Abstract
The generation of kinematic models of the heart using 3D echocardiography (echo) can be difficult due to poor image contrast and signal dropout, particularly at the epicardial surface. 2D echo images generally have a better contrast-to-noise ratio compared to 3D echo images, thus wall thickness (WT) estimates from 2D echo may provide a reliable means to constrain model fits to 3D echo images. WT estimates were calculated by solving a pair of differential equations guided by a vector field, which is constructed from the solution of Laplace’s equation on binary segmentations of the left ventricular myocardium. We compared 2D echo derived WT estimates against values calculated using gold-standard cardiac cine magnetic resonance imaging (MRI) to assess reliability. We found that 2D echo WT estimates were higher compared to WT values from MRI at end-diastole with a mean difference of 1.3 mm (95% CI: 0.74–1.8 mm), 1.5 mm (95% CI: 0.91–2.1 mm) and 2.1 mm (95% CI: 1.6–2.6 mm) for basal, mid-ventricular and apical segments respectively. At end-systole, the WT estimates from MRI were higher compared to those derived from 2D echo with a mean difference of 2.6 mm (95% CI: 2.0–3.1 mm), 2.1 mm (95% CI: 1.5–2.7 mm) and 1.1 mm (95% CI: 0.49–1.7 mm) for basal, mid-ventricular and apical segments, respectively. The quantitative WT comparison in this study will contribute to the ongoing efforts to better translate kinematic modelling analyses from gold-standard cardiac MRI to the more widely accessible echocardiography.
The authors gratefully acknowledge the Health Research Council of NZ for funding this project (17/608). This would not have been possible without the dedicated work of our research support staff, Craig McDougal, and the MRI technologists at Center of Advanced MRI, University of Auckland.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Kinematic modelling of the heart is important to the understanding of the underlying mechanisms of heart failure, which is a complex, multifactorial disease with divergent characteristics [1]. Cardiovascular disease is the world’s leading cause of mortality and morbidity. It is estimated that 23 million people worldwide are suffering from heart failure, which has a mortality rate of 50% within five years [1]. Recent advances in pharmacologic therapy and implantable cardiac devices have improved survival, however continued advancements in diagnostics and therapeutics are needed to further reduce the economic burden of heart failure as the current therapies rarely prove curative [1]. The new knowledge that can be gathered from kinematic modelling can be key for the development of novel treatments that are patient specific [12].
In order to create a model of the heart, an accurate representation of its geometry is needed from cardiac imaging techniques. Magnetic resonance imaging (MRI) is the gold standard imaging technique for assessing cardiac function, due to its high contrast-to-noise ratio, resolution and reproducibility compared to other imaging techniques [4]. However MRI is expensive, time consuming and often not suitable for patients with implantable devices [9]. Echocardiography (echo) remains the work-horse for diagnostic imaging in hospitals, due to its portability and relative low cost. It is also widely available and deeply embedded in clinical decision making for patients with heart disease [9].
3D echo evaluations of left ventricular (LV) mass and volume are known to be systematically different to those acquired from MRI, due to shadowing artefacts in patients with poor acoustic windows and a lower contrast-to-noise ratio, which makes the quantification of LV shapes from 3D echo images difficult [14]. Previous research has shown the feasibility of building cardiac statistical shape atlasses using MRI and 3D echo, but also using 3D echo alone by applying a multi-view subspace learning algorithm to establish the discrepancy between the image modalities [11].
In this study, both 2D and 3D echo images, as well as MRI acquisitions from the same person were gathered. 3D echo images have poor epicardial surface definition which increases difficulty in accurately defining wall thickness from these images. 2D echo images generally have a better image quality compared to 3D echo images, thus 2D echo may be used to constrain kinematic model generation from 3D echo. This can be done using wall thickness (WT) estimates from 2D echo to obtain a more accurate geometry as well as wall mass.
We quantified regional WT from 2D echo data by solving a pair of differential equations guided by a vector field, constructed from the solution of Laplace’s equation on binary segmentation’s of the LV myocardium. These WT measurements were compared against values calculated using gold-standard cardiac cine MRI taken from the same participants to assess the reliability of WT estimates from 2D echo.
2 Methods
2.1 Data Collection
18 participants underwent cardiac MRI and echocardiographic examinations within 1 h of each other. Ethical approval for this study was obtained from the health and disability ethics committee of New Zealand (17/CEN/226). All participants gave written informed consent to participate in the study. The MRI was performed on a 1.5T Siemens AvantoFit. All cine images were acquired in long and short axis orientations with a steady-state free precession (SSFP) sequence with the following standard imaging parameters: echo time (TE) of 1.6 ms, repetition time (TR) of 3.7 ms, flip angle of 45\(^\circ \), field of view of 360 cm \(\times \) 360 cm, temporal resolution of 48 ms, voxel size of 1.5 mm \(\times \) 1.5 mm, and slice thickness of 6 mm. The echocardiographic examination was performed using a Siemens Acuson SC2000 ultrasound system. A standard 2D echo examination was performed in line with guidelines [6], and the cardiac cycle was captured three times for each acquisition. Four 3D echo datasets were acquired using a 4Z1c 3D transducer which captures the cardiac geometry in one heart beat. The field of view and frame rate were varied to fit the patient geometry, and optimise image quality. All apical two chamber (A2C), apical four chamber (A4C) and apical long axis (ALX) views with the best image quality were chosen for the WT measurements.
2.2 Segmentation of the LV Myocardium from 2D Echo Images and Cardiac Cine MRI
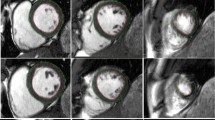
Segmentation of the LV myocardium in 2D echo images was performed manually in ITK-SNAP 3.8.0Footnote 1 [18]. Both end-diastolic (ED) and end-systolic (ES) frames in each of the three views were manually segmented, resulting in six annotated images for each of the 18 participants. Each of the manual contours were free drawn by hand, with no image processing techniques utilized. The ED and ES frames were selected according to the recommendation of the American Society of Echocardiography and the European Association of Cardiovascular Imaging [6]. The segmentations of the endocardial and epicardial borders were closed at the level of the mitral valve. Trabeculae and papillary muscles were excluded from the LV wall mass. 3D geometric models of the LV were made for all points in the cardiac cycle using CIM (v8.1.7), and the protocol defined in [17]. An example pair of contours on a 2D echo image and a 3D model overlayed on a MR image of the same patient is shown in Fig. 1. The triangular mesh defining the endo- and epicardial surfaces was converted into a mask of the myocardium using a voxelization methodology based on a ray intersection method similar to that described by [8]. The voxel size in the resulting mask was set to 0.75 mm \(\times \) 0.75 mm \(\times \) 0.75 mm.
2.3 Estimation of Regional Wall Thickness
The LV WT estimates from 2D echo and MRI were calculated using Laplace’s equation. This is an established method to calculate regional WT, for example, to identify cortical thickness [5]. It has also been used to calculate regional WT of the myocardium from MRI [10]. Solving Laplace’s equation over the myocardial domain provided a gradient field indicating the correspondence trajectories between the endocardial and epicardial surfaces of the domain. These correspondence trajectories have desirable properties as they are orthogonal to each surface, do not intersect, and are nominally parallel. The WT is then defined as the arc length of these correspondence trajectories, and is defined at each point (pixel) [16]. For the numerical implementation of Laplace’s equation (\(\varDelta \phi = 0\)), Dirichlet boundary conditions with values of 300 and 100 were applied on the endocardial (\(\varGamma _{endo}\)) and epicardial (\(\varGamma _{epi}\)) borders of the domain, respectively. The Laplace solver was implemented in MATLAB (R2018a) and adjusted from [15]. Solving for \(\phi \) by a finite difference scheme, the gradient field \(\varvec{T}\) is calculated according to Eq. 1.
Regional WT was calculated by solving two partial differential equations (PDEs) that determine two length functions: \(L_0 (x)\) and \(L_1 (x)\) (see Eqs. 2 and 3).
Given a point x on the correspondence trajectory, \(L_0 (x)\) gives the arc length between the endocardial border and x and \(L_1 (x)\) returns the arc length of the correspondence trajectory between the epicardial border and x. The WT is then defined by Eq. 4 [16].
2.4 Intra- and Inter-observer Variability Analyses
To assess the reproducibility of cardiac labelling on 2D echo images, both intra- and inter-observer variability analyses were performed on a randomly generated 10% sample (12 images) of the dataset. Each of the images was segmented three times with 2–3 days between measurements by an observer that had received some basic training in echo image segmentation. Furthermore, the most recent segmentations were chosen for evaluation against an independent segmentation provided by an observer with one year experience in image analysis. The intra- and inter-observer variabilities were evaluated using the Dice score (DSC) and Hausdorff distance (HD) and were also assessed on the derived WT estimates, after which paired t-tests were performed to test for significant differences.
2.5 Comparison of Regional WT Estimates
The WT was compared for each region in the American Heart Association (AHA) 17-segment model [6] using the clinical guidelines described in [2] for 2D echo. For cardiac cine MRI, the same guidelines were used to divide the 3D participant specific model derived from the cines at ED and ES into regions so that regional comparison of WT estimates could be performed. The AHA regions are shown in Fig. 2.
The mean of all the pixels/voxels in each surface region for 2D echo and each volume region for MRI were taken for comparison. First, WT estimates were compared using a correlation and an unpaired t-test at the levels of base, mid-ventricular and apex at ED and ES separately. Then, a two-way ANOVA was performed to assess the dependency of the WT estimates on the imaging modality and the different regions from the AHA 17-segment model. If significant terms were found in the ANOVA model, a Tukey multiple comparison of means (Tukey HSD) was performed to investigate the importance of each of the individual terms. The ANOVA model was analysed for the basal, mid-ventricular and apical segments at ED and ES separately, to be able to examine the differences between the individual segments at each level. All analyses were performed with R 3.4.3 and R Commander 2.4-4 [3].
3 Results
Results from the Laplace solver for the calculation of the WT are displayed on a model fitted to cardiac cine MRI in Fig. 3.
The intra- and inter-observer variability analyses is shown in Table 1. The intra-observer differences in WT were not significant when assessed using a paired t-test, while the inter-observer differences were found to be significant.
Results for the regional comparison of WT estimates from 2D echo to cardiac cine MRI are shown in Table 2. At ED, the WT was found significantly higher for 2D echo compared to cardiac cine MRI. At ES, this relationship is reversed. There was no correlation between the WT estimates from the two modalities.
Two-way ANOVA showed significant terms for the main effects, which were the imaging modality and the AHA region. Furthermore, there was a significant interaction term between the AHA region and the modality. The only exception was encountered for the two-way ANOVA model at the basal level at ES. The mean WT derived from 2D echo and cardiac cine MRI are shown in Figs. 4 and 5. Post-hoc analysis (Tukey multiple comparison of means) showed significant differences for regions 4, 5, 10, 11, 13, 15 and 16 at ED and regions 2, 3, 6, 8, 9 and 14 at ES.
Mean ED values of WT estimates calculated in each region of the AHA 17-segment model in both 2D echo (red) and cardiac cine MRI (black) for the 18 participants. The bars on each point display the standard errors on the means.The asterisks indicate the regions that were found to be significantly different in WT estimates using Tukey multiple comparison of means. (Color figure online)
4 Discussion
From the regional comparison of WT estimates, there was a clear difference in WT estimations for segmentations of ED and ES. At ED, the estimated WT from 2D echo is larger than when derived from cardiac MRI. At ES, this relationship is reversed. In cardiac MRI during systole, the trabeculae appear to combine with the compact myocardium, making the boundary between these structures difficult to distinguish, whereas in 2D echo this boundary is well defined [7].
Tukey HSD indicated that several segments differed significantly in WT derived from 2D echo and MRI at ED and ES. At ED, the segments with the largest differences in WT between the modalities were located in the inferior and inferolateral side of the heart. At ES, significant differences in WT were found on the septal side of the heart. Also within each modality, variations in WT across the different AHA regions were found. Whether these differences are consistent needs to be evaluated on a larger dataset and could be used to provide better translation in clinical indices between echo and MRI.
Mean ES values of WT estimates calculated in each region of the AHA 17-segment model in both 2D echo (red) and cardiac cine MRI (black) for the 18 participants. The bars on each point display the standard errors on the means. The asterisks indicate the regions that were found to be significantly different in WT estimates using Tukey multiple comparison of means. (Color figure online)
WT estimates were calculated using Laplace’s equation and PDE constraints. There are alternative techniques for calculating regional WT, such as the centerline method [13]. It has been reported that the use of the Laplace solver is more accurate than the centerline method because the latter carries the implicit assumption that the myocardial wall is always perpendicular to the acquisition plane [10]. Some limitations in the WT estimation include the fact that part of the basal myocardium needed to be excluded from the solution analysis. This was because an edge artefact occurred here due to the way the boundary conditions were defined. This could have been solved by adjusting the geometry only for the computation, although it would have required more computation time.
While the accuracy of the Laplace solver is good (in the order of 1 pixel), the use of manual segmentations for the delineation of the myocardial domain reduces the accuracy of the WT estimates on 2D echo, as there are inter- and intra-observer errors. The intra-observer error was relatively small (about 5%), which indicates a good consistency. The inter-observer error was much larger at about 35%. However, with a more detailed protocol for the segmentations and more training of the observers, the error is expected to decrease. It is important to investigate how these uncertainties in the 2D echo WT estimates can influence kinematic model predictions.
The comparison of regional WT estimates was performed based on the mean values found across one AHA region. These mean values come from a distribution of WT values and could be considered for future research.
5 Conclusions
This study has shown that WT estimates derived from 2D echo images of the heart are significantly different to WT estimates derived from 3D geometric models of the LV fitted to cardiac cine MRI using Laplace’s equation. As a next step, the WT estimates from 2D echo will be used to constrain model fits to 3D echo images. This knowledge contributes towards ongoing efforts to better translate kinematic modelling analyses from gold-standard cardiac MRI to the more widely accessible echocardiography.
Notes
References
Alpert, C., et al.: Symptom burden in heart failure: assessment, impact on outcomes, and management. Heart Fail. Rev. 22(1), 25–39 (2017)
Badano, L.P., Picano, E.: Standardized myocardial segmentation of the left ventricle. Stress Echocardiography, pp. 105–119. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-20958-6_7
Fox, J.: The R commander: a basic-statistics graphical user interface to RJ Stat. Software 14, 42 (2005)
Gonzalez, J.A., Kramer, C.M.: Role of imaging techniques for diagnosis, prognosis and management of heart failure patients: cardiac magnetic resonance. Curr. Hear. Fail. Rep. 12(4), 276–283 (2015)
Jones, S.E., et al.: Three-dimensional mapping of cortical thickness using Laplace’s equation. Hum. Brain Mapp. 11(1), 12–32 (2000)
Lang, R.M., et al.: Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American society of echocardiography and the European association of cardiovascular imaging. Eur. Hear. J.-Cardiovasc. Imaging 16(3), 233–271 (2015)
Park, E.A., et al.: Effect of papillary muscles and trabeculae on left ventricular measurement using cardiovascular magnetic resonance imaging in patients with hypertrophic cardiomyopathy. Korean J. Radiol. 16(1), 4–12 (2015)
Patil, S., Ravi, B.: Voxel-based representation, display and thickness analysis of intricate shapes. In: Ninth International Conference on Computer Aided Design and Computer Graphics (CAD-CG 2005) (2005)
Ponikowski, P., et al.: 2016 ESC guidelines for the diagnosis and treatment of acute and chronic heart failure: the task force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC). Developed with the special contribution of the heart failure association (HFA) of the ESC. Eur. J. Hear. Fail. 18(8), 891–975 (2016)
Prasad, M., et al.: Quantification of 3D regional myocardial wall thickening from gated magnetic resonance images. J. Magn. Reson. Imaging 31(2), 317–327 (2010)
Puyol-Antón, E., et al.: A multimodal spatiotemporal cardiac motion atlas from MR and ultrasound data. Med. Image Anal. 40, 96–110 (2017)
Sengupta, P.P., et al.: The new wave of cardiovascular biomechanics. JACC: Cardiovasc. Imaging 12(7), 1297–1299 (2019)
Sheehan, F.H., Bolson, E.L., Dodge, H.T., Mathey, D.G., Schofer, J., Woo, H.: Advantages and applications of the centerline method for characterizing regional ventricular function. Circulation 74(2), 293–305 (1986)
Shimada, Y.J., Shiota, T.: A meta-analysis and investigation for the source of bias of left ventricular volumes and function by three-dimensional echocardiography in comparison with magnetic resonance imaging. Am. J. Cardiol. 107(1), 126–138 (2011)
Wang, Y., et al.: A robust computational framework for estimating 3D bi-atrial chamber wall thickness. Trans. Biomed. Eng. (2019, under Revision)
Yezzi, A.J., Prince, J.L.: An eulerian PDE approach for computing tissue thickness. IEEE Trans. Med. Imaging 22(10), 1332–1339 (2003)
Young, A.A., et al.: Left ventricular mass and volume: fast calculation with guide-point modeling on MR images. Radiology 216(2), 597–602 (2000)
Yushkevich, P.A., et al.: User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31(3), 1116–1128 (2006)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
van Hal, V.H.J. et al. (2020). Comparison of 2D Echocardiography and Cardiac Cine MRI in the Assessment of Regional Left Ventricular Wall Thickness. In: Pop, M., et al. Statistical Atlases and Computational Models of the Heart. Multi-Sequence CMR Segmentation, CRT-EPiggy and LV Full Quantification Challenges. STACOM 2019. Lecture Notes in Computer Science(), vol 12009. Springer, Cham. https://doi.org/10.1007/978-3-030-39074-7_6
Download citation
DOI: https://doi.org/10.1007/978-3-030-39074-7_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-39073-0
Online ISBN: 978-3-030-39074-7
eBook Packages: Computer ScienceComputer Science (R0)