Abstract
The segmentation and classification of atherosclerotic plaque (AP) are of great importance in the diagnosis and treatment of coronary artery disease. Although the constitution of AP can be assessed through a contrast-enhanced coronary computed tomography angiography (CCTA), the interpretation of CCTA scans is time-consuming and tedious for radiologists. Automation of AP segmentation is highly desired for clinical applications and further researches. However, it is difficult due to the extreme unbalance of voxels, similar appearance between some plaques and background tissues, and artefacts. In this paper, we propose a vessel-focused 3D convolutional network for automatic segmentation of AP including three subtypes: calcified plaques (CAP), non-calcified plaques (NCAP) and mixed calcified plaques (MCAP). We first extract the coronary arteries from the CT volumes; then we reform the artery segments into straightened volumes; finally, a 3D vessel-focused convolutional neural network is employed for plaque segmentation. The proposed method is trained and tested on a dataset of multi-phase CCTA volumes of 25 patients. We further investigate the effect of artery straightening through a comparison experiment, in which the network is trained on original CT volumes. Results show that by artery extraction and straightening, the training time is reduced by 40% and the segmentation performance of non-calcified plaques and mixed calcified plaques gains significantly. The proposed method achieves dice scores of 0.83, 0.73 and 0.68 for CAP, NCAP and MCAP respectively on the test set, which shows potential value for clinical application.
J. Liu and C. Jin—These two authors contribute equally to this study.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Coronary artery disease (CAD) is one of the biggest causes of mortality in the world. It is usually caused by atherosclerosis, of which the plaques are divided into three subtypes: calcified (CAP), non-calcified (NCAP) and mixed calcified (MCAP). The plaque composition is an important indicator for the diagnosis and treatment of CAD [1].
Contrast-enhanced coronary CT angiography (CCTA) allows assessment of AP composition, while it is demanding to interpret CCTA scans due to the large volume of data and the numerous segments of coronary arteries. Therefore, automatic segmentation and classification of AP is highly desirable. It can not only facilliate the interpretation of CCTA scans, but also provides a quantitative measurement of AP. However, AP segmentation on voxel level is difficult due to extreme unbalance of voxels (much more background voxels than plaque voxels), similar appearance between some plaques and background tissues, and artefacts.
Many studies have focused on CAP segmentation in 3D CT volumes. CAP is characterized by bright appearance in CT images, which can be easily discerned in both non-contrast-enhanced and contrast-enhanced cardiac CT. Wolterink et al. [2] summarized CAP segmentation methods. In non-contrast cardiac CT scans, CAP can be detected by a threshold of 130 HU [3] and subsequent classifiers, while in CCTA the detection threshold may vary depending on protocols, scanners and contrast agents, and thus special techniques are needed to determine the threshold. CNN-based methods have emerged recently, which typically consist of two network: the first performs a rough segmentation to restrict the area of interest and the second performs a meticulous segmentation [4, 5].
Although fewer studies have covered the segmentation of NCAP and MCAP, the detection of them is important because they are more prone to rupture and result in acute coronary syndromes such as stroke and sudden death. In contrast to CAP, NCAP and MCAP show similar intensity with surrounding tissues, which makes segmentation of NCAP and MCAP more challenging (Fig. 1). [6] adopts a two-fold methodology for NCAP segmentation, in which the first step extracts the coronary arteries and NCAP is detected based on the extracted arteries. To our knowledge, there is no existing method that segments all three subtypes of AP at the same time.
Recently, fully convolutional neural networks (FCNs) have demonstrated state-of-the-art performance on many challenging image segmentation tasks. 3D U-Net [7] is one example that is especially suitable for medical image analysis.
In this paper, we propose a robust method based on 3D FCNs for automatic coronary artery plaques segmentation including all three types. To begin with, a bounding box encasing the coronary arteries are automatically generated for the purpose of reducing computation. Then we extract the coronary arteries and reform artery segments into straightened volumes as inputs of the network using multi-planar reformation (MPR) technique. Finally, a vessel-focused 3D convolutional network with attention layers [8] is trained to segment subtypes of AP. We further investigate the effect of artery straightening through a comparison experiment in which the network is trained on original data.
2 Method
The main challenges for the segmentation of artery plaques include:
-
class imbalance (a lot more background voxels than plaque voxels)
-
high variability of the plaque appearance
-
high similarity between non-calcified plaques and background
To address the first two problems, we first extract and straighten vessel segments along artery centerlines as inputs, which restricts the volume-of-interest that alleviates the class imbalance and simplifies the analysis of lumen curvature variation and surrounding tissues. We also use a multi-class Dice loss function that increases the cost of segmentation mistakes on the plaques.
To better distinguish plaques from background tissues, we design a U-Net [9] like encoder-decoder network with residual blocks that preserve the signals from shallow layers, deep supervision that encourages multi-scale segmentation and attention layers [8] that helps to locate the pathologies.
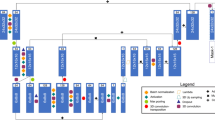
Figure 2 shows the workflow of the proposed method. The network architecture is described in detail in Sect. 2.3.
(a) Auto bounding box generation. Three convolutional neural networks (ConvNets) are trained on axial, sagittal and coronal planes respectively and determine the boundary of coronary artery on each axis. (b) Illustration of the 3D MPR technique. Cross-sectional planes are extracted along the centerline and stacked into a rectangular volume.
2.1 Auto Bounding Box Detection
To restrict the volume-of-interest for computation reduction, three ConvNets are employed to determine the bounding box around the heart for the comparison method (Fig. 3(a)). Slices along different directions are given a label (1 for slice with coronary). We use 100 additional volumes to train the ConvNets. Each ConvNet is a pretrained AlexNet [10], while we added an additional neuron with sigmoid activation on top of the original AlexNet output for the binary classification and fine-tuned the networks on three orthogonal planes (axial, sagittal, coronal). We chose the utmost predicted foreground slices as the boundary of the bounding box to make sure all the coronary arteries were enclosed.
2.2 Coronary Artery Extraction and Straightening
The coronary artery trees are extracted by MSCAR-DBT [11] which only requires two manually identified seed points. Vessels with diameter less than 2 mm are left out. The centerlines is then extracted based on the coronary artery extraction results.
We reform the artery segments into straightened volumes using 3D multi-planar reformation technique, which is illustrated in Fig. 3(b). We first subdivide the centerline into n segments with a equal length of 0.5 mm, and obtain \( n+1 \) evenly spaced control points \( P_m \,(1\le m \le n) \) along the centerline. Then we extract cross-sectional planes with a size of 32 \( \times \) 32 pixels at each control points and stack them into a rectangular volume. We define the reference vector  as a unit vector parallel to the y-axis of \( \sum _P \). Let
as a unit vector parallel to the y-axis of \( \sum _P \). Let  to be the normalized tangent to the centerline at \( P_m \). The bases of the cross-sectional plane at \( P_m \) are calculated by:
to be the normalized tangent to the centerline at \( P_m \). The bases of the cross-sectional plane at \( P_m \) are calculated by:

Equation 2 defines the mapping of point P(i, j, k) in the straightened space \( \sum _S\) to the original physical coordinate \( \sum _P\).

The reformed voxel values are obtained by bicubic interpolation. We cut the volumes that are longer than 128 voxels into several overlapping segments. The segmentation results for overlapped areas are obtained by averaging the output of the overlapping segments.
2.3 Network Architecture
The network is an encoder-decoder network shown in Fig. 4. The left part shows the encoding path. In each stage, one to three convolutional layers extract abstract context information. Each layer is with kernels of 3 \( \times \) 3 \( \times \) 3, symmetric padding, instance normalization and PRelu non linearity. We formulate each stage as a residual block, that is the input of each stage is added to the output of the last layer of the stage. Then convolutions with stride two are applied to halve the resolution and double the number of feature channels. The right part of the network is the decoding path. Each stage has a similar structure with the left but consists of a concatenation with the corresponding feature maps from the attention layers of the contracting path. De-convolution is applied to increase feature map resolution and halve the number of feature channels.
The idea of attention layer is to use attention gates to implicitly learn to suppress irrelevant regions in the input while highlighting salient features useful for plaque segmentations, which is described detailedly in [8].
We employ deep supervision in the expansive path by combining output of different stages via element-wise summation to form the final network output, which forces the network to produce an accurate segmentation in an early stage.
To address the extreme class unbalance in our data, we use a multiclass dice loss function [12] instead of the conventional categorical crossentropy loss:
where u is the output of the network and v is a one hot encoding of the ground truth. For both u and v, i is the number of pixels and \(k\in K\) being the classes.
3 Experiment Results
3.1 Dataset
We collected ECG-gated 4D-CTA scans by Philip 256-iCT from 25 patients, among which 13 were diagnosed with AP. The 4D-CT data sets are constructed in 20 phases: 5%, 10%,..., 100%. The size of each slice is 512 \(\times \) 512 pixels with an isotropic resolution of 0.414 mm. The number of slices in each volume ranged from 213 to 358 with thickness of 0.335 mm. We pick 4 phases (25%, 45%, 55% and 75%) from each patient and obtain a dataset consist of 100 scans, which allows the network to learn the variance introduced by cardiac motion and enhances the generalization ability. We select 80 scans as training set and the rest as test set.
The APs were annotated and classified by 5 trained radiologists, each scan is only annotated by one radiologist and then examined by a second one. The annotations serve as ground truth.
3.2 Comparison Experiment
To investigate the effect of artery straightening, we train a second network with the same architecture on the original data. Figure 5 shows the workflow of the method for comparison experiment.
3.3 Training Procedure
We implemented the proposed network in Keras with TensorFlow backend. For the proposed method, the input size is \(64\times 64\times 128\); for the comparison method, the input size is \(128\times 128\times 128\). The initial learning rate was 0.0001, and we reduced the learning rate by 50% if the validation loss did not improve after 10 epochs. The weights were updated by stochastic gradient descent with Adam optimizer. We ran more than 200 epochs on four NVIDIA 1080 GPUs.
Segmentation Result. (a)–(c): CAP; (d)–(f): NCAP; (g)–(i): MCAP. CAP is easily segmented by both methods. However, NCAP shows similar intensity with surrounding tissues, and comparison method fails to distinguish a majority of the plaque from background in (f), while proposed method successfully segments the whole structure in (e). MCAP is a mixture of bright and gray area while comparison method only captures the bright part in (i).
We use extensive data augmentation techniques including rotation, scaling, flipping, and smooth dense deformations on both data and ground truth labels.
3.4 Results
We remapped the output of the 3D network to the original space with nearest interpolation for visualization. Figures 6 and 7 show some examples. We evaluate the performance of the proposed method by true positive rate (sensitivity), positive predictive value (PPV) and dice score. The dice score of two sets A and B is evaluated as \( {2|A \cap B|}/{(|A| + |B|)}. \)
The proposed method achieved dice scores of 0.83, 0.73 and 0.68 for CAP, NCAP and MCAP respectively on the test set. Table 1 compares the performance of the proposed method and comparison experiment. It is shown that both methods perform well on the segmentation of CAP while proposed achieves significant gains in the segmentation of NCAP and MCAP. In addition, the training time for proposed method is around 40% less than the comparison method because of the smaller input size, which also demonstrates the benefits of artery straightening. It is also observed during training that the comparison method is more prone to overfit due to the small dataset while the proposed method benefits from the MPR transformation that makes the data more uniform and reduces the gap between training and testing data.
Existing methods only segment one type of the plaques and mostly reports sensitivity. For calcified plaques(CAP), [13] reports a sensitivity of 91.24%. For non-calcified plaques(NCAP), [14] reports a sensitivity of 92.5%. The result of proposed method is comparable to existing results while generalizes to three subtypes.
4 Discussion and Conclusion
Studies has shown that multi-planar reconstructions are useful in the evaluation of CT scans of coronary arteries for human radiologists [15]. The comparison experiment shows that MPR also contributes to a better segmentation performance for convolutional neural networks.
The detection and segmentation of artery plaques is very challenging due to class imbalance, high similarity between non-calcified plaques and high variability of the plaque appearance. The proposed method achieved dice scores of 0.83, 0.73 and 0.68 for CAP, NCAP and MCAP respectively on the test set. However, there are several drawbacks of this study. First, the current dataset only consists of 100 scans from 25 patients, which may not be representative considering the large variability of plaques. Second, the label for each scan is annotated by only one radiologist, and thus the network learns the segmentation regarding inter-observer error, which may hinder the network performance. Third, the location priors of the plaques are not utilized. In addition, although the MPR helps to boost the segmentation performance, it relies on the centerline of coronary artery which can be difficult to extract with the presence of plaques; and the coronary artery extraction method we are using now is semi-automatic and can be replaced with recent fully automatic state-of-the-art coronary artery segmentation algorithms.
To summarize, in this study we propose a pipeline for automatic plaque segmentation, which shows potential value in clinical use. Results show that by artery extraction and straightening, the training time is reduced by 40% and the segmentation performance of NCAP and MCAP gains significantly. The proposed method remains to be improved and tested on larger datasets.
References
Pasterkamp, G., Falk, E., Woutman, H., Borst, C.: Techniques characterizing the coronary atherosclerotic plaque: influence on clinical decision making? J. Am. Coll. Cardiol. 36(1), 13–21 (2000)
Wolterink, J.M., Leiner, T., de Vos, B.D., van Hamersvelt, R.W., Viergever, M.A., Išgum, I.: Automatic coronary artery calcium scoring in cardiac CT angiography using paired convolutional neural networks. Med. Image Anal. 34, 123–136 (2016)
Agatston, A.S., Janowitz, W.R., Hildner, F.J., Zusmer, N.R., Viamonte, M., Detrano, R.: Quantification of coronary artery calcium using ultrafast computed tomography. J. Am. Coll. Cardiol. 15(4), 827–832 (1990)
Wolterink, J.M., Leiner, T., Viergever, M.A., Išgum, I.: Automatic coronary calcium scoring in cardiac CT angiography using convolutional neural networks. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9349, pp. 589–596. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24553-9_72
Lessmann, N., et al.: Automatic calcium scoring in low-dose chest CT using deep neural networks with dilated convolutions. IEEE Trans. Med. Imaging 37, 615 (2017)
Renard, F., Yang, Y.: Coronary artery extraction and analysis for detection of soft plaques in MDCT images. In: 2008 15th IEEE International Conference on Image Processing, pp. 2248–2251. IEEE (2008)
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Oktay, O., et al.: Attention U-Net: learning where to look for the pancreas. arXiv preprint arXiv:1804.03999 (2018)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Krizhevsky, A., Sutskever, I., Hinton, G.E.: Imagenet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems, pp. 1097–1105 (2012)
Zhou, C., et al.: Pulmonary vessel segmentation utilizing curved planar reformation and optimal path finding (crop) in computed tomographic pulmonary angiography (CTPA) for CAD applications. In: Medical Imaging 2012: Computer-Aided Diagnosis, vol. 8315, p. 83150N. International Society for Optics and Photonics (2012)
Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., Maier-Hein, K.H.: Brain tumor segmentation and radiomics survival prediction: contribution to the BRATS 2017 challenge. In: Crimi, A., Bakas, S., Kuijf, H., Menze, B., Reyes, M. (eds.) BrainLes 2017. LNCS, vol. 10670, pp. 287–297. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-75238-9_25
Santini, G., et al.: An automatic deep learning approach for coronary artery calcium segmentation. In: Eskola, H., Visnen, O., Viik, J., Hyttinen, J. (eds.) EMBEC & NBC 2017. EMBEC 2017, NBC 2017. IFMBE Proceedings, vol. 65, pp. 374–377. Springer, Singapore (2018). https://doi.org/10.1007/978-981-10-5122-7_94
Wei, J., et al.: Computerized detection of non-calcified plaques in coronary CT angiography: evaluation of topological soft gradient prescreening method and luminal analysis. In: Medical Physics, vol. 41, no. 8Part1 (2014)
AChenbaCh, S., Moshage, W., Ropers, D., BaChmann, K.: Curved multiplanar reconstructions for the evaluation of contrast-enhanced electron beam CT of the coronary arteries. Am. J. Roentgenol. 170(4), 895–899 (1998)
Acknowledgement
This work is supported by the National Natural Science Foundation of China under Grant 61622207.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Liu, J., Jin, C., Feng, J., Du, Y., Lu, J., Zhou, J. (2019). A Vessel-Focused 3D Convolutional Network for Automatic Segmentation and Classification of Coronary Artery Plaques in Cardiac CTA. In: Pop, M., et al. Statistical Atlases and Computational Models of the Heart. Atrial Segmentation and LV Quantification Challenges. STACOM 2018. Lecture Notes in Computer Science(), vol 11395. Springer, Cham. https://doi.org/10.1007/978-3-030-12029-0_15
Download citation
DOI: https://doi.org/10.1007/978-3-030-12029-0_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-12028-3
Online ISBN: 978-3-030-12029-0
eBook Packages: Computer ScienceComputer Science (R0)