Abstract
The multi-level organization of nature is self-evident: proteins do interact among them to give rise to an organized metabolism while in the same time each protein (a single node of such interaction network) is itself a network of interacting amino-acid residues allowing coordinated motion of the macromolecule and systemic effect as allosteric behaviour.
Similar situations hold for ecology, anatomy, organ physiology. The most diffuse approach to such situation is to give for granted that causally relevant events pertain to the most fundamental level (the molecular one) in the form of regularities (or perturbations in the case of pathological situations), that ‘climb up’ the hierarchy reaching the ultimate layer of macroscopic behaviour.
Such causative model, is not the only one: we observe top-down, bottom-up as well as middle-out perturbation/control trajectories.
The recent complex network studies allow to go further the pure qualitative observation of the existence of both non-linear and non-bottom-up processes and to uncover the deep nature of multi-level organization. Here, taking as paradigm protein science, we will give an account of how the information travelling across a network can create meaning so offering a more realistic frame of causation in complex systems.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Allosteric effect
- Biodynamic interfaces
- Causative models
- Complex networks
- Complexity
- Multi-level organization
- Proteins
1 Introduction
The network formalism is probably the most natural way to represent biological systems. Even if in the last decades the analysis of complex networks became a very widespread paradigm to face problems going from macromolecular structures (Di Paola et al., 2013) to genetic regulation circuits (Demongeot et al., 2003), neuroscience (Sporns, 2018) and ecological systems (Mendonça et al., 2018) this is not a new idea. In 1948 Warren Weaver (1948) one of the fathers of mathematical information theory sketched a very intriguing synthetic tripartite description of science into problems of ‘organized simplicity’, ‘disorganized complexity’ and ‘organized complexity’ with biology located in the last class.
The first class (simplicity) refers to the case of very few elements interacting among them with largely invariant relations, being its paradigm classical mechanics. Class 1 problems allow for an extreme abstraction (e.g. a planet can be thought as a dimensionless ‘material point’). The possibility to take into consideration only very few basic features like mass and distance makes this approach largely object independent (this is the basic reason of the use of examples of the same physical law based upon cars, cannonballs or skiers).
Problems of Disorganized Complexity (class 2) have as paradigm classical thermodynamics and reach a great generalization power by means of a very different style of reasoning with respect to Class 1. In Class 2 problems, the predictive power stems from giving up the analysis of system fundamental aggregation level (e.g. the single molecules) preferring a statistical knowledge corresponding to gross averages (like pressure, volume, temperature are) on a transfinite number of atomic elements. Both the approaches must fulfil very stringent constraints. Class 1 approach asks for few involved elements interacting in a stable way, class 2 style needs a very large number of identical particles with only negligible (or very stable and invariant) interactions among them. Biological systems, only in very few cases do satisfy these constraints, so we step into Weaver’s third class (Organized Complexity). Organized Complexity arises whenever many (even if not so many as in class 2) non-identical elements each other interact by means of links endowed with time-varying correlation strength.
The interaction of ‘non identical elements’ with ‘varying correlation strengths’ corresponds to a network of links (edges, correlations) with variable strength, connecting different nodes that in turn are ‘non identical’ being themselves networks with variable wiring structure.
Weaver (1948) commented that while science was at home (relying on the usual repertoire of laws and boundary conditions deciding for their application) in both Class 1 and Class 2 phenomena, the overwhelming importance of contextual information with respect to lawful invariant behaviour, of Class 3 systems, makes the situation much more uncomfortable. After more than 70 years from Weaver’s paper, we made some steps ahead in Organized Complexity studies and the present work deals with these advancements.
The paper is organized as follows: in the first chapter (biodynamic interfaces) we will discuss the basic principles of the interaction between complex systems, with an emphasis on the need of an intermediate layer shared by the two interacting systems with a partially independent nature with respect to the two interactors. In the second chapter (the middle way) we will introduce the concept of mesoscopic or ‘middle-out’ organization demonstrating why the ‘network representation’ allows for a natural, hypothesis free formalization of the meso-scale. The third chapter will be devoted to the transit of information across a network system and the consequent discrimination from noise of the relevant (signal) perturbations able to ‘climb-up’ or ‘stepping-down’ the multilevel organization using allosteric effect in proteins as model system.
2 Biodynamic Interfaces
There is no interaction without information exchange and there is no information exchange without an efficient communication channel. This ‘channel’ is exactly what we call ‘Interface’. If Mary calls Peter by means of her smartphone, the establishing of a contact strictly depends on the existence of an electromagnetic field endowed with a band of frequencies devoted to cell phone communication. Peter smartphone corresponds to a very specific frequency modulation of the field that is elicited by the digits Mary composes on her phone and sends on the specific band of frequencies, consequently Peter’s smartphone rings and the communication begins. We do not enter into the actual content of communication (that only pertains to Mary and Peter), instead we focus on two crucial points of the process:
-
1.
The existence of a medium (the field) that cannot be considered as a discrete entity with a specific location in both space and time but as a ‘global feature’ covering the space and assuming different values in different locations. The interactors (here the Mary and Peter phones) are causally linked in both directions only because they share the same field. From basic physics we know that a point charge embedded into an electromagnetic field both ‘senses’ (i.e. is influenced by the field) and modifies (i.e. influences) the field. This is exactly what happens in human-environment interaction in which environment influences physiology (e.g. toxic effects, sensory information..) and is in turn influenced by humans. Both human beings and environment are complex systems and, for their interaction, they need a shared interface (Arora et al., 2020).
-
2.
The interface (field) oscillates with a specific frequency, this implies it has both a ‘spatial’ and a ‘temporal’ structure, it is a dynamic interface. The frequency of oscillation is not independent from the spatial features of the interface, more in general, any network system (even a field can be imagined as a grid with some focal points, the ‘cells’ in the case of mobile phones) has characteristic oscillation modes originating from its wiring structure. We will go back on this point when dealing with protein structures ‘resonating’ with specific modes that are the carriers of across levels information.
Both these issues are at work in multi-level organization and, more in general, in biological regulation.
3 The Middle Way
The most common style of explanation follows an IF-THEN style in which what happens at a given level influences (or determines depending on the relative importance of stochasticity embedded in the link). These linear fluxes of implications give for granted the existence of a fundamental ‘explanatory layer’ located at the most microscopic level that, thanks to a sort of domino effect, ends up into a macroscopic consequence.
This view is in sharp contrast with what we know about complex structured systems, where a multi-layer (and bi-directional) causality is at work. One of the most clear falsifications of the obliged ‘bottom-up’ character of biological causation, comes from a 1945 paper (Fankhauser, 1945) by the German (but USA based) embryologist Gerhard Fankhauser. He considered cell size in polyploid triton larvae that have a doubled chromosome number with respect to their diploid counterpart. The polyploid individuals have a doubled cell size with respect to the diploid ones, on the contrary, they have exactly the same dimension of organs and ducts. Arora et al. (2020) with respect to the diploid counterpart. This comes from the fact that the polyploid organism uses half the number of cells, though each cell was itself double in size, to build up its organs. This is crucial for life: the optimization of the calibre of a biological structure (the duct) is fine tuned to fit with the flow of biological fluids (a top-down constraint) and cannot be established by more fundamental levels like its constituent cells or the genome. While this is an intuitive tenet for a ‘designed’ or ‘teleological’ process (after all, we do not decide the size of our house based solely on the size of the bricks!), the Fankhauser finding was considered as a largely unexpected finding in a natural system. This is why Albert Einstein (a colleague of Fankhauser at Princeton) told he was expecting the double size cells should give rise to double size organs, concluding that the Fankhauser observation pointed to still largely unknown principles. The brilliant Fankhauser experiment was largely overlooked and obscured by the successes of molecular biology in the years to come, but it is a clear example of a top-down causative model in which a ‘high-level’ constraint ‘slaved’ the microscopic cellular/genomic level.
It is important to stress that the ‘bottom-up only’ obsession is not shared by all the biological fields of investigation, Ecologists recognized since many years that the ‘most microscopic’ level of organization is not necessarily the place where ‘the most relevant facts do happen’.
On the contrary, the most fruitful scale of investigation is where ‘non-trivial determinism is maximal’ (Pascual & Levin, 1999). That is to say, the scale more ‘rich’ in meaningful correlations between features pertinent to micro and macroscale that directly recalls the above sketched concept of ‘Interface’.
Non-trivial determinism can be defined in terms of prediction error as:
In the above formula, E is the mean prediction error and S the standard deviation.
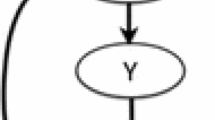
In the case of a simple linear regression in which a dependent variable Y must be predicted by an independent variable X, the non-trivial determinism is nothing else than the usual squared Pearson correlation between the two X and Y variables.
The formula can be extended to any other situation in which we wish to predict a system feature Y, both X and Y do not need to represent single variables but any suitable set of information at any definition scale.
The ‘non-trivial’ attribute of determinism stands for the need of ‘explaining the variance’ of the system at hand (the statistic r2 corresponds to the proportion of variance explained by a model) and not its ‘average’ pattern. The aim is to get rid of the actual behaviour of the system in both space and time and not to describe a ‘frozen’ ideal configuration.
The individuation of ‘mesoscopic principles’ largely independent from the material constitution of the studied system and only dependent from their relational structure was faced by 1998 Nobel prize in Physics Robert Laughlin and colleagues. A paper appeared in year 2000 (Laughlin et al., 2000) entitled ‘The Middle Way’ that aptly individuated in the discovery of universal mesoscopic principles the next frontier of science.
As pointed out by Nicosia et al. (2014): ‘Networks are the fabric of complex systems’ and this tells us that network formalism is probably the ideal instrument in the search for such principles. The basic idea of complex network style of reasoning is that shared organization rules (i.e. similar wiring patterns) give rise to similar phenomenology, independently of the nature of the constituting elements. In other words, complex network invariants promise to be the place where to look for universal mesoscopic principles, for the simple fact that they have not different regularities and laws for the different levels, this promises to be the viewpoint that maximizes ‘non-trivial determinism’ (Pascual & Levin, 1999) favoring the emergence of between-level correlations.
In Mickulecki (2001) paper, the author demonstrates the neat separation of the laws governing the internal functioning of the nodes of a network (constitutive laws) from the laws and regularities only dependent from the wiring structure of the system (relational laws). This allows to build an electrical analogue of a mechanical or physiological system only based on conservation principles of both potential and flux across a network analogous to Kirchoff’s laws. The flux does not need to be an electrical current and the same holds for the potential: a system represented by a set of nodes linked by edges with a given topology has similar emerging properties independently of the physical nature of nodes and edges. This opens the way to a ‘network thermodynamics’, whose principles are strictly dependent from wiring architecture while largely independent of the constitutive laws governing the single elements. Still more important, this provokes a shift from the founding of the unitary character of science from the consideration that ‘all the entities are made of the same fundamental building blocks’ to the recognition that ‘all the entities can be represented by a set of relations among their parts’. These relations can be formalized in terms of graph (network) invariants catching different aspects of the wiring structure of the system at hand.
Complex network invariants catch the essence of multi-level organization for the simple fact their estimation merges different level of definition of the system at hand without the need of any strong hypothesis.
Mathematically speaking, a network corresponds to a graph whose entire information is caught by its adjacency matrix (see Fig. 1): a binary matrix having as rows and columns the nodes and at each i, j position a unit value if the i and j nodes have a direct link between them and 0 otherwise.
The figure reports the adjacency matrix (left panel) correspondent to the wiring diagram on the right. The presence of a direct link between two nodes corresponds to a unit value of the corresponding element of the matrix on the left. Here all the links are supposed to have the same strength, in other cases we can substitute the unit values with a quantitative estimate of the correlation strength. The represented graph is bi-directional
Graph invariants are relative to local (single nodes), global (entire network), and mesoscopic (clusters of nodes, optimal paths) levels respectively. The “degree” (how many links are attached to a given node) is a local descriptor, the “average shortest path” (characteristic length) is the average length of minimal paths connecting all the node pairs, and can be considered as a mesoscopic feature, while the general connectivity of the network (density of links) is a global property (Giuliani, 2019). All these descriptors (and many others) are strictly intermingled across different organization layers. Thus, characteristic length inherits from the ‘bottom’ the information of the single node degree (higher degree nodes have an higher probability to enter into shortest paths). In turn, betweeness of a node (the number of shortest paths passing by a node, thus a microscopic feature of the network) inherits from the ‘top’ (mesoscopic level) the existence of clusters (modules) of nodes.
In this way, a node in between two different A, B clusters is traversed by all the shortest paths linking the A, B node pairs so scoring an high betweeness (Fig. 2).
The figure reports schematically the most common graph invariants. Each index concentrates on a particular aspect of network wiring, shortest paths, participation coefficient and betweeness centrality are particularly important for describing fluxes across the network, clustering coefficient and modularity point to the existence of ‘structural domain’ within the network
Describing a system by network formalism implies a multi-level structural representation without the need of ‘imposing’ a particular bottom-up or top-down causative pattern.
4 Information Fluxes Within Networks
Proteins are the smallest objects that have all the features typical of complex systems, it is not without reason that the title of a seminal work on protein structure and dynamics (Frauenfelder & Wolynes, 1994) is ‘Biomolecules: where the physics of complexity and simplicity meet’ .
Proteins ‘sense’ the environment, can acquire different stable state configurations, have an emergent behaviour not predictable from the accurate knowledge of their composition and perform complex ‘actions’ relevant for the system that host them. In addition the structural and compositional knowledge we have on protein molecules is order of magnitudes mere detailed and reliable than for any other complex system. This makes protein sciences a perfect playground for complexity studies.
Probably the most straightforward paradigm of information transfer through a network is the allosteric effect. Allostery is a neologism coming from Greek language, which has to do with the ability of proteins to transmit a signal from one site to another in response to environmental stimuli. The sensing (and consequent adaptation) of relevant information from the microenvironment is crucial for protein physiological role. This ability relates to the transmission of information across the protein molecule from a sensor (allosteric) site to the effector (binding) site (Di Paola & Giuliani, 2015). The protein molecule, hence, perceives ligand binding (or any other micro-environmental perturbation) at distance from the active site (where in turn the effective action takes place, e.g. where two small molecules are put together in order to catalyse their chemical reaction), and adapts its configuration accordingly. Thus, haemoglobin molecule senses at the allosteric site the partial pressure of oxygen (p[O2]): when p[O2] is high the affinity of haemoglobin for oxygen increases and the protein binds oxygen molecules at active site, on the contrary when p[O2] is low, affinity decreases and bound oxygen is released to cells. This process is crucial for life: in lungs there is a very high oxygen pressure and the haemoglobin present in red blood cells must catch oxygen molecules that in turn must be released in peripheral tissues (low p[O2]) so to make oxidative metabolism possible. How the protein molecule can discriminate such a relevant signal from the continuous perturbations of its structure coming from thermal noise and transmit the information at distance so to reach the active site.
To answer this question is useful to consider a protein molecule as a network (Fig. 3) having as nodes the aminoacid residues and as edges the intermolecular non-covalent bonds between residues generated by the 3D folding of the molecule. These networks are called Protein Contact Network (PCN) (Di Paola et al., 2013).
In this figure, the left panel the 3D structure of a small protein (recovering) follows the usual ‘ribbon’ style: the polypeptidic chain is represented in terms of contiguous segments of ‘secondary structures’ namely helices, random coils, and beta sheets. The right panel represents the same protein in terms of the adjacency matrix of the corresponding network (PCN = Protein Contact Network) whose nodes are the constituent amino-acids while the darkened pixels mark the relevant between amino-acid residues contacts (the unit values of Fig. 1)
In Fig. 3 the aminoacid residues are ordered along the protein sequence from the left to the right in the X axis of the adjacency matrix and from the top to the bottom on Y. The ‘trivial’ contacts between aminoacids adjacent along the chain are not considered. This implies the scored contacts (links of the PCN) correspond to non-covalent intermolecular bonds putting different parts of the molecule into close contact (see Fig. 4, where a protein molecule is represented as a bracelet having aminoacid residues as pearls and PCN relevant contacts as red dashed lines).
The blue line sequentially connecting the different aminoacid residues (pearls) corresponds to the covalent bond that generates the primary structure (sequence) of the macromolecule. In solution the protein molecule acquires its ‘native’ form by a folding process that generates its 3D structure responsible of its physiological role. The folding process puts aside residues otherwise distant along the sequence creating contacts (dashed red lines) among them. These contacts allow for a direct communication of the interacting aminoacid residues
In PCNs the shortest paths passing by the network edges mediate concerted motions and energy transmission upon stimulation of allosteric site (Di Paola & Giuliani, 2015). The topological metrics of shortest paths (minimum number of links separating two residues) is thus the actual metrics for signalling. Recently it was demonstrated (Poudel et al., 2020) that this purely topological metrics is coincident with the dynamical modes of protein molecule. This creates a spatio-temporal link of ‘sustained modes’ fulfilling the stable oscillation constraint we set for biodynamic interfaces. Thus we can say we are in presence of a ‘fine tuned’ grid deciding of the fate of external stimuli across the system.
The discrimination between relevant signals to be transmitted at distance without loss of information and non-informative perturbations to be dissipated without relevant changes in the 3D structure, relies upon two very important mesoscopic network descriptors: ‘Guimera and Amaral’ z and P indexes (Guimera & Amaral, 2005). The index z quantifies the number of contacts a given node (aminoacid residue in this case) has with other nodes of its own cluster (local contacts), while P scales with the number of edges linking the node to aminoacid residues pertaining to different clusters.
A perturbation affecting specifically an ‘high P’ node travels a long distance across the network passing by subsequent ‘high P’ nodes and arriving at destination supporting allosteric effects, on the contrary generic (noisy) thermal motion rapidly dissipates distributing across non-directional cycles thru intra-module motions.
High P nodes create a ‘fast lane’ for relevant information neatly separated by noise. This is exactly the role of biodynamic interfaces: some proteins, called multimeric, consist of distinct chains held together by intermolecular contacts. This is the case of haemoglobin made of four distinct polypetidic chains: the allosteric effect ends up into a different re-arrangement of the relative positions of the four chains that go back and forth between two different patterns (R and T for Relaxed and Tense) with high and low affinity for oxygen. The interface between these four chains is made of high P aminoacid residues that allow for the among chains concerted motions. Figure 5 gives a pictorial description of the situation.
From Fig. 5 it is evident the presence of ‘displaced contacts’ in the form of residues that, while pertaining to a given chain (module of the network) have the majority of their contacts with residues pertaining to different chains. These ‘displaced contacts’ are the long ‘whiskers’ contacting zones different from their own cluster (e.g. the pale blue line pertaining to the first chain (1–150)) that is in contact with the orange (second chain) module. These whiskers correspond to high P nodes that generate ‘something in between’ the interacting systems with a ‘shared ontology’ across the interacting systems (polypeptide chains).
Perturbations relevant for the allosteric effect (signals) enter the fast lane passing by high P residues and arriving at destination, on the contrary, not relevant (noisy) perturbations instead dissipate along futile within-module circuits. The presence of both fast (directional low loss) and slow (no-directional high loss) lanes of communication is shared by all natural networks (Kohestani et al., 2018) even if in protein molecules is much more evident than in other natural networks.
The discrimination between relevant and irrelevant stimuli is a form of ‘meaning creation’ by purely structural means that allow for a causative process embedded (and not imposed from the external) in the relational structure of the system at hand. This kind of causation makes obsolete the bottom-up top-down distinction and asks for a different explanation style in terms of ‘attractor-like’ dynamics spanning different layers of organization.
References
Arora, M., Giuliani, A., & Curtin, P. (2020). Biodynamic interfaces are essential for human–environment interactions. BioEssays, 42(11), e2000017.
Demongeot, J., Aracena, J., Thuderoz, F., Baum, T. P., & Cohen, O. (2003). Genetic regulation networks: Circuits, regulons and attractors. Comptes Rendus Biologies, 326(2), 171–188.
Di Paola, L., De Ruvo, M., Paci, P., Santoni, D., & Giuliani, A. (2013). Protein contact networks: An emerging paradigm in chemistry. Chemical Reviews, 113(3), 1598–1613.
Di Paola, L., & Giuliani, A. (2015). Protein contact network topology: A natural language for allostery. Current Opinion in Structural Biology, 31, 43–48.
Fankhauser, G. (1945). Maintenance of normal structure in heteroploid salamander larvae, through compensation of changes in cell size by adjustment of cell number and cell shape. Journal of Experimental Zoology, 100(3), 445–455.
Frauenfelder, H., & Wolynes, P. G. (1994). Biomolecules: Where the physics of complexity and simplicity meet. Physics Today, 47(2), 58–64.
Giuliani, A. (2019). The actual status of quantitative approaches in biology: Problems and perspectives. Rend Mat Appl, 40(7), 165–176.
Guimera, R., & Amaral, L. A. N. (2005). Functional cartography of complex metabolic networks. Nature, 433(7028), 895–900.
Kohestani, H., Totonkuban, M., Di Paola, L., Todde, V., & Giuliani, A. (2018). The basic principles of topology-dynamics relations in networks: An empirical approach. Physica A: Statistical Mechanics and its Applications, 508, 584–594.
Laughlin, R. B., Pines, D., Schmalian, J., Stojković, B. P., & Wolynes, P. (2000). The middle way. Proceedings of the National Academy of Sciences, 97(1), 32–37.
Mendonça, V., Madeira, C., Dias, M., Vermandele, F., Archambault, P., Dissanayake, A., & Vinagre, C. (2018). What’s in a tide pool? Just as much food web network complexity as in large open ecosystems. PLoS One, 13(7), e0200066.
Mickulecki, D. (2001). Network thermodynamics and complexity: A transition to relational systems theory. Computers & Chemistry, 25, 369–391.
Nicosia, V., De Domenico, M., & Latora, V. (2014). Characteristic exponents of complex networks. Europhysics Letters, 106(5), 58005.
Pascual, M., & Levin, S. A. (1999). From individuals to population densities: Searching for the intermediate scale of nontrivial determinism. Ecology, 80(7), 2225–2236.
Poudel, H., Reid, K. M., Yamato, T., & Leitner, D. M. (2020). Energy transfer across nonpolar and polar contacts in proteins: Role of contact fluctuations. The Journal of Physical Chemistry B, 124(44), 9852–9861.
Sporns, O. (2018). Graph theory methods: Applications in brain networks. Dialogues in Clinical Neuroscience, 20(2), 111.
Weaver, W. (1948). Science and complexity. American Scientist, 36, 536.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Giuliani, A. (2021). Information from Structure: How Networks Face Biological Complexity. In: Minati, G. (eds) Multiplicity and Interdisciplinarity. Contemporary Systems Thinking. Springer, Cham. https://doi.org/10.1007/978-3-030-71877-0_5
Download citation
DOI: https://doi.org/10.1007/978-3-030-71877-0_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-71876-3
Online ISBN: 978-3-030-71877-0
eBook Packages: Business and ManagementBusiness and Management (R0)