Abstract
Brain tumor segmentation by computer computing is still an exciting challenge. UNet architecture has been widely used for medical image segmentation with several modifications. Attention blocks have been used to modify skip connections on the UNet architecture and result in improved performance. In this study, we propose the development of UNet for brain tumor image segmentation by modifying its contraction and expansion block by adding Attention, adding multiple atrous convolutions, and adding a residual pathway that we call Multiple Atrous convolutions Attention Block (MAAB). The expansion part is also added with the formation of pyramid features taken from each level to produce the final segmentation output. The architecture is trained using patches and batch 2 to save GPU memory usage. Online validation of the segmentation results from the BraTS 2021 validation dataset resulted in dice performance of 78.02, 80.73, and 89.07 for ET, TC, and WT. These results indicate that the proposed architecture is promising for further development.
Supported by Ministry of Education and Culture, Indonesia.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Segmentation of brain tumors using computer computing is still an exciting challenge. Several events have been held to get the latest methods with the best segmentation performance. One event that continues to invite researchers to innovate related to the segmentation method is the Brain Tumor Segmentation Challenge (BraTS Challenge). This BraTS Challenge has been held every year, starting in 2012 until now in 2021 [4].
The BraTS 2021 challenge is held by providing a larger dataset than the previous year. Until now, the dataset provided consists of training data accompanied by a label with a total of 1251 data and validation data that is not accompanied by a label with a total of 219 data. This validation data can be checked for correctness of labeling using the online validation tool provided on the https://www.synapse.org site [5,6,7, 12].
Among the many current architectures, UNet has become the widely used architecture as a medical image segmentation model. Starting with use in segmenting neuronal structures in the EM Stack by [14], this architecture has been developed for segmenting 3D medical images. The development of UNet includes modifying existing blocks at each level, both in the expansion and decoder parts, modifying skip connections, and adding links in the decoder section by adding some links to form pyramid features.
One of the developments of the UNet architecture is to modify the skip connection part. Modifications are made by adding an attention gate which is intended to be able to focus on the target segmentation object. This attention-gate model is taught to minimize the influence of the less relevant parts of the input image while still focusing on the essential features for the segmentation target [15].
Other UNet architecture developments are block modification as done in [1] by creating two paths in one block. One path uses convolution with kernel size 5\(\,\times \,\)5 followed by normalization and relu. The other path uses convolution with a kernel size of 3\(\,\times \,\)3 followed by residual blocks. Merging the output of each path is done by concatenating the output features of each path. On the other hand, some modify the block from UNet by using atrous convolution to get a wider reception area [17].
The merging of feature maps which are the outputs of each level in the UNet decoder section, to form a feature pyramid is also carried out to improve segmentation performance as was done in [13]. The formation of this pyramid feature was inspired by the [10] research which was used to carry out the object detection process. This pyramid feature is also used in several studies to segment brain tumors [18, 21, 22].
In this study, a modification of the UNet architecture was proposed for processing brain tumor segmentation from 3D MRI images. The modifications include modifying each block with multiple atrous convolutions, adding an attention gate accompanied by a residual path to keep accelerating the convergence of the model. The skip connection portion of UNet was modified by adding an attention gate connected to the output of the lower expansion block. Moreover, the last modification is using pyramid features by combining the feature outputs from each level in the expansion section, which is connected to a convolution block to produce segmented outputs. The segmentation performance obtained is promising.
2 Methods
2.1 Dataset
The datasets used in this study are the BraTS 2021 Training dataset and the BraTS 2021 validation dataset. Each dataset was obtained with different clinical protocols and from different MRI scanners from multiple providing institutions. The BraTS 2021 Training dataset contains 1251 patient data with four modalities, T1, T1Gd, T2, and T2-Flair, accompanied by one associated segmentation label. There are four types of segmentation labels with a value of 1 indicating Necrosis/non-enhancing tumor, 2 representing edema, a value of 4 indicating tumor enhancing, and 0 for non-tumor and background. The labels provided are annotated by one to four annotation officers and are checked and approved by expert neuro-radiologists.
The BraTS 2021 Validation dataset, on the other hand, is a dataset that does not come with a label. The segmentation results must be validated online by submitting it to the provided online validation siteFootnote 1 to obtain the correctness of labeling. This BraTS 2021 validation dataset contains 219 patient data with the same four modalities as the BraTS 2021 Training dataset.
2.2 Preprocessing
The 3D images of the BraTS 2021 training dataset and the BraTS 2021 validation dataset were obtained from a number of different scanners and multiple contributing institutions. The value of the voxel intensity interval of each 3D image produced will be different. So these values need to be normalized so that they are in the same interval. Each of these 3D images was normalized using the Eq. 1 similar to that done in [2].
where \(I_{norm}\) and \(I_{orig}\) are the normalized image and the original image, while \(\mu \) and \(\sigma \) are the average value and standard deviation of all non-zero voxels in the 3D image. The normalization process was carried out for each patient data and each modality-both for the BraTS 2021 training dataset during training and the BraTS 2021 validation dataset during inference.
2.3 Proposed Architecture
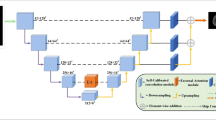
The architecture proposed in this study is developing the UNet architecture with a 3D Image processing approach. The proposed architecture used is shown in Fig. 1.
All modalities are used in this study, followed by a dropout layer as regularization-the use of dropout as one of the regularization models as proposed by [16]. The use of dropout as regularization is also used in several studies with a rate that varies between 0.1 to 0.5 [3, 8, 9, 11, 19, 20]. In this paper, the dropout rate value used is 0.2 with the placement at the beginning of the layer.
The next layer is the Multi Atrous Attention Block (MAAB). There are several levels in this block, starting with levels 1, 2, 3 and 4. Details of the internal visualization within the block are shown in Fig. 2.
This MAAB block processes feature maps equipped with atrous convolutions with different dilatation factors according to their level. The atrous convolution function expands the receptive field area of the feature map without increasing the number of parameters that must be studied. The deeper the downsampling level, the greater the level of the MAAB block to increase the receptive field area that can be covered and increase architectural performance in studying feature maps.
In the first level, the MAAB block contains one convolution layer with a pre-activation strategy. For the second level, in addition to containing the first level layer, one atrous convolution layer is also added with a factor of 2. The following blocks contain the previous blocks with an increasing convolution atrous layer-the order of the dilatation factors in the convolution layers 1, 2, 4, and 8. The residual path is connected from the convolution results at the beginning of the block with the combined output of the levels used in this MAAB block by using the feature addition function. At the end of the block, an attention sub-block is added to keep the focus on relevant features.
The skip connection is modified by adding an attention block before being connected to the expansion section feature. This attention block is used to keep the model focused on relevant features such as the initiative in [15]. The attention diagram used in this study is shown in the Fig. 3. G in the figure is a feature that comes from the expansion level before being upsampled, while X is a feature of the skip connection of the contraction section. The output of this attention block is combined with the upsampling feature at an equivalent level for subsequent processing.
In the expanding section, the feature maps at each level are concatenated together before being inserted into the last MAAB level 1 block. The feature map at the lowest level is upsampled by a factor of four, while the second level is upsampled by a factor of two to equal the size of the feature map at level one. This connection forms a feature map of the pyramid and the supervision of each lower level. The output of the last MAAB block is convoluted into three channels representing the segmentation target (ET, WT, and TC).
2.4 Loss Function
The loss function used during the training process is diceloss with the formula expressed in the Eq. 2. The objects detected in the image consist of 3 types, namely Enhanced Tumor, Tumor Core, a combination of Enhanced Tumor and Necrotic objects, and Whole Tumor, which is a combination of all tumor objects. So that the loss function used uses the combination of the three areas with the weighting as stated in the Eq. 3.
where P represents the predicted result, Y represents the segmentation target, \(\epsilon \) is filled with a small value to avoid dividing by zero. Furthermore, ET, TC, and WT represent Enhanced Tumor, Tumor Core, and Whole Tumor areas.
2.5 Experiment Settings
The hardware used in this study includes an Nvidia RTX 2080i 11GB, 64GB RAM, and a Core I7 processor. While the Deep Learning framework software used is Tensorflow/Keras version 2.5.
The training was carried out using the BraTS 2021 training dataset, which contained 1251 patient data with four modalities (T1, T1Gd, T2, T2-Flair) and one ground-truth file for each patient. The data is split into two parts, with 80% as training data and 20% as local validation data. To minimize variation in training, a 5-fold cross-validation strategy is used.
The model was trained using Adam’s optimizer with a learning rate of 1e-4 for 300 epochs for each fold. Data augmentation techniques used include random crop, three-axis random permutation, random replace channel with gaussian distribution, and random mirroring of each axis.
Data is trained with patches of size \(72\times 72\times 72\) and batch size of 2 to minimize GPU memory requirements. The 3d image patches were taken from the area containing the tumor at random. During the inference process, the data is processed at size \(72\times 72\times 72\) but with a shift of 64 voxels to each axis. Voxels from the overlapping segmentation results are averaged to get the final segmentation result.
3 Results
The time required for training and inference model using the five-fold strategy as shown in the Table 1. From the Table 1 it can be seen that the average time required for a 5-fold training with 300 epochs is 104408 s. Alternatively, per-epoch, it takes 348,027 s. This time is needed for training 1001 data and local validation for 250 data. The average inference time required is 1530 s seconds as shown in Table 1. This time is used to segment the data as much as 219 data. So that processing for each data takes an average of 6.99 s. Meanwhile, if using a combination of 5 models, it will take 10054 s so that the processing of an ensemble of 5 models for each data takes an average of 45.91 s.
Loss obtained during training for each fold as shown in Fig. 4. From the figure, the most stable is the 3rd fold and the 5th fold with no spikes in value in the graph. While in others, there is a spike in value at certain times. As in the 1st fold, there was a spike value at the epoch between 50–100 for both training and validation loss. Likewise, in the 2nd fold and fourth fold. This condition is possible because this training uses random patches. When taking a random patch, there may not be an object, but the model detects an object so that the loss value will approach the value of 1.
From Fig. 4(f), it can be seen that the overall training of this model is convergent. The spikes in value do not exceed the initial loss value. At the end of the epoch, the loss values for training and validation also converge. In all graphs (a-e), the existing convergence pattern is close to the convergent value. The validation loss value is also not much different from the training loss value, so it can be said that the model is not overfitting.
The results of the dice score performance during training are congruent with the loss value. Assuming that the loss function used is \(1-dice\). However, because there are three objects counted in the dice, the loss value is an amalgamation of the dice scores of each object with a weight determined in the Eq. 3. The average dice value of each object during training for all folds as shown in Fig. 5. The validation scores for ET and TC objects have a good pattern, with values increasingly outperforming the training score near the end of the epoch. In comparison, the validation score for the WT object is always below the training score of the WT. However, the score pattern of each object increases until the end of the epoch.
Online validation of segmentation results using the 1st to fifth fold model is displayed in Table 2. Five models of training results ensembled using the average method can also be seen in the table.
This architecture is also tested with the BraTS 2021 testing dataset for the challenge. The ground truth for this dataset is not provided. We only send the codes that form the architecture and the mechanism for segmenting one patient data individually along with the weight file of the model in a docker format. We use five models that are ensembled into one with the same averaging method as the ensemble model used in the Table 2. The performance results of the 5 model ensemble applied to the BraTS 2021 testing dataset are outstanding, as shown in the Table 3.
4 Discussion
In this study, we propose a modified Unet3D architecture for brain tumor segmentation. Modifications include modification of each block with atrous convolution, attention gate, and the addition of residual path. The skip connection section is modified by adding an attention gate that combines the features of the contraction section with the expansion section one level below its equivalent level. The pyramid feature is also added to get better segmentation performance results. Checking using the combination of 5 models on the validation dataset resulted in segmentation performance of 78.02, 80.73, and 89.07 for ET, TC, and WT objects.
In Fig. 4 especially in parts (a), (b), and (d) there is a spike in loss value in certain epochs. The alleged cause of this incident is that random patch picking will result in a volume that has no object, either ET, TC, or WT, but the model still gets its predictions, causing the loss value to spike suddenly. However, the exact cause needs further investigation.
Notes
References
Aghalari, M., Aghagolzadeh, A., Ezoji, M.: Brain tumor image segmentation via asymmetric/symmetric UNet based on two-pathway-residual blocks. Biomed. Signal Process. Control 69, 102841 (2021). https://doi.org/10.1016/j.bspc.2021.102841
Akbar, A.S., Fatichah, C., Suciati, N.: Simple myunet3d for brats segmentation. In: 2020 4th International Conference on Informatics and Computational Sciences (ICICoS), pp. 1–6 (2020). https://doi.org/10.1109/ICICoS51170.2020.9299072
Akbar, A.S., Fatichah, C., Suciati, N.: Modified mobilenet for patient survival prediction. In: Crimi, A., Bakas, S. (eds.) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, pp. 374–387. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-72087-2_33
Baid, U., Ghodasara, S., Bilello, M., et al.: The RSNA-ASNR-MICCAI BraTS 2021 Benchmark on Brain Tumor Segmentation and Radiogenomic Classification, July 2021. http://arxiv.org/abs/2107.02314
Bakas, S., Akbari, H., Sotiras, A., et al.: Segmentation labels for the pre-operative scans of the tcga-gbm collection (2017). https://doi.org/10.7937/K9/TCIA.2017.KLXWJJ1Q, https://wiki.cancerimagingarchive.net/x/KoZyAQ
Bakas, S., Akbari, H., Sotiras, A., et al.: Segmentation labels for the pre-operative scans of the tcga-lgg collection (2017). https://doi.org/10.7937/K9/TCIA.2017.GJQ7R0EF, https://wiki.cancerimagingarchive.net/x/LIZyAQ
Bakas, S., Akbari, H., Sotiras, A., et al.: Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Scientific Data 4(1), September 2017. https://doi.org/10.1038/sdata.2017.117
Chang, J., Zhang, L., Gu, N., et al.: A mix-pooling CNN architecture with FCRF for brain tumor segmentation. J. Visual Commun. Image Representation 58, 316–322 (2019). https://doi.org/10.1016/j.jvcir.2018.11.047
Kabir Anaraki, A., Ayati, M., Kazemi, F.: Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms. Biocybern. Biomed. Eng. 39(1), 63–74 (2019). https://doi.org/10.1016/j.bbe.2018.10.004
Lin, T.Y., Dollár, P., Girshick, R., et al.: Feature pyramid networks for object detection. In: Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017. vol. 2017-Janua, pp. 936–944. IEEE, July 2017. https://doi.org/10.1109/CVPR.2017.106. http://ieeexplore.ieee.org/document/8099589/
Liu, L., Wu, F.X., Wang, J.: Efficient multi-kernel DCNN with pixel dropout for stroke MRI segmentation. Neurocomputing 350, 117–127 (2019). https://doi.org/10.1016/j.neucom.2019.03.049
Menze, B.H., Jakab, A., Bauer, S., et al.: The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2015). https://doi.org/10.1109/tmi.2014.2377694
Moradi, S., Oghli, M.G., Alizadehasl, A., et al.: MFP-Unet: a novel deep learning based approach for left ventricle segmentation in echocardiography. Phys. Medica 67, 58–69 (2019). https://doi.org/10.1016/J.EJMP.2019.10.001. https://www.sciencedirect.com/science/article/pii/S1120179719304508
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Schlemper, J., Oktay, O., Schaap, M., et al.: Attention gated networks: Learning to leverage salient regions in medical images. Med. Image Anal. 53, 197–207 (2019). https://doi.org/10.1016/j.media.2019.01.012https://www.sciencedirect.com/science/article/pii/S1361841518306133
Srivastava, N., Hinton, G., Krizhevsky, A., et al.: Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15(1), 1929–1958 (2014)
S. V. and I. G.: Encoder enhanced atrous (EEA) unet architecture for retinal blood vessel segmentation. Cogn. Syst. Res. 67, 84–95 (2021). https://doi.org/10.1016/j.cogsys.2021.01.003
Wang, J., Gao, J., Ren, J., et al.: DFP-ResUNet: convolutional neural network with a dilated convolutional feature pyramid for multimodal brain tumor segmentation. Comput. Methods Programs Biomed., 106208, May 2021. https://doi.org/10.1016/j.cmpb.2021.106208.https://linkinghub.elsevier.com/retrieve/pii/S0169260721002820
Xie, H., Yang, D., Sun, N., et al.: Automated pulmonary nodule detection in CT images using deep convolutional neural networks. Pattern Recogn. 85, 109–119 (2019). https://doi.org/10.1016/j.patcog.2018.07.031
Yang, T., Song, J., Li, L.: A deep learning model integrating SK-TPCNN and random forests for brain tumor segmentation in MRI. Biocybern. Biomed. Eng. 39(3), 613–623 (2019). https://doi.org/10.1016/J.BBE.2019.06.003. https://www.sciencedirect.com/science/article/pii/S0208521618303292
Zhou, Z., He, Z., Jia, Y.: AFPNet: a 3D fully convolutional neural network with atrous-convolution feature pyramid for brain tumor segmentation via MRI images. Neurocomputing 402, 235–244 (2020). https://doi.org/10.1016/j.neucom.2020.03.097. https://www.sciencedirect.com/science/article/pii/S0925231220304847
Zhou, Z., He, Z., Shi, M., et al.: 3D dense connectivity network with atrous convolutional feature pyramid for brain tumor segmentation in magnetic resonance imaging of human heads. Comput. Biol. Med. 121, 103766 (2020). https://doi.org/10.1016/j.compbiomed.2020.103766
Acknowledgements
This work was supported by the Ministry of Education and Culture, Indonesia. We are deeply grateful for BPPDN (Beasiswa Pendidikan Pascasarjana Dalam Negeri) and PDD (Penelitian Disertasi Doktor) 2020–2021 Grant, which enabled this research could be done.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Akbar, A.S., Fatichah, C., Suciati, N. (2022). Unet3D with Multiple Atrous Convolutions Attention Block for Brain Tumor Segmentation. In: Crimi, A., Bakas, S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2021. Lecture Notes in Computer Science, vol 12962. Springer, Cham. https://doi.org/10.1007/978-3-031-08999-2_14
Download citation
DOI: https://doi.org/10.1007/978-3-031-08999-2_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-08998-5
Online ISBN: 978-3-031-08999-2
eBook Packages: Computer ScienceComputer Science (R0)