Abstract
Objectives
Volumetric evaluation of coronary artery disease (CAD) allows better prediction of cardiac events. However, CAD segmentation is labor intensive. Our objective was to create an open-source deep learning (DL) model to segment coronary plaques on coronary CT angiography (CCTA).
Methods
Three hundred eight individuals’ 894 CCTA scans with 3035 manually segmented plaques by an expert reader (considered as ground truth) were used to train (186/308, 60%), validate (tune, 61/308, 20%), and test (61/308, 20%) a 3D U-net model. We also evaluated the model on an external test set of 50 individuals with vulnerable plaques acquired at a different site. Furthermore, we applied transfer learning on 77 individuals’ data and re-evaluated the model’s performance using intra-class correlation coefficient (ICC).
Results
On the test set, DL outperformed the currently used minimum cost approach method to quantify total: ICC: 0.88 [CI: 0.85–0.91] vs. 0.63 [CI: 0.42–0.76], noncalcified: 0.84 [CI: 0.80–0.88] vs. 0.45 [CI: 0.26–0.59], calcified: 0.99 [CI: 0.98–0.99] vs. 0.96 [CI: 0.94–0.97], and low attenuation noncalcified: 0.25 [CI: 0.13–0.37] vs. −0.01 [CI: −0.13 to 0.11] plaque volumes. On the external dataset, substantial improvement was observed in DL model performance after transfer learning, total: 0.62 [CI: 0.01–0.84] vs. 0.94 [CI: 0.87–0.97], noncalcified: 0.54 [CI: −0.04 to 0.80] vs. 0.93 [CI: 0.86–0.96], calcified: 0.91 [CI:0.85–0.95] vs. 0.95 [CI: 0.91–0.97], and low attenuation noncalcified 0.48 [CI: 0.18–0.69] vs. 0.86 [CI: 0.76–0.92].
Conclusions
Our open-source DL algorithm achieved excellent agreement with expert CAD segmentations. However, transfer learning may be required to achieve accurate segmentations in the case of different plaque characteristics or machinery.
Key Points
• Deep learning 3D U-net model for coronary segmentation achieves comparable results with expert readers’ volumetric plaque quantification.
• Transfer learning may be needed to achieve similar results for other scanner and plaque characteristics.
• The developed deep learning algorithm is open-source and may be implemented in any CT analysis software.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Coronary artery disease (CAD) is responsible for the majority of avoidable deaths among elderly individuals, and the prevalence of CAD is still increasing [1, 2]. Therefore, more accurate estimates of diseases burden are needed to identify individuals at risk of later major adverse cardiac events (MACE). Coronary computed tomography angiography (CCTA) is an effective noninvasive tool to visualize and quantify CAD. Recent societal guidelines have endorsed CCTA as the primary tool to assess CAD in symptomatic patients [3, 4]. However, despite the isovolumetric image acquisition properties of CCTA allowing for volumetric quantification of CAD, in clinical practice, visual assessment is used to describe the composition and quantity of CAD [5, 6]. This is mostly due to the time-consuming and labor-intensive nature of quantitative plaque assessment as it requires trained and experienced readers to do the segmentations [7].
Volumetric plaque assessment, especially with regard to compositional measurements, has the potential to improve MACE prediction and overall clinical outcomes [8,9,10]. Furthermore, these segmentations can also be used for radiomics and machine learning research, which have shown promising results and may shape future evaluation of CAD [11, 12]. However, without robust and accurate automated segmentation algorithms, widespread adaption of volumetric analysis in clinical practice is not possible. Previously, various gradient-based methods like the variants of model-guided minimum cost approach (MCA) have been used to delineate vessel inner and outer contours [13]. However, these techniques provide moderate accuracy, as the segmentation of the outer vessel wall is challenging as Hounsfield unit (HU) differences between noncalcified plaque components and perivascular fat tissue are modest [14]. Recently, deep learning (DL) algorithms have been used to automate the segmentation of medical images [15]. Several promising results in the area of cardiovascular imaging show that the U-net DL architecture is especially well suited to accurately define given organs or abnormalities on radiological images [16, 17]. However, there is no information on whether DL may outperform previously used techniques to delineate coronary atherosclerotic lesions.
Therefore, our aim was to develop and evaluate the performance of an open-source DL model for the segmentation of CAD on CCTA. Furthermore, we wished to assess the performance of the model on an external test set containing larger vulnerable plaques imaged using different machinery, and to assess whether transfer learning may increase the accuracy of the model.
Materials and methods
Study design, participants, and scanning parameters of the internal dataset
Between May 2004 and August 2015, 1,429 individuals without cardiovascular symptoms were prospectively enrolled in an epidemiological observational study investigating the effects of human immunodeficiency virus infection and associated factors on CAD in the USA [18, 19]. Previously, randomly selected 308 study participants’ 894 CCTAs were manually segmented to infer the effects of human immunodeficiency virus, cocaine use, and cardiovascular risk factors on CAD [12, 20, 21]. The resulting dataset is referred to as the internal dataset. In the present investigation, we use the manual segmentation masks to train and validate an automatic DL algorithm for the segmentation of coronary atherosclerotic plaques.
Inclusion, exclusion criteria, and patient characteristics have been published previously [12, 18,19,20]. The Institutional Review Board approved the study protocol, and all study participants provided written informed consent. All procedures were in accordance with HIPAA, local and federal regulations, and the Declaration of Helsinki.
Detailed imaging parameters have been published [12, 18,19,20] and are available in the supplemental material.
Study design, participants, and scanning parameters of the external validation dataset
We identified 127 vulnerable plaques of 127 consecutive individuals who underwent CCTA between August 1st and December 31st of 2017 at a European tertiary cardiac center. This dataset was previously used to assess how image quality and segmentation affect volumetric and radiomic parameters of coronary plaques [14, 22]. The resulting dataset is referred to as the external dataset and is used to validate our results. Detailed inclusion, exclusion criteria, and scanning parameters have been published previously [14, 22, 23] and are detailed in the supplementary material.
Segmentation and volumetric quantification of CAD
Coronary plaque was defined as any discernible structure that could be assigned to the coronary artery wall and could be identified in at least 2 independent planes resulting in at least a 10% reduction in lumen caliber [24]. All segmentations were done using a dedicated software (QAngioCT software v3.1.3.13; Medis Medical Imaging Systems), which determined the inner and outer vessel contours [13]. Plaque volumes resulting from these automatic segmentations are referred to as MCA plaque volumes. The MCA segmentations were then manually corrected if needed by a level-3 certified specialist in cardiac CT imaging (M.K. with 7 years of experience) for both the external and internal datasets. Plaque volumes resulting from manual segmentations are referred to as manual plaque volumes.
We stratified total plaque volume based on HU values to noncalcified plaque volume: −100 to 350 HU, calcified plaque volume: ≥ 351 HU, and low attenuation noncalcified plaque volume: −100 to 30 HU [25].
DL architecture
Detailed methodology can be found in the supplementary material. In brief, after the determination of the coronary centerlines, a dedicated software tool (QAngioCT software v3.1.3.13; Medis Medical Imaging Systems) generated the straight multiplanar reconstruction (MPR) images which were used as inputs to the DL models. The images were then converted to patches of 24 × 32 × 32 voxels with a stride of 12 pixels along the long axis. We implemented an attention U-net DL model for the segmentation task [26]. U-net models have previously been proven to be suitable for automatic cardiac/coronary segmentation [27, 28].
The DL network was implemented in Python, using Keras (v2.3.1) with TensorFlow (v2.1.0) backend. Hyperopt (v0.2.5) package was used to optimize hyperparameters of the network during training. All codes developed, including the fitted models, are available at https://github.com/balinthomonnay/coronary_cta_prediciton.
DL training and testing design
In the internal dataset, the individuals were randomly divided into three separate groups. Sixty percent of the study participants was the training set, 20% was the validation set (tuning set), and the remaining 20% was used for testing the algorithm. If an individual had multiple CCTA datasets, then for training and validation all available data was used, while for testing a single CCTA scan was selected randomly.
The external dataset contained 127 coronary plaques from 127 individuals’ 127 CT scans. The data was split into training (50% of the plaques: 64/127), validation (tuning set, 10% of the plaques: 13/127), and test set (40% of the plaques: 50/127). The external dataset contained fewer plaques than the internal; therefore, a different split percentage was used on the external dataset in order to have enough plaques for transfer learning.
Statistical analysis
Data are represented as medians and interquartile ranges (IQR) or frequencies and percentages as appropriate. Kruskal-Wallis one-way ANOVA with Dunn’s nonparametric all-pairs comparison test with Holm’s p value correction was used for multiple comparisons. In the case of categorical variables, we used the chi-square test, with pairwise comparisons corrected using Holm’s p value correction for multiple comparisons. To compare the resulting plaque volumes, we first compared them using the Friedman rank sum test with Durbin’s (Conover-Iman) all-pairs comparison test with Holm’s p value correction for multiple comparisons. Then, we quantified the accuracy of the segmentations using the intra-class correlation coefficient (ICC, two-way random, single measures, absolute agreement) with 95% confidence intervals (CI). To assess potential biases between the resulting plaque volumes for the different segmentations, we conducted Bland-Altman analyses between the different segmentations. All statistical calculations were performed in the R environment (v4.0.0) [29]. Methodologies and results are reported in accordance with the Reporting Diagnostic Accuracy Studies (STARD) guidelines [30].
Results
Participant population — internal dataset
Altogether, 308 individuals’ (age at first CCTA: 49.1±6.9 years, 69.2% males) 894 CCTA scans (average time between scans: 3.6±2.7 years) contained a total of 3035 coronary plaques. The individuals were split into training (60%, 186/308 of individuals’ 532 scans, 1693 plaques), validation (tuning) (20%, 61/308 participants’ 177 CCTA scans, 625 plaques), and testing (20%, 61/308 of individuals’ 185 scans). In the case of the testing dataset, for individuals with multiple scans, we randomly selected one single case resulting in 61 scans of 61 individuals with 219 plaques.
Plaque volumes distributions — internal dataset
Based on manual segmentation, the median total plaque volumes were 27.8 mm3 (IQR: 12.6–56.4 mm3; range: 0.2–588.3 mm3). Noncalcified, calcified, and low attenuation noncalcified plaque volumes were 22.9 mm3 (IQR: 10.0–45.8 mm3; range: 0.2–583.3 mm3), 0.4 mm3 (IQR: 0.0–8.5 mm3; range: 0.0–363.8 mm3), and 0.1 mm3 (IQR: 0.0–0.8 mm3; range: 0.0–79.0 mm3) respectively. The only significant difference between the three groups regarding plaque volumes was in calcified plaque volumes. Distribution of plaque volumes in the three groups can be found in Supplementary Table 1.
Training of the DL model
The DL model was trained on the 1693 plaques of the 532 CCTA scans from 186 individuals in the training dataset. The optimal hyperparameters are reported in the supplementary material. The model architecture is shown in Fig. 1. All codes developed, including the fitted models, are available at https://github.com/balinthomonnay/coronary_cta_prediciton.
Block diagram of the 3D U-net model architecture for coronary plaque segmentation. In each box, the numbers denote the various convolutional filter sizes [24 × 32 × 32, 12 × 16 × 16, 6 × 8 × 8], and the number of channels [1, 64, 128]. The input layer is 24 × 32 × 32 voxels in size. All of the convolutions were identically padded. Each of the 3 outputs is in the same dimensions as the input layer, representing the inner mask, outer mask, and the plaque. The inner mask consists of all voxels surrounded by the inner arterial luminal wall; the outer mask represents all voxels encompassed by the whole vessel; and the plaque is defined as a subtraction of the inner and outer masks
Results on the internal test set: statistical comparisons and ICC analysis
The median total plaque volume was 25.9 mm3 (IQR: 12.1–56.5 mm3) in the test set based on manual segmentations. The MCA algorithm resulted in significantly lower plaque volumes (11.4 mm3 [IQR: 0.0–33.0 mm3]) as compared to the manual segmentations (p < 0.0001), while the DL model achieved comparable plaque volumes which were not statistically different from the manual segmentation (24.4 mm3 [IQR: 10.5–57.1 mm3], p = 0.74). Similar results were observed for noncalcified and calcified plaque volumes, where the MCA algorithm resulted in significantly different values, while the DL-based method provided comparable results. In the case of low attenuation noncalcified plaque volume, all three segmentations were statistically different (manual: 0.1 mm3 [IQR: 0.0–1.0 mm3]; MCA: 0.0 mm3 [0.0–0.0 mm3]; DL: 0.0 mm3 [IQR: 0.0–0.5 mm3]; p < 0.0001 for omnibus test, respectively, pairwise tests: manual vs. MCA: p < 0.0001, Manual vs. DL: p = 0.03, MCA vs. DL-based: p < 0.0001). Results are presented in Table 1.
The DL-based segmentation method achieved excellent ICC values (ICC = 0.88 [CI: 0.85–0.91]) for total plaque volumes, higher than the MCA method (ICC = 0.63 [CI: 0.42–0.76]). Similar results were true for noncalcified (ICC: 0.84 [CI: 0.80–0.88] vs. 0.45 [CI: 0.26–0.59] respectively) and calcified plaque volumes (ICC: 0.99 [CI: 0.98–0.99] vs. 0.96 [CI: 0.94–0.97] respectively). In the case of low attenuation noncalcified plaque volumes, the DL-based algorithm outperformed the MCA method; however, both algorithms had poor accuracies (ICC: 0.25 [CI: 0.13–0.37] vs. −0.01 [CI: −0.13 to 0.11] respectively). Scatter plots of the plaque volumes are presented in Fig. 2.
Scatter plots and ICC values of the plaque volumes from the internal test set. For each plaque component, the deep learning algorithm provided superior ICC values as compared to the conventional MCA method. However, in the case of low attenuation noncalcified plaque volume, both algorithms provided poor agreement. DL, deep learning; ICC, intra-class correlation coefficient; MCA, minimum cost approach
Results on the internal test set: Bland-Altman analysis
The DL-based segmentation algorithm produced near 0 bias for total, noncalcified, and calcified plaque volumes (mean difference: −0.7 mm3, −0.9 mm3, and 0.2 mm3, respectively), while the MCA method had higher bias (mean difference: −18.7 mm3, −17.1 mm3, and −1.6 mm3, respectively). Furthermore, the DL model had smaller variance regarding total, noncalcified, and calcified plaque volumes (95% limits of agreement: −48.5 to 47.0 mm3, −47.2 to 45.4 mm3, and −5.1 to 5.4 mm3, respectively) as compared to the MCA method (95% limits of agreement: −86.9 to 49.5 mm3, −84.3 to 50.1 mm3, and −9.4 to 6.2 mm3, respectively). In the case of low attenuation noncalcified plaque volume, both algorithms had relative high bias and variance as compared to the absolute values themselves (mean difference: −1.1 mm3 vs. −1.8 mm3, 95% limits of agreement: −12.1 to 9.8 mm3 vs. −13.6 to 10.1 mm3, respectively). Bland-Altman plots are presented in Fig. 3.
Results on the external test set prior to transfer learning
All plaque volumes were higher in the external test (plaque n = 50) set as compared to the test set of the initial cohort (plaque n = 219, total plaque volume: 163.0 mm3 (IQR: 105.1–243.9 mm3; range: 50.8–610.8 mm3), noncalcified plaque volume: 146.6 mm3 (IQR: 91.2–215.4 mm3; range: 32.8–596.6 mm3), calcified plaque volume: 14.5 mm3 (IQR: 3.6–28.0 mm3; range: 0.2–151.5 mm3), and low attenuation noncalcified plaque volume: 30.1 mm3 (IQR: 12.1–57.9 mm3; range: 1.1–193.1 mm3, respectively, p < 0.0001 for all).
Prior to transfer learning, the MCA and DL segmentation algorithms only provided comparable results to the ground truth in the case of calcified plaque volumes, while for total and noncalcified plaque volumes the two methods produced similar values. In the case of low attenuation noncalcified plaque volume, the DL model provided significantly better results than the MCA method; however, this was still significantly different than the ground truth values. Detailed results are presented in the supplementary material, Table 2, and Figs. 4, 5.
Scatter plots and ICC values of the plaque volumes from the external test set. Prior to transfer learning, both the DL and the MCA segmentations showed moderate agreement with the manual segmentations. However, after applying transfer learning to the DL model, it showed excellent ICC values for all plaque components. DL, deep learning; ICC, intra-class correlation coefficient; MCA, minimum cost approach
Bland-Altman plots of the external test set. Agreements are shown between the ground truth manual segmentation and the MCA and the DL-based model before and after applying transfer learning. Except for the calcified plaque volume, there is a substantial bias for all plaque type subcomponents, in both the MCA segmentation and the DL segmentation before transfer learning. After transfer learning, the DL model’s performance substantially improved, resulting in low bias and variance. DL, deep learning; 95% LOA, 95% limit of agreement; MCA, minimum cost approach
Results on the external test set after transfer learning
After transfer learning the fitted model using 64 plaques for training and 13 for validation (tuning), we observed a substantial improvement in the predicted volumes on the external test set (n = 50) (Table 2). This resulted in excellent ICC values for all plaque components, total plaque volume: ICC = 0.94 [CI: 0.87–0.97], noncalcified plaque volume: ICC = 0.93 [CI: 0.86–0.96], calcified plaque volume: ICC = 0.95 [CI: 0.91–0.97], and low attenuation noncalcified plaque volume: ICC = 0.86 [CI: 0.76–0.92] (Fig. 4). Furthermore, there was a substantial improvement resulting in lower biases and variance (Bland-Altman analysis results are presented in Fig. 5).
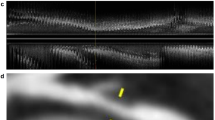
Dice coefficients for both internal and external datasets are provided in Supplementary Table 2. Also, a representative example showing plaque cross-sections where the DL algorithm correctly identified the plaque while the MCA did not, and an example where both failed, is presented in Fig. 6.
Representative examples of MCA and DL segmentations. The top row depicts a coronary plaque cross-section in which case the DL algorithm correctly identified the atherosclerotic lesion, while the MCA algorithm did not. The bottom row shows a cross-section where both algorithms failed to correctly identify the plaque. DL, deep learning; MCA, minimum cost approach
Discussion
We developed an open-source DL algorithm for the segmentation of coronary plaques on CCTA which resulted in excellent agreement with manual segmentations. Our open-source DL algorithm outperformed the conventionally available MCA algorithm. However, agreement with manually measured low attenuation noncalcified plaque volumes was moderate. To further validate the model, we obtained an external dataset, which consisted of significantly larger vulnerable plaques scanned using different machinery. Initially, our model had better accuracies than a conventional MCA method, but agreement was moderate. After transfer learning our model using images from the same machinery, we observed significantly better results with excellent agreement as compared to manual measurements. To facilitate the implementation of volumetric plaque quantification in daily clinical practice, we have made our fitted models publicly available at https://github.com/balinthomonnay/coronary_cta_prediciton.
Quantitative plaque assessment beyond the evaluation of coronary artery stenosis severity has proven to have incremental value in the evaluation of CAD [8, 9, 31,32,33]. However, contouring a single vessel can take up to 30 min, which greatly limits its application in the daily clinical routine [34]. Nevertheless, if we wish to utilize the additional information present in plaque volumes, we need a fast, robust method for the delineation of atherosclerotic lesions.
Several machine learning–based approaches focus on coronary calcium scoring or stenosis severity classification. By employing a recurrent convolutional neutral network (which is the combination of a 3D convolutional neutral network and a recurrent neutral network), Zreik et al have demonstrated the feasibility of automatic plaque characterization (no plaque, noncalcified, mixed, calcified) and qualitative stenosis assessment by achieving an accuracy of 0.77 and 0.80, respectively [35]. Liu et al used a vessel-focused 3D convolutional network to label coronary plaques as calcified, mixed, and noncalcified with a dice score of 0.83, 0.68, and 0.73, respectively [36]. However, there is limited data on automated plaque volume assessment using DL methods.
Our results show that despite the subtle differences in atherosclerotic and perivascular tissue attenuation and the small size of the coronary lesions, DL is able to provide accurate estimates of plaque volumes. Total, noncalcified, and calcified plaque volumes of the DL model resulted in comparable results with manual segmentation, unlike the MCA algorithm, which provided significantly different values on the internal test set. Also, the limits of agreements had smaller ranges for DL segmentations as compared to the MCA algorithm, but they were still substantial considering the average volumes of the plaques. Furthermore, both for DL and MCA, the ICC values for low attenuation noncalcified plaque volumes were poor. Differentiating between low attenuation noncalcified plaques and its surrounding epicardial fat tissue might be the most challenging for both human readers and automatic algorithms. The number of plaques with low attenuation noncalcified volumes was lower in our training set, whereas in the external test set they were artificially overrepresented. These circumstances may explain the initial poor performance of quantifying low attenuation noncalcified plaques, which significantly improved after transfer learning. Nevertheless, the reproducibility of this plaque component has been shown to be poor among experts, therefore questioning whether this specific plaque component can actually be accurately measured [14].
When testing the generalizability of our model on an external dataset of vulnerable plaques showing significantly larger plaque volumes imaged on different machinery, we found that our model did not generalize well. The DL model outperformed the MCA model, but the ICC values were only moderate. However, after applying transfer learning, we found that the DL model again achieved excellent accuracies for all plaque compositions. These results indicate that creating DL segmentation models which generalizes well to all situations is difficult. Nevertheless, our results indicate that if there is an available model trained on sufficient amount of data, then fine tuning it to new data is easy even with limited data, and the results significantly outperform the general model. However, both the internal and external test set results highlight that in some cases manual editing of the contours may be needed as there are cases when the DL automatic segmentation is insufficient.
Our study has some limitations. The inputs of the 3D U-net were single plaques, not whole coronary arteries; hence, manual marking of the plaques is necessary. Furthermore, the proposed DL model only enables volumetric plaque quantification, and it was not trained for stenosis assessment. Also, despite our external validation set, the datasets only contain images from two companies; therefore, generalizability to other scanners may be different. A further limitation is that all datasets were manually segmented by the same level-3 certified specialist in cardiac CT imaging. However, the lack of inter-reader variability also allows more accurate comparisons between the algorithms as the observed differences are not the result of possible differences in manual segmentations. Also, both the internal and external datasets had a low prevalence of calcified plaques. However, we focused on noncalcified plaques as currently available algorithms yield a high accuracy on segmenting calcified lesions, unlike noncalcified ones. Furthermore, in several cases, repeated CCTA scans of the same patient were used to train the model, potentially limiting the accuracy of the model.
In conclusion, our DL model achieved comparable results to manual segmentations and significantly outperformed currently used MCA methods for volumetric plaque quantification. On an external test set consisting of larger vulnerable plaques imaged on different hardware, we showed that applying our model only resulted in moderate accuracy. However, transfer learning on a small dataset was able to fine-tune the model to later have excellent accuracy. We have made our fitted models publicly available to the community to facilitate the implementation of volumetric plaque quantification in daily clinical practice. We encourage the scientific community to provide transfer learned models on other machinery and software providers to implement these models into their CAD analysis software, so that we may all increase the quality of care and help reduce the burden of coronary atherosclerosis.
Abbreviations
- CAD:
-
Coronary artery disease
- CCTA:
-
Coronary CT angiography
- CI:
-
Confidence intervals
- DL:
-
Deep learning
- HU:
-
Hounsfield unit
- ICC:
-
Intra-class correlation coefficient
- MACE:
-
Major adverse cardiac events
- MCA:
-
Minimum cost approach
- MPR:
-
Multiplanar reconstruction
References
Mathers CD, Stevens GA, Boerma T, White RA, Tobias MI (2015) Causes of international increases in older age life expectancy. Lancet 385:540–548
GBD 2017 Causes of Death Collaborators (2018) Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980-2017: a systematic analysis for the Global Burden of Disease Study 2017
Knuuti J, Wijns W, Saraste A et al (2020) 2019 ESC Guidelines for the diagnosis and management of chronic coronary syndromes. Eur Heart J 41:407–477
Moss AJ, Williams MC, Newby DE, Nicol ED (2017) The updated NICE guidelines: cardiac CT as the first-line test for coronary artery disease. Curr Cardiovasc Imaging Rep 10:15
Leipsic J, Abbara S, Achenbach S et al (2014) SCCT guidelines for the interpretation and reporting of coronary CT angiography: a report of the Society of Cardiovascular Computed Tomography Guidelines Committee. J Cardiovasc Comput Tomogr 8:342–358
Kolossvary M, Szilveszter B, Merkely B, Maurovich-Horvat P (2017) Plaque imaging with CT-a comprehensive review on coronary CT angiography based risk assessment. Cardiovasc Diagn Ther 7:489–506
Williams MC, Earls JP, Hecht H (2021) Quantitative assessment of atherosclerotic plaque, recent progress and current limitations. J Cardiovasc Comput Tomogr. https://doi.org/10.1016/j.jcct.2021.07.001
Lee SE, Sung JM, Rizvi A et al (2018) Quantification of coronary atherosclerosis in the assessment of coronary artery disease. Circ Cardiovasc Imaging 11:e007562
Ferencik M, Mayrhofer T, Puchner SB et al (2015) Computed tomography-based high-risk coronary plaque score to predict acute coronary syndrome among patients with acute chest pain--results from the ROMICAT II trial. J Cardiovasc Comput Tomogr 9:538–545
Williams MC, Kwiecinski J, Doris M et al (2020) Low-attenuation noncalcified plaque on coronary computed tomography angiography predicts myocardial infarction: results from the multicenter SCOT-HEART trial (Scottish Computed Tomography of the HEART). Circulation 141:1452–1462
Kolossváry M, Karády J, Szilveszter B et al (2017) Radiomic features are superior to conventional quantitative computed tomographic metrics to identify coronary plaques with napkin-ring sign. Circ Cardiovasc Imaging 10:e006843
Kolossvary M, Gerstenblith G, Bluemke DA et al (2021) Contribution of risk factors to the development of coronary atherosclerosis as confirmed via coronary CT angiography: a longitudinal radiomics-based study. Radiology 299:97–106
Boogers MJ, Broersen A, van Velzen JE et al (2012) Automated quantification of coronary plaque with computed tomography: comparison with intravascular ultrasound using a dedicated registration algorithm for fusion-based quantification. Eur Heart J 33:1007–1016
Kolossvary M, Javorszky N, Karady J et al (2021) Effect of vessel wall segmentation on volumetric and radiomic parameters of coronary plaques with adverse characteristics. J Cardiovasc Comput Tomogr 15:137–145
Chen C, Qin C, Qiu H et al (2020) Deep learning for cardiac image segmentation: a review. Front Cardiovasc Med 7:25
Zeleznik R, Foldyna B, Eslami P et al (2021) Deep convolutional neural networks to predict cardiovascular risk from computed tomography. Nat Commun 12:715
van Velzen SGM, Lessmann N, Velthuis BK et al (2020) Deep learning for automatic calcium scoring in CT: validation using multiple cardiac CT and chest CT protocols. Radiology 295:66–79
Lai S, Fishman EK, Lai H et al (2008) Long-term cocaine use and antiretroviral therapy are associated with silent coronary artery disease in African Americans with HIV infection who have no cardiovascular symptoms. Clin Infect Dis 46:600–610
Lai H, Moore R, Celentano DD et al (2016) HIV infection itself may not be associated with subclinical coronary artery disease among African Americans without cardiovascular symptoms. J Am Heart Assoc 5:e002529
Kolossvary M, Fishman EK, Gerstenblith G et al (2021) Cardiovascular risk factors and illicit drug use may have a more profound effect on coronary atherosclerosis progression in people living with HIV. Eur Radiol 31:2756–2767
Kolossvary M, Celentano D, Gerstenblith G et al (2021) HIV indirectly accelerates coronary artery disease by promoting the effects of risk factors: longitudinal observational study. Sci Rep 11:23110
Kolossvary M, Szilveszter B, Karady J, Drobni ZD, Merkely B, Maurovich-Horvat P (2019) Effect of image reconstruction algorithms on volumetric and radiomic parameters of coronary plaques. J Cardiovasc Comput Tomogr 13:325–330
Karady J, Panajotu A, Kolossvary M et al (2017) The effect of four-phasic versus three-phasic contrast media injection protocols on extravasation rate in coronary CT angiography: a randomized controlled trial. Eur Radiol 27:4538–4543
Newby DE, Adamson PD, Berry C et al (2018) Coronary CT angiography and 5-year risk of myocardial infarction. N Engl J Med 379:924–933
Inoue K, Motoyama S, Sarai M et al (2010) Serial coronary CT angiography-verified changes in plaque characteristics as an end point: evaluation of effect of statin intervention. JACC Cardiovasc Imaging 3:691–698
Oktay O, Schlemper J, Folgoc LL et al (2018) Attention U-Net: learning where to look for the pancreas. arXiv preprint arXiv:180403999
Pan LS, Li CW, Su SF, Tay SY, Tran QV, Chan WP (2021) Coronary artery segmentation under class imbalance using a U-Net based architecture on computed tomography angiography images. Sci Rep 11:14493
Siriapisith T, Kusakunniran W, Haddawy P (2021) A 3D deep learning approach to epicardial fat segmentation in non-contrast and post-contrast cardiac CT images. Peer J Comput Sci 7:e806
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, 4.0.0
Bossuyt PM, Reitsma JB, Bruns DE et al (2015) STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. Radiology 277:826–832
Diaz-Zamudio M, Dey D, Schuhbaeck A et al (2015) Automated quantitative plaque burden from coronary CT angiography noninvasively predicts hemodynamic significance by using fractional flow reserve in intermediate coronary lesions. Radiology 276:408–415
Park HB, Heo R, B Óh et al (2015) Atherosclerotic plaque characteristics by CT angiography identify coronary lesions that cause ischemia: a direct comparison to fractional flow reserve. JACC Cardiovasc Imaging 8:1-10
Nakazato R, Shalev A, Doh JH et al (2013) Aggregate plaque volume by coronary computed tomography angiography is superior and incremental to luminal narrowing for diagnosis of ischemic lesions of intermediate stenosis severity. J Am Coll Cardiol 62:460–467
Hell MM, Motwani M, Otaki Y et al (2017) Quantitative global plaque characteristics from coronary computed tomography angiography for the prediction of future cardiac mortality during long-term follow-up. Eur Heart J Cardiovasc Imaging 18:1331–1339
Zreik M, van Hamersvelt RW, Wolterink JM, Leiner T, Viergever MA, Isgum I (2019) A recurrent CNN for automatic detection and classification of coronary artery plaque and stenosis in coronary CT angiography. IEEE Trans Med Imaging 38:1588–1598
Liu J, Jin C, Feng J, Du Y, Lu J, Zhou J (2019) A vessel-focused 3D convolutional network for automatic segmentation and classification of coronary artery plaques in cardiac CTA. Springer International Publishing, Cham, pp 131–141
Acknowledgements
The authors thank the study participants. Without their participation and commitment, this study would not have been possible.
Funding
This study has received funding by Shenghan Lai.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Shenghan Lai.
Conflict of interest
The authors declare no competing interests.
Statistics and biometry
One of the authors has significant statistical expertise.
Informed consent
Written informed consent was obtained from all subjects (patients) in this study.
Ethics approval
Institutional Review Board approval was obtained.
Methodology
• diagnostic or prognostic study
• observational
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 25 kb)
Rights and permissions
About this article
Cite this article
Jávorszky, N., Homonnay, B., Gerstenblith, G. et al. Deep learning–based atherosclerotic coronary plaque segmentation on coronary CT angiography. Eur Radiol 32, 7217–7226 (2022). https://doi.org/10.1007/s00330-022-08801-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-08801-8