Abstract
Tumor delineation is critical for the precise diagnosis and treatment of glioma patients. Since manual segmentation is time-consuming and tedious, automatic segmentation is desired. With the advent of convolution neural network (CNN), tremendous CNN models have been proposed for medical image segmentation. However, the small size of kernel limits the shape of the receptive view, omitting the global information. To utilize the intrinsic features of brain anatomical structure, we propose a modified U-Net with an attention block (AttU-Net) to tract the complementary information from the whole image. The proposed attention block can be easily added to any segmentation backbones, which improved the Dice score by 5%. We evaluated our approach on the dataset of BraTS 2021 challenge and achieved promising performance on this dataset. The Dice scores of enhancing tumor, tumor core, and whole tumor segmentation are 0.793, 0.819, and 0.879, respectively.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Glioma is the most common tumor of the central nerves system in adults and glioblatoma is the most aggressive one, with nonspecific signs and symptoms. Early diagnosis and promote therapy are main determinants of prognosis. Magnetic resonance images (MRI) is wildly utilized in clinical practice for tumor localization, diagnosis, risk stratification and precise resection.
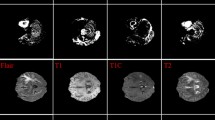
Therefore, tumor delineation is quite important but manual segmentation is rather time-consuming. Automatic and precise tumor segmentation is desired. This year, the Brain Tumor Segmentation (BraTS) challenge is held to encourage the development of brain tumor segmentation [2, 3]. Consistent with clinical practice, four common MRI sequences, a native pre-contrast (T1), a post-contrast T1-weighted (T1Gd), a T2-weighted (T2) and a T2 Fluid Attenuated Inversion Recovery (T2-FLAIR), are provided for segmentation. Figure 1 shows an example of image set. The tumor tissue shown in MRI could be categorized into 3 sub-regions, i.e., the enhancing tumor (ET), the tumor core (TC) and the whole tumor (WT). The TC entails the ET and the necrotic (NCR), while the whole tumor (WT) describes the whole tumor region, consisting TC and the peritumoral edematous/invaded tissue (ED) [4, 5, 15]. There is an intersection between ET, TC and WT, for prediction convenience, the independent regions, NCR, ED and ET, are set to segmentation labels. As shown in the Fig. 1, there is clear inclusion relation between target labels, i.e., \(ED \notin \) ET, NCR.
Recently, tremendous deep learning approaches for brain tumor segmentation have been proposed. U-Net [16] is a prevailing backbone in medical image segmentation, with symmetric encoder and decoder structure. Kayalibay et al. proposed a successful U-Net variant for BraTS 2015, and achieved state of art results that year [12]. Later, Fabian et al. enhanced the U-Net’s skip-connection strategy, which was added after convolution layer in each encode and decode stage, to maintain previous extracted feature, and achieve promising performance in BraTS 2017 [10]. Moreover, Isensee et al. proposed a compositive and automatic segmentation framework with preprocessing, network and post-processing, which could automatically configures itself and suit for the most of tasks [8], and this network achieved top performance in BraTS2020 [9].
However, there are intrinsic drawbacks lying behind CNN. For brain datasets, its symmetric anatomical structure information would dramatically facilitate the pathology segmentation, i.e., doctors will highly concern about the asymmetric structure. Whereas, the size of CNN kernel is rather small, generally \(3 \times 3\), leading to local receptive view and omitting global information while extracting feature [1, 18]. In this way, the introduction of attention mechanism for global relation extraction, could alleviate the aforementioned drawback. Attention mechanism was first proposed for natural language processing task to perceive context [17]. Its promising performance boosted mass related research in the filed of computer vision [6, 14]. Hu et al. first combined attention mechanism with CNN for classification. The attention map weighted the feature map, as the output of the attention block [7]. In [13], a lightweight attention mechanism were proposed, which applied to the feature maps of decoder, for retinal vessel image segmentation.
Inspired by the aforementioned works, we proposed a 3D attention U-Net, namely AttU-Net for brain tumor segmentation. The contribution of our approach can be summarized as follows.
-
1)
We propose a segmentation model which could segment pathology with complementary information from the intrinsic anatomical brain structure. The attention maps are utilized for the enhancement of the determinant feature for segmentation.
-
2)
We modify a light-weight attention block for volume data and combine it with U-Net organically. The proposed attention block could easily adapt for any common backbone.
-
3)
We propose a fully automatic 3D segmentation framework for brain tumor, and validate it with a public dataset from BraTS 2021.
2 Methods
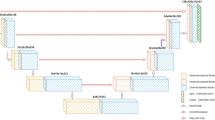
The overall structure of AttU-Net consists five components, i.e., an encoder, a symmetric decoder, attention blocks, multi-scale supervision and skip-connection strategy, shown in Fig. 2. We will formulate the backbone first in the Sect. 2.1 and then elaborate the structure of the attention block in Sect. 2.2.
2.1 Network Architecture
The backbone is modified from the typical U-Net [16]. The encoder is composed of five convolution blocks, connected by the maximum pooling layer. Each of convolution block consists of three \(3 \times 3 \times 3\) convolution layers with stride 2 to reduce feature resolution, and followed by a Leaky-ReLu Activation Layer. The decoding process is symmetric with encoder, also with five block but connected by up-sampling layer. The shape of feature map in each encoder convolution blocks are 16, 32, 64, 128 and 256, while the order in decoder is opposite. The attention blocks (named A) are added to encoding process, to track feature correlation, which will be elaborated later. The last convolution layer in decoder is a \(1 \times 1 \times 1\) convolution, and the number of output channel is 4, representing background, NCR, ET and ED. Moreover, we leverage multi-scale supervision for details segmentation. The output of each stage will go over a \(1 \times 1 \times 1\) convolution layer to predict multi-scale segmentation map, followed by up-sampling. The reshaped predicted map will be added to the final segmentation map (Fig. 3).
2.2 Attention Block
As stated previously, the anatomic structure of brain is intrinsic symmetric. We assume that the region which is heterogeneous compared with the other side has higher probability to be tumor [11]. In order to extract global feature, we adopt a pixel-wise attention block to weight the middle layer feature map. We propose a slice-self attention block, to extract the asymmetric regions, which can indicate areas at risk.
An attention block consists three stages, feature mapping, similarity calculation and skip-connection. Feature mapping stage is to map the three origin feature map to the space K, Q, V for following calculation. K, Q are calculated from feature of decoder (\(F_D\)) and the feature from encoder in the same stage (\(F_E\)), respectively, while V are calculated from the feature after skip-connection (\(F_{D+E}\)). The target feature are first go over a convolution layer with \(1 \times 1 \times 1\) kernel, to reduce dimension. \(F_D\) and \(F_E\) \(\in R^{D\times C\times H\times W}\) while \(F_{D+E}\) \(\in R^{D\times C\times H\times W}\), and the outputs of the convolution layer are all \(\in R^{D/2\times C\times H\times W}\). Since it’s more convenient to compute similarity matrix in two dimension, we flatten the feature map from \(D/2 \times C \times H \times W\) to \(D/2 \times C \times N\), where \(N=H\times W\). Then, we compute attention map with K and Q, using
where A presents attention map \(\in R^{D/2\times N \times N}\). Since similarity matrix is a coefficient matrix whose value should be within [0,1], so we apply softmax on the previous result. As mentioned before, the attention map indicates the pixel-wide similarity, the \(a_{ji}\) represents the similarity between \(k_{j}\) and \(q_{i}\). Since we compute the feature with encoder feature and decoder feature, the attention map highlights model attention, where is determinant to segmentation results. Then we utilize the attention map to weight V, calculated it by function \(f(\cdot ) = V \cdot A\), \(\in R^{D/2 \times C \times N}\). Finally, we reshape the previous result to \(D/2 \times C \times N\), and concatenate which with original feature \(F_{D+E}\), using
where Y represents output of attention block, X stands for \(F_{D+E}\) after first convolutional layer and \(\lambda \) is a hyper parameter. The input of attention map \(F_{D+E}\) \(\in R^{2D\times C\times H\times W}\) while the output \(\in R^{D\times C\times H\times W}\).
The Dice loss is utilized as loss function. Since, it’s a four classes segmentation task, while training, we noticed that NCR (label 1) was the most difficult part. Hence we utilized weighted Dice score, that is,
where Lab0 means value equal to 0, etc. Based on experiments, we set \(\lambda _1=1.2\) while \(others=1\).
3 Experiments
3.1 Dataset
The proposed model was evaluated in the BraTS2021 challenge dataset which contains 1000 training instances and 216 validation instances, each of which consists four sequence, i.e., T1, T2, T1-Gd and FLAIR. The training sets is consisted of labeled image for supervised learning. The depth, width and length of 3D brain MR images are 155, 240, 240, respectively.
3.2 Pre-processing
To reduce the modeling difficulty, we omitted noisy information from original image, by cropping the image into 128\(\,\times \,\)128 \(\,\times \,\) 128 size. Later, data augmentation techniques, including random crop, flipping, rotation, were utilized to improve model generalization ability. Four sequences were concatenated into four channel and the shape of input was 128\(\,\times \,\)128 \(\,\times \,\) 128.
3.3 Implementations
We treated brain MRI as volume data with the depth, width and length were 155, 240, 240, respectively. Four sequences would be concatenated as four dimensions data, [4, 155, 240, 240], as a whole input. The origin images have been cropped into [4, 128, 128, 128], which could roughly contain all the brain tissue. The training process adopted the batch iteration method, with the batch size as 8, 1000 epochs performed. The model was implemented in Pytorch and optimized by the Adam algorithm. The initial learning rate was set to 0.004, and decay with epoch growing, updated by the equation,
The all model was performed on four NVIDIA GTX 3080Ti grahics cards. Training time was about 12 h per model using 4 GPUs.
The segmentation results. Three Prediction samples for randomly were selected slices from middle, top and bottom, respectively. AttU-Net obtained more precise prediction, especially on the edge and tiny regions, than U-Net. The predicited edge of AttU-Net is smoother and suit for origin image, represented by arrows.
3.4 Results
Our approach achieves final Dice score (on the testing dataset) 0.793, 0.879, 0.819 and Hausdorff Distance (HD) 23.1 mm, 7.80 mm and 20.7 mm on label ET, WT and TC respectively, as shown in Table 1. The sensitivity and specificity are also presented in Table 2. In general, the performance of the delineation of WT is much better than the other two labels. The reason for dramatic gap between the performance among labels may lie in the different size among them. Generally, the size of the of ET is dramatically smaller than WT region and with more variant shape, which would be more challenging for automatic segmentation.
In order to compare the performance of different strategies, we also present the quantitative results of ablation study on the validation dataset, shown in the Table 3, since the ground-truth of test dataset is not public.
The proposed network achieved the best value in almost every metrics, except for HD of TC. The performance of attention block can be clearly indicated by the metrics value’s gap between Att-UNet and baseline U-Net. The baseline U-Net shares the same convolution backbone, but without attention block, achieving 0.748, 0.767 and 0.871 Dice scores on label ET, TC and WT, respectively. Attention block strategy improved the segmentation results by nearly 5%. Moreover, the multi-scale segmentation strategy could dramatically benefit the prediction, especially for edge and tiny regions, shown in Fig. 4.
The predicted results are presented in Fig. 4. Our algorithm is capable to detect large tumor regions as well as small details. The predicted edge of Att-Unet is smoother than baseline U-Net. However, there are drawback of segmenting the slices near top. Since the proposed model did not consider spatial information. We tried to flatten the last dimension (C, H, W) to calculate spatial similarity, but the results was under desired and additional computing consumption was introduced. It may be due to the lose of position after flatten.
3.5 Conclusion
In this paper, we designed an AttU-Net for the BraTS2021 challenge. We leveraged attention blocks to extract symmetric shape information. Moveover, we modified segmentation backbone from U-Net, adapting for volume datasets. On the validation set, we achieved Dice score 0.808, 0.818 and 0.913 on label ET, TC and WT, respectively. Due to the time limits, we omitted the number of architectural variants. As mentioned before, there were some outliers prediction on the slices near the top of brain. It may be due to the lacks of spatial information, since it just calculates the similarity within one slice. Moreover, there are intrinsic relation between targets regions, we could constrains the prediction using prior relation in the future.
References
Albawi, S., Mohammed, T.A., Al-Zawi, S.: Understanding of a convolutional neural network. In: 2017 International Conference on Engineering and Technology (ICET), pp. 1–6. IEEE (2017)
Baid, U., et al.: The rsna-asnr-miccai brats 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv preprint arXiv:2107.02314 (2021)
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the tcga-gbm collection. the cancer imaging archive. Nat. Sci. Data 4, 170117 (2017)
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the tcga-lgg collection. The cancer imaging archive 286 (2017)
Bakas, S., Akbari, H., Sotiras, A., Bilello, M., Rozycki, M., Kirby, J.S., Freymann, J.B., Farahani, K., Davatzikos, C.: Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 4(1), 1–13 (2017)
Chen, J., et al.: Transunet: transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306 (2021)
Hu, J., Shen, L., Sun, G.: Squeeze-and-excitation networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 7132–7141 (2018)
Isensee, F., Jaeger, P.F., Kohl, S.A., Petersen, J., Maier-Hein, K.H.: nnu-net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18(2), 203–211 (2021)
Isensee, F., Jäger, P.F., Full, P.M., Vollmuth, P., Maier-Hein, K.H.: nnU-Net for brain tumor segmentation. In: Crimi, A., Bakas, S. (eds.) BrainLes 2020. LNCS, vol. 12659, pp. 118–132. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-72087-2_11
Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., Maier-Hein, K.H.: Brain tumor segmentation and radiomics survival prediction: contribution to the BRATS 2017 challenge. In: Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M. (eds.) BrainLes 2017. LNCS, vol. 10670, pp. 287–297. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-75238-9_25
Kalavathi, P., Senthamilselvi, M., Prasath, V.: Review of computational methods on brain symmetric and asymmetric analysis from neuroimaging techniques. Technologies 5(2), 16 (2017)
Kayalibay, B., Jensen, G., van der Smagt, P.: CNN-based segmentation of medical imaging data. arXiv preprint arXiv:1701.03056 (2017)
Li, X., Jiang, Y., Li, M., Yin, S.: Lightweight attention convolutional neural network for retinal vessel image segmentation. IEEE Trans. Industr. Inf. 17(3), 1958–1967 (2020)
Liang, J., Homayounfar, N., Ma, W.C., Xiong, Y., Hu, R., Urtasun, R.: Polytransform: deep polygon transformer for instance segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 9131–9140 (2020)
Menze, B.H., Jakab, A., Bauer, S., Kalpathy-Cramer, J., Farahani, K., Kirby, J., Burren, Y., Porz, N., Slotboom, J., Wiest, R., et al.: The multimodal brain tumor image segmentation benchmark (brats). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2014)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Vaswani, A., et al.: Attention is all you need. In: Advances in Neural Information Processing Systems, pp. 5998–6008 (2017)
Zheng, H., Fu, J., Mei, T., Luo, J.: Learning multi-attention convolutional neural network for fine-grained image recognition. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 5209–5217 (2017)
Acknowledgement
This work was funded by the National Natural Science Foundation of China (grant no. 61971142, 62111530195 and 62011540404), the development fund for Shanghai talents (no. 2020015) and the Fujian Province Joint Funds for the Innovation of Science and Technology (2019Y9070).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Wang, S., Li, L., Zhuang, X. (2022). AttU-NET: Attention U-Net for Brain Tumor Segmentation. In: Crimi, A., Bakas, S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2021. Lecture Notes in Computer Science, vol 12963. Springer, Cham. https://doi.org/10.1007/978-3-031-09002-8_27
Download citation
DOI: https://doi.org/10.1007/978-3-031-09002-8_27
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-09001-1
Online ISBN: 978-3-031-09002-8
eBook Packages: Computer ScienceComputer Science (R0)