Abstract
Homeostasis is a regulatory mechanism that keeps some specific variable close to a set value as other variables fluctuate, and is of particular interest in biochemical networks. We review and investigate a reformulation of homeostasis in which the system is represented as an input-output network, with two distinguished nodes ‘input’ and ‘output’, and the dynamics of the network determines the corresponding input-output function of the system. Interpreting homeostasis as an infinitesimal notion—namely, the derivative of the input-output function is zero at an isolated point—we apply methods from singularity theory to characterise homeostasis points in the input-output function. This approach, coupled to graph-theoretic ideas from combinatorial matrix theory, provides a systematic framework for calculating homeostasis points in models, classifying different types of homeostasis in input-output networks, and describing all small perturbations of the input-output function near a homeostasis point.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
1 Introduction
Homeostasis is an important concept, occurring widely in biology, especially biochemical networks, and in many other areas including control engineering. A system exhibits homeostasis if some output variable remains constant, or almost constant, when an input variable or parameter changes by a relatively large amount. In the control theory literature, mathematical models of homeostasis are often constructed by requiring the output to be constant when the input lies in some range. That is, the derivative of the input-output function is identically zero on that interval of input values. Such models have perfect homeostasis or perfect adaptation [17, 41].
An alternative approach is introduced and studied in [22, 23, 25, 37, 42], using an ‘infinitesimal’ notion of homeostasis—namely, the derivative of the input-output function is zero at an isolated point—to introduce singularity theory into the study of homeostasis. From this point of view, perfect homeostasis is an infinite-codimension phenomenon, hence highly non-generic. It is also unlikely to occur exactly in a biological system. Nonetheless, perfect homeostasis can be a reasonable modeling assumption for many purposes.
The singularity-theoretic analysis leads to conditions that are very similar to those that occur in bifurcation theory when recognizing and unfolding bifurcations (see [20, 24]). These conditions have been used to organize the numerical computation of bifurcations in nonlinear systems, for example in conjunction with continuation methods. See for example Dellnitz [9,10,11], Dellnitz and Junge [12], Dellnitz et al. [13], Jepson and Spence [27], Jepson et al. [28], and Moore et al. [31]. It might be possible to adapt some of these methods to homeostasis. Donovan [15, 16] has used the singularity-theoretic framework to adapt such numerical methods to homeostasis. As well as organizing the numerical calculations, singularity theory and homeostasis matrix techniques may help to simplify them.
Mathematically, homeostasis can be thought of as a network concept. One variable (a network node) is held approximately constant as other variables (other nodes) vary (perhaps wildly). Network systems are distinguished from large systems by the desire to keep track of the output from each node individually. If we are permitted to mix the output from several nodes, then homeostasis is destroyed, since the sum of a constant variable with a wildly varying one is wildly variable. Placing homeostasis in the general context of network dynamics leads naturally to the methods reviewed here.
Summary of Contents
Section 2 opens the discussion with a motivational example of homeostasis: regulation of the output ‘body temperature’ in an opossum, when the input ‘environmental temperature’ varies. The graph of body temperature against environmental temperature \({\mathcal I}\) is approximately linear, with nonzero slope, when \({\mathcal I}\) is either small or large, while in between is a broad flat region, where homeostasis occurs. This general shape is called a ‘chair’ by Nijhout and Reed [34] (see also [33, 35]), and plays a central role in the singularity theory discussion. This example is used in Sect. 4 to motivate a reformulation of homeostasis in terms of the derivative of an output variable with respect to an input being zero at some point, hence approximately constant near that point. We discuss this mathematical reformulation in terms of singularities of input-output functions.
Section 5 introduces input-output networks – networks that have input and output nodes. In such networks the observable is just the value of the output node as a function of the input that is fed into the input node. This simplified form of the observable and the input-output map allows us to use Cramer’s rule to simplify the search for infinitesimal homeostasis points. See Lemma 5.2.
As it happens, many nodes and arrows in input-output networks may have no effect on the existence of homeostasis. The end result is that when looking for infinitesimal homeostasis in the original network, we may first reduce that network to a ‘core’ network. The definition of and reduction to the core are given in Sect. 6. These reductions allow us to discuss three different types of infinitesimal homeostasis in three-node input-output networks. The first is that there are only three core networks in three-node input-output networks (even though there are 78 possible input-output three-node networks) and there are three types of infinitesimal homeostasis (Haldane, null-degradation, and structural) distinguished by the mathematics. The mathematics of three-node input-output networks is presented in Sect. 8, and the relation to the biochemical networks that motivated the mathematics is given in Sect. 3.
Section 9 discusses the relationship between infinitesimal homeostasis and singularity theory—specifically elementary catastrophe theory [19, 36, 43]. The two simplest singularities are simple homeostasis and the chair. We characterize these singularities, discuss their normal forms (the simplest form into which the singularity can be transformed by suitable coordinate changes), and universal unfoldings, which classify all small perturbations as other system parameters vary. We relate the unfolding of the chair to observational data on two species of opossum and the spiny rat, Fig. 2. Section 9 also provides a brief discussion of how chair points can be calculated analytically by implicit differentiation, and considers a special case with extra structure, common in biochemical applications, where the calculations simplify.
Catastrophe theory enables us to discuss how infinitesimal homeostasis can arise in systems with an extra parameter. In Sect. 10 we see that the simplest such way for homeostasis to evolve is through a chair singularity. This observation gives a mathematical reason for why infinitesimal chairs are important and complements the biological reasons given by Nijhout, Reed, and Best [33, 35].
Until this point the paper has dealt with input-output functions having one input variable. This is the most important case; however multiple input systems are also important. We follow [23] and discuss two input systems in Sect. 11. We argue that the hyperbolic umbilic of elementary catastrophe theory plays the role of the chair in systems with two inputs.
The paper ends with a discussion of a possible singularity theory description of housekeeping genes in Sect. 12. Here we emphasize how both the homeostasis network theory and the network singularity theory intertwine. The details of this application are given in Antoneli et al. [1].
2 Thermoregulation: A Motivation for Homeostasis
Homeostasis occurs when some feature of a system remains essentially constant as an input parameter varies over some range of values. For example, in thermoregulation the body temperature of an organism remains roughly constant despite variations in its environment. (See Fig. 1 for such data in the brown opossum where body temperature remains approximately constant over a range of \(18\,^\circ \)C in environmental temperature [32, 33].) Or in a biochemical network the equilibrium concentration of some important biochemical molecule might not change much when the organism ingests food.
Homeostasis is almost exactly opposite to bifurcation. At a bifurcation, the state of the system undergoes a change so extensive that some qualitative property (such as number of equilibria, or the topological type of an attractor) changes. In homeostasis, the state concerned not only remains topologically the same: some feature of that state does not even change quantitatively. For example, if a steady state does not bifurcate as a parameter is varied, that state persists, but can change continuously with the parameter. Homeostasis is stronger: the steady state persists, and in addition some feature of that steady state remains almost constant.
Homeostasis is biologically important, because it protects organisms against changes induced by the environment, shortage of some resource, excess of some resource, the effect of ingesting food, and so on. The literature is extensive [44]. However, homeostasis is not merely the existence (and persistence as parameters vary) of a stable equilibrium of the system, for two reasons.
First, homeostasis is a stronger condition than ‘the equilibrium varies smoothly with parameters’, which just states that there is no bifurcation. In the biological context, approximately linear variation of the equilibrium with nonzero slope as parameters change is not normally considered to be homeostasis, unless the slope is very small. For example, in Fig. 1, body temperature appears to be varying linearly when the environmental temperature is either below \(10\,^\circ \)C or above \(30\,^\circ \)C and is approximately constant in between. Nijhout et al. [33] call this kind of variation (linear, constant, linear) a chair.
Second, some variable(s) of the system may be homeostatic while others undergo larger changes. Indeed, other variables may have to change dramatically to keep some specific variable roughly constant.
We assume that there is an input-output function, which we consider to be the product of a system black box. Specifically, we assume that for each input \({\mathcal I}\) there is an output \(x_o({\mathcal I})\). For opossums, \({\mathcal I}\) is the environmental temperature from which the opossum body produces an internal body temperature \(x_o({\mathcal I})\).
Nijhout et al. [33] suggest that there is a chair in the body temperature data of opossums [32]. We take a singularity-theoretic point of view and suggest that chairs are better described locally by a homogeneous cubic function (that is, like \(x_o({\mathcal I})\approx {\mathcal I}^3\)) rather than by the previous piecewise linear description. Figure 2(a) shows the least-squares fit of a cubic function to data for the brown opossum, which is a cubic with a maximum and a minimum. In contrast, the least squares fit for the eten opossum, Fig. 2(b), is monotone.
These results suggest that in ‘opossum space’ there should be a hypothetical type of opossum that exhibits a chair in the system input-output function of environmental temperature to body temperature. In singularity-theoretic terms, this higher singularity acts as an organizing center, meaning that the other types of cubic can be obtained by small perturbations of the homogeneous cubic. In fact, data for the spiny rat have a best-fit cubic very close to the homogeneous cubic, Fig. 2(c). We include this example as a motivational metaphor, since we do not consider a specific model for the regulation of opossum body temperature.
This example, especially Fig. 2, motives a formulation of homeostasis in a way that can be analyzed using singularity theory. The first step in any discussion of homeostasis must be the formulation of a model that defines, perhaps only implicitly, the input-output function \(x_o\). Our singularity theory point of view suggests defining infinitesimal homeostasis as an input \({\mathcal I}_0\) where the derivative of output \(x_o\) with respect to the input vanishes at \({\mathcal I}_0\); that is, \(x_o'({\mathcal I}_0) = 0\).
3 Biochemical Input-Output Networks
We provide context for our results by first introducing some of the biochemical models discussed by Reed in [37]. In doing so we show that input-output networks form a natural category in which homeostasis may be explored.
There are many examples of biochemical networks in the literature. In particular examples, modelers decide which substrates are important and how the various substrates interact. Figure 3 shows a network resulting from the detailed modeling of the production of extracellular dopamine (eDA) by Best et al. [3] and Nijhout et al. [33]. These authors derive a differential equation model for this biochemical network and use the results to study homeostasis of eDA with respect to variation of the enzyme tyrosine hydroxylase (TH) and the dopamine transporters (DAT).
In another direction, relatively small biochemical network models are often derived to help analyse a particular biochemical phenomenon. We present four examples; three are discussed in Reed et al. [37] and one in Ma et al. [29]. These examples belong to a class that we call biochemical input-output networks (Sect. 5) and will help to interpret the mathematical results.
3.1 Feedforward Excitation
The input-output network corresponding to feedforward excitation is in Fig. 4. This motif occurs in a biochemical network when a substrate activates the enzyme that removes a product. The standard biochemical network diagram for this process is shown in Fig. 4a. Here \(\mathbf {X}\), \(\mathbf {Y}\), \(\mathbf {Z}\) are the names of chemical substrates and their concentrations are denoted by lower case x, y, z. Each straight arrow represents a flux coming into or going away from a substrate. The differential equations for each substrate simply state that the rate of change of the concentration is the sum of the arrows going towards the substrate minus the arrows going away (conservation of mass). The curved line indicates that substrate is activating an enzyme.
Feedforward excitation: (a) Motif from [37]; (b) Input-output network with two paths from \(\iota \) to o corresponding to the motif in (a).
Both diagrams in Fig. 4 represent the same information, but in different ways. The framework employed in this paper for the mathematics focuses on the structure of the model ODEs. Figure 4b uses nodes to represent variables, and arrows to represent couplings. In other areas, conventions can differ, so it is necessary to translate between the two representations. The simplest method is to write down the model ODEs.
In this motif, one path consists of two excitatory couplings: \(g_1(x) > 0\) from \(\mathbf{X}\) to \(\mathbf{Y}\) and \(g_2(y) >0\) from \(\mathbf{Y}\) to \(\mathbf{Z}\). The other path is an excitatory coupling \(f(x) > 0\) from \(\mathbf{X}\) to the synthesis or degradation \(g_3(z)\) of \(\mathbf{Z}\) and hence is an inhibitory path from \(\mathbf{X}\) to \(\mathbf{Z}\) having a negative sign.
The equations are the first column of:
It is shown in [37] (and reproduced using this theory in [25]) that the model system (1) (left) for feedforward excitation leads to infinitesimal homeostasis at \(X_0\) if
where \(X_0 = (x_0,y_0,z_0)\) is a stable equilibrium.
Figure 4b redraws the diagram in Fig. 4a using the math network conventions of this paper, together with some extra features that are crucial to this particular application. We consider x to be a distinguished input variable, with z as a distinguished output variable, while y is an intermediate regulatory variable. Accordingly we change notation and write
The second column in (1) shows which variables occur in the components of the model ODE for each of \(x_\iota , x_\rho , x_o\). In Fig. 4b these variables are associated with three nodes \(\iota , \rho , o\). Each node has its own symbol, here a square for \(\iota \), circle for \(\rho \), and triangle for o. Here these symbols are convenient ways to show which type of variable (input, regulatory, output) the node corresponds to. Arrows indicate that the variables corresponding to the tail node occur in the component of the ODE corresponding to the head node. For example, the component for \(\dot{x}_o\) is a function of \(x_\iota \), \(x_\rho \), and \(x_o\). We therefore draw an arrow from \(\iota \) to o and an arrow from \(\rho \) to o. We do not draw an arrow from o to itself, however: by convention, every node variable can appear in the component for that node. In a sense, the node symbol (circle) represents this ‘internal’ arrow.
The mathematics described here shows that infinitesimal homeostasis occurs in the system in the second column of (1) if and only if
at the stable equilibrium \(X_0\).
Here Fig. 4b incorporates some additional information. The arrow from \({\mathcal I}\) to node \(\iota \) shows that \({\mathcal I}\) occurs in the equation for \(\dot{x}_\iota \) as a parameter. Similarly the arrow from node o to \({\mathcal O}\) shows that node o is the output node. Finally, the ± signs indicate which arrows are excitatory or inhibitory. This extra information is special to biochemical networks and does not appear as such in the general theory.
3.2 Product Inhibition
Here substrate \(\mathbf{X}\) influences \(\mathbf{Y}\), which influences \(\mathbf{Z}\), and \(\mathbf{Z}\) inhibits the flux \(g_1\) from \(\mathbf{X}\) to \(\mathbf{Y}\). The biochemical network for this process is shown in Fig. 5a.
This time the model equations for Fig. 5a are in the first column of (2)
and the input-output equations in the second column of (2) can be read directly from the first column. The input-output network in Fig. 5b then follows.
Product inhibition: (a) Motif from [37]; (b) Input-output network with one path from \(\iota \) to o corresponding to the motif in (a).
Reed et al. [37] discuss why the model equations for product inhibition also satisfy
Our general mathematical results show that the system in the second column of (2) exhibits infinitesimal homeostasis at a stable equilibrium \(X_0\) if and only if either
It follows from (3) and (4) that the model equations cannot satisfy infinitesimal homeostasis. Nevertheless, Reed et al. [37] show that these bichemical network equations do exhibit homeostasis; that is, the output z is almost constant for a broad range of input values \({\mathcal I}\).
3.3 Substrate Inhibition
The biochemical network model for substrate inhibition is given in Fig. 6a, and the associated model system is given in the first column of (5). This biochemical network and the model system are discussed in Reed et al. [37]. In particular, this paper provides justification for taking \(g_1'(x) > 0\) for all relevant x, whereas the coupling (or kinetics term) \(g_2'(y)\) can change sign.
Substrate inhibition: (a) Motif from [37]; (b) Input-output network corresponding to the motif in (a).
The equations are the first column of:
That model system of ODEs is easily translated to the input-output system in the second column of (5). Our theory shows that the equations for infinitesimal homeostasis are identical to those given in (4) for product inhibition. Given the assumption on \(g_1'\) infinitesimal homeostasis is possible only if the coupling is neutral (that is, if \(f_{o,x_\rho } = g_2' = 0\) at the equilibrium point). This observation agrees with the observation in [37] that \(\mathbf{Z}\) can exhibit infinitesimal homeostasis in the substrate inhibition motif if the infinitesimal homeostasis is built into the kinetics term \(g_2\) between \(\mathbf{Y}\) and \(\mathbf{Z}\).
Reed et al. [37] note that neutral coupling can arise from substrate inhibition of enzymes, enzymes that are inhibited by their own substrates. See the discussion in [38]. This inhibition leads to reaction velocity curves that rise to a maximum (the coupling is excitatory) and then descend (the coupling is inhibitory) as the substrate concentration increases. Infinitesimal homeostasis with neutral couplings arising from substrate inhibition often has important biological functions and has been estimated to occur in about \(20\%\) of enzymes [38].
3.4 Negative Feedback Loop
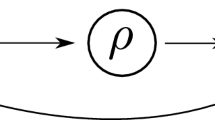
The input-output network in Fig. 7b corresponding to the negative feedback loop motif in Fig. 7a has only one simple path \(\iota \rightarrow o\). Our results imply that infinitesimal homeostasis is possible in the negative feedback loop if and only if the coupling \(\iota \rightarrow o\) is neutral (Haldane) or the linearized internal dynamics of the regulatory node \(\rho \) is zero (null-degradation).
Negative feedback loop: (a) Motif adapted from [29]. Unlike the arrows in Figs. 4, 6 and 5 that represent mass transfer between substrates, positive or negative arrows between enzymes in this negative feedback motif indicate the activation or inactivation of an enzyme by a different enzyme. (b) Input-output network corresponding to the motif in (a).
The equations are:
where \(k_{{\mathcal I}x}, K_{{\mathcal I}x}, F_x, k'_{F_x}, K'_{F_x}, k_{zy}, F_y, k'_{F_y}, k_{xz}, K_{xz}, k'_{yz}, K'_{yz}\) are 12 constants.
Each enzyme \(\mathbf{X}, Y, Z\) in the feedback loop motif (Fig. 7a) can have active and inactive forms. In the kinetic equations (6, left) the coupling from \(\mathbf{X}\) to \(\mathbf{Z}\) is non-neutral according to [29]. Hence, in this model only null-degradation homeostasis is possible. In addition, in the model the \(\dot{y}\) equation does not depend on y and homeostasis can only be perfect homeostasis. However, this model is a simplification based on saturation in y [29]. In the original system \(\dot{y}\) does depend on y and we expect standard null-degradation homeostasis to be possible in that system.
Stability of the equilibrium in this motif implies negative feedback. The Jacobian of (6, right) is
At null-degradation homeostasis (\(f_{\rho , x_\rho } = 0\)) it follows from linear stability that
Conditions (7) imply that both the input node and the output node need to degrade and the couplings \(\rho \rightarrow o\) and \(o\rightarrow \rho \) must have opposite signs. This observation agrees with [29] that homeostasis is possible in the network motif Fig. 7a if there is a negative loop between \(\mathbf{Y}\) and \(\mathbf{Z}\) and when the linearized internal dynamics of \(\mathbf{Y}\) is zero.
Another biochemical example of null-degradation homeostasis can be found in [17, Fig. 2].
4 Infinitesimal Homeostasis
In applications, homeostasis is often a property of an observable on a many-variable system of ODEs. Specifically, consider a system of ODEs
in a vector of variables \(X= (x_1, \ldots , x_m) \in {\mathbf{R}}^m\) that depends on an input parameter \({\mathcal I}\in {\mathbf{R}}\). Although not always valid in applications we assume that F is infinitely differentiable. Suppose that (8) has a linearly stable equilibrium at \((X_0,{\mathcal I}_0)\). By the implicit function theorem there exists a family of linearly stable equilibria \(X({\mathcal I})=(x_1({\mathcal I}),\ldots ,x_m({\mathcal I}))\) near \({\mathcal I}_0\) such that \(X({\mathcal I}_0)=X_0\) and
By assumption, we are interested in homeostasis of a chosen observable \(\varphi :{\mathbf{R}}^m\rightarrow {\mathbf{R}}\). The input-output function is
This system exhibits homeostasis if the input-output function \(x_o({\mathcal I})\) remains roughly constant as \({\mathcal I}\) is varied.
Often times the observable is just one coordinate of the ODE system; that is, \(\varphi (X) = x_j\), which we denote as the output variable \(x_o.\) This formulation of homeostasis is often a network formulation. The output variable is just a choice of output node and the input parameter can be assumed to affect only one node—the input node \(x_\iota \).
We now introduce a formal mathematical definition of infinitesimal homeostasis, one which opens up a potential singularity-theoretic approach that we discuss later.
Definition 4.1
The equilibrium \(X_0\) is infinitesimally homeostatic at \({\mathcal I}_0\) if
where \('\) indicates differentiation with respect to \({\mathcal I}\).
By Taylor’s theorem, infinitesimal homeostasis implies homeostasis, but the converse need not be true. See [37] and the discussion of product inhibition in Sect. 3.
5 Input-Output Networks
We now apply the notion of infinitesimal homeostasis to input-output networks—a natural formulation in biochemical networks that we discussed in detail in Sect. 3. We assume that one node \(\iota \) is the input node, a second node o is the output node, and the remaining nodes \(\rho = (\rho _1,\ldots , \rho _n)\) are the regulatory nodes. Our discussion of network infinitesimal homeostasis follows [25]. Input-output networks equations have the form \(F = (f_\iota , f_\rho , f_o)\) where each coordinate function \(f_\ell \) depends on the state variables of the nodes coupled to node \(\ell \) in the network graph. We assume that only the input node coordinate function \(f_\iota \) depends on the external input variable \({\mathcal I}\).
As shown in [25] there are 13 distinct three-node fully inhomogeneous networks and six choices of input and output nodes for each network. Thus, in principle, there are 78 possible ways to find homeostasis in three-node input-output networks. The number of input-output four-node networks increases dramatically: there are 199 fully inhomogeneous networks and more than 2000 four-node input-output networks.
Further motivated by biochemical networks, we assume:
-
(a)
The state space for each node is 1-dimensional and hence the state space for an input-output network system of differential equations is \({\mathbf{R}}^{n+2}\).
-
(b)
The coordinate functions \(f_\ell \) are usually distinct functions, so the network is assumed to be fully inhomogeneous.
-
(c)
Generically
$$\begin{aligned} f_{\iota , {\mathcal I}} \ne 0 \end{aligned}$$(11)is valid everywhere, where the notation \(f_{\ell , y}\) denotes the partial derivative of the coordinate function \(f_\ell \) with respect to y.
Cramer’s Rule and Infinitesimal Homeostasis
The equilibria of an input-output system satisfy the system
The assumption of a stable equilibrium \(X_0\) at \(I_0\) implies that the Jacobian
has eigenvalues with negative real part at \((X_0,{\mathcal I}_0)\), so J is invertible.
To state the next result we first need:
Definition 5.1
The homeostasis matrix is:
Lemma 5.2
The input-output function for the input-output network (12) satisfies
Infinitesimal homeostasis occurs at a stable equilibrium \(X_0 = X({\mathcal I}_0)\) if and only if
Proof
Implicit differentiation of (12) with respect to \({\mathcal I}\) yields the matrix system
Cramer’s rule implies that
Since \(f_{\iota , {\mathcal I}} \ne 0\) by genericity assumption (11), \(X_0\) is a point of infinitesimal homeostasis if and only if \(x_o' = 0\), if and only if (15), as claimed. \(\square \)
6 Core Networks
The results in Sects. 6, 7 and 8 will appear in Wang et al. [42].
Definition 6.1
A node \(\rho \) is downstream from a node \(\tau \) if there is a path from \(\tau \) to \(\rho \) and upstream if there is a path from \(\rho \) to \(\tau \). An input-output network is a core network if every node is downstream from \(\iota \) and upstream from o.
A core network \({\mathcal G}_c\) can be associated to any given input-output network \({\mathcal G}\) as follows. The nodes in \({\mathcal G}_c\) are the nodes in \({\mathcal G}\) that lie on a path from \(\iota \) to o. The arrows in \({\mathcal G}_c\) are the arrows in \({\mathcal G}\) that connect nodes in \({\mathcal G}_c\).
Reduction to the Core
In this section we discuss why every network that exhibits infinitesimal homeostasis can be reduced to a core network in such a way that the core has essentially the same input-output function as the original network. This reduction is performed in two stages.
-
(a)
Homeostasis implies that the output node o is downstream from the input node \(\iota \).
-
(b)
Nodes that are not upstream from the output node, and nodes that are not downstream from the input node, may be deleted.
We show that if infinitesimal homeostasis occurs in the original network, then that infinitesimal homeostasis can be computed in the smaller core network.
Lemma 6.2
In an input-output network, the existence of (generic) infinitesimal homeostasis implies that the output node o is downstream from the input node \(\iota \).
Heuristically, if the input node is not upstream from the output node, then changes in the input node cannot affect the dynamics of the output node. So the input-output map must satistfy \(x_o'({\mathcal I}) \equiv 0\) and the set value \(x_o({\mathcal I})\) is constant (and not generic).
We assume that there is a path from the input node to the output node and show that nodes that are not upstream from o and nodes that are not downstream from \(\iota \) can be deleted without changing the existence of homeostasis.
Proposition 6.3
Let \({\mathcal G}\) be a connected input-output network where there is a path from the input node \(\iota \) to the output node o. Divide the regulatory nodes \(\rho \) into three classes \(\rho = (u,\sigma ,d)\), where
-
nodes in u are not upstream from o,
-
nodes in d are not downstream from \(\iota \), and
-
regulatory nodes \(\sigma \) are both upstream from o and downstream from \(\iota \).
Then all nodes u, d and all arrows into nodes in u and out of nodes in d can be deleted to form a core network \({\mathcal G}_c\) without affecting the existence of infinitesimal homeostasis.
Again, heuristically the proof is straightforward. If a node is not upstream from the output node, than its value cannot affect the output node and if a node is not downstream from the input node than its value cannot be affected by the value of the input node. So deleting these nodes should not affect the input-output map.
Core Equivalence
Definition 6.4
Two core networks are core equivalent if the determinants of their homeostasis matrices are identical.
The general result concerning core equivalence is given in Theorem 7.2. Here we give an example of arrows that do not affect the homeostasis matrix and therefore the input-output function.
Definition 6.5
A backward arrow is an arrow whose head is the input node \(\iota \) or whose tail is the output node o.
Proposition 6.6
If two core networks differ from each other by the presence or absence of backward arrows, then the core networks are core equivalent.
Proof
Backward arrows are not present in the homeostasis matrix (14). \(\square \)
Therefore, backward arrows can be ignored when computing infinitesimal homeostasis from the homeostasis matrix \(H\). However, backward arrows cannot be completely ignored, since they can be involved in the existence of both equilibria of (12) and their stability.
7 Types of Infinitesimal Homeostasis
Infinitesimal homeostasis is found in an input-output network \({\mathcal G}\) by simultaneously solving two equations: Find a stable equilibrium of an admissible system \(\dot{X} = F(X,{\mathcal I})\) and find a zero of the determinant of the homeostasis matrix \(H\). In this section, we discuss the different types of zeros \(\det (H)\) can have and (for the most part) ignore the question of finding an equilibrium and its stability.
The homeostasis matrix \(H\) of an admissible system has three types of entries: linearized coupling strengths \(f_{k,x_\ell }\) where node \(\ell \) is connected to node k, linearized internal dynamics \(f_{k,x_k}\) of node k, and 0. We emphasize that the entries that are forced to be 0 depend specifically on network architecture.
Assume that the input-output network has \(n+2\) nodes: the input \(\iota \), the output o, and the n regulatory nodes \(\rho = (\rho _1,\ldots ,\rho _n)\). It follows that \(\det (H)\) is a homogeneous polynomial of degree \(n+1\) in the variables \(f_{k,x_\ell }\). It is discussed in [42], based on Frobenius-König theory (see [40] for a historical account), that the homeostasis matrix \(H\) can be put in block upper triangular form. Specifically, there exist two constant \((n+1)\times (n+1)\) permutation matrices P and Q such that
where the square matrices \(H_1,\ldots ,H_m\) are unique up to permutation, that is, individually the blocks cannot be brought into the form (16) by permutation of their rows and columns.
Moreover, when \(\det (H)\) is viewed as a homogeneous polynomial in the entries of the matrix \(H\) there is a factorization
into irreducible homogeneous polynomials \(\det (H_1),\ldots ,\det (H_m)\). That is, the irreducible blocks of the decomposition (16) correspond to the irreducible components in the factorization (17) (this follows from Theorem 4.2.6 (pp. 114–115) and Theorem 9.2.4 (p. 296) of [5]). We note that the main nontrivial result that allows us to write Eq. (17)—proved in [5, Theorem 9.2.4 (p. 296)]—is that \(\det (H_j)\) is irreducible as a polynomial if and only if the matrix \(H_j\) is irreducible in the sense that \(H_j\) cannot be brought to the form (16) by permutation of \(H_j\)’s rows and columns.
Low Degree Irreducible Factors of det( H )
Wang et al. [42] show that there can be two types of degree 1 factors (Haldane and null-degradation) and two types of degree 2 factors (structural and appendage). The principal result in [42] is the assertion that these four irreducible factors of \(\det (H)\) can be associated with topological characteristics of the network \({\mathcal G}\) that in turn defines a type of homeostasis. The connection between the form of a factor \(\det (H_j)\) and the topology of the network is given by certain determinant formulas that are reminiscent of the connection between a directed graph and its adjacency matrix and has been rediscovered by many authors [7, 8, 14, 26] (see [6] for a modern account). Before stating the classification we introduce some graph theoretic terminology.
Definition 7.1
Let \({\mathcal G}\) be an input-output network.
-
(a)
A directed path between two nodes is called a simple path if it visits each node on the path at most once. An \(\iota o\)-simple path is a simple path connecting the input node \(\iota \) to the output node o.
-
(b)
A node in an input-output network \({\mathcal G}\) is simple if the node is on an \(\iota o\)-simple path and appendage if the node is not simple.
-
(c)
The appendage subnetwork \({\mathcal A}_{\mathcal G}\) of \({\mathcal G}\) is the subnetwork consisting of appendage nodes and arrows in \({\mathcal G}\) that connect appendage nodes.
-
(d)
The complementary subnetwork corresponding to an \(\iota o\)-simple path S is the network \({\mathcal C}_S\) consisting of all nodes not in S and all arrows in \({\mathcal G}\) between nodes in \({\mathcal C}_S\).
Given these definitions we can state necessary and sufficient conditions for core equivalence:
Theorem 7.2
Two core networks are core equivalent if and only if they have the same set of \(\iota o\)-simple paths and the Jacobian matrices of the complementary subnetworks to any simple path have the same determinant up to sign.
We isolate four types of homeostasis.
-
(A)
Haldane homeostasis is associated with the arrow \(\ell \rightarrow k\), where \(k\ne \ell \), if homeostasis is caused by the vanishing of the degree 1 irreducible factor \(f_{k,x_\ell }\) of \(\det (H)\).
Theorem 7.3. Haldane homeostasis associated with an arrow \(\ell \rightarrow k\) can occur if and only if the arrow \(\ell \rightarrow k\) is contained in every \(\iota o\) -simple path.
-
(B)
Null-degradation homeostasis is associated with a node \(\tau \) if homeostasis is caused by the vanishing of the degree 1 irreducible factor \(f_{\tau ,x_\tau }\) of \(\det (H)\).
Theorem 7.4. Null-degradation homeostasis associated with a node \(\tau \) can occur if and only if for every \(\iota o\) -simple path S
-
(a)
\(\tau \) belongs to the complementary subnetwork \({\mathcal C}_S\) and
-
(b)
\(\tau \) is not contained in a cycle of \({\mathcal C}_S\).
-
(a)
-
(C)
Structural homeostasis of degree 2 is caused by the vanishing of a degree 2 irreducible factor of \(\det (H)\) that has the form
$$ f_{\rho _2,x_{\rho _1}} f_{\rho _3,x_{\rho _2}} - f_{\rho _3,x_{\rho _1}}f_{\rho _2,x_{\rho _2}} $$that is, the determinant of the homeostasis matrix of a feedforward loop motif defined by two \(\iota o\)-simple path snippets: one snippet is \(\rho _1\rightarrow \rho _2\rightarrow \rho _3\) and the other snippet is \(\rho _1\rightarrow \rho _3\). A snippet of a path is a connected subpath.
Theorem 7.5. Structural homeostasis of degree 2 can occur if and only if
-
(a)
two \(\iota o\)-simple path snippets form a feedforward loop motif and
-
(b)
all \(\iota o\)-simple paths contain one of the two snippets of the feedforward loop motif.
Structural homeostasis of degree 2 is exactly the structural homeostasis considered in [25] for 3-node core networks; it often arises in biochemical networks associated with the mechanism of feedforward excitation.
-
(a)
-
(D)
Appendage homeostasis of degree 2 is caused by the vanishing of a degree 2 irreducible factor of \(\det (H)\) that has the form
$$ f_{\tau _1,x_{\tau _1}}f_{\tau _2,x_{\tau _2}} - f_{\tau _2,x_{\tau _1}} f_{\tau _1,x_{\tau _2}} $$where the two node cycle \({\mathcal A}=\{\tau _1\smash {{}^\leftarrow _\rightarrow }\tau _2\}\) consists of appendage nodes.
Theorem 7.6. Appendage homeostasis of degree 2 associated with a two-node cycle \({\mathcal A}\subset {\mathcal A}_{\mathcal G}\) can occur if and only if for every \(\iota o\)-simple path S
-
(a)
\({\mathcal A}\) belongs to the complementary subnetwork \({\mathcal C}_S\) and
-
(b)
nodes in \({\mathcal A}\) do not form a cycle with other nodes in \({\mathcal C}_S\).
-
(a)
The four types of infinitesimal homeostasis (A)–(D) correspond to the only possible factors of degree \(\leqslant 2\). More precisely:
Theorem 7.7
Any factor of degree 1 is of type (A) or (B) and any irreducible factor of degree 2 is of type (C) or (D).
Homeostasis can also occur in blocks of degree 3 or higher. There are three types of such blocks: structural (all couplings are between simple nodes), appendage (all couplings are between appendage nodes), and mixed (both simple and appendage nodes appear in the block). Theorem 7.6 generalizes to higher degree appendage homeostasis. Specifically:
Theorem 7.8
Let \({\mathcal G}\) be a network with appendage subnetwork \({\mathcal A}\subset {\mathcal A}_{\mathcal G}\). Appendage homeostasis associated with \({\mathcal A}\) can occur if and only if for every \(\iota o\)-simple path S
-
(a)
\({\mathcal A}\) belongs to the complementary subnetwork \({\mathcal C}_S\) and
-
(b)
nodes in \({\mathcal A}\) do not form a cycle with other nodes in \({\mathcal C}_S\).
8 Low Degree Homeostasis Types
The homeostasis matrix \(H\) of a three-node input-output network is a \(2\,\times \, 2\) matrix. It follows that a homeostasis block is either \(1\times 1\) or \(2\times 2\). If the block is \(2\times 2\), it must be structural. For if it were appendage, the network would need to have two appendage nodes and one simple node. If the network had only one simple node, then the input node and the output node would be identical and that is not permitted.
Examples of Haldane, Structural of Degree 2, and Null Degradation
The admissible systems of differential equations for the three-node networks in Fig. 8 are:

The determinants of the \(2\times 2\) homeostasis matrices are:
A vanishing determinant in (19)(a) leads to two possible instances of Haldane homeostasis. A vanishing determinant in (19)(b) leads to balancing of two simple paths and structural homeostasis. Finally, a vanishing determinant in (19)(c) leads to null-degradation or Haldane homeostasis. These types of homeostasis were classified in [25] where it was also noted that Haldane occurs in product inhibition, structural occurs in feedforward excitation, and null-degradation occurs in a negative feedback loop.
Appendage Homeostasis of Degree 2
The admissible systems of differential equations for the four-node network in Fig. 9 have the form:
The homeostasis matrix is
and
It follows that \(\det (H) = 0\) can lead either to Haldane homeostasis or appendage homeostasis of degree 2.
9 Singularity Theory of Input-Output Functions
As discussed in Sect. 2, Nijhout et al. [33, 35] observe that homeostasis appears in many applications through the notion of a chair. Golubitsky and Stewart [23] observed that a chair can be thought of as a singularity of the input-output function, one where \(x_o({\mathcal I})\) ‘looks like’ a homogeneous cubic \(x_o({\mathcal I}) \approx {\mathcal I}^3\). More precisely, the mathematics of singularity theory [19, 36] replaces ‘looks like’ by ‘up to a change of coordinates.’
Definition 9.1
Two functions \(p,q:{\mathbf{R}}\rightarrow {\mathbf{R}}\) are right equivalent on a neighborhood of \({\mathcal I}_0\in {\mathbf{R}}\) if
where \(\Lambda :{\mathbf{R}}\rightarrow {\mathbf{R}}\) is an invertible change of coordinates on a neighborhood of \({\mathcal I}_0\) and \(K\in {\mathbf{R}}\) is a constant.
The simplest singularity theory theorem states that \(q:{\mathbf{R}}\rightarrow {\mathbf{R}}\) is right equivalent to \(p({\mathcal I}) = {\mathcal I}^3\) on a neighborhood of the origin if and only if \( q'({\mathcal I}_0) = q''({\mathcal I}_0) = 0\) and \(q'''({\mathcal I}_0)\ne 0\). Hence we call a point \({\mathcal I}_0\) an infinitesimal chair for an input-output function \(x_o\) if
A simple result is:
Lemma 9.2
An input-output map \(x_o\) has an infinitesimal chair at \({\mathcal I}_0\) if and only if
where \(h({\mathcal I}) = \det (H)\).
Proof
Suppose that \(x'_o({\mathcal I}) = k({\mathcal I}) h({\mathcal I})\) where \(k({\mathcal I})\) is nowhere zero. Then \(h({\mathcal I}_0) = h'({\mathcal I}_0) = 0\) if and only if \(x'_0({\mathcal I}_0) = x_0''({\mathcal I}_0) = 0\) because \(x_o'' = k'h + kh'\). Moreover, if \(h = h' = 0\), then \(x_o'' = k h''\). Finally, it follows from the Cramer’s rule calculation in Lemma 5.2 that
Hence, \(k({\mathcal I})\) is nowhere zero. \(\square \)
A simpler result states the following. The input-output function defines simple infinitesimal homeostasis if
which is equivalent to \(h = 0\) and \(h'\ne 0\). The graph of \(x_o\) ‘looks like’ a parabola near a point of simple infinitesimal homeostasis.
9.1 Chair Points for Blocks of Degree 1 and 2
Lemma 9.2 gives necessary and sufficient conditions for the existence of infinitesimal homeostasis using the function \(h = \det (H)\). In general, the homeostasis function can be simplified by recalling from (16) that the homeostasis matrix \(PHQ\) is block upper triangular. It follows that if homeostasis stems from block j, then \(\det (H)\) is a nonzero multiple of \(\det (H_j)\). The results in Sect. 7 imply
Theorem 9.3
Given an input-output network. Then, the defining conditions for infinitesimal chair homeostasis are given by \(h_j = h_j' = 0\) where \(h_j\) is defined by (21).
We now calculate chair equations for the two degree 1 three-node examples.
Lemma 9.4
-
(a)
If the arrow \(\rho \rightarrow o\) has Haldane homeostasis in the network \(\iota \rightarrow \rho \rightarrow o\), then
$$ h = h' = 0 \quad \Longleftrightarrow \quad f_{o,x_\rho } = f_{o,x_\rho x_\rho } = 0 $$ -
(b)
If the node \(\tau \) has null-degradation homeostasis in the network \(\iota \rightarrow o\), \(o\rightarrow \tau \), \(\tau \rightarrow \iota \), then
$$ h = h' = 0 \quad \Longleftrightarrow \quad f_{\tau ,x_\tau } = f_{\tau ,x_\tau x_\tau } = 0 $$
Proof
Suppose \(h({\mathcal I}) = h_j({\mathcal I}) k({\mathcal I})\), where \(k({\mathcal I}_0)\) is nonzero at \(I_0\), then \(h({\mathcal I}_0) = h'({\mathcal I}_0) = 0\) if and only if \(h_j({\mathcal I}_0) = h_j'({\mathcal I}_0) = 0\). The proof proceeds in two parts.
-
(a)
Observe that
$$ h_j = f_{o,x_\rho }(x_\rho ,x_o)=0 $$is one equation for a Haldane chair and the second equation is
$$ h_j' = f_{o,x_\rho x_\rho }x_\rho ' + f_{o,x_\rho x_o}x_o' = 0 $$Since \(h_j'\) is evaluated at a point of homeostasis, \(x_o' = 0\). It follows that either \(f_{o,x_\rho x_\rho } = 0\) or \(x_\rho ' = 0\). We can use Cramer’s rule to solve for \(x_\rho '\); it is a nonzero multiple of \(f_{\rho ,x_\iota } f_{o,x_o}\). If \(f_{\rho ,x_\iota } = 0\), then we would have a second Handane in the \(\iota \rightarrow \rho \) arrow - a codimension 2 homeostasis. So, generically, we can assume \(f_{\rho ,x_\iota }\ne 0\). By computing the Jacobian at the assumed Haldane point we see that \(f_{o,x_o}\) is an eigenvalue and therefore negative by the assumed stabilty.
-
(b)
We use the admissible system equilibrium equations from (18) (c) to see that null-degradation is defined by \(h_j = f_{\tau ,x_\tau }(x_\tau ,x_o) = 0\) and a chair by
$$ h_j' = f_{\tau ,x_\tau x_\tau }x_\tau ' + f_{\tau ,x_\tau x_o}x_o' = 0 $$Since \(x_o'=0\) and \(x_\tau '\ne 0\) at the generic homeostasis point, it follows that \( f_{\tau ,x_\tau x_\tau } = 0\) is the chair equation, as claimed. \(\square \)
9.2 Elementary Catastrophe Theory and Homeostasis
The transformations of the input-output map \(x_o({\mathcal I})\) given in Definition 9.1 are just the standard change of coordinates in elementary catastrophe theory [19, 36, 43]. We can therefore use standard results from elementary catastrophe theory to find normal forms and universal unfoldings of \(x_o({\mathcal I})\), as we now explain.
Because \(x_o({\mathcal I})\) is 1-dimensional, we consider singularity types near the origin of a 1-variable function \(g({\mathcal I})\). Such singularities are determined by the first nonvanishing \({\mathcal I}\)-derivative \(g^{(k)}(0)\) (unless all derivatives vanish, which is an ‘infinite codimension’ phenomenon that we do not discuss further). Informally, the codimension of a singularity is the number of conditions on derivatives that determine it. This is also the minimum number of extra variables required to specify all small perturbations of the singularity, up to changes of coordinates. These perturbations can be organized into a family of maps called the universal unfolding, which has that number of extra variables.
Definition 9.5
\(G({\mathcal I}, a)\) is an unfolding of \(g({\mathcal I})\) if \(G({\mathcal I},0)=g({\mathcal I})\). G is a universal unfolding of g if every unfolding of \(H({\mathcal I},b)\) factors through G. That is,
It follows that every small perturbation \(H(\cdot , b)\) is equivalent to a perturbation \(G(\cdot ,A(b))\) of g in the G family.
If such k exists, the normal form is \(\pm {\mathcal I}^k\). Simple infintesimal homeostasis occurs when \(k = 2\), and an infinitesimal chair when \(k=3\). When \(k \ge 3\) the universal unfolding for catastrophe theory equivalence is
for parameters \(a_j\) and when \(k = 2\) the universal unfolding is \(\pm {\mathcal I}^2\). The codimension in this setting is therefore \(k\,-\,2\). See [4] Example 14.9 and Theorem 15.1; [18] chapter IV (4.6) and chapter VI (6.3); and [30] chapter XI Sect. 1.1 and chapter XII Sects. 3.1, 7.2.
To summarize: the normal form of the input-output function for simple infinitesimal homeostasis is
and no unfolding parameter is required. Similarly,
is the normal form of the input-output function for a chair, and
is a universal unfolding.
10 Evolving Towards Homeostasis
Control-theoretic models of homeostasis often build in an explicit ‘target’ value for the output, and construct the equations to ensure that the input-output function is exactly flat over some interval. Such models are common, and provide useful information for many purposes. In singularity theory an exactly flat input-output function has ‘infinite codimension’, so our approach is not appropriate for models of this type.
However, in biology, homeostasis is an emergent property of biochemical networks, not a preset target value, and the input-output function is only approximately flat, for example as in Fig. 2 (left). Many of the more recent models of homeostasis do not assume a preset target value; instead, this emerges from the dynamics of a biochemical network. Here we expect typical singularities to have finite codimension, and our approach is then potentially useful. For example, in [21, Section 8] we proved that for one such model, of feedforward inhibition [33, 39], the input-output map has a ‘chair’ singularity, with normal form \(x^3+\lambda x\). Other examples of chair singularities are given in [37].
A key question is: In a mathematical sense, how does a biological system evolve towards homeostasis? Imagine a system of differential equations depending on parameters. Suppose that initially the parameters are set so that the associated input-output function has no regions of homeostasis. Now vary the parameters so that a small region of homeostasis appears in the input-output function. Since this region of homeostasis is small, we can assume that it is spawned by a singularity associated with infinitesimal homeostasis. How can that happen?
Singularities Organizing Evolution Towards Homeostasis
A plausible answer follows from the classification of elementary catastrophes. If there is one input and one output, the assumption of no initial homeostasis implies that the input-output function \(x_o:{\mathbf{R}}\rightarrow {\mathbf{R}}\) is strictly increasing (or strictly decreasing). Generically, evolving towards infinitesimal homeostasis can occur in only one way. As a parameter \(\beta \) is varied, at some point \({\mathcal I}_0\) the function \(x_o({\mathcal I})\) approaches a singularity, so there is a point \({\mathcal I}_0\) where \(x_o'({\mathcal I}_0)=0\). This process can happen only if \(x_o''({\mathcal I}_0)=0\) is also satisfied. That is, from a singularity-theoretic point of view, the simplest way that homeostasis can evolve is through an infinitesimal chair.
This process can be explained in the following way. The system can evolve towards infinitesimal homeostasis only if the universal unfolding of the singularity has a parameter region where the associated function is nonsingular. For example, simple homeostasis (\(x_o({\mathcal I})={\mathcal I}^2\), which is structurally stable) does not have this property. All small perturbations of \({\mathcal I}^2\) have a Morse singularity. The simplest (lowest codimension) singularity that has nonsingular perturbations is the fold singularity \(x_o({\mathcal I}) = {\mathcal I}^3\); that is, the infinitesimal chair.
At least two assumptions underlie this discussion. First, we have assumed that all perturbations of the input-output function can be realized by perturbations in the system of ODEs. This is true; see Lemma 10.1. Second, we assume that when evolving towards homeostasis the small region of homeostasis that forms is one that could have grown from a point of infinitesimal homeostasis.
When \(x_o\) depends on one parameter, generically the infinitesimal chair is the only possible singularity that can underlie the formation of homeostasis.
Lemma 10.1
Given a system of ODEs \(\dot{x}=F(x,{\mathcal I})\) whose zero set is defined by
and a perturbation \(\tilde{X}({\mathcal I}) = X({\mathcal I}) + P({\mathcal I})\) of that zero set. Then \(\tilde{X}\) is the zero set of the perturbation
Therefore any perturbation of the input-output function \(x_o({\mathcal I})\) can be realized by perturbation of F.
Proof
Clearly
If we write \(P({\mathcal I})=(0,P_o({\mathcal I}))\) where \(P_o({\mathcal I})\) is a small perturbation of \(x_o({\mathcal I})\), then we can obtain the perturbation \(x_o+P_o\) of \(x_o\) by the associated perturbation of F. \(\square \)
Theorem 10.2
Consider input-output functions with one input and one output. Then the only singularities of codimension \(\le 3\) that have perturbations with no infinitesimal homeostasis are the fold (chair) and the swallowtail.
Proof
It is easy to see that perturbations of \({\mathcal I}^k\) always have a local minimum when k is even. So the only normal forms with perturbations that have no infinitesimal homeostasis occur when k is odd. Those that have codimension at most 3 are the fold (\(k=3\)) and the swallowtail (\(k=5\)). \(\square \)
We remark that folds occur in the unfoldings of swallowtails and that the generic non-homeostatic approach to a swallowtail would also give a non-homeostatic approach to a fold (or chair).
11 Input-Output Maps with Two Inputs
Suppose now that the input \({\mathcal I}\) consists of several variables. In general terms, consider a parametrized family of ODEs
where \(X=(x_1,\ldots ,x_m) \in {\mathbf{R}}^m\), \({\mathcal I}\in {\mathbf{R}}^k\), and F is infinitely differentiable. We assume that (26) stems from an input-output network where one of the nodes (or coordinates of X) is the output node that is denoted, as before, by o. We also assume that (26) has a stable equilibrium at \(X_0\) when \({\mathcal I}={\mathcal I}_0\).
The equilibria of (26) are given by:
By the implicit function theorem, we can solve (27) near \((X_0,{\mathcal I}_0)\) to obtain a map \(X:{\mathbf{R}}^k \rightarrow {\mathbf{R}}^m\) such that
where \(X({\mathcal I}_0)=X_0\). Let
Definition 11.1
The input-output map of (27) near \((X_0,{\mathcal I}_0)\) is \(x_o: {\mathbf{R}}^k \rightarrow {\mathbf{R}}\).
Definition 11.2
The point \({\mathcal I}_0\) is an infinitesimal homeostasis point of \(x_o\) if the derivative
In particular, \({\mathcal I}_0\) is a singularity—that is, the derivative of \(x_o\) is singular there—but the vanishing of all first derivatives selects a special subclass of singularities, said to have ‘full corank’.
The interpretation of an infinitesimal homeostasis point is that \(x_o({\mathcal I})\) differs from \(x_o({\mathcal I}_0)\) in a manner that depends quadratically (or to higher order) on \(|{\mathcal I}-{\mathcal I}_0 |\). This makes the graph of \(x_o({\mathcal I})\) flatter than any growth rate with a nonzero linear term. This condition motivates for the condition (29) rather than merely \(D_{\mathcal I}x_o({\mathcal I}_0)\) being singular.
Definition 11.2 places the study of homeostasis in the context of singularity theory, and we follow the standard line of development in that subject. A detailed discussion of singularity theory would be too extensive for this paper. A brief summary is given in [21] in the context of homeostasis, accessible descriptions can be found in [36, 43], and full technical details are in [18, 30] and many other sources.
Following Nijhout et al. [33] we define:
Definition 11.3
A plateau is a region of \({\mathcal I}\) over which \(X({\mathcal I})\) is approximately constant.
Remark 11.4
Universal unfolding theory implies that small perturbations of \(x_o\) (that is, variation of the suppressed parameters) change the plateau region only slightly. This point was explored for the chair singularity in [21]. It follows that for sufficiently small perturbations plateaus of singularities depend mainly on the singularity itself and not on its universal unfolding.
Remark 11.5
In this section we focus on how singularities in the input-output map shape plateaus, and we use the normal form and unfolding theorems of elementary catastrophe theory to do this. We remark that typically the variables other than \(x_o\), the manipulated variables Y, can vary substantially while the output variable is held approximately constant. See, for example, Fig. 3 in [1].
11.1 Catastrophe Theory Classification
The results of [21] reduce the classification of homeostasis points for a single node to that of singularities of input-output maps \({\mathbf{R}}^k \rightarrow {\mathbf{R}}\). As mentioned in Sect. 9.2, this is precisely the abstract set-up for elementary catastrophe theory [4, 18, 36, 43]. The case \(k=1\) is discussed there.
We now consider the next case \(k = 2\). Table 1 summarizes the classification when \(k = 2\), so \({\mathcal I}= ({\mathcal I}_1, {\mathcal I}_2) \in {\mathbf{R}}^2\). Here the list is restricted to codimension \(\le 3\). The associated geometry, especially for universal unfoldings, is described in [4, 18, 36] up to codimension 4. Singularities of much higher codimension have also been classified, but the complexities increase considerably. For example Arnold [2] provides an extensive classification up to codimension 10 (for the complex analog).
Remark 11.6
Because \(k = 2\), the normal forms for \(k = 1\) appear again, but now there is an extra quadratic term \(\pm {\mathcal I}_2^2\). This term is a consequence of the splitting lemma in singularity theory, arising here when the second derivative \(D^2 x_o\) has rank 1 rather than rank 0 (corank 1 rather than corank 2). See [4, 36, 43]. The presence of the \(\pm {\mathcal I}_2^2\) term affects the range over which \( x_o({\mathcal I})\) changes when \({\mathcal I}_2\) varies, but not when \({\mathcal I}_1\) varies.
11.2 Normal Forms and Plateaus
The standard geometric features considered in catastrophe theory focus on the gradient of the function \(x_o({\mathcal I})\) in normal form. In contrast, what matters here is the function itself. Specifically, we are interested in the region in the \({\mathcal I}\)-plane where the function \(x_o\) is approximately constant.
More specifically, for each normal form \(x_o({\mathcal I})\) we choose a small \(\delta >0\) and form the set
This is the plateau region on which \(x_o({\mathcal I})\) is approximately constant, where \(\delta \) specifies how good the approximation is. If \(x_o({\mathcal I})\) is perturbed slightly, \(P_\delta \) varies continuously. Therefore we can compute the approximate plateau by focusing on the singularity, rather than on its universal unfolding.
This observation is important because the universal unfolding has many zeros of the gradient of \(x_o({\mathcal I})\), hence ‘homeostasis points’ near which the value of \(x_o({\mathcal I})\) varies more slowly than linear. However, this structure seems less important when considering the relationship of infinitesimal homeostasis with homeostasis. See the discussion of the unfolding of the chair summarized in [21, Figure 3].
The ‘qualitative’ geometry of the plateau—that is, its differential topology and associated invariants—is characteristic of the singularity. This offers one way to infer the probable type of singularity from numerical data; it also provides information about the region in which the system concerned is behaving homeostatically. We do not develop a formal list of invariants here, but we indicate a few possibilities.
The main features of the plateaus associated with the six normal forms are illustrated in Table 1. Figure 10 plots, for each normal form, a sequence of contours from \(-\delta \) to \(\delta \); the union is a picture of the plateaus. By unfolding theory, these features are preserved by small perturbations of the model, and by the choice of \(\delta \) in (30) provided it is sufficiently small. Graphical plots of such perturbations (not shown) confirm this assertion. Again, we do not attempt to make these statements precise in this paper.
11.3 The Hyperbolic Umbilic
As we have discussed, homeostasis can occur when one variable is held approximately constant on variation of two or more input parameters. For example, body temperature can be homeostatic with respect to both external temperature and amount of exercise. A biological network example is Fig. 3, where there is homeostasis of extracellular dopamine (eDA) in response to variation in the activities of the enzyme tyrosine hydroxylase (TH) and the dopamine transporters (DAT), Best et al. [3]. These authors derive a differential equation model for this biochemical network. They fix reasonable values for all parameters in the model with the exception of the concentrations of TH and DAT. Figure 11 (left) shows the equilibrium value of eDA as a function of TH and DAT in their model. The white dots indicate the predicted eDA values for the observationally determined values of TH and DAT in the wild type genotype (large white disk) and the polymorphisms observed in human populations (small white disks). Their result is scientifically important because almost all of the white disks lie on the plateau (the region where the surface is almost horizontal) that indicates homeostasis of eDA. Note that the flat region contains a line from left to right at about eDA = 0.9. In this respect the surface graph in Fig. 11 (left) appears to resemble that of a nonsingular perturbed hyperbolic umbilic (see Table 1) in Fig. 11 (right). See also the level contours of the hyperbolic umbilic in Fig. 10. This figure shows that the hyperbolic umbilic is the only low codimension singularity that contains a single line in its zero set.
(Left): Nijhout et al. [33, Fig. 8] and Reed et al. [37, Fig. 14]. At equilibrium there is homeostasis of eDA as a function of TH and DAT. There is a plateau around the wild-type genotype (large white disk). Smaller disks indicate positions of polymorphisms of TH and DAT found in human populations. (Right): Graph of surface of perturbed hyperbolic umbilic without singularities: \(Z({\mathcal I}_1,{\mathcal I}_2) = {\mathcal I}_1^3+{\mathcal I}_2^3+{\mathcal I}_1+{\mathcal I}_2/2\).
The number of curves (‘whiskers’) forming the zero-level contour of the plateau is a characteristic of the plateau. For example, Fig. 11 appears to have one curve in the plateau. This leads us to conjecture that the hyperbolic umbilic is the singularity that organizes the homeostatic region of eDA in the example discussed in [3]. It may be the case however, that there is no infinitesimal homeostasis in this example, and the cause is more global. We have discussed in Sect. 10 why the chair and the hyperbolic umbilic are the singularities that might be expected to organize two output homeostasis.
Theorem 11.7
Consider input-output functions with two inputs and one output. Then the only singularities of codimension \(\le 3\) that have perturbations with no infinitesimal homeostasis are the fold (chair), swallowtail, and hyperbolic umbilic.
The proof of this theorem is in [23].
Remark 11.8
In Sect. 10 we note that a system of equations that evolves toward infinitesimal homeostasis does so by transitioning through a singularity that has unfolding parameters with no infinitesimal homeostasis. It follows from Theorems 10.2 and 11.7 that the most likely ways to transition to homeostasis in systems with one input variable is through the chair and in systems with two input variables the hyperbolic umbilic and the two variable chair.
12 Gene Regulatory Networks and Housekeeping Genes
Antoneli et al. [1] use infinitesimal homeostasis to find regions of homeostasis in a differential equation model for the gene regulatory network (GRN) that is believed to regulate the production of the protein PGA2 in Escherichia coli and yeast. Specifically, in this model the input parameter is an external parameter \({\mathcal I}\) that represents the collective influence of other gene proteins on this specific GRN. We find regions of homeostasis that gives a plausible explanation of how the level of the PGA2 protein might be held approximately constant while other reactions are taking place.
Gene expression is a general name for a number of sequential processes, the most well known and best understood being transcription and translation. These processes control the level of gene expression and ultimately result in the production of a specific quantity of a target protein.
The genes, regulators, and the regulatory connections between them forms a gene regulatory network (GRN). A gene regulatory network can be represented pictorially by a directed graph where the genes correspond to network nodes, incoming arrows to transcription factors, and outgoing arrows to levels of gene expression (protein concentration).
12.1 Gene Regulatory Networks and Homeostasis
Numerous terms are used to describe types of genes according to how they are regulated. A constitutive gene is a gene that is transcribed continually as opposed to a facultative gene that is transcribed only when needed. A housekeeping gene is a gene that is required to maintain basic cellular function and so is typically expressed in all cell types of an organism. Some housekeeping genes are transcribed at a ‘relatively constant rate’ in most non-pathological situations and are often used as reference points in experiments to measure the expression rates of other genes.
Even though this scheme is more or less universal among all life forms, from uni-cellular to multi-cellular organisms, there are some important differences according to whether the cell possesses a nucleus (eukaryote) or not (prokaryote). In single-cell organisms, gene regulatory networks respond to changes in the external environment adapting the cell at a given time for survival in this environment. For example, a yeast cell, finding itself in a sugar solution, will turn on genes to make enzymes that process the sugar to alcohol.
Recently, there has been an ongoing effort to map out the GRNs of some the most intensively studied single-cell model organisms: the prokaryote E. coli and the eukaryote Saccharomyces cerevisiae, a species of yeast. A hypothesis that has emerged from these efforts is that the GRN has evolved into a modular structure in terms of small sub-networks appearing as recurrent patterns in the GRN, called network motifs. Moreover, experiments on the dynamics generated by network motifs in living cells indicate that they have characteristic dynamical functions. This suggests that network motifs may serve as building blocks in modeling gene regulatory networks.
Much experimental work has been devoted to understanding network motifs in gene regulatory networks of single-cell model organisms. The GRNs of E. coli and yeast, for example, contain three main motif families that make up almost the entire network. Some well-established network motifs and their corresponding functions in the GRN of E. coli and yeast include negative (or inhibitory) self-regulation, positive (or excitatory) self-regulation and several types of feedforward loops. Nevertheless, most analyses of motif function are carried out looking at the motif operating in isolation. There is, however, mounting evidence that network context, that is, the connections of the motif with the rest of the network, are important when drawing inferences on characteristic dynamical functions of the motif.
In this context, an interesting question is how the GRN of a single-cell organism is able to sustain the production rates of the housekeeping genes and at same time be able to quickly respond to environmental changes, by turning on and off the appropriate facultative genes. If we assume that the dynamics of gene expression is modeled by coupled systems of differential equations then this question can be formulated as the existence of a homeostatic mechanism associated to some types of network motifs imbedded in the GRN.
Latest estimates on the number of feedforward loops in the GRN of S. cerevisiae assert that there are least 50 feedforward loops (not all of the same type) potentially controlling 240 genes. One example of such a feedforward loop is shown in Fig. 12. The three genes in this network are considered constitutive.
An example of feedforward regulation network from the GRN of S. cerevisiae, involving the genes SFP1, CIN5 and PGA2. The PGA2 gene produces an essential protein involved in protein trafficking (null mutants have a cell separation defect). The CIN5 gene is a basic leucine zipper (bZIP) transcription factor. The SPF1 gene regulates transcription of ribosomal protein and biogenesis genes.
12.2 Basic Structural Elements of GRNs
The fundamental building block or node in a gene regulatory network is a gene that is composed of two parts: transcription and translation. The transcription part produces messenger RNA (mRNA) and the translation part produces the protein. The system of ODEs associated to one gene has the form
where \(x^R\) is the mRNA concentration, \(x^P\) is the protein concentration and the \(t_J\) are the coupling protein concentrations of transcription factors that regulate the gene and are produced by other genes in the network. The parameter \({\mathcal I}\) represents the effect of upstream transcription factors that regulate the gene but are not part of the network. The vector field f has the form
where \(f^R\) models the dynamics of mRNA concentration and \(f^P\) models the dynamics of the protein concentration.
When the gene is not self-regulated the system has the form
and when the gene is self-regulated the system of two scalar equations has the form
In both cases the gene output is the scalar variable \(x_P\).
12.3 The Gene Regulatory Network for PGA2
Consider the network consisting of three genes (and six nodes) shown in Fig. 13, where the dashed lines represent inhibitory coupling (repression or negative control) and the solid lines represent excitatory coupling (activation or positive control).
Observe that the six-node network in Fig. 13 has two simple paths:
There are two possible Haldane homeostasis arrows \(x^R\rightarrow x^P\) and \(z^R\rightarrow z^P\), and one structural homeostasis of degree three consisting of two paths \(x^P\rightarrow y^R\rightarrow y^P\rightarrow z^R\) and \(x^P\rightarrow z^R\). To verify this we compute the homeostasis matrix.
The steady-state equations associated with the network in Fig. 13 have the form:
where the input parameter \({\mathcal I}\) represents the action of all upstream transcription factors that affect the x-gene and do not come from the y- and z-genes. Our goal is to find regions of homeostasis in the steady-state protein concentration \(z^P\) as a function of the input parameter \({\mathcal I}\). To do this we compute \(\det (H)\), where H is the \(5\times 5\) homeostasis matrix.
A short calculation shows that
Therefore structural homeostasis is found by solving \(h = h' = 0\), where
This equation is analysed in Antoneli et al. [1], who show that standard ODE models for gene regulation, when inserted into a feedforward loop motif, do indeed lead to chair structural homeostasis in the output protein housekeeping genes. In [1] this cubic expression was obtained by direct calculation and its appearance was somewhat mysterious; here it emerges from the general theory of homeostasis matrices in Sect. 7.
References
Antoneli, F., Golubitsky, M., Stewart, I.: Homeostasis in a feed forward loop gene regulatory network motif. J. Theor. Biol. 445, 103–109 (2018). https://doi.org/10.1016/j.jtbi.2018.02.026
Arnold, V.I.: Local normal forms of functions. Invent. Math. 35, 87–109 (1976)
Best, J., Nijhout, H.F., Reed, M.: Homeostatic mechanisms in dopamine synthesis and release: a mathematical model. Theor. Biol. Med. Model. 6 (2009). https://doi.org/10.1186/1742-4682-6-21
Bröcker, Th., Lander, L.: Differentiable Germs and Catastrophes. LMS Lect. Notes, vol. 17. Cambridge University Press, Cambridge (1975)
Brualdi, R.A., Ryser, H.J.: Combinatorial Matrix Theory. Cambridge University Press, Cambridge (1991)
Brualdi, R.A., Cvetkoić, D.M.: A Combinatorial Approach to Matrix Theory and its Applications. Chapman & Hall/CRC Press, Boca Raton (2009)
Coates, C.L.: Flow graph solutions of linear algebraic equations. IRE Trans. Circuit Theory CT–6, 170–187 (1959)
Cvetković, D.M.: The determinant concept defined by means of graph theory. Mat. Vesnik 12(27), 333–336 (1975)
Dellnitz, M.: Hopf-Verzweigung in Systemen mit Symmetrie und deren Numerische Behandlung. Uni. Diss, Hamburg (1988)
Dellnitz, M.: A computational method and path following for periodic solutions with symmetry. In: Roose, D., De Dier, B., Spence, A. (eds.) Continuation and Bifurcations: Numerical Techniques and Applications, pp. 153–167. Kluwer, Dordrecht (1990)
Dellnitz, M.: Computational bifurcation of periodic solutions in systems with symmetry. IMA J. Numer. Anal. 12, 429–455 (1992)
Dellnitz, M., Junge, O.: On the approximation of complicated dynamical behavior. SIAM J. Numer. Anal. 36, 491–515 (1999)
Dellnitz, M., Junge, O., Thiere, B.: The numerical detection of connecting orbits. Discret. Continuous Dyn. Syst. - B 1, 125–135 (2001)
Desoer, C.A.: The optimum formula for the gain of a flow graph or a simple derivation of Coates’ formula. Proc. IRE 48, 883–889 (1960)
Donovan, G.M.: Biological version of Braess’ paradox arising from perturbed homeostasis. Phys. Rev. E 98, 062406-1 (2018)
Donovan, G.M.: Numerical discovery and continuation of points of infinitesimal homeostasis. Math. Biosci. 311, 62–67 (2019)
Ferrell, J.E.: Perfect and near perfect adaptation in cell signaling. Cell Syst. 2, 62–67 (2016)
Gibson, C.: Singular Points of Smooth Mappings. Research Notes in Math, vol. 25. Pitman, London (1979)
Golubitsky, M.: An introduction to catastrophe theory and its applications. SIAM Rev. 20(2), 352–387 (1978)
Golubitsky, M., Schaeffer, D.G.: Singularities and Groups in Bifurcation Theory I. Applied Mathematics Series, vol. 51. Springer, New York (1985)
Golubitsky, M., Stewart, I.: Symmetry methods in mathematical biology. São Paulo J. Math. Sci. 9, 1–36 (2015)
Golubitsky, M., Stewart, I.: Homeostasis, singularities and networks. J. Math. Biol. 74, 387–407 (2017). https://doi.org/10.1007/s00285-016-1024-2
Golubitsky, M., Stewart, I.: Homeostasis with multiple inputs. SIAM J. Appl. Dyn. Syst. 17, 1816–1832 (2018)
Golubitsky, M., Stewart, I., Schaeffer, D.G.: Singularities and Groups in Bifurcation Theory II. Applied Mathematics Series, vol. 69. Springer, New York (1988)
Golubitsky, M., Wang, Y.: Infinitesimal homeostasis in three-node input-output networks. J. Math. Biol. 80, 1163–1185 (2020). https://doi.org/10.1007/s00285-019-01457-x
Harary, F.: The determinant of the adjacency matrix of a graph. SIAM Rev. 4(3), 202–210 (1962)
Jepson, A.D., Spence, A.: The numerical solution of nonlinear equations having several parameters, I: scalar equations. SIAM J. Numer. Anal. 22, 736–759 (1985)
Jepson, A.D., Spence, A., Cliffe, K.A.: The numerical solution of nonlinear equations having several parameters, III: equations with \({\mathbf{Z}}_{2}\) symmetry. SIAM J. Numer. Anal. 28, 809–832 (1991)
Ma, W., Trusina, A., El-Samad, H., Lim, W.A., Tang, C.: Defining network topologies that can achieve biochemical adaptation. Cell 138, 760–773 (2009)
Martinet, J.: Singularities of Smooth Functions and Maps. London Mathematical Society Lecture Notes Series, vol. 58. Cambridge University Press, Cambridge (1982)
Moore, G., Garratt, T.J., Spence, A.: The numerical detection of Hopf bifurcation points. In: Roose, D., De Dier, B., Spence, A. (eds.) Continuation and Bifurcations: Numerical Techniques and Applications, pp. 227–246. Kluwer, Dordrecht (1990)
Morrison, P.R.: Temperature regulation in three Central American mammals. J. Cell Comp. Physiol. 27, 125–137 (1946)
Nijhout, H.F., Best, J., Reed, M.C.: Escape from homeostasis. Math. Biosci. 257, 104–110 (2014)
Nijhout, H.F., Reed, M.C.: Homeostasis and dynamic stability of the phenotype link robustness and plasticity. Integr. Comp. Biol. 54(2), 264–275 (2014). https://doi.org/10.1093/icb/icu010
Nijhout, H.F., Reed, M., Budu, P., Ulrich, C.: A mathematical model of the folate cycle: new insights into folate homeostasis. J. Biol. Chem. 226, 55008–55016 (2004)
Poston, T., Stewart, I.: Catastrophe Theory and Its Applications. Surveys and Reference Works in Math, vol. 2. Pitman, London (1978)
Reed, M., Best, J., Golubitsky, M., Stewart, I., Nijhout, F.: Analysis of homeostatic mechanisms in biochemical networks. Bull. Math. Biol. 79, 2534–2557 (2017). https://doi.org/10.1007/s11538-017-0340-z
Reed, M.C., Lieb, A., Nijhout, H.F.: The biological significance of substrate inhibition: a mechanism with diverse functions. BioEssays 32, 422–429 (2010)
Savageau, M.A., Jacknow, G.: Feedforward inhibition in biosynthetic pathways: inhibition of the aminoacyl-tRNA synthetase by intermediates of the pathway. J. Theor. Biol. 77, 405–425 (1979)
Schechter, M.: Modern Methods in Partial Differential Equations. McGraw-Hill, New York (1977)
Tang, Z.F., McMillen, D.R.: Design principles for the analysis and construction of robustly homeostatic biological networks. J. Theor. Biol. 408, 274–289 (2016)
Wang, Y., Huang, Z., Antoneli, F., Golubitsky, M.: The structure of infinitesimal homeostasis in input-output networks, preparation
Zeeman, E.C.: Catastrophe Theory: Selected Papers 1972–1977. Addison-Wesley, London (1977)
www.biology-online.org/4/1_physiological_homeostasis.htm (2000, updated)
Acknowledgments
We thank Janet Best, Tony Nance, and Mike Reed, for helpful conversations. This research was supported in part by the National Science Foundation Grant DMS-1440386 to the Mathematical Biosciences Institute. FA and MG were supported in part by the OSU-FAPESP Grant 2015/50315-3.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 The Editor(s) (if applicable) and The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Golubitsky, M., Stewart, I., Antoneli, F., Huang, Z., Wang, Y. (2020). Input-Output Networks, Singularity Theory, and Homeostasis. In: Junge, O., Schütze, O., Froyland, G., Ober-Blöbaum, S., Padberg-Gehle, K. (eds) Advances in Dynamics, Optimization and Computation. SON 2020. Studies in Systems, Decision and Control, vol 304. Springer, Cham. https://doi.org/10.1007/978-3-030-51264-4_2
Download citation
DOI: https://doi.org/10.1007/978-3-030-51264-4_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-51263-7
Online ISBN: 978-3-030-51264-4
eBook Packages: EngineeringEngineering (R0)