Abstract
Evidence-based medicine has received increasing attention. This type of medicine would have the benefit of using large data sets to investigate clinical–laboratory associations and validate hypotheses grounded on data. Pathology is one area that has been benefited from large data sets of images, having advances leveraged by computational pathology, which in turn relies in the advances of the methods conceived by the computational intelligence and the computer vision fields. This type of medicine would benefit of using large. By particularly considering kidney biopsies, computational nephropathology seeks to identify renal lesions from primary computer vision tasks that involve classification and segmentation of renal structures on histology images. In this context, this chapter aims at discussing some advances in computational nephropathology, contextualizing them in the scope of the PathoSpotter project. We also address current achievements and challenges, as well as dig in future prospects to the field.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
16.1 Introduction
Kidney is among the human organs with the most clearly demarcated compartments at the histological level of glomeruli, tubules, interstitium, and blood vessels (arteries and veins). These structures are organized in functional units, called nephrons, based on the flow of the blood through afferent arterioles, glomeruli, efferent arterioles, and the flow of the liquid ultra-filtrated from the blood, through the tubules (proximal convoluted tubule, Henle’s loop, and distal convoluted tubule). Diseases affecting any of these compartments progressively extend to the other structures. Most of the known nephrologic diseases affect primarily the glomeruli with inflammation, fibrosis, anomalous deposits, or degenerative changes. A continuous effort has been made by pathologists toward establishing a consensus on histological markers to be used in renal disease classification and to predict the outcome of renal diseases. Although most of these markers have shown to be useful in defining diagnosis and prognosis of renal diseases, the criteria adopted to define these markers are time consuming, labor intensive, and sometimes have poor agreement among pathologists. Automatizing the process of renal disease classification would contribute to improve the accuracy and reduce the time spent with this process.
The dissemination of digital image acquisition systems, for collecting either snapshot images or large whole slide image (WSI), opened possibilities to facilitate the transit of the information about the biopsies among pathologists. In this sense, a new field of research has arisen, which was coined as digital pathology. With advances of hardware and software to digitize WSIs, large histological image libraries emerge to improve diagnostic performance and the course of consensus. One perceived soon that the information contained in these images would remain largely unexplored without a proper effort to automatize the analysis of the histological images. Such large collection of digital histological images, gathered in different laboratories with different hardware devices, provided resources for supervised learning, directly considering the training of increasingly accurate intelligent systems able to recognize lesions in WSIs. This made possible the emergence of a new area of research called computational pathology. Computational pathology systems rely on the extraction and recognition of visual patterns from images and are used in multiple tasks. In the nephrology context, intelligent systems are applied, for instance, in the segmentation of renal structures and lesion classification.
16.2 Digital × Computational Pathology
The use of computers as a tool for assisting in cell analysis is not new. Tolles [1] introduced some basic tools and techniques for image acquisition and image analysis. Henceforth an expressive evolution of computational techniques for this field has driven the rise of digital pathology, which is a broad term that encompasses tools and systems to digitize pathology slides and associated metadata, their storage, review, analysis, and infrastructure [2].
The increasing application of artificial intelligence (AI) and machine learning (ML) techniques for image analysis in digital pathology drove the emergence of a new field called computational pathology, which guides its efforts to the pattern recognition from digitized histology images and their associated meta-data. Although in the pathology community there are no formal definitions neither for digital pathology, nor for computational pathology, we could define the main tasks for the first as to acquire, process, storage, and distribute digital histology images, while for the second as to analyze digital histology images with the purpose of recognizing image patters and ultimately aid pathologist with the diagnosis. From the information technology perspective, digital pathology infrastructure and methods are related to the quality of acquired images, the reliability of the stored images, and the image distribution and security. Besides, methods for computational pathology are related to the detection, segmentation, labeling, retrieval, and classification of structures presented in the images.
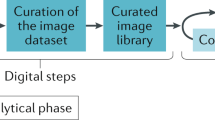
Figure 16.1 illustrates the digital and computational pathology procedures and tasks. Activities in digital pathology encompass the tissue sampling and slide preparation, which is usually followed by scanning to turn them into high-resolution images that must be stored in secure repositories. Complementarily, the computational pathology exploits the image collections to perform the many artificial intelligence tasks for medical decision support. A common method relies on visual feature extraction and representation through image descriptors.
Digital and computational pathology procedures. Digital pathology is a broad term that encompasses tools and systems to digitize pathology slides and associated meta-data, their storage, review, analysis, and infrastructure. Computational pathology guides its efforts to the pattern recognition from digitized histology images and their associated meta-data across tasks of image classification, object detection, image and per-instance segmentation, labeling and retrieval
16.3 How Do Computers Find Patterns in Histology Images?
The importance of finding patterns in several human activities has stimulated the development of computational systems that have the goal of automatizing tasks, ultimately reducing classification errors prone to be achieved when done exclusively by humans.
Computational techniques have been a commonplace to aid in diagnoses based on radiological images [3,4,5] and dermatology [6,7,8], while the use of such techniques on biopsies is rapidly increasing, particularly in nephropathology [9,10,11,12,13,14].
A computational system used for histology image classification is usually built upon the pipeline depicted in Fig. 16.1. Although image acquisition takes place via any proper device to the considered task, the pre-processing stage is specifically responsible to make the image appropriately uniform to the next stage, where features sufficiently representative to help the image classification task are extracted. Broadly speaking, image classification should be understood here either as object classification where an image is labeled to pertain to a specific class, or as semantic or per-instance segmentation where a pixel or a set of pixels are classified to belong to a specific class, respectively. Each stage presents its own set of challenges to be overcome, and classification error may occur individually or as a result of interaction among all phases.
In computational nephropathology, the acquisition phase captures an entire slide that is converted into a digital image using some type of optical-electronic transductor. Aspects such as image resolution (number of pixels), noise filtering, scale adjustments, and color spectrum are strongly influenced by the quality of the lamp used to light the slide, as well as the accuracy of the transductor. The result of this phase is a digital colored image composed by the combination of three channels (R-red, G-green and B-blue), computationally represented as a matrix of integer numbers associated to the color intensity at each image point (pixel), which was captured by the transductor. Once the image has been digitized, the next step is to guarantee image uniformity regarded lighting, bright, and other essential aspects inherited to histology images. Image classification can be regarded to the level of a pixel, a set of pixels, or an object. The first two are related to semantic or instance segmentation, respectively, while the last one is the traditional object classification where a box is labeled as a specific known class. Image segmentation is the process of partitioning a digital image into multiple segments in order to separate the parts of interest from all other parts. For the sake of computer algorithms, image segmentation is the process of assigning a label to every pixel in an image, such that pixels with the same label share certain characteristics. Figure 16.2 illustrates an example of how image segmentation works over a cropped image containing a glomerulus. Feature extraction and image classification are very interdependent and critical to the overall quality of the system. Given a specific image classification problem, the most difficult challenge is accurately selecting and extracting the best features, and then choosing a classification supervised model that is capable of correctly separating the classes. Due to the complexity in translating which features are the most appropriate and how they best represent the object of interest in computer language is that the whole task is challenging. Several algorithms have been developed to extract features based on color, edges, corners, shape, and textures when analyzing images with different performance levels.
To be able to make predictions, ML-based algorithms must first rely on data in order to build a mathematical model [15]. This stage is called training, which is by itself a complex, time-consuming task, as several models and parameters must be tuned to reach the best classifier. Each model built must be “trained” with a sample data set in order to “learn” how to classify samples with an acceptable degree of error, in the prediction phase. For each context, a specific set of features will be relevant, and a certain model of the classifier will yield optimal results when analyzing these features. Since there is no way to a priori determine either the best feature set or the best classifier model, for many years, computer scientists struggled with the development of mathematical functions that could represent images as features in order to obtain the best result possible in the classification stage.
For several years, computer scientists attempted to tackle those limitations until a major breakthrough occurred in the field of the computer vision: The use of convolutional neural networks (CNN) to automatize the feature extraction process during the training stage. A CNN is a combination of convolutional filters designed to achieve trainable feature extractors, followed by a fully connected artificial neural network (ANN) to perform image classification. Figure 16.3 illustrates an earlier form of a CNN architecture, which is comprised of a backbone and a top layer, the latter formed by a multi-layer perceptron (MLP). In the 1990s, this architecture was shown to work well for recognizing hand-written digits [16]. In recent years, thanks to the increasing use of graphics processing units (GPU) to speed up computational processes, and the rise of large image data sets [17], it has been possible to develop larger, more complex computational vision models based on CNNs. These models favor the creation of a new ML approach, called deep learning (DL) [18], which led to the widespread use of deeper networks [2] in computer vision, mainly after the success of the AlexNet model [19], which won the 2012 edition of ImageNet Large Scale Visual Recognition Challenge [20].
An example of an earlier CNN architicture [16] that is comprised of a backbone and a top layer. The backbone is formed by convolutional layers followed by pooling layer, this latter is in charge of dimensionality reduction and consequently control of the parameter degree-of-freedom in the process of training. The top layer is usually laid out by a multilayer perceptron (MLP). Before feeding the fully connected (FC) MLP, data from backbone is vectorized through 1D transformations; after passing through the MLP, 𝐾 neurons are outputted according to the number of classes to be classified
In computational pathology, the conception of CNN architectures in association with the increasing availability of digital biopsy data sets, stimulated by the adoption of slide scanners, has created momentum in the development of computational systems capable of assisting pathologists. A very useful feature of CNN is the possibility of transfer learning [21]. In this technique, a CNN is initially trained with as large data set as possible, enabling it to adjust its filters to correctly extract the best features from images. Then, using the previously trained feature extractor, a new data set of interest (usually with a few number of training image samples) is submitted to the network, which will then quickly converge on the classification. Transfer learning has proven to be a useful technique for developing effective CNN models even using small training data sets.
There has been a steady increase in the use of computational image classification systems in computational pathology, mainly in the detection of cellular lesions to assist in diagnosis [22,23,24]. Every day new techniques and methods are being developed to improve the quality of these systems. We believe that in the near future, computational systems capable of performing automatic image analysis will become an important tool for pathologists, and these will foster significant evolution in evidence-based medicine.
16.4 The PathoSpotter Project
Histological images bear a variety of information about patient disease. Frequently, patients with similar forms of clinical disease have different histological presentations and differing prognoses (see Fig. 16.4). Take, for example, a situation frequently faced by pediatricians: A child with high proteinuria leading to hypoalbuminemia, edema and dyslipidemia presents a condition known as nephrotic syndrome. If the renal biopsy of this patient reveals normal glomeruli, as shown in Fig. 16.4a, the pathologist knows that child has a benign disease and will completely recover under treatment with corticosteroids [25]. However, if a child with similar clinical presentation shows scars in the glomeruli, as seen in Fig. 16.4b, this patient has a progressive disease that will not respond to corticosteroid treatment [26]. Then histological lesions contain information that is relevant to defining a diagnosis and, consequently, to disease prognosis. Fig. 16.4c depicts a kidney with diffuse interstitial widening due to fibrosis, as well as tubular atrophy. Interstitial fibrosis and tubular atrophy are considered among the most relevant histological markers of renal disease chronicity [27]. Histological images also contain information regarding the activity, chronicity, and progression of diseases [25].
Although one is able to recognize many signs that predict the course of disease, pathologists are constantly revising the histological criteria used to define disease activity, chronicity, progression, and even diagnosis [28]. It is not uncommon that pathologists find variations in the characteristics of ordinary lesions in their day-to-day practice or encounter lesions that are rare or of unknown relevance [9].Footnote 1 Due to the relevance of images used in anatomopathological diagnoses, pathologists typically will maintain records of the images of lesions received in their routines. The emergence of digital image collection and storage has allowed for the creation of huge data sets consisting of histology images. Despite that, no tools have been fully developed that permit the exploration of the information contained in these images. Accordingly, the elaboration of a system capable of automatically detecting and comparing histological lesions would be of great interest and could be used to:
-
Support pathologists in their routines, mainly those residing in remote areas with limited access to specialists in different areas of anatomical pathology;
-
Teach young pathologists by providing access to a variety of images of the same histological lesion across different cases;
-
Perform large-scale clinical-pathological correlations, accelerating research into new treatments and the definition of consensuses.
Hence several research groups around the world have been working in the development of intelligent systems for automatic classification of histological lesions of breast, prostate, skin, and other organs using different approaches [3, 10, 23].Footnote 2 In 2014, a group of pathologists and computer science experts started the PathoSpotter project [3] in an effort to build a system to perform automatic identification of histological lesions. The most significant developments produced by PathoSpotter to date have been in the area of renal pathology, due to the availability of a robust digital image library of histological renal lesions. This library, stored at the Gonçalo Moniz Institute of the Oswaldo Cruz Foundation, [4] began in 1997 and contains all biopsies received for diagnosis from all public referral nephrology services in the municipality of Salvador (Bahia, Brazil). The digital image library was built by Dr. Washington LC dos-Santos. Unlike most of the data sets built for experimental research, this library possesses a diversity that likely reflects that found in everyday life of pathologists. The library contains more than 110,000 images of more than 3000 biopsies, mostly of native kidneys stained with hematoxylin and eosin (H&E), periodic acid–Schiff (PAS), periodic acid-methenamine silver stain (PAMS), Mallory’s trichrome (AZAN) or picrosirius red stain, as well as immunofluorescence for IgA, IgG, IgM, kappa and lambda chains of immunoglobulins, C1q, C3, and fibrinogen, in addition to images obtained by transmission electron microscopy. Lesions are the main focus of the images, which employ different magnifications. These images were generated using at least five different digital image capture systems. Most are in JPEG format with a resolution of 1024 × 768 pixels. Additionally, the library contains 400 WSIs. The clinical characteristics of the patients from whom the biopsies originated have been previously reported [29]. Most patients were adults, with similar male vs. female distribution, and 50% had nephrotic syndrome. The main biopsy diagnoses were focal and segmental glomerulosclerosis, lupus nephritis, and membranous glomerulopathy. The library also contains about 2000 images donated by colleagues from four other laboratories at the Federal Universities of Piaui (Teresina, Brazil) and Minas Gerais (Belo Horizonte, Brazil), the Kidney Hospital of the Federal University of São Paulo (São Paulo, Brazil), and Imagepat, a private service in Salvador, Bahia.
16.4.1 A Map of Kidney Histological Lesions
As mentioned before, histological images bear a variety of information about the patient’s disease. Having a way of organizing and visualizing the images under different points of view is essential for pathologists and computer science specialists to glance through the histological patterns of renal diseases and to grasp the distinctive presentation of the same category of the lesion in different diseases. To solve this problem, we developed an interactive web-based visualization tool that accesses a database of images and displays the information in a circular hierarchical form (see Fig. 16.5).
The compact visualization of the hierarchy is built on the fly based on the available information for each image: Staining, nosological diagnosis, pathological anatomical diagnosis, renal compartment, and morphological changes. By using this tool, the pathologists can navigate the hierarchy, including zooming and panning, collapse and expand branches, search for a specific keyword, and change the hierarchy order. Additionally, the pathologists can see and change the details of each image displayed on the right panel. This flexibility allows the pathologists to reorganize the images based on different aspects and explore how the images are related.
16.4.2 A Content-Based Image Retrieval System
Anatomopathological diagnosis is performed through visual findings of lesions in WSIs. A type of lesion can emerge with variations in visual presentation, which are diagnostically relevant. Quite often, the pathologists need to find previous cases that are similar to the one under analysis. This process is usually conducted by manually inspecting the previous cases to search for similar lesion variations, which is a laborious, time-consuming task, especially considering large image repositories. Therefore a system capable of automatically retrieving histological images that present similar characteristics to a target image is of great importance for the refinement of the anatomopathological diagnosis and for the research of histological markers for diagnostic purposes. In computer science, such task is known as content-based image retrieval (CBIR). A typical CBIR system performs two primary tasks: (a) extraction of visual features for the representation of the images and (b) computation of the similarity between the query image and the other images in the database using their feature descriptors, which is followed by ranking them accordingly (Fig. 16.6).
Typical CBIR system. A feature extraction method creates image descriptors that are stored in a database. Such descriptors allow comparison and indexing of 𝑡ℎ images. When a query image is submitted, a descriptor is built and compared to the stored ones. Finally, the images are ranked according to a similarity score and the most similar are presented to user
The effectiveness of a CBIR system strongly bears in the quality of the features adopted to describe the images [30]. To select and extract representative features, traditional CBIR systems have to deal with several specificities of image objects, such as scale and rotation variances. Some algorithms like SIFT [31], SURF [32], and HOG [33] have been traditionally used to deal with these issues. For medical purposes, the development of effective feature extractors has been considered a challenging problem [4, 34].
The evolution of the CNN-based architectures facilitated the feature extraction by automatizing the learning and extraction of invariant features [35], while allowing its use as feature extractors for CBIR systems [34, 36,37,38,39], including some works in histopathology [40, 41].
In CBIR systems applied in nephropathology, the images may be indexed by their visual content (color, texture, shape) and by metadata representative of the histopathological findings (characterization of renal structures). Both information can be used for image indexing and retrieval. Although many systems used in medical practice are capable of comparing images based on their global appearance, subsequently list them according to some similarity score, they are not capable of finding relevant images based on the particular features presented in specific nephrological findings. A system able to perform such specialized retrieval depends on the development of methods for extracting specific discriminative visual patterns in images from renal biopsies.
The PathoSpotter SearchFootnote 3 service is the first initiative for a CBIR system devoted to nephropathology. It uses a feature extraction algorithm based on CNNs (trained with the nephropathology images) to compute visual descriptors for the set of images that will be used as the retrieval repository and save them in a database for future querying. To search for images in this set that are similar to a query image, the pathologist submits this query image to the system and select the database to be used for retrieval. The system uses the same feature extraction algorithm to extract a descriptor for the query image and compare such descriptors to all descriptors in the database. Each comparison yields a number that express the similarity between the query image and the compared image (similarity score). Finally, the system ranks the images in the database according this similarity score and shows the ones that present the higher similarity score with the query image.
16.4.3 How PathoSpotter Finds Patterns in Histology Images
Detection of renal lesions in histology images is the ultimate goal of nephropathology. When considering the analyzed renal structure along with the clinical data, the pathologist and the nephrologist are capable to provide a nosological diagnosis as accurate as possible. Likewise, the main goal of PathoSpotter is to pursue precision in the detection of lesions over biopsy images by using ML and computer vision techniques. The PathoSpotter system is fed with WSIs or cropped images, and to accomplish that goal, our team has been working with two fundamental tasks: image classification and segmentation. We consider segmentation as pixel classification when dealing with the semantic or the per-instance forms of this task. Here however we differentiate both tasks as to label an object (considering a bounding box) or to label a pixel, respectively, inside the specific concepts of object classification and image segmentation.
The first classification system developed in PathoSpotter was based on handcrafted features and a K-nearest neighborhood classifier to label images of no-lesion or with hypercellularity glomerulus [9]. The input is a cropped image containing a glomerulus, and the data set was comprised of digitized images fixed in formalin (to preserve their histological structure), embedded in paraffin, cut into 2–3 μm thick sections, and finally stained using one of the following techniques: H&E, PAS, PAMS, AZAN, or picrosirius red stain. In total, 811 images were used, considering 300 images of no-lesion (normal) glomeruli and 511 images of glomeruli from kidneys with hypercelullarity. A binary classification was performed, yielding 88% of accuracy in small image samples used to assess the performance of the system.
Lately an evolved system was conceived, [11] grounded on an own CNN architecture and support vector machine (SVM). As mentioned before, CNNs provide features that are found in the process of training the classifier. After training, these features are appropriated to be used in any other classifier rather than the MLP on top of the CNN; in our case, an SVM was used. Over the same data set used in the work of Barros et al. [9], using a tenfold cross-validation evaluation procedure, this new classification method found near perfect results in hypercellularity/normal binary problem. Considering four classes—endocapillary, mesangial, both, and normal–this new classifier achieved 82% of mean F1-score and accuracy.
CNN-based classifiers commonly output the probability of classifying an object via a softmax function placed in the last layer. The softmax function assures that the scores of all classes sum up to 1, then guaranteeing that the highest score is given to the “most probable” class. Since these softmax scores are also nonnegative, by definition, we can say they represent probabilities, although these cannot be interpreted as reliable confidence scores. Hein et al. [42] showed that CNNs almost always return high confidence predictions even for inputs far away from the training distribution. Since the scores tend to be high regardless of the input, they are not a suitable measure of uncertainty.
An uncertainty metric should indicate whether the model is “certain” or “uncertain” about its prediction, being mostly useful for high-risk applications such as autonomous driving or computer-aided diagnoses (CAD). Ideally, when evaluating a model, the incorrect predictions should have higher uncertainty scores than correct predictions. This way, when a model is uncertain about a given image, we can consider that the prediction is probably wrong. As the classification cannot be fully trusted, the sample should be properly assessed by specialists. Recently research on uncertainty estimation is gaining more relevance [43]. Specifically for CAD applications, Begoli et al. [44] highlight the need for uncertainty measurements in machine-assisted, medical systems. Indeed several works studied how uncertainty can be applied in medical imaging classification approaches [45, 46] even for nephropathology [47].
Diving into this issue, Chagas et al. [12] proposed to evolve the PathoSpotter classification system by combining CNN architectures with an uncertainty estimation method for membranous nephropathy classification. Besides achieving competitive classification results (average F1-score of 93.6%), their uncertainty scores showed high relation with correctness on predictions (higher uncertainty scores represented mostly incorrect predictions). For practical applications, the uncertainty score works as additional support information for the pathologist. If a model returns results with high uncertainty, the pathologist can reevaluate, ignore the prediction, or mark the image for further inspection.
Starting from the uncertainty estimation, Chagas et al. [13] also increased PathoSpotter potential by tackling the problem of out-of-distribution (OOD) detection for membranous nephropathy classification. OOD detection aims to determine whether the input image belongs or not to the training distribution. Considering the overconfidence problem of neural networks, it might be troublesome to analyze the output of a model when the target image belongs to a different domain (e.g., a non-glomerulus image) or to an unknown or novel class of the same domain (e.g., a glomerulus with an unknown or novel lesion). This OOD detection was based on an unbounded open-set setup, i.e., when there are no constraints to the unknown classes (novel glomerular classes and non-glomerular classes are considered OOD in the same way) [48]. OOD detection ensures the safety and robustness of the evaluation pipeline, as OOD samples might represent an outlier or noisy data.
A whole pipeline can be depicted considering uncertainty estimation and OOD detection, as Fig. 16.7 suggests. An ideal pipeline is based on a model that performs class prediction, uncertainty score, and OOD detection. Figure 16.7 details how we propose using these model outcomes. Firstly, one must determine whether the input image is an OOD sample or not. Depending on the OOD method and implementation, this step could be achieved automatically or via threshold. If the image is classified as OOD data, the sample could represent a non-glomerulus or a novel class, thus should be assessed in another pipeline defined by specialists. We propose discarding the image, reevaluate it, or mark it for further analysis, but this phase depends on the application and specialists involved. Alternatively, if the sample is not classified as OOD data, one must determine how reliable the class prediction is. With the uncertainty score, we can define (automatically or via threshold) whether the model is “certain” or “uncertain” about the class prediction. If the model is “uncertain,” the sample should be investigated, using an assessment similar to the OOD case. If the model is “certain,” we can trust the class prediction and use it as our final classification label. Knowing when the model “does not know” is of underlying importance because not only the image might not be related to the data domain, but also the model could return an overconfidence score for any given image. Regardless, in a safe evaluation pipeline, the pathologists should be aware of when any of those situations occur.
Uncertainty-aware open-set classification pipeline proposal. Instead of predicting a single label, the model performs class prediction, uncertainty score, and OOD detection. Each model outcome is used as input (represented by dashed lines) for other steps. Depending on whether the input image is an OOD data and whether the model is “uncertain,” a different end for the pipeline can be reached (represented by dark blue boxes): Discarding the image or further assessment or using regular class prediction for glomerular classification
In the classification works by Barros et al. [9] and Chagas et al. [11], the glomeruli were cut manually by the pathologist, making the classification task still with some degree of human interference. Thus, with the aim of improving the injury classification capabilities of the PathoSpotter system, Rehem et al. [14] propose a glomerulus detection method on renal histology images. For that, we evaluated two state-of-the-art deep-learning techniques: single-shot multi-box detector with InceptionV2 (SI2) and faster region-based convolutional neural network with InceptionV2 (FRI2). As a result, we reached 0.88 of mAP and 0.94 of F1-score, when using SI2, and 0.87 of mAP and 0.97 of F1-score, when using FRI2. On average, to process each image, FRI2 required 30.91s, while SI2 just 0.79s. In the experiments, we found that SI2 model is the best detection method for the particular task as it is 64% faster in the training stage and 98% faster to detect the glomeruli in each image.
16.5 Achievements, Challenges, and Future Prospects
We have seen several groundbreaking achievements in computational pathology and nephropathology in the literature, mainly due to the advances in the DL methods to classify and segment images. Some examples can be found in detecting skin cancer [23] or classifying glomerular lesions in WSIs [11, 49]. Although much has been done toward solving fundamental problems in the field, unfortunately very few studies exploit large and heterogeneous data sets with substantial cohorts that might validate the clinical usefulness of some pre-clinical prospective works. This situation may result in some skewed opinions from what is hype and what is effectively promised in the state-of-the-art works. This is so because many challenges are involved in the development of systems capable of automatically integrating expert knowledge pertaining to specific histological lesions, such as:
-
The number of existing histological lesions in a given organ. It is very difficult to precisely define the exact number of existing lesions in a given organ. For instance, in renal pathology, the concept of a histological lesion is vague. These are considered to be discrete, specific, and defined as elementary changes in a histological structure (e.g., mesangial hypercellularity), but can also be a combination of distinct lesions, also referred as a lesion pattern (e.g., membranous, membranoproliferative). The denominations used are sometimes misleading, due to associations with nosological entities [50]. In some cases, due to the origin and progression of a structural change in a tissue, it will not always be possible to stratify a complex pattern of structural changes into different elementary lesions. Some elementary lesions may become evident only through the use of histochemical techniques that highlight specific structures or chemical components [50]. Although this strategy is helpful for the pathologist to perform a diagnosis, it also expands numerous possible representations of a given lesion. Additionally, the histochemical staining technique used to highlight specific structures varies greatly among laboratories. Although H&E is the most widely used stain, it is also used together with various other stain combinations. Thus, efforts to establish a comprehensive classification of all lesions within a given organ may prove unsuccessful.
-
Lack of agreement in lesion definition. In their daily routine, pathologists must decide whether a given histological structure is normal or if it presents a lesion. Most of the time these decisions are easily made, and most pathologists would share the same opinion. Difficulty emerges when faced with early stage or not fully-developed lesions. Decision-making can be further complicated by the existence of lesions that, at early stages, bear some resemblance to artifacts that appear during histological preparation (sectioning or staining for instance). In fact, these issues are a frequent topic of debate among specialists, with consensus generally achieved through the exclusion of borderline lesions. For instance, the definition of glomerular mesangial hypercellularity varies between the MEST-C classification of IgA nephropathy and the lupus nephritis classification revised by Bajema et al. [27] and Markowitz [51]. Furthermore, the definition of mesangial hypercellularity used in MEST-C classification excludes nonclustered adjoining mesangial cell nuclei [52].
-
Low frequency of lesions. Reports on the frequency of biopsy-confirmed glomerular diseases attribute more than 50% of cases to focal and segmental glomerulosclerosis, membranous glomerulopathy IgA nephropathy or lupus nephritis, while amyloidosis and Alport syndrome are less frequent (about 1% each) and fibrillary glomerulopathy and fabry glomerulopathy are rarely observed [29, 53]. Consequently, the elementary lesions associated with these diseases are represented differently in most histological image libraries. Since the current supervised approaches used in computer vision require large amounts of images, it may prove difficult to find suitable sets of images for analysis.
-
The emergence of new histological lesions. One of the most interesting observations in pathology meetings is shifting relevance attributed to structural changes in tissues. These fluctuations profoundly impact disease classification and patient treatment. Great effort is expended in the identification and validation of the relevance attributed to a given lesion. For instance, tubulitis attained high relevance in the context of kidney transplant rejection [28].
-
Tissue processing. Although a trend exists toward the use of phosphate-buffered formalin as a preserving medium for renal biopsies, the choice of fixative varies among pathology laboratories. In practice, formalin acetic alcohol and Bouin’s fluid are used by many laboratories. These fixatives preserve molecular residues differently, affecting tissue morphology and staining properties. At least four different staining techniques are used for highlighting different structures. Although most pathology laboratories commonly use H&E, PAS, and PAMS, staining and counter-staining techniques vary widely among pathology laboratories [54].
-
Image capture and processing. Although much emphasis has been given to whole slide scanners, systems based on portable image capture devices, such as smartphones, could facilitate rapid consultations among pathologists. This practice requires an adequate normalization step prior to conducting image analysis.
-
The obtainment of a diversified, annotated data set of images, validated by pathologists, presents a major challenge for computational pathology. This is largely due to a shortage of professionals with high levels of expertise in specific areas or diagnostic pathology.
-
One of the expected uses of a system capable of identifying histological lesions is to perform clinical-pathological correlations on large scale. This approach may require flexibility in order to quickly learn new lesions as well as the capability to combine images acquired by different sources and methods (immunofluorescence, electron microscopy) with meta-data obtained clinically.
Based on the challenges cast before, we can dig in some future prospects to the field of computational nephropathology until pathologists are able to effectively use CAD-based systems in their daily laboratory routines. They are:
-
Classification systems must provide true reliability when examine WSIs in the wild. For that, methods should provide a consistent uncertainty score.
-
Supervised methods must provide appropriated generalization either to accomplish any kind of classification or even to segment renal structures. Due to limited availability of data for all types of renal lesions, it is hard to guarantee broad generalization in the process of training supervised methods. An interesting topic of research that is gaining enough attention lately is the self-learning methods, which should be capable to recognize patterns even if it was not trained for this purpose.
-
AI/ML-based methods should work on stain-free biopsy images or learn how to generalize from one stain over the others. Each stain highlight characteristics of the image edges, which usually skew the generalization performance of an ML technique. Working on stain-free biopsy images, it is possible to subsequently stain the histology image according to generalization purposes; another way could be to conceive powerful methods that could generalize from a unique stain to rule images stained by other methods.
-
Integrate clinical data with histology images. One way to provide more information to ML-based systems is across specialized text, which would create more powerful workflows in computational nephropathology. Broadly speaking, textual data would bring large possibilities to build systems to recognize timely patterns, rather than spatial patterns as it is commonly done by DL methods, nowadays.
-
The computational nephropathology would benefit considerably if there was an integration of data from clinical sources and images, but particularly with data from genomics and proteomics. These two types of information would favor, for instance, the identification of new histological markers of disease previously unrecognized by pathologists.
To reach these future milestones, there is an avenue to improve current research in computational nephropathology, considering since data set gathering to how researchers measure the performance and generalizability of the proposed methods.
16.6 Concluding Remarks
Computational pathology is a fast-developing area with a wide scope and research agenda full of opportunities and challenges that must be overcome to allow its full adoption by pathologists in their daily practice. The new techniques and tools yielded by computational pathology may improve the way that the pathologists perform their tasks, allowing them to dedicate more time to the integration and analysis of clinical, morphological, and molecular information collected from tissue specimens. Even though much has been done in the field, the proposed solutions in the scientific literature are far from the point where they can be applied in preclinical trials. Weaknesses in terms of analytical and clinical validation still need to be bridged so that such solutions are robust and reliable enough to be deployed to be used by the pathologists in their daily routine.
Notes
- 1.
- 2.
The Brazilian Health Ministry Research Agency. https://portal.fiocruz.br/en/
- 3.
References
Tolles WE. Section of biology: the cytoanalyzer—an example of physics in medical research. Trans N Y Acad Sci. 1955;17(3 Series II):250–6.
Abel J, Ouillette P, Williams C, Blau J, Cheng J, Yao K, Lee W, Cornish T, Balis U, McClintock D. Display characteristics and their impact on digital pathology: a current review of pathologists future microscope. J Pathol Inform. 2020;11:23. https://doi.org/10.4103/jpi.jpi_38_20.
Jader G, Fontineli J, Ruiz M, Abdalla K, Pithon M, Oliveira L. Deep instance segmentation of teeth in panoramic X-ray images. In: 2018 31st SIBGRAPI conference on graphics, patterns and images (SIBGRAPI). Piscataway, NJ: IEEE; 2018. p. 400–7.
Silva G, Oliveira L, Pithon M. Automatic segmenting teeth in X-ray images: trends, a novel data set, benchmarking and future perspectives. Expert Syst Appl. 2018;107:15–31.
Silva B, Pinheiro L, Oliveira L, Pithon M. A study on tooth segmentation and numbering using end-to-end deep neural networks. In: 2020 33rd SIBGRAPI conference on graphics, patterns and images (SIBGRAPI). Piscataway, NJ: IEEE; 2020. p. 164–71.
Abhishek D, Aarti S, Sachi G, Sudip D. Use of artificial intelligence in dermatology. Indian J Dermatol. 2020;65:352–7.
Abraham A, Sobhanakumari K, Mohan A. Artificial intelligence in dermatology. J Skin Sex Transm Dis. 2021;3(1):99–102.
Young AT, Xiong M, Pfau J, Keiser MJ, Wei ML. Artificial intelligence in dermatology: a primer. J Invest Dermatol. 2020;140(3):1504–12.
Barros GO, Navarro B, Duarte A, dos Santos WLC. PathoSpotter-K: a computational tool for the automatic identification of glomerular lesions in histological images of kidneys. Sci Rep. 2017;7:1–8. https://doi.org/10.1038/srep46769.
Ginley B, Tomaszewski JE, Yacoub R, Chen F, Sarder P. Unsupervised labeling of glomerular boundaries using Gabor filters and statistical testing in renal histology. J Med Imaging. 2017;4(2):021102. https://doi.org/10.1117/1.jmi.4.2.021102.
Chagas P, Souza L, Araújo I, Aldeman N, Duarte A, Angelo M, dos Santos WLC, Oliveira L. Classification of glomerular hypercellularity using convolutional features and support vector machine. Artif Intell Med. 2020;103:101808. https://doi.org/10.1016/j.artmed.2020.101808.
Chagas P, Souza L, Calumby R, Duarte A, Angelo M, dos Santos WLC, Oliveira L. Deep-learningbased membranous nephropathy classification and Monte-Carlo dropout uncertainty estimation. In: Simpósio Brasileiro de Computação Aplicada à Saúde 𝑆𝐵𝐶𝐴𝑆; 2021. https://doi.org/10.5753/sbcas.2021.
Chagas P, Souza L, Calumby R, Pontes I, Araújo S, Duarte A, Pinheiro N, dos Santos WLC, Oliveira L. Toward unbounded open-set recognition to say “I don’t know” for glomerular multi-lesion classification. In: International symposium on medical information processing and analysis (SIPAIM); 2021.
Rehem JMC, Santos WLC, Duarte AA, Oliveira LR, Angelo MF. Automatic glomerulus detection in renal histological images. In: Proceedings–SPIE medical imaging 11603; 2021. https://doi.org/10.1117/12.2582201.
Tan P-N, Steinbach M, Karpatne A, Kumar V. Introduction to data mining. London: Pearson; 2019.
Lecun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE. 1998;86:2278–323. https://doi.org/10.1109/5.726791.
Rajaraman A, Ullman JD. Mining of massive datasets. Cambridge: Cambridge University Press; 2011. https://doi.org/10.1017/CBO9781139058452.
Goodfellow I, Bengio Y, Courville A. Deep learning. Cambridge, MA: MIT Press; 2016. http://www.deeplearningbook.org.
Krizhevsky ISA, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM. 2017;60(6):84–90. https://doi.org/10.1145/3065386.
Russakovsky O, Deng J, Hao S, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, Fei-Fei L. ImageNet large scale visual recognition challenge. Int J Comput Vis. 2015;115:211–52. https://doi.org/10.1007/s11263-015-0816-y.
Pratt LY, Pratt LY, Hanson SJ, Giles CL, Cowan JD. Discriminability-based transfer between neural networks. In: Advances in neural information processing systems; 1993. p. 204–11.
Prewitt JMS, Mendelsohn ML. The analysis of cell images. Ann N Y Acad Sci. 1966;128(3):1035–53. https://doi.org/10.1111/j.1749-6632.1965.tb11715.x.
Hekler A, Utikal JS, Enk AH, Solass W, Schmitt M, Klode J, Schadendorf D, Wiebke S, Franklin C, Bestvater F, Flaig MJ, Krahl D, von Kalle C, Fröhling S, Brinker TJ. Deep learning outperformed 11 pathologists in the classification of histopathological melanoma images. Eur J Cancer. 2019;118:91–6. https://doi.org/10.1016/j.ejca.2019.06.012.
Arvaniti E, Fricker KS, Moret M, Rupp N, Hermanns T, Fankhauser C, Wey N, Wild PJ, Rüschoff JH, Claassen M. Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci Rep. 2018;8(12054):1–11. https://doi.org/10.1038/s41598-018-30535-1.
Fogo AB, Lusco MA, Najafian B, Alpers CE. AJKD atlas of renal pathology: minimal change disease. Am J Kidney Dis. 2015;66(2):376–7. https://doi.org/10.1053/j.ajkd.2015.04.006.
Fogo AB, Lusco MA, Najafian B, Alpers CE. AJKD atlas of renal pathology: focal segmental glomerulosclerosis. Am J Kidney Dis. 2015;66(2):e1–2. https://doi.org/10.1053/j.ajkd.2015.04.007.
Bajema IM, Wilhelmus S, Alpers CE, Bruijn JA, Colvin RB, Terencecook H, D’Agati VD, Ferrario F, Haas M, Jennette JC, Joh K, Nast CC, Noël LH, Rijnink EC, Roberts ISD, Seshan SV, Sethi S, Fogo AB. Revision of the International Society of Nephrology/Renal Pathology Society classification for lupus nephritis: clarification of definitions, and modified National Institutes of Health activity and chronicity indices. Kidney Int. 2018;93(4):789–96. https://doi.org/10.1016/j.kint.2017.11.023.
Bhowmik DM, Dinda AK, Mahanta P, Agarwal SK. The evolution of the Banff classification schema for diagnosing renal allograft rejection and its implications for clinicians. Indian J Nephrol. 2010;20(1):2–8. https://doi.org/10.4103/0971-4065.6208.
dos Santos WLC, Sweet GMM, Azevêdo LG, Tavares MB, Soares MFS, de Melo CVB, Carneiro MFM, de Souza Santos RF, Conrado MC, Braga DTL, Bessa MC, de Freitas Pinheiro Junior N, Bahiense-Oliveira M. Current distribution pattern of biopsy-proven glomerular disease in Salvador, Brazil, 40 years after an initial assessment. J Bras Nefrol. 2017;39(4):376–83. https://doi.org/10.5935/0101-2800.20170069.
Zhou XS, Huang TS. CBIR: from low-level features to high-level semantics. In: Image and video communications and processing, vol. 3974. Bellingham: International Society for Optics and Photonics; 2000. p. 426–31.
Mikolajczyk K, Schmid C. Scale & affine invariant interest point detectors. Int J Comput Vis. 2004;60(1):63–86.
Bay H, Ess A, Tuytelaars T, Van Gool L. Speededup robust features (SURF). Comput Vis Image Underst. 2008;110(3):346–59.
Dalal N, Triggs B. Histograms of oriented gradients for human detection. In: 2005 IEEE computer society conference on computer vision and pattern recognition (CVPR’05), vol. 1; 2005. p. 886–93.
Rinaldi AM, Russo C. A content based image retrieval approach based on multiple multimedia features descriptors in E-health environment. In: 2020 IEEE international symposium on medical measurements and applications (MeMeA); 2020. p. 1–6.
LeCun Y, Kavukcuoglu K, Farabet C. Convolutional networks and applications in vision. In: Proceedings of 2010 IEEE International symposium on circuits and systems. Piscataway, NJ: IEEE; 2010. p. 253–6.
Shah A, Naseem R, Iqbal S, Shah MA, et al. Improving cbir accuracy using convolutional neural network for feature extraction. In: 2017 13th international conference on emerging technologies (ICET). Piscataway, NJ: IEEE; 2017. p. 1–5.
Azizpour H, Razavian AS, Sullivan J, Maki A, Carlsson S. From generic to specific deep representations for visual recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition workshops; 2015. p. 36–45.
Kruthika KR, Maheshappa HD, Initiative A’s DN, et al. CBIR system using capsule networks and 3D CNN for Alzheimer’s disease diagnosis. Inform Med Unlock. 2019;14:59–68.
Sezavar A, Farsi H, Mohamadzadeh S. Content-based image retrieval by combining convolutional neural networks and sparse representation. Multimed Tools Appl. 2019;78(15):20895–912.
Ozen Y, Aksoy S, Kösemehmetoğlu K, Önder S, Üner A. Self-supervised learning with graph neural networks for region of interest retrieval in histopathology. In: 2020 25th International conference on pattern recognition (ICPR); 2021. p. 6329–34. https://doi.org/10.1109/ICPR48806.2021.9412903.
Zheng Y, Jiang Z, Xie F, Shi J, Zhang H, Huai J, Cao M, Yang X. Diagnostic regions attention network (DRA-net) for histopathology WSI recommendation and retrieval. IEEE Trans Med Imaging. 2021;40(3):1090–103.
Hein M, Andriushchenko M, Bitterwolf J. Why ReLU networks yield high-confidence predictions far away from the training data and how to mitigate the problem. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition; 2019. p. 41–50.
Kendall A, Gal Y. What uncertainties do we need in Bayesian deep learning for computer vision? In: Proceedings of the 31st international conference on neural information processing systems. NIPS’17. Long Beach, CA: Curran Associates; 2017. p. 5580–90.
Begoli E, Bhattacharya T, Kusnezov D. The need for uncertainty quantification in machine-assisted medical decision making. Nat Mach Intell. 2019;1(1):20–3.
Leibig C, Allken V, Ayhan MS, Berens P, Wahl S. Leveraging uncertainty information from deep neural networks for disease detection. Sci Rep. 2017;7(1):1–14.
Combalia M, Hueto F, Puig S, Malvehy J, Vilaplana V. Uncertainty estimation in deep neural networks for dermoscopic image classification. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition workshops; 2020. p. 744–5.
Cicalese PA, Mobiny A, Shahmoradi Z, Yi X, Mohan C, Van Nguyen H. Kidney level lupus nephritis classification using uncertainty guided Bayesian convolutional neural net-works. IEEE J Biomed Health Inform. 2020;25(2):315–24.
Roady R, Hayes TL, Kemker R, Gonzales A, Kanan C. Are open set classification methods effective on large-scale datasets? PLoS One. 2020;15(9):e0238302.
Ginley B, Lutnick B, Jen KY, Fogo A, Jain S, Rosenberg A, Walavalkar V, Wilding G, Tomaszewski JE, Yacoub R, Rossi GM, Sarder P. Computational segmentation and classification of diabetic glomerulosclerosis. J Am Soc Nephrol. 2019;30(10):1953–67.
D’Agati VD, Fogo AB, Bruijn JA, Charles Jennette J. Pathologic classification of focal segmental glomerulosclerosis: a working proposal. Am J Kidney Dis. 2004;43(2):368–82. https://doi.org/10.1053/j.ajkd.2003.10.024.
Markowitz G. Updated Oxford classification of IgA nephropathy: a new MEST-C score. Nat Rev Nephrol. 2017;13(7):385–6.
Roberts ISD, Cook HT, Troyanov S, Alpers CE, Amore A, Barratt J, Berthoux F, Bonsib S, Bruijn JA, Cattran DC, Coppo R, D’Agati V, D’Amico G, Emancipator S, Emma F, Feehally J, Ferrario F, Fervenza SFFC, Geddes Agnes Fogo CC, Groene H-J, Andrew M, Haas HM, Hill PA, Hogg RJ, Hsu SI, Jennette JC, Joh K, Julian BA, Kawamura T, Lai FM, Li L-S, Li PKT, Liu Z-H, Mackinnon B, Mezzano S, Schena FP, Tomino Y, Walker HWPD, Weening JJ, Yoshikawa N, Zhang H. The Oxford classification of IgA nephropathy: pathology definitions, correlations, and reproducibility. Kidney Int. 2009;76(5):546–56. https://doi.org/10.1038/ki.2009.168.
Polito MG, De Moura LAR, Kirsztajn GM. An overview on frequency of renal biopsy diagnosis in Brazil: clinical and pathological patterns based on 9617 native kidney biopsies. Nephrol Dial Transplant. 2010;25:490–6. https://doi.org/10.1093/ndt/gfp355.
Alturkistani HA, Tashkandi FM, Mohammedsaleh ZM. Histological stains: a literature review and case study. In: Global journal of health science. Richmond Hill, ON: Canadian Center of Science and Education; 2015. p. 72–9. https://doi.org/10.5539/gjhs.v8n3p72.
Acknowledgments
The PathoSpotter project is partially sponsored by the Fundação de Amparo à Pesquisa do Estado da Bahia (FAPESB), grants TO-P0008/15 and TO-SUS0031/2018, and by the Inova FIOCRUZ grant. Washington dos Santos and Luciano Oliveira are research fellows of Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), grants 306779/2017 and 307550/2018-4, respectively.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Oliveira, L. et al. (2022). PathoSpotter: Computational Intelligence Applied to Nephropathology. In: Bezerra da Silva Junior, G., Nangaku, M. (eds) Innovations in Nephrology. Springer, Cham. https://doi.org/10.1007/978-3-031-11570-7_16
Download citation
DOI: https://doi.org/10.1007/978-3-031-11570-7_16
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-11569-1
Online ISBN: 978-3-031-11570-7
eBook Packages: MedicineMedicine (R0)