biofilm inhibition and pathogenicity attenuation in bacteria by Proteus mirabilis. Royal Society Open Science, 5, 170702. https://doi.org/10.1098/rsos.170702
Zheng, L. Y., Congdon, R. B., Sadik, O. A., Marques, C. N. H., Davies, D. G., Sammakia, B. G., Lesperance, L. M., & Turner, J. N. (2013). Electrochemical measurements of biofilm development using polypyrrole enhanced flexible sensors. Sensors and Actuators B: Chemical, 182, 725–732. https://doi.org/10.1016/j.snb.2013.03.097
Zhong, N., Zhao, M., & Li, Y. (2016). U-shaped, double-tapered, fiber-optic sensor for effective biofilm growth monitoring. Biomedical Optics Express, 7(2), 335–351. https://doi.org/10.1364/BOE.7.000335
Zhong, N., Wu, Y., Wang, Z., Chang, H., Zhong, D., Xu, Y., Hu, X., & Huang, L. (2019). Monitoring microalgal biofilm growth and phenol degradation with fiber-optic sensors. Analytical Chemistry, 91(23), 15155–15162. https://doi.org/10.1021/acs.analchem.9b03923
Zinke, M., Schröder, G. F., & Lange, A. (2022). Major tail proteins of bacteriophages of the order Caudovirales. Journal of Biological Chemistry, 298. https://doi.org/10.1016/j.jbc.2021.101472
Zupančič, J., Raghupathi, P. K., Houf, K., Burmølle, M., Sørensen, S. J., & Gunde-Cimerman, N. (2018). Synergistic interactions in microbial biofilms facilitate the establishment of opportunistic pathogenic fungi in household dishwashers. Frontiers in Microbiology, 9. https://www.frontiersin.org/articles/10.3389/fmicb.2018.00021
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Firoze, S., Sami, H., Azhar, A., Asaad, M., Khan, P.A., Khan, H.M. (2024). Microbial Biofilms and the Role of Biotechnology as a Solution. In: Ahmad, F., Mohammad, Z.H., Ibrahim, S.A., Zaidi, S. (eds) Microbial Biotechnology in the Food Industry. Springer, Cham. https://doi.org/10.1007/978-3-031-51417-3_9
Download citation
DOI: https://doi.org/10.1007/978-3-031-51417-3_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-51416-6
Online ISBN: 978-3-031-51417-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)