Abstract
Lignocellulose-derived sugars and other biorefinery by-product streams such as glycerol and acetic acid are useful carbon feedstocks for microbes that produce lipids. Lipids have high energy density and are easily converted into versatile biofuels and valuable oleochemicals. Common, robust yeasts such as Saccharomyces cerevisiae and Yarrowia lipolytica have been the most successfully exploited as cell factories for lipid production, and excellent progress has been made in productivity with the implementation of synthetic biology tools and metabolic engineering strategies. Accumulation and storage of standard fatty acids as triacylglycerols or secretion of free fatty acids has been enhanced by modification of metabolic pathways yielding maximal fatty acid titers above 100 g L−1 and productivity of 0.8 g L−1 h−1. Production of higher-value exotic fatty acids that are not native to yeast, such as short chain, hydroxylated, and cyclopropane, has great potential but requires more research into lipid synthesis pathways and new metabolic engineering strategies to achieve similar productivities as achieved for standard fatty acids. In addition, monitoring of cell viability and health, balancing cofactor demands, and minimizing stress are important strategies to avoid or reduce metabolic burden caused by engineering of cells.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Metabolic engineering
- Yeast
- Fatty acids
- Saccharomyces cerevisiae
- Yarrowia lipolytica
- Synthetic biology
- Hydroxylated
- Cyclopropane
- Metabolic burden
- Biofactories
9.1 Introduction
The economic, environmental, and social sustainability problems caused by the dependency on petroleum have motivated a global shift to renewable, sustainable, and green alternative energy sources [1]. Replacement of crude oil-derived fuels and chemicals by the production of biofuels and bioproducts can be an effective strategy to reduce pollution and carbon dioxide emissions [2]. In particular, a useful feedstock for biofuels and bioproducts are lipids, consisting mainly of triacylglycerols, as they have high energy density and are readily converted to mono-alkyl esters for use as a diesel substitute. While lipids have excellent commercial utility, they are relatively expensive and in short supply, as there are important applications for lipids in food processing and oleochemical manufacture. In 2018, the natural fatty acid global market was valued at nearly $13.5 billion and expected to reach $17.5 billion in 2023 with a compound annual growth rate (CAGR) of 5.4% [3] (BCC Research LLC, 2019). Therefore, new sources of cost-effective lipids for the production of fuels and chemicals will be in increasingly high demand.
Lipids produced by microbes have huge potential to satisfy the growing demand for bio-based energy-dense hydrocarbons and related natural products [4], especially where their production is based on non-food carbon sources such as lignocellulosic sugars or by-product streams from biorefineries. Recent advances in microbial metabolic engineering and process technologies have brought us closer to cost-effective yields and diversity of oleaginous products that can support this growing market [5].
Compared with the ubiquitous bacterium Escherichia coli, yeasts like Saccharomyces cerevisiae and Yarrowia lipolytica are more effective hosts for lipid production because they synthesize C16-18 carbon chain fatty acids very efficiently requiring just two fatty acid synthases, whereas E. coli requires ten enzymes to reach the same endpoint [1, 6]. Furthermore, yeast can store large quantities of fatty acid internally as triacylglycerol. S. cerevisiae is a widely used industrial yeast due to its robustness and good tolerance of harsh industrial conditions [7] and its long history of use in large-scale fermentation to produce ethanol and beverages [8]. The oleaginous yeast Y. lipolytica has also had wide use in biotechnology and has several advantages over S. cerevisiae in that it naturally stores substantially more lipid within the cell and utilizes a broad range of low-cost feedstocks such as glycerol. The yeast holds generally recognized as safe (GRAS) status for the production of citric acid and has been explored for the production of sugar derivatives and nonnative, lipid products such as β-carotene and lycopene [9,10,11,12,13,14,15].
Another active area of research is in the production of exotic fatty acids and derivatives in yeasts as feedstocks in the production of fine chemicals, medicines, detergents and soaps, lubricants, cosmetics, and skin care products [16]. These lipids are not naturally present in yeast but are produced through the introduction of genes sourced from other organisms. Exotic fats include fatty alcohols and esters and unusual fatty acids such as those with modifications to fatty acid chain length, polyunsaturation, or added functional groups.
In recent years, the emerging synthetic biology field has brought new vitality into the development of microbial cell factories providing more powerful tools and methods to modify the microbial metabolic pathways [17]. To date, both natural lipids and lipid derivatives have been successfully produced through the benefits of synthetic biology and metabolic engineering in impressive yields. While most of the basic research in yeast lipid engineering have used purified sugars as carbon feedstocks, it has also been shown that lignocellulosic-derived sugars and other biorefinery by-products will also be effective substrates for these organisms.
In one concept of a biorefinery, cheap, plentiful biomass can be deconstructed to produce lignocellulosic sugars that are used as feedstocks for microbial oil production leaving lignin and hemicellulose sugars which can be further converted into products. For example, Wei et al. tested loblolly pine and sweetgum autohydrolysates after detoxification as feedstocks for lipid production via the oleaginous bacterium, Rhodococcus opacus, and achieved 0.25–0.31 g/L lipid titer [18]. Slininger et al. screened and identified three oleaginous yeasts that could utilize raw enzyme hydrolysates of ammonia fiber expansion (AFEX)-pretreated corn stover and acid-pretreated switchgrass as feedstocks for lipid production, and the lipid titer reached 25–30 g/L (39–45% of the theoretical yield) [19]. Here, we review recent progress in the application of synthetic biology and metabolic engineering focusing on yeasts S. cerevisiae and Y. lipolytica, as cell factories to produce lipids and higher-value fatty acid derivatives as part of a biorefinery.
9.2 Microbial Lipids from Lignocellulose-Derived Substrates
Microorganisms that can use lignocellulosic-derived substrates such as glucose, xylose, glycerol, and acetic acid for the production of microbial lipids are important for the utilization of biorefinery streams. Here, we review recent promising microbial lipid production research featuring the yeasts S. cerevisiae and Y. lipolytica cultured using lignocellulose-derived substrates.
Apart from glucose, the other two major sugars from lignocellulosic biomass are xylose and arabinose. For S. cerevisiae to be a more competitive chassis for the biotechnology industry, it is important to extend its growth substrates beyond glucose. The Pronk group has adopted metabolic engineering strategies, laboratory evolution, and co-culture approaches to enable S. cerevisiae to use xylose as a carbon source and improved ethanol fermentation performance using mixed sugars including glucose-xylose-arabinose [20, 21]. Ionic-liquid-pretreated switchgrass and sorghum were used as feedstocks for fatty alcohol production by S. cerevisiae engineered with 11 genetic modifications compared with the parent BY4741 strain, and the fatty alcohol titer reached 0.7 g/L in shaker flasks [22].
Also, a series of lignocellulose substrates were assessed for growth of engineered Y. lipolytica for microbial lipid production in bioreactors. For example, Li and Alper used xylose as carbon source bringing lipid production to 15 g/L [23]; Rakicha et al. improved lipid titer to 24.2 g/L using molasses/glycerol as feedstocks [24], Ledesma-Amaro et al. further improved lipid titer to 50.5 g/L using xylose and glycerol as substrates [25], and Niehus et al. achieved a very high lipid titer of 16.5 g/L and showed Y. lipolytica was tolerant to the toxicity of xylose-rich agave bagasse hydrolysate [26]. Furthermore, in a semicontinuous system, the high-density cell culture of Y. lipolytica was assessed using 3% acetic acid as a carbon source. The acetic acid was consumed completely, and yeast achieved a lipid titer of 115 g/L, yield of 0.16 g/g, and productivity of 0.8 g·L−1·h−1, respectively [27]. In further examples, Slininger et al. screened and identified three oleaginous yeasts that could use non-detoxified enzyme hydrolysates of ammonia fiber expansion (AFEX)-pretreated corn stover and acid-pretreated switchgrass as feedstocks for lipid production. The highest lipid yield reached 25–30 g/L, 39–45% of the theoretical yield [19].
As these successful attempts in production of microbial lipids show, biorefinery by-product streams such as non-glucose sugars, glycerol, and acetic acid are feasible cheap substrates for microbial lipids production. An ongoing challenge of the biorefinery concept is to develop robust microbial cell factories with greater productivity and cost-effectiveness. Recent research toward this aim is reviewed in the following section focusing on progress with S. cerevisiae and Y. lipolytica.
9.3 Metabolic Engineering Strategies and Recent Progress Toward Improved Yeast Lipid Production
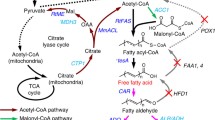
In general, natural lipid production with yeast can be enhanced by (1) increasing fatty acid (FA) biosynthesis , such as by “pushing” carbon flux toward precursor acetyl-CoA and malonyl-CoA pools; (2) “blocking” competing pathways that consume lipids or free fatty acids, such as beta-oxidation; (3) balancing cofactor requirements and enzyme activity to deliver a steady NADPH supply to support fatty acid synthase activity; and (4) secreting free fatty acids into culture media or sequestering nascent lipids within lipid droplet to avoid toxicity [28,29,30]. This section highlights the recent advances in engineering efforts to increase lipid production in S. cerevisiae and Y. lipolytica yeast. The key genes/enzymes that have been engineered to improve lipid production and their cellular locations are shown in Fig. 9.1.
Main metabolic pathways, control points, and organelles from sugars to lipid in yeast S. cerevisiae. The key genes/enzymes that have been engineered to impact lipid production have been highlighted in red text (ADH alcohol dehydrogenase, ALD6 cytosolic aldehyde dehydrogenase 6, ACS acetyl-coA synthetase, SeACSL641p acetyl-CoA synthetase with L641P mutation, derived from Salmonella enterica, ACC1 acetyl-CoA carboxylase, ACC1** acetyl-CoA carboxylase carrying two mutations ser659ala and ser1157ala, RtFAS fatty acid synthetase from R. toruloides, FAS1, 2 fatty acid synthetase, ACL1, 2 ATP-citrate synthase subunit 1, 2, MmACL ACL from Mus musculus, FAA2 medium-chain fatty acyl-CoA synthetase, PXA1 subunit of heterodimeric peroxisomal ABC transport complex, POX1 fatty-acyl coenzyme A oxidase, MEF1 peroxisomal multifunctional enzyme, PEX10 peroxisome biogenesis factor 10, FAA1, 4 long-chain fatty acyl-CoA synthetase, FAT1 very-long-chain fatty acyl-CoA synthetase, ‘tesA truncated E. coli thioesterase, TGL3-5 triacylglycerol lipase genes, GUT2 glycerol-3-phosphate dehydrogenase gene, GPAT glycerol 3-phosphate acyltransferase, LPAT lysophosphatidate acyltransferase, PAP phosphatidate phosphatase, DGA1, LRO1 diacylglycerol acyltransferase, ARE1, 2 sterol O-acyltransferase genes, AtCLO1 caleosin, lipid droplet stabilization protein from Arabidopsis thaliana)
9.3.1 Lipid Metabolic Engineering of S. cerevisiae
Storage lipids make up no more than 10% dry cell weight (DCW) in wild-type S. cerevisiae, while they are accumulated to a much higher degree in some oleaginous yeast like Y. lipolytica [31]. Well-targeted single gene or pathway modifications in yeast normally lead to increased lipid content though improvement is limited. For more considerable improvement in lipid production, it is necessary to combine multiple approaches including synthetic biology, metabolic engineering, protein/enzyme engineering, adaptive laboratory evolution, machine learning, etc. Several successful attempts have been undertaken by researchers to enhance extracellular and intracellular lipid production in S. cerevisiae , and here, only recent examples with promising lipid yield are summarized.
Successful approaches to enhancing fatty acid (FA) biosynthesis include the overexpression of ACC1 (or ACC1**) [32, 33], ACS1, FAS1, and FAS2 [6, 34,35,36], blocking FA competing pathways by deleting genes in beta-oxidation such as POX1 and POX2 [37, 38] and assessing the effects of lipid accumulation and storage genes such as DGAT1 and PDAT1 [39,40,41]. In terms of intracellular free fatty acid (FFA) accumulation, as opposed to esterified fatty acids, Valle-Rodriguez et al. (2014) deleted DGA1, LRO1, ARE1, and ARE2 to block formation of neutral lipid and deleted POX1 to avoid FA degradation whereby the engineered S. cerevisiae reached 1.5% intracellular FFA by DCW, fivefold higher than control [37].
S. cerevisiae BY4742 produced 17% DCW TAG was produced by overexpressing genes coding for FAS, ACC, and DGA [6]. Then, introducing ATP-citrate lyase (ACL) from the metabolism of an oleaginous microorganism to S. cerevisiae and disrupting isocitrate dehydrogenase genes IDH1 and IDH2, they could increase the total fatty acids to 21% [42]. Peng et al. (2018) strengthened three steps of lipid production including FA biosynthesis (Ald6-SEACSL641P, ACC1**), lipid accumulation (DGAT1), and lipid sequestration (ΔTGL3, AtCLO1) and achieved 8.0% DCW (2.6-fold than control) and 0.3 g/L lipid (4.6-fold than control) in a two-stage bioprocess [30]. Notably, the Nielsen group implemented a comprehensive strategy to increase TAG accumulation and reached 254 mg TAG/g DCW in S. cerevisiae. The strategy included increasing acetyl-CoA supply (ACC1**), improving lipid accumulation (PAH1 and DGA1), blocking lipid degradation (ΔTGL3, 4, 5, ΔPOX1, ΔPXA1), sterol synthesis (ΔARE1), glycerol-3-phosphate utilization (ΔGUT2) [43].
For secreted free fatty acid (FFA) production , Li et al. (2014) disrupted β-oxidation, deleted acyl-CoA synthetase, and overexpressed thioesterases and ACC1 in S. cerevisiae to achieve 140 mg/L [44]. The Da Silva group achieved 2.2 g/L extracellular FFAs through disrupted neutral lipid recycle in S. cerevisiae including disruption of β-oxidation (△FAA2, PXA1, POX1), acyl-CoA synthetase genes (FAA1, FAA4, FAT1), and coexpression of DGA1 and TGL3 [45]. Zhou et al. (2016) reached 10.4 g/L extracellular FFAs by enhancing acetyl-CoA supply, malonyl-CoA pathway, and fatty acid synthase expression and blocking fatty acid activation and degradation. The Nielsen group further engineering efforts to reprogram yeast metabolism from alcohol fermentation to lipogenesis whereby they constructed an impressive FFA-producing yeast delivering up to 33.4 g/L FFAs. The metabolic engineering included increasing cytosolic acetyl-CoA and NADPH supplies, redistributing carbon flux toward fatty acid biosynthesis, abolishing ethanol production pathway, mutating pyruvate kinase, and directing evolution [46].
9.3.2 Lipid Metabolic Engineering of Y. lipolytica
Due to the similarity of lipid metabolism between yeasts S. cerevisiae and Y. lipolytica, general metabolic strategies to enhance lipid production are transferable. Similar to the effectiveness of lipid pathway engineering in S. cerevisiae, there has been much progress in increasing lipid production in Y. lipolytica. Here, recent successful examples with promising lipid yields or addressing the key bottleneck metabolic issues have been addressed.
The Stephanopoulos group used lipid pathway engineering in Y. lipolytica to markedly improve production; their strategies have ranged the introduction of multiple gene combinations to the analysis of cellular physiological issues. Tai and Stephanopoulos (2013) firstly identified a more efficient promoter (intron-containing TEF) to assist heterologous gene expression by 17-fold and then improved ACC1 and DGA1 expression to increase lipid to 61.7% DCW, 0.270 g/g lipid yield, and 0.253 g L−1 h−1 lipid productivity [47]. Qiao et al. (2015) successfully identified the Δ9 stearoyl-CoA desaturase (SCD), which was overexpressed to avoid the repression of acetyl-CoA carboxylase via increasing fatty-acyl-CoA desaturation. Meanwhile, simultaneous overexpression of SCD, ACC1, and DGA1 in Y. lipolytica achieved improved cell growth and increased tolerance to sugars plus a high-level lipid titer of 55 g/L and high carbon to lipid conversion yield (84.7% of theoretical maximal yield) [48]. Further, Qiao et al. (2017) successfully demonstrated that redox engineering via the modulation of the NADPH recovery pathway in Y. lipolytica increased lipid accumulation to 98.9 g/L measured as fatty acid methyl ester (FAME) [49]. Furthermore, Xu et al. (2017) employed a semicontinuous fermentation mode to bring the lipid titer of 115 g/L with an engineered Y. lipolytica (PO1g: ACC1, DGA1) and acetic acid as substrates [27].
Cellular oxidative stress defense pathways were investigated in Y. lipolytica to determine their impact on lipid production. Additional glutathione disulfide reductase to reduce oxidative stress, glucose-6-phosphate dehydrogenase for NADPH recycling and an engineered aldehyde dehydrogenase with broad substrate range were introduced into the yeast which proved to be efficient solutions to combat reactive oxygen and aldehyde stress in Y. lipolytica. The lipid titer reached 72.7 g/L and oil content 84.4% [50].
A comprehensive overexpression strategy in Y. lipolytica was adopted by the Alper group to improve lipid production. Blazeck et al. (2014) improved lipid production titer to 25 g/L using metabolic engineering strategies that included enhancing TAG biosynthesis (DGA1, 2), increasing acetyl-CoA (ACL1, 2), increasing NADPH cofactor supply (MAE), inhibiting the TCA cycle, increasing the citric acid level (ΔAMPD), and preventing beta-oxidation and peroxisome biogenesis (knockout of mfe1, pex10) [51]. Based on the engineered strains, Liu et al. (2015) identified a mutant Mga2p regulator in Y. lipolytica, which increased unsaturated fatty acid biosynthesis, possibly due to reduced feedback inhibition of ACC or reduced degradation of the stearoyl-CoA desaturase. Also, the mutant strain containing Mga2p maintained a high lipid titer (25 g/L) [52]. Furthermore, Liu et al. adopted a laboratory adaptive evolution approach to further screen for a super lipid producer strain with 87.1% DCW and 39.1 g/L lipid production [53].
Further examples of Y. lipolytica metabolic engineering with promising lipid yield include Ledesma-Amaro et al. (2016) who tested two synthetic approaches, firstly redirecting carbon flux to neutral lipids and, secondly, by mimicking a bacterial system to produce free FFAs. One optimal strain engineered to overexpress lipases that convert lipids to FFAs, and prevented the formation of CoA esters and β-oxidation of fats, produced up to 20.8 g/L lipids in a 5 L bioreactor [54]. Meanwhile, Ledesma-Amaro et al. (2016b) engineered PO1d strain with the following interventions: Δpox1–6, ΔTGL4, GDP1, DGA2, ssXR, ssXDH, and ylXK. Using xylose/glycerol as substrates, the lipid titer reached 50.5 g/L [25]. Friedlander et al. (2016) enhanced lipid accumulation and sequestration in Y. lipolytica by overexpression of both DGA1 from Rhodosporidium toruloides and DGA2 from Claviceps purpurea, plus deleted a key lipase (TGL3). The final engineered strain NS432 achieved 77% lipid content and 0.21 g lipid per g glucose yield in batch fermentation and 85 g/L lipid in fed-batch glucose fermentation [55]. Besides, 13C-metabolic flux analysis was employed to understand whether the malic enzyme contributes to lipogenic NADPH production in Y. lipolytica, and the oxidative pentose phosphate pathway was proved to be the primary source of NADPH for lipid overproduction from glucose [56] (Table 9.1).
9.4 Exotic Fatty Acid/Alcohol Production in Engineered Yeast
9.4.1 Short- and Medium-Chain Fatty Acids
Short-chain fatty acids (SCFAs) , where the carbon chain length is less than 10, are important industrial products as they can be used as gasoline and jet fuel precursors and intermediates in the synthesis of alkenes [59]. Producing SCFAs in common biotechnological organisms is challenging as they do not natively produce short-chain fatty acids but prefer chain length range between C14 and C18 as these are primarily precursors for the formation of cellular membranes to support cell homeostasis [60]. Beyond the challenge of producing substantial SCFA within the cells, the potential cytotoxicity due to SCFAs’ capacity to damage cell membranes needs to be addressed [61].
The first challenge is that the acyl carrier protein (ACP) and a phosphopantetheine transferase (PPT) are too large for the natural fatty acid synthase (FAS) of S. cerevisiae to passively diffuse into for elongation [62,63,64]. Also, the size of the short-chain thioesterases (TE) cleaving the elongating fatty acid is more than 9 kDa [65, 66]. To overcome these issues, Leber and Da Silva (2014) [67] expressed the FAS from Homo sapiens (hFAS); two heterologous TEs from Cuphea palustris, a plant that naturally produces SCFA , and Rattus norvegicus; and PTTs from E. coli and Bacillus subtilis in S. cerevisiae, respectively. Compared with native yeast, C8 levels were increased by 17-fold by overexpression of hFAS. Linking hFAS with heterologous TEs further improved the yield of C8 by four- and nine-fold. After introducing heterologous PPTs, total SCFA titers and C8 titers could reached 111 mg/L and 82 mg/L, respectively. In 2015, the freestanding thioesterase (HTEII) in H. sapiens was found to have a primary chain length selectivity for octanoic acid. HTEII was fused to hFAS and PTTs from H. sapiens was expressed in S. cerevisiae. Also, β-oxidation was fully disrupted. Finally, hexanoic and octanoic acid levels were increased by eight- and 79-fold over the parent strain with hFAS only [68].
Zhu et al. achieved the production of >1 g/L extracellular SCFA (C6-C12) in S. cerevisiae, a more than 250-fold improvement over the original strain. To achieve this, they engineered both the endogenous FAS and an orthogonal bacterial type I FAS and performed directed evolution on the membrane transporter Tpo1. They further developed the strain via adaptive laboratory evolution and metabolic flux control to markedly improve the SCFA production [69].
Meanwhile, Xu et al. (2016) demonstrated the specific structure of fungal type I FAS in Y. lipolytica. Then, they swapped malonyl/palmitoyl transacylase domain in FAS1 and fused the truncated FAS1 with smaller TE to improve medium-chain fatty acid production, which resulted in remarkably increasing C12 and C14 portions of fatty acids to 29.2% and 7.5%, respectively [70] .
9.4.2 Fatty Acid Esters and Alcohols
Fatty acid ethyl esters (FAEEs) are an attractive diesel oil alternative with high energy density and low toxicity to the production host (Zhang et al., 2012; Zhou et al., 2014). Acyl-CoAs formed within the cell can be condensed by wax ester synthase/acyl-CoA:diacylglycerol acyltransferase with ethanol to synthesize FAEEs. In order to improve FAEE yield, the pathway for the intermediate acyl-CoAs is enhanced by metabolic engineering. Shi et al. screened five wax ester synthases for FAEE biosynthesis; a candidate obtained from Marinobacter hydrocarbonoclasticus gave 6.3 mg/L FAEE titer (Shi et al., 2012). With integration of this wax synthetase into the S. cerevisiae genome, FAEE yield improved to 34 mg/L (Shi et al., 2014b).
In addition, reducing competition for acyl-CoAs from non-lipid pathways was shown to improve FAEE productivity . For example, Valle Rodriguez et al. blocked β-oxidation, sterol esters, and TAG biosynthesis in S. cerevisiae to yield 17.2 mg/L in the mutant strain, threefold higher than the wild-type strain (Valle-Rodríguez et al., 2014). As NADPH and acetyl-CoA are required to synthesize acyl-CoA, De Jong et al. 2014 upregulated ethanol degradation and constructed a phosphoketolase pathway to increase flux of acetyl-CoA and NADPH, which can improve the pool of acyl-CoA. Alcohol dehydrogenase Adh2, the Salmonella enterica acetyl-CoA synthetase variant SeACS (L641P), and acetaldehyde dehydrogenase Ald6 were overexpressed to accelerate ethanol degradation, which improved threefold FAEE yield (Starai et al., 2005). The overexpression of ACC1 also contributed to the accumulation of acetyl-CoA, whereby FAEE production reached 8.2 mg/L (Shi et al., 2012). Y. lipolytica has also been developed as a host for FAEE production by similar metabolic engineering strategies. An efficient FAEE biosynthetic pathway was constructed by expression of heterologous wax ester synthase gene with codon optimization for Y. lipolytica and under strong promoters. In addition, carbon flux was redirected toward the FAEE biosynthesis pathway by modifying the acetyl-CoA node, and β-oxidation was deleted by PEX10 knockout. Finally, the engineered strains coupled with the exogenous optimized ethanol concentration can produce an extracellular FAEE yield of 1.18 g/L via shake-flask fermentation [71] .
Fatty alcohols have applications in detergents, medicine, cosmetics, and biofuels (Beller et al., 2015). In yeast, fatty alcohol can be obtained by the reduction of a fatty aldehyde intermediate or directly synthesized by fatty acyl-CoAs that undergo reduction via the action of a bifunctional fatty acyl-CoA reductase (Willis et al., 2011). The expression of fatty acyl-CoA reductase from mouse in S. cerevisiae resulted in 47.4 mg/L of fatty alcohols (Sangwallek et al., 2013). To further improve fatty alcohol yield, a mouse fatty acid reductase MmFar1p (NADPH-dependent) with high activity was expressed in S. cerevisiae. Also, diacylglycerol acyltransferase1 DGA1, fatty aldehyde dehydrogenase HFD1, and medium-chain alcohol dehydrogenase ADH6 were deleted to redirect carbon flux toward fatty alcohols instead of toward TAG, FFA, and ethanol. Further, a mutant acetyl-CoA carboxylase was overexpressed to increase acetyl-CoA flux . The Δ9-desaturase OLE1 was overexpressed to increase membrane fluidity and access of MmFar1p to the substrate. The final strain containing 11 genetic modifications than parent BY4741 strain produced 1.2 g/L fatty alcohols in shake flasks from glucose (d’Espaux et al., 2017).
9.4.3 Ricinoleic Fatty Acids
Ricinoleic acid (RA) accounts for around 90% of the total fatty acid in castor seeds [72]. Because of its specific structure, RA can be a substrate for double bond and hydroxyl-group reactions and, therefore, an important natural raw material for the chemical industry [73]. RA and its derivatives have broad commercial applications, including food, textile, paper, plastics, perfumes, cosmetics, paints, inks and lubricants, and biofuels [74, 75]. Although RA is the major component of castor seeds, the castor plant has many serious challenges in its production. In addition, the process of extracting RA from the castor seeds is complicated [76].
To date, RA biosynthesis has been most successful in Y. lipolytica although a major challenge is that the hydroxylated ricinoleic acid is formed at the sn-2 position of phosphatidylcholine (PC) in membranes when the Δ12 hydroxylase (FAH12) from castor is expressed. As Y. lipolytica accumulates high amounts of oleic acid, the substrate for FAH12, it provides a direct precursor for RA synthesis. Bressy et al. (2014) [77] expressed the castor FAH12 in Y. lipolytica which resulted in 7% RA of the total fatty acid; however, when two copies of the Claviceps purpurea hydroxylase CpFAH12 were expressed in a modified strain, RA content increased to 35% of the total lipids. Next, they deleted six POX genes to prevent β-oxidation of fatty acids, the native Δ12-desaturase which converts oleic acid to linoleic acid and DGA1 and DGA2 which form TAG via the addition of acyl-CoA to the glycerol backbone. In the final version, the native Y. lipolytica PDAT acyltransferase (Lro1p) was overexpressed, and RA yield reached 43% of total fatty acid and over 60 mg/g of dry cell weight in small scale-cultures and up to 12 g/L and 60% of total lipids when supplemented with 24 g/L of oleic acid at 10 L bioreactor scale (Fig. 9.2).
9.4.4 Long-Chain Polyunsaturated Fatty Acids
The most common carbon chain length of yeast fatty acids is 16–18, whereas a group of valuable long-chain polyunsaturated fatty acids (LC-PUFAs) has carbon chain lengths of 20–24 and includes multiple double bonds in a methylene interrupted pattern. Two main categories of desaturation of the fatty acid carbon chain are known as omega-6 (n-6) and omega-3 (n-3), and the numbering is determined by the position of the first double bond from the methyl end group of the fatty chain [78]. Omega-6 LC-PUFA can be precursors to the eicosanoids, a group of powerful bioactive molecules that include prostaglandins and thromboxane. The omega-3 PUFAs are important human dietary fatty acids that can regulate the immune system, blood clots, neurotransmitters, and cholesterol metabolism and adjust membrane phospholipids of both the brain and the retina [79]. Although LC-PUFA can have a positive effect on health, these LC-PUFAs cannot be synthesized in the human body and so are required to be taken via the diet [80]. Currently, dietary omega-3 LC-PUFAs, especially eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA), are obtained mainly from fish oil, and due to fish stock depletion and an increasing demand, obtaining alternative sources is becoming necessary [76]. Both S. cerevisiae and Y. lipolytica have been engineered to produce LC-PUFA.
LC-PUFAs are biosynthesized in cells by a series of alternating fatty acid desaturations and carbon chain elongation. In yeast, the elongation step occurs in the acyl-CoA pool but the special desaturases introduced into the cells undertake desaturation of the phospholipid-linked fatty acids, which causes an acyl exchange bottleneck and reduces yield [81]. Also, in the final step, a double bond is introduced between carbon 5 and 6 in dihomo-γ-linolenic acid (DHGLA, 20:3ω6) and eicosatetraenoic acid (ETA 20:4ω3) (Fig. 9.3a) by a Δ5-desaturase belonging to “front-end” desaturase family. Key genes relating to the pathway of DHGLA (20:3ω6) and ETA (20:4ω3), containing acyl-CoA-dependent Δ6-desaturase from the microalga O. tauri; Δ9-desaturase, Δ12-desaturase, and Δ6-elongase from M. alpina; and ω3-desaturase from S. kluyveri, in S. cerevisiae were constructed (Fig. 9.3). The Δ6-desaturase from the microalga O. tauri can use CoA-bound substrates to avoid transferring the Δ6-desaturated fatty acid from phospholipid to acyl-CoA and directly pass on the substrate to Δ6-elongation, which could overcome the bottleneck of Δ6-elongation [82]. Finally, DHGLA (20:3ω6) and ETA (20:4ω3) were obtained in the engineered strain. Subsequently, through further engineering, EPA was synthesized. In subsequent research, the efficiency of 5-desaturase from P. tetraurelia was found to be higher than that from other organisms [83].
Compared with engineered S. cerevisiae, the yield of EPA in Y. lipolytica was much higher, and a different metabolic engineering strategy was taken [84]. Starting with linoleic acid (C18:2 n-6) which is naturally synthesized in Y. lipolytica wild-type strain, genes introduced included a Δ6-desaturase, C18/20 elongase, Δ5-desaturase, and Δ17-desaturase resulting in 3% EPA of total fatty acids. Subsequently, overexpression of a C16/18 elongase from M. alpina, introducing a Δ12 desaturase from Fusarium moniliforme, increased gene copy numbers, and promoter optimization resulted in 40% EPA of total fatty acids. However, a large amount of γ-linolenic acid (C18:3ω6) was also accumulated via this strategy, which showed that the conversion of GLA to DHGLA (C20:3ω6) was rate limiting. Therefore, Δ9 pathway was constructed by introducing Δ9-elongase, Δ8-desaturase, Δ5-desaturase, and a Δ17 desaturase sourced from a range of organisms to avoid the buildup of GLA (Fig. 9.3). In the same way, they integrated multiple copies of the genes after codon optimization with strong promoters. In order to reduce the consumption of LC-PUFA by β-oxidation, PEX10 was deleted. The final strains contained 30 copies of nine different genes, and the yield of EPA was 56.6% of the total, which can be used as a commercial product produced by metabolically engineered yeast to take the place of that derived from fish [85].
9.4.5 Cyclopropane Fatty Acids
Cyclopropane fatty acids (CFAs) are naturally occurring saturated fatty acids that possess a strained three-membered ring within the fatty acid chain. They have been found in bacteria [86, 87], some fungi [88], plants [89, 90], and parasites [91]. This fatty acid has potential high value as an equivalent compound to isostearic acid which has industrial application in the lubrication and oleochemical industries [92]. Cyclopropane fatty acids have unique characteristics such as ring opening by hydrogenation to produce methyl branched-chain fatty acid, which combines the chemical and physical properties of unsaturated fatty acid with oxidative stability of saturated fatty acids [93]. There has been recent interest and research into building microbial cell factories for the production of CFAs, including both S. cerevisiae and Y. lipolytica.
Peng et al. (2018) expressed the E. coli CFA gene in S. cerevisiae that had been engineered for higher fatty acid (FA) biosynthesis, lipid production, and sequestration. TGL3, encoding triglyceride lipase 3, the main enzyme responsible for hydrolyzing CFA from TAG, was knocked out to block CFA loss from the lipid droplet. The highest CFA yield was 12 mg/g dry cell weight (DCW) which was four-fold above the strain expressing E. coli CFA gene only and up to 68.3 mg/L in a two-stage bioprocess [30, 94]. Y. lipolytica has also been engineered for the production of CFAs. E. coli was the preferred candidate from among a range of CFA genes screened from bacteria and selected plants for expression as it provided good yield and both C17 and C19 cyclopropane products [95]. Blocking β-oxidation by knocking out PEX10 and MFE1, overexpression of DGA1, and increasing the genomic copy number of the E. coli CFA gene were successful strategies to produce cyclopropane fatty acids in Y. lipolytica [96]. A further strain was constructed by mutating regulatory protein encoded by MGA2 paired with DGA1 overexpression and CFA expression, which produced 200 mg/L of C19:0 CFA in small-scale fermentation. Moreover, more than 3 g/L of C19:0 CFA was achieved in bioreactor fermentation, which accounted for up to 32.7% of total lipids [96].
9.5 Conclusion and Outlook
With increasing interest globally toward sustainable industrial production, microbial lipids are attracting significant attention due to their energy density and versatility plus the prospect of obtaining microbial lipids with a broad range of functionalities. Microbes require carbon as a key feedstock for growth, and lignocellulose is a rich and sustainable resource that has enormous potential as a substrate for microbial lipid production [97]. Within biorefineries, microbial lipid factories can play a central role in converting renewable substrates into versatile lipid products. Regarding microbial lipid factories, yeasts such as S. cerevisiae and Y. lipolytica have been the most widely investigated. With advances in synthetic biology and metabolic engineering, more tools and approaches have become available to support and enhance the introduction of gene modifications [76]. Although high performing strains with good tolerance to stressful environments and efficient lipid conversion rates can be obtained via synthetic biology and metabolic engineering strategies, the quality and availability of the sugar feedstock remain one of the important limiting factors for microbial factories. Despite the development of successful lignocellulosic sugar production from raw materials in biorefineries, the volume is not currently sufficient to meet the growing demand for bioproduction. Therefore, further research and development for low-cost and efficient production of non-food sugar sources needs to be undertaken to ensure these supplies [98,99,100].
There are some limitations to what can be achieved in yeast biofactories through metabolic engineering and synthetic biology. Cellular metabolic burden is a long-standing problem in biotechnology which was first noticed by metabolic engineers in the 1970s and 1980s when they attempted to overexpress proteins for desired products, and they found the cell growth reduced and mutation rates increased after the overexpression of protein [101,102,103]. The metabolic burden can be caused by an imbalance of energy molecules (e.g., NAD(P)H and ATP) or redirection of carbon building blocks away from essential cellular processes, for example. A cell’s carbon and energy resource distribution have been optimized to reach equilibrium states by a natural evolution [101] and modification and manipulation of these via metabolic engineering and synthetic biology alter the natural balance. There are a number of effective strategies available and in development to address these specific issues of carbon and energy imbalance in engineered strains [104].
In short, much progress in metabolic engineering of yeast for enhanced lipid production has been made, and while natural fatty acid production levels are reportedly close to commercial realization, there is more research needed for exotic lipid production to improve productivity and purity. Furthermore, the engineering strategies and modifications that have been shown to be highly effective in laboratory strains now need to undergo development and translation to their industrial environments. The fast development of stable strains useful for the industrial environment is still challenging [7]. Therefore, further research is needed to overcome the many challenges to bring microbial lipid factories to commercial reality. With more technologies and strategies becoming integrated in the future, such as systems biology, protein engineering, and “omics” analysis, these can enrich the progress to date and help obtain the goals for producing fatty acid-derived biofuels and bioproducts in an affordable and sustainable manner.
Abbreviations
- ‘tesA:
-
Truncated E. coli thioesterase
- ACC1 :
-
Acetyl-CoA carboxylase
- ACC1** :
-
Acetyl-CoA carboxylase carrying two mutations ser659ala and ser1157ala
- ACL1,2 :
-
ATP-citrate synthase subunit 1,2
- ACS:
-
Acetyl-coA synthetase
- ADH:
-
Alcohol dehydrogenase
- ALD6 :
-
Native aldehyde dehydrogenase isoform 6
- ARE1, 2:
-
Sterol O-acyltransferase
- AtCLO1 :
-
Caleosin, lipid droplet stabilization protein from Arabidopsis thaliana
- DGAT :
-
acyl-CoA: Diacylglycerol acyltransferase
- FA:
-
Fatty acid
- FAA1, 4 :
-
Long-chain fatty acyl-CoA synthetase
- FAA2 :
-
Medium-chain fatty acyl-CoA synthetase
- FAME:
-
Fatty acid methyl ester
- FAS1 :
-
Fatty acid synthase subunit β
- FAS2 :
-
Fatty acid synthase subunit α
- FAT1 :
-
Very-long-chain fatty acid transport protein
- FFA(s):
-
Free fatty acid(s)
- GPAT:
-
Glycerol 3-phosphate acyltransferase
- GUT2 :
-
Glycerol-3-phosphate dehydrogenase gene
- LPAT:
-
Lysophosphatidate acyltransferase
- MAE:
-
Malic acid transport protein
- MFE1 :
-
Peroxisomal multifunctional enzyme
- MmACL :
-
ACL from Mus musculus
- PAP:
-
Phosphatidate phosphatase
- PEX10:
-
Peroxisome biogenesis factor 10
- POX :
-
Peroxisomal β-oxidation
- PXA1 :
-
Subunit of heterodimeric peroxisomal ABC transport complex
- RtFAS :
-
Fatty acid synthetase from R. toruloides
- SCD :
-
Acyl-CoA desaturase
- SeACS L641p :
-
Acetyl-CoA synthetase with L641P mutation, derived from Salmonella enterica
- TAG:
-
Triacylglycerol
- TGL3–5 :
-
Triacylglycerol lipase 3–5
- WT:
-
Wild type
References
Zhang, F., Rodriguez, S., & Keasling, J. D. (2011). Metabolic engineering of microbial pathways for advanced biofuels production. Current Opinion in Biotechnology, 22(6), 775–783.
Fortman, J., Chhabra, S., Mukhopadhyay, A., Chou, H., Lee, T. S., Steen, E., & Keasling, J. D. (2008). Biofuel alternatives to ethanol: Pumping the microbial well. Trends in Biotechnology, 26(7), 375–381.
BBC Publishing (2019). Oleochemical fatty acids: Global markets to 2023, report highlights.
Karmee, S. K., Linardi, D., Lee, J., & Lin, C. S. K. (2015). Conversion of lipid from food waste to biodiesel. Waste Management, 41, 169–173.
Zhou, Y. J., Kerkhoven, E. J., & Nielsen, J. (2018). Barriers and opportunities in bio-based production of hydrocarbons. Nature Energy, 3, 925.
Runguphan, W., & Keasling, J. D. (2014). Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metabolic Engineering, 21, 103–113.
Hong, K.-K., & Nielsen, J. (2012). Metabolic engineering of Saccharomyces cerevisiae: A key cell factory platform for future biorefineries. Cellular and Molecular Life Sciences, 69(16), 2671–2690.
Mussatto, S. I., Dragone, G., Guimarães, P. M., Silva, J. P. A., Carneiro, L. M., Roberto, I. C., Vicente, A., Domingues, L., & Teixeira, J. A. (2010). Technological trends, global market, and challenges of bio-ethanol production. Biotechnology Advances, 28(6), 817–830.
Abdel-Mawgoud, A. M., Markham, K. A., Palmer, C. M., Liu, N., Stephanopoulos, G., & Alper, H. S. (2018). Metabolic engineering in the host Yarrowia lipolytica. Metabolic Engineering, 50, 192–208.
Kamzolova, S. V., & Morgunov, I. G. (2017). Metabolic peculiarities of the citric acid overproduction from glucose in yeasts Yarrowia lipolytica. Bioresource Technology, 243, 433–440.
Rymowicz, W., Rywińska, A., & Marcinkiewicz, M. (2009). High-yield production of erythritol from raw glycerol in fed-batch cultures of Yarrowia lipolytica. Biotechnology Letters, 31(3), 377–380.
Janek, T., Dobrowolski, A., Biegalska, A., & Mirończuk, A. M. (2017). Characterization of erythrose reductase from Yarrowia lipolytica and its influence on erythritol synthesis. Microbial Cell Factories, 16(1), 118.
Carly, F., Vandermies, M., Telek, S., Steels, S., Thomas, S., Nicaud, J.-M., & Fickers, P. (2017). Enhancing erythritol productivity in Yarrowia lipolytica using metabolic engineering. Metabolic Engineering, 42, 19–24.
Kildegaard, K. R., Adiego-Pérez, B., Belda, D. D., Khangura, J. K., Holkenbrink, C., & Borodina, I. (2017). Engineering of Yarrowia lipolytica for production of astaxanthin. Synthetic and Systems Biotechnology, 2(4), 287–294.
Ma, T., Shi, B., Ye, Z., Li, X., Liu, M., Chen, Y., Xia, J., Nielsen, J., Deng, Z., & Liu, T. (2019). Lipid engineering combined with systematic metabolic engineering of Saccharomyces cerevisiae for high-yield production of lycopene. Metabolic Engineering, 52, 134–142.
Liu, R., Zhu, F., Lu, L., Fu, A., Lu, J., Deng, Z., & Liu, T. (2014). Metabolic engineering of fatty acyl-ACP reductase-dependent pathway to improve fatty alcohol production in Escherichia coli. Metabolic Engineering, 22, 10–21.
Kondo, A., Ishii, J., Hara, K. Y., Hasunuma, T., & Matsuda, F. (2013). Development of microbial cell factories for bio-refinery through synthetic bioengineering. Journal of Biotechnology, 163(2), 204–216.
Wei, Z., Zeng, G. M., Huang, F., Kosa, M., Sun, Q. N., Meng, X. Z., Huang, D. L., & Ragauskas, A. (2015). Microbial lipid production by oleaginous Rhodococci cultured in lignocellulosic autohydrolysates. Applied Microbiology and Biotechnology, 99(17), 7369–7377.
Slininger, P. J., Dien, B. S., Kurtzman, C. P., Moser, B. R., Bakota, E. L., Thompson, S. R., O’Bryan, P. J., Cotta, M. A., Balan, V., Jin, M., Sousa, L. d. C., & Dale, B. E. (2016). Comparative lipid production by oleaginous yeasts in hydrolyzates of lignocellulosic biomass and process strategy for high titers. Biotechnology and Bioengineering, 113(8), 1676–1690.
Verhoeven, M. D., de Valk, S. C., Daran, J. G., van Maris, A. J. A., & Pronk, J. T. (2018). Fermentation of glucose-xylose-arabinose mixtures by a synthetic consortium of single-sugar-fermenting Saccharomyces cerevisiae strains. FEMS Yeast Research, 18(8), 1–12.
Papapetridis, I., Verhoeven, M. D., Wiersma, S. J., Goudriaan, M., van Maris, A. J. A., & Pronk, J. T. (2018). Laboratory evolution for forced glucose-xylose co-consumption enables identification of mutations that improve mixed-sugar fermentation by xylose-fermenting Saccharomyces cerevisiae. FEMS Yeast Research, 18(6), foy056.
d’Espaux, L., Ghosh, A., Runguphan, W., Wehrs, M., Xu, F., Konzock, O., Dev, I., Nhan, M., Gin, J., Apel, A. R., Petzold, C. J., Singh, S., Simmons, B. A., Mukhopadhyay, A., Martin, H. G., & Keasling, J. D. (2017). Engineering high-level production of fatty alcohols by Saccharomyces cerevisiae from lignocellulosic feedstocks. Metabolic Engineering, 42, 115–125.
Li, H., & Alper, H. S. (2016). Enabling xylose utilization in Yarrowia lipolytica for lipid production. Biotechnology Journal, 11(9), 1230–1240.
Rakicka, M., Lazar, Z., Dulermo, T., Fickers, P., & Nicaud, J. M. (2015). Lipid production by the oleaginous yeast Yarrowia lipolytica using industrial by-products under different culture conditions. Biotechnology for Biofuels, 8, 104.
Ledesma-Amaro, R., Lazar, Z., Rakicka, M., Guo, Z., Fouchard, F., Coq, A. C., & Nicaud, J. M. (2016). Metabolic engineering of Yarrowia lipolytica to produce chemicals and fuels from xylose. Metabolic Engineering, 38, 115–124.
Niehus, X., Crutz-Le Coq, A. M., Sandoval, G., Nicaud, J. M., & Ledesma-Amaro, R. (2018). Engineering Yarrowia lipolytica to enhance lipid production from lignocellulosic materials. Biotechnology for Biofuels, 11, 1.
Xu, J. Y., Liu, N., Qiao, K. J., Vogg, S., & Stephanopoulos, G. (2017). Application of metabolic controls for the maximization of lipid production in semicontinuous fermentation. Proccedings of the National Academy of Sciences of the United States of America, 114(27), E5308–E5316.
Pfleger, B. F., Gossing, M., & Nielsen, J. (2015). Metabolic engineering strategies for microbial synthesis of oleochemicals. Metabolic Engineering, 29, 1–11.
Yan, Q., & Pfleger, B. F. (2019). Revisiting metabolic engineering strategies for microbial synthesis of oleochemicals. Metabolic Engineering, 58, 35–46.
Peng, H., He, L., & Haritos, V. S. (2018). Metabolic engineering of lipid pathways in Saccharomyces cerevisiae and staged bioprocess for enhanced lipid production and cellular physiology. Journal of Industrial Microbiology & Biotechnology, 45(8), 707–717.
Li, Y., Zhao, Z. K., & Bai, F. (2007). High-density cultivation of oleaginous yeast Rhodosporidium toruloides Y4 in fed-batch culture. Enzyme and Microbial Technology, 41(3), 312–317.
Shi, S., Chen, Y., Siewers, V., & Nielsen, J. (2014). Improving production of Malonyl coenzyme A-Derived metabolites by abolishing Snf1-dependent regulation of Acc1. MBio, 5(3), e01130-14.
Choi, J. W., & Da Silva, N. A. (2014). Improving polyketide and fatty acid synthesis by engineering of the yeast acetyl-CoA carboxylase. Journal of Biotechnology, 187, 56–59.
Chen, Y., Daviet, L., Schalk, M., Siewers, V., & Nielsen, J. (2013). Establishing a platform cell factory through engineering of yeast acetyl-CoA metabolism. Metabolic Engineering, 15, 48–54.
Chen, F., Zhou, J., Shi, Z., Liu, L., Du, G., & Chen, J. (2010). Effect of acetyl-CoA synthase gene overexpression on physiological function of Saccharomyces cerevisiae. Wei Sheng Wu Xue Bao = Acta Microbiologica Sinica, 50(9), 1172–1179.
Shiba, Y., Paradise, E. M., Kirby, J., Ro, D.-K., & Keasling, J. D. (2007). Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae for high-level production of isoprenoids. Metabolic Engineering, 9(2), 160–168.
Valle-Rodriguez, J. O., Shi, S. B., Siewers, V., & Nielsen, J. (2014). Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid ethyl esters, an advanced biofuel, by eliminating non-essential fatty acid utilization pathways. Applied Energy, 115, 226–232.
Chen, L., Zhang, J., & Chen, W. N. (2014). Engineering the Saccharomyces cerevisiae β-oxidation pathway to increase medium chain fatty acid production as potential biofuel. PLoS One, 9(1), e84853: 1-10.
Greer, M. S., Truksa, M., Deng, W., Lung, S. C., Chen, G. Q., & Weselake, R. J. (2015). Engineering increased triacylglycerol accumulation in Saccharomyces cerevisiae using a modified type 1 plant diacylglycerol acyltransferase. Applied Microbiology and Biotechnology, 99(5), 2243–2253.
Dahlqvist, A., Stahl, U., Lenman, M., Banas, A., Lee, M., Sandager, L., Ronne, H., & Stymne, H. (2000). Phospholipid: Diacylglycerol acyltransferase: An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proccedings of the National Academy of Sciences of the United States of America, 97(12), 6487–6492.
Peng, H., Moghaddam, L., Brinin, A., Williams, B., Mundree, S., & Haritos, V. S. (2018). Functional assessment of plant and microalgal lipid pathway genes in yeast to enhance microbial industrial oil production. Biotechnology and Applied Biochemistry, 65(2), 138–144.
Tang, X., Feng, H., & Chen, W. N. (2013). Metabolic engineering for enhanced fatty acids synthesis in Saccharomyces cerevisiae. Metabolic Engineering, 16, 95–102.
Ferreira, R., Teixeira, P. G., Gossing, M., David, F., Siewers, V., & Nielsen, J. (2018). Metabolic engineering of Saccharomyces cerevisiae for overproduction of triacylglycerols. Metabolic Engineering Communications, 6, 22–27.
Li, X., Guo, D., Cheng, Y., Zhu, F., Deng, Z., & Liu, T. (2014). Overproduction of fatty acids in engineered Saccharomyces cerevisiae. Biotechnology and Bioengineering, 111(9), 1841–1852.
Leber, C., Polson, B., Fernandez-Moya, R., & Da Silva, N. A. (2015). Overproduction and secretion of free fatty acids through disrupted neutral lipid recycle in Saccharomyces cerevisiae. Metabolic Engineering, 28, 54–62.
Yu, T., Zhou, Y. J., Huang, M., Liu, Q., Pereira, R., David, F., & Nielsen, J. (2018). Reprogramming yeast metabolism from alcoholic fermentation to lipogenesis. Cell, 174, 1549.
Tai, M., & Stephanopoulos, G. (2013). Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metabolic Engineering, 15, 1–9.
Qiao, K., Imam Abidi, S. H., Liu, H., Zhang, H., Chakraborty, S., Watson, N., Kumaran Ajikumar, P., & Stephanopoulos, G. (2015). Engineering lipid overproduction in the oleaginous yeast Yarrowia lipolytica. Metabolic Engineering, 29(0), 56–65.
Qiao, K., Wasylenko, T. M., Zhou, K., Xu, P., & Stephanopoulos, G. (2017). Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nature Biotechnology, 35(2), 173–177.
Xu, P., Qiao, K., & Stephanopoulos, G. (2017). Engineering oxidative stress defense pathways to build a robust lipid production platform in Yarrowia lipolytica. Biotechnology and Bioengineering, 114(7), 1521–1530.
Blazeck, J., Hill, A., Liu, L., Knight, R., Miller, J., Pan, A., Otoupal, P., & Alper, H. S. (2014). Harnessing Yarrowia lipolytica lipogenesis to create a platform for lipid and biofuel production. Nature Communications, 5, 3131.
Liu, L. Q., Markham, K., Blazeck, J., Zhou, N. J., Leon, D., Otoupal, P., & Alper, H. S. (2015). Surveying the lipogenesis landscape in Yarrowia lipolytica through understanding the function of a Mga2p regulatory protein mutant. Metabolic Engineering, 31, 102–111.
Liu, L., Pan, A., Spofford, C., Zhou, N., & Alper, H. S. (2015). An evolutionary metabolic engineering approach for enhancing lipogenesis in Yarrowia lipolytica. Metabolic Engineering, 29, 36–45.
Ledesma-Amaro, R., Dulermo, R., Niehus, X., & Nicaud, J.-M. (2016). Combining metabolic engineering and process optimization to improve production and secretion of fatty acids. Metabolic Engineering, 38, 38–46.
Friedlander, J., Tsakraklides, V., Kamineni, A., Greenhagen, E. H., Consiglio, A. L., MacEwen, K., Crabtree, D. V., Afshar, J., Nugent, R. L., & Hamilton, M. A. (2016). Engineering of a high lipid producing Yarrowia lipolytica strain. Biotechnology for Biofuels, 9(1), 1.
Wasylenko, T. M., Ahn, W. S., & Stephanopoulos, G. (2015). The oxidative pentose phosphate pathway is the primary source of NADPH for lipid overproduction from glucose in Yarrowia lipolytica. Metabolic Engineering, 30, 27–39.
Zhou, Y. J., Buijs, N. A., Zhu, Z., Qin, J., Siewers, V., & Nielsen, J. (2016). Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nature Communications, 7, 11709.
Xu, P., Qiao, K. J., Ahn, W. S., & Stephanopoulos, G. (2016). Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals. Proccedings of the National Academy of Sciences of the United States of America, 113(39), 10848–10853.
Peralta-Yahya, P. P., Zhang, F., Del Cardayre, S. B., & Keasling, J. D. (2012). Microbial engineering for the production of advanced biofuels. Nature, 488(7411), 320.
Beld, J., Lee, D. J., & Burkart, M. D. (2015). Fatty acid biosynthesis revisited: Structure elucidation and metabolic engineering. Molecular BioSystems, 11(1), 38–59.
Jarboe, L. R., Royce, L. A., & Liu, P. (2013). Understanding biocatalyst inhibition by carboxylic acids. Frontiers in Microbiology, 4, 272.
Lomakin, I. B., Xiong, Y., & Steitz, T. A. (2007). The crystal structure of yeast fatty acid synthase, a cellular machine with eight active sites working together. Cell, 129(2), 319–332.
Mootz, H. D., Finking, R., & Marahiel, M. A. (2001). 4′-Phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis. Journal of Biological Chemistry, 276(40), 37289–37298.
White, S. W., Zheng, J., Zhang, Y.-M., & Rock, C. O. (2005). The structural biology of type II fatty acid biosynthesis. Annual Review of Biochemistry, 74, 791–831.
Buchbinder, J. L., Witkowski, A., Smith, S., & Fletterick, R. J. (1995). Crystallization and preliminary diffraction studies of thioesterase II from rat mammary gland. Proteins: Structure, Function, and Bioinformatics, 22(1), 73–75.
Dehesh, K., Edwards, P., Hayes, T., Cranmer, A. M., & Fillatti, J. (1996). Two novel thioesterases are key determinants of the bimodal distribution of acyl chain length of Cuphea palustris seed oil. Plant Physiology, 110(1), 203–210.
Leber, C., & Da Silva, N. A. (2014). Engineering of Saccharomyces cerevisiae for the synthesis of short chain fatty acids. Biotechnology and Bioengineering, 111(2), 347–358.
Leber, C., Choi, J. W., Polson, B., & Da Silva, N. A. (2016). Disrupted short chain specific β-oxidation and improved synthase expression increase synthesis of short chain fatty acids in Saccharomyces cerevisiae. Biotechnology and Bioengineering, 113(4), 895–900.
Zhu, Z., Hu, Y., Teixeira, P. G., Pereira, R., Chen, Y., Siewers, V., & Nielsen, J. (2020). Multidimensional engineering of Saccharomyces cerevisiae for efficient synthesis of medium-chain fatty acids. Nature Catalysis, 3(1), 64–74.
Xu, P., Qiao, K., Ahn, W. S., & Stephanopoulos, G. (2016). Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals. Proceedings of the National Academy of Sciences, 113(39), 10848–10853.
Gao, Q., Cao, X., Huang, Y.-Y., Yang, J.-L., Chen, J., Wei, L.-J., & Hua, Q. (2018). Overproduction of fatty acid ethyl esters by the oleaginous yeast Yarrowia lipolytica through metabolic engineering and process optimization. ACS Synthetic Biology, 7(5), 1371–1380.
Yamamoto, K., Kinoshita, A., & Shibahara, A. (2008). Ricinoleic acid in common vegetable oils and oil seeds. Lipids, 43(5), 457–460.
Mander, L., & Liu, H.-W. (2010). Comprehensive natural products II: Chemistry and biology (Vol. 1). Boston: Elsevier.
Kılıç, M., Uzun, B. B., Pütün, E., & Pütün, A. E. (2013). Optimization of biodiesel production from castor oil using factorial design. Fuel Processing Technology, 111, 105–110.
Ogunniyi, D. S. (2006). Castor oil: A vital industrial raw material. Bioresource Technology, 97(9), 1086–1091.
Ledesma-Amaro, R., & Nicaud, J.-M. (2016). Yarrowia lipolytica as a biotechnological chassis to produce usual and unusual fatty acids. Progress in Lipid Research, 61, 40–50.
Béopoulos, A., Verbeke, J., Bordes, F., Guicherd, M., Bressy, M., Marty, A., & Nicaud, J.-M. (2014). Metabolic engineering for ricinoleic acid production in the oleaginous yeast Yarrowia lipolytica. Applied Microbiology and Biotechnology, 98(1), 251–262.
Venegas-Calerón, M., Sayanova, O., & Napier, J. A. (2010). An alternative to fish oils: Metabolic engineering of oil-seed crops to produce omega-3 long chain polyunsaturated fatty acids. Progress in Lipid Research, 49(2), 108–119.
Salas Lorenzo, I., Chisaguano Tonato, A. M., de la Garza Puentes, A., Nieto, A., Herrmann, F., Dieguez, E., Castellote, A. I., López-Sabater, M. C., Rodríguez-Palmero, M., & Campoy, C. (2019). The effect of an infant formula supplemented with AA and DHA on fatty acid levels of infants with different FADS genotypes: The COGNIS study. Nutrients, 11(3), 602.
Blondeau, N., Lipsky, R. H., Bourourou, M., Duncan, M. W., Gorelick, P. B., & Marini, A. M. (2015). Alpha-linolenic acid: An omega-3 fatty acid with neuroprotective properties—Ready for use in the stroke clinic? BioMed Research International, 2015, 519830.
Domergue, F., Abbadi, A., Ott, C., Zank, T. K., Zähringer, U., & Heinz, E. (2003). Acyl carriers used as substrates by the desaturases and elongases involved in very long-chain polyunsaturated fatty acids biosynthesis reconstituted in yeast. Journal of Biological Chemistry, 278(37), 35115–35126.
Domergue, F., Abbadi, A., Zähringer, U., Moreau, H., & Heinz, E. (2005). In vivo characterization of the first acyl-CoA Δ6-desaturase from a member of the plant kingdom, the microalga Ostreococcus tauri. Biochemical Journal, 389(2), 483–490.
Tavares, S., Grotkjær, T., Obsen, T., Haslam, R. P., Napier, J. A., & Gunnarsson, N. (2011). Metabolic engineering of Saccharomyces cerevisiae for production of eicosapentaenoic acid, using a novel Δ5-desaturase from Paramecium tetraurelia. Applied and Environmental Microbiology, 77(5), 1854–1861.
Xie, D., Jackson, E. N., & Zhu, Q. (2015). Sustainable source of omega-3 eicosapentaenoic acid from metabolically engineered Yarrowia lipolytica: From fundamental research to commercial production. Applied Microbiology and Biotechnology, 99(4), 1599–1610.
Xue, Z., Sharpe, P. L., Hong, S.-P., Yadav, N. S., Xie, D., Short, D. R., Damude, H. G., Rupert, R. A., Seip, J. E., & Wang, J. (2013). Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nature Biotechnology, 31(8), 734.
Barry, C., 3rd, Lee, R. E., Mdluli, K., Sampson, A. E., Schroeder, B. G., Slayden, R. A., & Yuan, Y. (1998). Mycolic acids: Structure, biosynthesis and physiological functions. Progress in Lipid Research, 37(2–3), 143.
Grogan, D. W., & Cronan, J. E. (1997). Cyclopropane ring formation in membrane lipids of bacteria. Microbiology and Molecular Biology Reviews, 61(4), 429–441.
Law, J. H. (1971). Biosynthesis of cyclopropane rings. Accounts of Chemical Research, 4(6), 199–203.
Bao, X., Katz, S., Pollard, M., & Ohlrogge, J. (2002). Carbocyclic fatty acids in plants: Biochemical and molecular genetic characterization of cyclopropane fatty acid synthesis of Sterculia foetida. Proceedings of the National Academy of Sciences, 99(10), 7172–7177.
Bao, X., Thelen, J. J., Bonaventure, G., & Ohlrogge, J. B. (2003). Characterization of cyclopropane fatty-acid synthase from Sterculia foetida. Journal of Biological Chemistry, 278(15), 12846–12853.
Rahman, M. D., Ziering, D. L., Mannarelli, S. J., Swartz, K. L., Huang, D. S., & Pascal, R. A., Jr. (1988). Effects of sulfur-containing analogs of stearic acid on growth and fatty acid biosynthesis in the protozoan Crithidia fasciculata. Journal of Medicinal Chemistry, 31(8), 1656–1659.
Schmid, K. M.. (1999). Cyclopropane fatty acid expression in plants. Google Patents.
Gontier, E., Thomasset, B., Wallington, E., & Wilmer, J. (2008) Plant cyclopropane fatty acid synthase genes and uses thereof. Google Patents.
Peng, H., He, L., & Haritos, V. S. (2019). Enhanced production of high-value cyclopropane fatty acid in yeast engineered for increased lipid synthesis and accumulation. Biotechnology Journal, 14(4), 1800487.
Czerwiec, Q., Idrissitaghki, A., Imatoukene, N., Nonus, M., Thomasset, B., Nicaud, J. M., & Rossignol, T. (2019). Optimization of cyclopropane fatty acids production in Yarrowia lipolytica. Yeast, 36(3), 143–151.
Markham, K. A., & Alper, H. S. (2018). Engineering Yarrowia lipolytica for the production of cyclopropanated fatty acids. Journal of Industrial Microbiology & Biotechnology, 45(10), 881–888.
Shields-Menard, S. A., Amirsadeghi, M., French, W. T., & Boopathy, R. (2018). A review on microbial lipids as a potential biofuel. Bioresource Technology, 259, 451–460.
Chandel, A. K., & Singh, O. V. (2011). Weedy lignocellulosic feedstock and microbial metabolic engineering: Advancing the generation of ‘biofuel’. Applied Microbiology and Biotechnology, 89(5), 1289–1303.
Elkins, J. G., Raman, B., & Keller, M. (2010). Engineered microbial systems for enhanced conversion of lignocellulosic biomass. Current Opinion in Biotechnology, 21(5), 657–662.
Wilson, D. B. (2011). Microbial diversity of cellulose hydrolysis. Current Opinion in Microbiology, 14(3), 259–263.
Glick, B. R. (1995). Metabolic load and heterologous gene expression. Biotechnology Advances, 13(2), 247–261.
Colletti, P. F., Goyal, Y., Varman, A. M., Feng, X., Wu, B., & Tang, Y. J. (2011). Evaluating factors that influence microbial synthesis yields by linear regression with numerical and ordinal variables. Biotechnology and Bioengineering, 108(4), 893–901.
Poust, S., Hagen, A., Katz, L., & Keasling, J. D. (2014). Narrowing the gap between the promise and reality of polyketide synthases as a synthetic biology platform. Current Opinion in Biotechnology, 30, 32–39.
Wu, G., Yan, Q., Jones, J. A., Tang, Y. J., Fong, S. S., & Koffas, M. A. (2016). Metabolic burden: Cornerstones in synthetic biology and metabolic engineering applications. Trends in Biotechnology, 34(8), 652–664.
Acknowledgments
WJ is financially supported by Monash University for Monash Graduate Scholarship (MGS) and Monash International Tuition Scholarships (MITS), and Graduate Research International Travel Award (GRITA). WJ thanks RLA for providing an opportunity to work within his group at Imperial College Centre for Synthetic Biology. RLA and HP received funding from BBSRC (BB/R01602X/1).
Authors’ Contributions
VH determined the book chapter design, WJ drafted Sects. 9.1, 9.4, and 9.5 and HP drafted Sects. 9.2 and 9.3. VH, WJ, HP, and RLA revised the manuscript, proofread, and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Jiang, W., Peng, H., Ledesma Amaro, R., Haritos, V. (2021). Metabolic Engineering of Yeast for Enhanced Natural and Exotic Fatty Acid Production. In: Liu, ZH., Ragauskas, A. (eds) Emerging Technologies for Biorefineries, Biofuels, and Value-Added Commodities. Springer, Cham. https://doi.org/10.1007/978-3-030-65584-6_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-65584-6_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-65583-9
Online ISBN: 978-3-030-65584-6
eBook Packages: EnergyEnergy (R0)