Abstract
Iron is an essential nutrient for all mammalian cells, and numerous proteins are dedicated to the uptake and distribution of iron within cells. As discussed in previous chapters, there are proteins that facilitate iron uptake, including transferrin receptors 1 and 2 (TfR1 and TfR2) [1] and DMT1 [2], proteins dedicated to iron storage, including ferritin H and L chains [3, 4], and an iron exporter, ferroportin [5, 6]. Most cellular proteins are synthesized in the cytosol and those that require an iron cofactor generally acquire their iron from the cytosolic iron pool. The nature of the cytosolic iron pool remains uncharacterized, as the iron is not associated with a specific known iron carrier molecule. The iron in this accessible iron pool may be bound with low affinity to abundant negatively charged molecules, including citrate and ATP. Because the cytosol of cells is a reducing environment, most cellular iron is likely in the ferrous (Fe2+) rather than in the ferric (Fe3+) state. Iron that enters the endosome in the transferrin cycle is initially in the ferric state, but an endosomal reductase, Steap3 [7], reduces it to ferrous iron, the form that is transported into cytosol by DMT1. When iron is taken up by ferritin, it is oxidized by ferroxidase activity of the ferritin H chain [3, 4], which facilitates precipitation and storage of iron in the ferritin heteropolymer.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Iron is an essential nutrient for all mammalian cells, and numerous proteins are dedicated to the uptake and distribution of iron within cells. As discussed in previous chapters, there are proteins that facilitate iron uptake, including transferrin receptors 1 and 2 (TfR1 and TfR2) [1] and DMT1 [2], proteins dedicated to iron storage, including ferritin H and L chains [3, 4], and an iron exporter, ferroportin [5, 6]. Most cellular proteins are synthesized in the cytosol and those that require an iron cofactor generally acquire their iron from the cytosolic iron pool. The nature of the cytosolic iron pool remains uncharacterized, as the iron is not associated with a specific known iron carrier molecule. The iron in this accessible iron pool may be bound with low affinity to abundant negatively charged molecules, including citrate and ATP. Because the cytosol of cells is a reducing environment, most cellular iron is likely in the ferrous (Fe2+) rather than in the ferric (Fe3+) state. Iron that enters the endosome in the transferrin cycle is initially in the ferric state, but an endosomal reductase, Steap3 [7], reduces it to ferrous iron, the form that is transported into cytosol by DMT1. When iron is taken up by ferritin, it is oxidized by ferroxidase activity of the ferritin H chain [3, 4], which facilitates precipitation and storage of iron in the ferritin heteropolymer.
To ensure that iron supplies are sufficient to support synthesis of new proteins and growth, cells coordinate iron uptake, storage, and export activities by regulating expression levels of the proteins responsible for these activities. Iron regulatory proteins 1 and 2 (IRP1 and IRP2) are homologous proteins that sense cytosolic iron levels and regulate expression of TfR1, one isoform of DMT1, ferritin H and L chains, ferroportin and several other iron metabolism proteins (reviewed in 8–10). In cells that are iron starved, IRP1 and IRP2 bind to conserved stem-loop structures found in the mRNAs that encode these proteins, which are known as iron-responsive elements (IREs). In the ferritin and ferroportin transcripts, the IREs are found in the 5′UTR near the cap-binding site where translation factors initially assemble. In TfR1 and DMT1, the IREs are found in the 3′UTR, a region of mRNA that is often important in determining the half-life of transcripts. The mechanism by which IRPs sense cytosolic iron levels and accordingly increase or decrease their capacity to bind to IREs is the major subject of this chapter.
2 The IRE–IRP Regulatory System
The IRE–IRP regulatory system is based on the ability of cytosolic IRPs to affect the stability of a previously synthesized mRNA, or to alter the efficiency with which a given transcript is translated. Unlike many cellular regulatory systems, most of the regulatory activity in the IRE–IRP system occurs post-transcriptionally [8–10]. An important key to the system is that target transcripts have evolved to contain one or more IREs. An IRE consists of conserved sequence and structural features [11–13]. It contains an upper stem composed of five base-paired residues that assume a helical structure, which is generally separated from a lower base-paired stem by an unpaired “bulged” cytosine near the 5′ end. The bulged C divides the upper from the lower stem, functioning as a hinge that enables the upper and lower stems to flexibly alter their relative alignments. In some IREs, such as those found in ferritin, the lower stem contains a second characteristic bulged “U” residue, which may further facilitate bending of the lower stem relative to the upper stem. In addition, IREs contain a six-membered loop, which contains a base-pair between residues 1 and 5 that structures the loop, enabling the A, G, and U residues at positions 2, 3, and 4 to protrude out (Fig. 3.1). The “bulged” C and the “AGU” pseudotriloop of the loop are distinctive molecular features that are separated from one another by a fixed distance and angle. They constitute the two major recognition features for binding by iron regulatory proteins [13, 14]. The bulged C fits into a binding pocket of domain 4 of IRP1, and the AGU residues fit into a binding pocket that opens between domains 2 and 3 of apo-IRP1.
The IRE is an RNA stem-loop structure. The upper and lower stems are composed of Watson–Crick base-pairs which are in a helical conformation. In the six-membered loop, the C at position 1 of the loop forms a base-pair with the G at position 5. The C–G base-pair structures the loop, allowing the A, G, and U residues (denoted by asterisks) to form a pseudotriloop of residues that can form multiple hydrogen bonds with protein(s). The upper and lower stems are separated by an unpaired “bulge” C that confers flexibility on the structure by interrupting the helix
Mammalian cells contain two iron regulatory proteins, IRP1 and IRP2, which differ in the mechanism by which they sense cellular iron status. IRP1 is a bifunctional protein, which, like IRP2, is expressed in virtually all mammalian cells. In cells that are iron replete, IRP1 contains a cubane iron–sulfur cluster and is a functional aconitase enzyme that interconverts citrate and isocitrate in cytosol, similar to the reaction catalyzed by mitochondrial aconitase in the citric acid cycle. Two distinct but homologous genes encode the cytosolic and mitochondrial aconitase genes. As there is also a distinct cytosolic isocitrate dehydrogenase, active cytosolic aconitase likely contributes to extra-mitochondrial metabolism in several ways, particularly perhaps by facilitating NADPH synthesis by cytosolic isocitrate dehydrogenase [15]. The iron–sulfur cluster of cytosolic aconitase is crucial for aconitase activity, as it is one of the ligands for the substrates citrate and isocitrate, along with over 23 amino acid residues that line that enzymatic active site cleft [16, 17].
The status of the iron–sulfur cluster is also the key to transformation of IRP1 into an IRE-binding protein, as the IRE-binding form lacks the cluster. The mechanism by which IRP1 converts into the apo-form devoid of an iron–sulfur cluster may involve the fundamental chemical properties of exposed iron–sulfur clusters. Iron–sulfur clusters are notoriously sensitive to oxidation events, which can cause spontaneous disassembly of the cluster. In the aconitases, iron–sulfur clusters are unusually sensitive to oxidation because the cluster is exposed to solvent [18]. Although disassembly of iron–sulfur clusters may depend on random oxidation events, it is also thought that disassembly of the cluster is facilitated when IRP1 is phosphorylated at serine 138 [19, 20]. In contrast to cluster disassembly, numerous proteins dedicated to assembly of iron–sulfur clusters are expressed in mammalian cells and it is likely that the iron–sulfur cluster of cytosolic aconitase is reconstituted by reassembly after spontaneous degradation in cells, but only when cells contain sufficient iron to resynthesize the cluster [21–23].
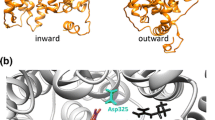
Loss of the iron–sulfur cluster of IRP1 permits IRP1 to undergo a conformational change that transforms it into an IRE-binding protein. Solution of the structure of the IRE bound to IRP1 confirmed that major conformational changes were required for acquisition of IRE-binding activity [13, 14] (Fig. 3.2). The bulged C fits into a pocket within domain 4 of IRP1, which the RNA can access because domain 4 of apo-IRP1 (apo-IRP1 refers to IRP1 devoid of an iron–sulfur cluster) rotates on a hinge linker that connects it to domains 1–3. Opening of the active site cleft allows the residues that normally face into the aconitase active-site cleft to interact with large molecules that would otherwise be excluded from the narrow cleft. Another important change occurs between domains 2 and 3 in apo-IRP1, where conformational changes create a pocket that accommodates the unpaired AGU residues of the loop. Most of the important RNA–protein interactions occur between the bulged C and its binding pocket, and the AGU pseudotriloop and its pocket, and the multiple hydrogen bonds at both sites (about 12 per site) account for the very high-affinity binding (in the picomolar range) that has been observed [13]. Thus, IRP1 functions as an aconitase when it contains an intact iron–sulfur cluster, whereas it functions as an IRE-binding protein when it lacks an iron–sulfur cluster.
IRP1 functions as a cytosolic aconitase or an IRE-binding protein. When IRP1 contains an iron–sulfur cluster, it functions as a cytosolic aconitase, whereas upon loss of the cluster, it undergoes a significant conformational change that enables it to bind IREs with high affinity. The bulged C of the IRE fits into a pocket in domain 4, whereas residues of the pseudotriloop reach into a pocket opened by movement of domain 3 relative to domain 2
IRP2 is a second IRE-binding protein that is highly homologous to IRP1 and binds with high affinity to IREs [24, 25]. However, unlike IRP1, IRP2 does not appear to have another function or to bind an iron–sulfur cluster. In addition, IRP2 is very difficult to find in iron-replete cells, because it undergoes iron-dependent degradation in vivo [26, 27]. Complicating matters further, IRP2 is subject to rapid degradation in lysates that contain iron and are exposed to oxygen, and analysis of IRP2 levels and binding activity can be misleadingly low unless assays are performed in lysates that contain a strong iron chelator such as desferrioxamine, or in lysates that are made and tested under anaerobic conditions [28]. The initial step of iron-dependent degradation of IRP2 in cells likely involves some type of iron-dependent oxidation, but its exact nature is not known [29]. Although there is agreement that the degradation pathway of IRP2 involves ubiquitination and proteasomal degradation, the initial identification of the ubiquitin ligase [30] has been disputed [31]. IRP2 contains a 73 amino acid insertion relative to IRP1 that was initially thought to be important for degradation [26], but it appears that features outside of this cysteine- and proline-rich domain are more important for iron-dependent degradation [32, 33]. Thus, much remains to be discovered about the intracellular iron-dependent degradation pathway of IRP2. Nevertheless, IRP2 levels and binding activity appear to accurately reflect cytosolic iron status, and since the intracellular iron pool that is sensed by IRP2 cannot be directly measured, IRP2 levels often are used to indirectly assess intracellular iron levels.
Thus, in iron-replete cells, IRE-binding activities of both IRP1 and IRP2 are reduced, either because insertion of an iron–sulfur cluster promotes a conformational change, or an iron-dependent modification promotes degradation by the ubiquitin–proteasome system. In each case, IRE-binding activity correlates with levels of iron in the cytosolic pool of iron. Both IRP1 and IRP2 bind to most IREs with similar affinity and they therefore each have the potential to provide backup to a system that relies on their combined binding activity. In iron-starved cells, IRP binding represses translation of mRNAs that contain IREs near the 5′ end of the transcript, including ferritin and ferroportin [34], whereas it stabilizes the transcript of TfR1 and thereby increases expression of TfR1 protein. These changes are appropriate for intracellular iron homeostasis, as expression of ferritin should be diminished in cells that are iron starved, whereas expression of TfR1 should be increased to bring more iron into the cell (Fig. 3.3).
IRP binding to IREs results in repression of translation of ferritin and stabilization of the TfR1 mRNA. In iron-depleted cells, IRPs bind to the ferritin IRE at the 5′ end of the transcript and interfere with assembly of translation initiation factors. Ferritin IREs typically have a bulged C, and an unpaired U, as depicted. In the TfR1 mRNA, there are five IREs, each of which can bind an IRP. Binding of IRPs protects the transcript from being cleaved by an endonuclease. Upon cleavage, the mRNA cleavage products are rapidly degraded
Many questions have been raised about the specificity of binding of each IRP for different IRE targets [12, 35], with the hypothesis advanced that IRP2 binds well to ferritin-type IREs that contain a complex bulge in the lower stem, but not to simple IREs in which helical base-pairs in the upper and lower stems are separated only by the bulged C. A major difference between IREs of ferritin and other IREs is that ferritin IREs have an unpaired U in a position two residues 5′ of the bulged C. Since nucleotides that are exposed and are not engaged in base-pairing mediate most contacts with RNA binding proteins, the unpaired U has the potential to be important in specificity [13]. IRP1 and IRP2 were equally efficacious in translationally repressing ferritin expression in one study [36], suggesting that both IRPs contribute to translational regulation of ferritin. Questions about specificity cannot be definitively resolved using in vitro studies, but target specificities can be assessed in animals engineered to lack either IRP1 or IRP2. These studies revealed that IRP2 has a major role in regulation not only of ferritin IREs, but also of other transcripts that contain simple IREs such as the TfR1 transcript [37] and the eALAS transcript [38], as is discussed below.
3 The Role of IRPs in Physiology
Based on experiments performed in cell lines, it was not clear what role each IRP played in physiology and it seemed likely that each IRP would have the ability to function as a backup for the other. However, targeted deletion of the IRP1 or IRP2 genes yielded very different phenotypes in mice. Somewhat unexpectedly, the IRP1 “knockout” mouse appeared to have fairly normal iron status in most tissues [28]. In contrast, the IRP2 “knockout” mouse (Irp2−/−) developed a mild microcytic anemia [38, 39] and progressive adult-onset neurodegeneration characterized by an abnormal gait and tremor [37]. The symptoms and neuropathologic changes in Irp2−/− animals are subtle and progressive. One group suggested that their mice did not develop “symptomatic” neurodegeneration, although they reported a significant compromise in motor abilities of their mouse based on testing with a rotarod, which indicated that their mice were not neurologically normal [40]. In the erythroid cells and brain tissue of Irp2−/− mice, ferritin levels are high, whereas Tfr1 levels are low, consistent with the results expected if there is a marked decrease in IRE-binding activity. Since Irp1 is present in the affected tissues in mice, it was interesting to observe that Irp1 is mainly in the cytosolic aconitase form in mammalian tissues, and it does not readily convert to the IRE-binding form in the tissues of iron-starved mice, even in the absence of Irp2 [28].
Since activation of the IRE-binding activity of IRP1 was repeatedly observed in cell lines that were treated with iron chelators or otherwise deprived of iron, it was initially difficult to reconcile the results of multiple tissue culture experiments with the phenotypes of Irp−/− animals. However, culture of cells under different oxygen concentrations yielded an important insight into IRP activities. Because oxygen must be transported over significant distances to various tissues by heme in red cells, mammalian tissues are exposed to oxygen concentrations far below those in the atmosphere. At oxygen concentrations similar to those of mammalian tissues (oxygen concentrations of 3–6% compared to atmospheric concentrations of 21%), it is difficult to induce IRE-binding activity of IRP1, even by starving cells of iron. It is likely that the inability to activate IRE-binding activity of IRP1 is related to the fact that the iron–sulfur cluster is more stable at lower oxygen concentrations. Conversely, levels of IRP2 increase significantly in cells that are maintained at low oxygen concentrations, perhaps because the degradation pathway of IRP2 appears to be initiated by an oxidation event. The contrary effects of low oxygen on the IRE-binding activities of IRP1 compared to IRP2 can explain much about the phenotypes of Irp−/− mice [41]. At normal tissue oxygen concentrations (3–6% O2), IRP1 mainly functions as an aconitase, contributing less than half of total IRE-binding activity in most tissues, whereas IRE-binding activity of IRP2 increases concomitantly with an increase in protein levels at low tissue oxygen concentrations.
Although the differential effects of low tissue oxygen concentrations on IRE-binding activities of the two IRPs explain much about the phenotypes of Irp1−/− and Irp2−/− animals, there is yet another feature that is crucial in developing iron misregulation and the phenotypes of these mice. In Irp1−/− animals, Irp2 levels increase such that total IRE-binding activity levels almost equal the IRE-binding activities contributed by both Irp1 and Irp2 in wild-type animals [41]. Increased Irp2 levels compensate for the loss of Irp1 and its IRE-binding activity. It is not clear how Irp2 levels increase, but it is possible that loss of Irp1 and its IRE-binding activity leads to a minor increase in ferritin synthesis and a concomitant decrease in Tfr1 levels, resulting in a subtle diminution of cytosolic iron pools. Since Irp2 levels appear to correlate directly with cytosolic iron levels, the increase in Irp2 levels may directly reflect and oppose this subtle iron depletion.
In animals that lack both Irp2 alleles and one functional Irp1 allele, the anemia and neurodegenerative symptoms previously observed in Irp2−/− are much more severe. In addition, ferritin levels are higher, and Tfr1 levels are lower than in Irp2−/− mice [42]. These results suggest that both IRP1 and IRP2 contribute to regulation of target transcripts, a conclusion supported by the fact that animals that are bred to lack both alleles of Irp1 and Irp2 die at the blastocyst stage of development, before embryos have developed sufficiently to implant in the uterus [43]. Thus, the IRE–IRP system is indispensable in animals at the earliest stages of development. To analyze the effects of IRPs in specific tissues, animals were generated in which IRP activity could be eliminated in specific tissues. Animals engineered to lack IRP activity in the intestinal mucosa died within weeks of birth and levels of ferroportin in the intestinal mucosa were abnormally high, indicating that ferroportin is a true target of the IRP regulatory system [44].
The reason that neurons and red cells are compromised by loss of IRPs is likely because these cells are functionally iron starved. When ferritin is over-expressed, it competes with other proteins for iron, and sequesters iron that could otherwise be used for synthesis of proteins and prosthetic groups such as heme [45]. The fact that ferritin competes with other proteins for iron and effectively sequesters iron may be the central reason that ferritin translation is repressed by IRPs in iron-deficient cells. In the absence of IRE-binding activity, not only does ferritin expression increase, but iron uptake simultaneously decreases, which contributes further to iron depletion. Thus, cells that lack sufficient IRE-binding activity can misallocate iron reserves, allowing ferritin to sequester iron that should be available to meet basic metabolic needs, even though incoming iron supplies are also diminished.
4 The Evolution and Scope of the IRE–IRP System
The IRE–IRP regulatory system coordinates iron metabolism in a sophisticated and highly sensitive manner in mammalian cells, but the regulatory system does not appear to be well developed in lower animal forms or simple eukaryotes. There is no cytosolic aconitase in Saccharomyces cerevisiae, and although there is a cytosolic aconitase in Caenorhabditis elegans, there is no evidence that the C. elegans aconitase can function as an IRE-binding protein [46]. Ferritin IREs are present in metazoans, including sea anemones and sponges, but these organisms do not contain IREs in other transcripts [47]. In Drosophila melanogaster, there are two cytosolic aconitases, one of which binds IREs [48], and there is evidence that IRE-binding activity regulates synthesis of ferritin and succinate dehydrogenase in flies [49]. Thus, flies contain a rudimentary IRE–IRP system.
Since the IRE–IRP system is highly developed in mammalian cells, many of its characteristics must have evolved in mammals. As cytosolic aconitase activity, but not IRE-binding activity, is found in worms [46], it is likely that the IRE–IRP system developed by exploiting the potential instability of the iron–sulfur cluster of cytosolic aconitase. The cubane iron–sulfur cluster of cytosolic aconitase can be readily oxidized by superoxide, oxygen, nitric oxide, and other oxidants (reviewed in [8, 9]). When these oxidants remove an electron, the entire structure of the cluster is destabilized and it spontaneously disintegrates. Proteins dedicated to the synthesis of iron–sulfur clusters likely provide replacement clusters to cytosolic aconitase when the iron–sulfur clusters undergo oxidative degradation [22, 23]. However, if these iron–sulfur cluster assembly proteins obtain their iron from the cytosolic iron pool, and the pool has become depleted of iron in the time that elapsed since initial synthesis and cluster degradation, cells may be unable to synthesize a replacement iron–sulfur cluster. Failure to replace the iron–sulfur cluster of IRP1 would result in a buildup of apoprotein. In the absence of the iron–sulfur cluster, the apoprotein form of IRP1 may assume new conformations, because the cluster constrains the conformational space. It does this by binding to three cysteines of cytosolic aconitase as well as the substrates citrate and isocitrate. The substrates bind both to the cluster and to residues on the opposite side of the active-site cleft. Accumulation of apoprotein in cells would be expected to correlate with development of iron depletion. Thus, if this apoprotein could be recruited into a process that would reverse the iron depletion, it could offer cells a competitive advantage [50].
How might accumulation of the apoprotein form of cytosolic aconitase work to reverse cellular iron depletion? Since the apoprotein accumulates in the cytosol, it could either act upon transcripts in the cytosol, or it could be transported to the nucleus to function as a transcriptional repressor or activator. Transport into the nucleus would require developing a specialized nuclear transport signal, whereas acting upon cytosolic transcripts would require no extra modifications in the apoprotein form of cytosolic aconitase. In addition, the apoprotein would be in the correct location to resume its enzymatic activities if the iron–sulfur cluster was repaired or replaced. To transform the apoprotein into an important regulatory molecule, targets that would fit the conformation of the apoprotein likely had to evolve.
In eukaryotic mRNAs, the 5′ and 3′UTRs can evolve rapidly, as these portions of the mRNA are not constrained by the specific sequence requirements of protein-coding regions. If mutations in the 5′UTR produced an RNA stem-loop that could be bound tightly by the apo form of cytosolic aconitase, that RNA stem-loop might be retained in evolution. For example, ferritin H and L chains contain an IRE near the 5′ end of the transcript, and binding by apo-IRP1 inhibits translation by interfering with assembly of the translation machinery. Repression of ferritin synthesis produces a desirable result for cells because ferritin does not compete with other proteins to bind iron. Similarly, an IRE could have evolved near the 5′ end in ferroportin, since cells that are iron depleted need to hoard iron rather than allow their iron to be exported from the cell. As 3′UTRs tend to contain elements that determine the rate of mRNA decay, binding of apoprotein in the 3′UTR could interfere with degradation processes and could lengthen the mRNA half-life in transcripts such as TfR1. Increasing the levels of TfR1 mRNA results in increased TfR1 biosynthesis, which reverses cellular iron depletion by enhancing iron uptake.
As IREs in different transcripts differ from one another, particularly in the sequences that provide base-paired stems and structure, it is reasonable to suggest that each IRE arose as an independent evolutionary selection event. This scenario is compatible with the discovery of IREs in many different transcripts in many different species [47]. A functional IRE present in the mitochondrial aconitase transcript reduces synthesis of this iron–sulfur protein when cells have diminished iron stores [51–53]. IREs that likely affect mRNA half-life have been found in one transcript of DMT1 [54] and in the cell cycle protein Cdc14a [55]. In addition, a functional IRE is present in the 5′UTR of the hypoxia inducible factor 2 α (HIF2 α) transcript [56]. The discovery of an IRE in the HIF2 α transcript is important, because HIF1 and 2 α are important oxygen sensors that regulate transcription of numerous genes involved in the switch from normoxia to hypoxia, including glycolysis enzymes and the production of erythropoietin, the major growth factor that regulates erythropoiesis [57].
5 Diseases and the IRE–IRP System
Several diseases in humans and mice are caused by problems with the IRE–IRP regulatory system. Humans that have mutations in the ferritin L chain develop hereditary hyperferritinemia and bilateral cataract syndrome [58]. They have markedly elevated serum ferritin levels and cataracts, but they do not have other more serious symptoms. The reason for high serum ferritins is that these patients have mutations in the ferritin L chain IRE that reduce the ability of IRPs to bind and to repress translation. The severity of disease correlates with the reduction of IRE-binding ability [9].
Mice that lack Irp2 develop microcytic anemia, erythropoietic protoporphyria, and adult-onset neurodegeneration. These phenotypes arise as a result of increased ferritin, decreased TfR1 expression, and misregulation of other known IRP targets such as eALAS [38], ferroportin [44], and perhaps HIF2 α. The IRP2 gene is located on human chromosome 15q25, and thus far, no mutations of IRP2 have been described that cause human disease. However, it is fairly likely that mutations or deletions of IRP2 will prove to be a cause of human disease, as many diseases characterized by anemia and neurodegeneration are not yet well understood. The incentive to find such diseases is high, because studies in the mouse Irp2−/− mouse have demonstrated that the neurologic compromise of Irp2−/− animals can be prevented. When animals are treated with the stable nitroxide, Tempol, normal regulation of ferritin and Tfr1 is restored in the brain. Tempol can be supplied by supplementation of food, as Tempol is absorbed in the duodenum and crosses the blood-brain barrier. In cells, it causes disassembly of the iron–sulfur cluster of IRP1, which converts IRP1 into the IRE-binding form and counteracts the deficiency of IRE-binding activity caused by loss of IRP2 [59].
In cancer syndromes such as Von-Hippel Lindau (VHL) disease, many of the features of the renal cancer are thought to be attributable to high levels of HIF2 α, which is normally ubiquitinated by the VHL complex in normoxic cells [60]. HIF2 α is a transcription factor that increases expression of glycolysis enzymes, erythropoietin, VEGF, and numerous other genes. In normoxic cells, an iron-dependent prolyl hydroxylase modifies HIF residues, which then enables the VHL ubiquitination complex to bind to HIF and mark it for degradation. The fact that HIF2 α has an IRE in the 5′UTR [56] links its expression to the IRE–IRP system and indicates that reagents such as Tempol might be useful in treatment of VHL renal cancer.
6 Transcriptional Regulation of Iron Metabolism Genes
Although posttranscriptional regulation of TfR1, ferritin, and other IRP targets is very important in intracellular iron homeostasis, transcription of these genes is also regulated. In TfR1, several HIF-binding sites are important in transcription [61, 62], with transcription of TfR1 increasing in cells that over-express HIF1α [63]. More recently, it has been reported that HIF2α is required to transcriptionally activate DMT1 and Dcytb in response to iron deficiency [64]. Thus, HIF1 and 2α increase transcription of targets such as TfR1, DMT1, and Dcytb. Interestingly, HIFα inhibits transcription of hepcidin [65].
In addition to the emerging role of HIF α transcription factors, other transcription factors important in iron metabolism include Stat 5, Nrf2, and SMAD4. Both TfR1 and IRP2 are transcriptional targets of the signal transducer and activator of transcription, Stat5, in erythroid cells, which enables signaling through the erythropoietin receptor to increase expression of both TfR1 and IRP2 [66]. Increased transcription of both ferritin H and L chains is mediated by the transcription factor Nrf2 in response to oxidants and prooxidant xenobiotics [67, 68], which helps cells to avert some of the damage associated with oxidative stress by sequestering iron and reducing its participation in Fenton chemistry. Much remains to be learned about transcriptional regulation of genes that are important in iron metabolism. For instance, transcriptional regulation of ferroportin is clearly important, particularly in the intestinal mucosa [69], but this is poorly understood. Studies of the hepcidin promoter demonstrate important roles for Stat 3 [70, 71], for SMAD4 [72] and for HIFα [65].
7 Posttranscriptional Modifications of IRPs
Both IRP1 and IRP2 have been observed to undergo phosphorylation and the phosphorylation status potentially affects function. In IRP1, phosphorylation at serine 138 destabilizes the iron–sulfur cluster and facilitates conversion to the IRE-binding form [19, 20]. Phosphorylation of IRP1 at serine 711 inhibits the conversion of citrate to isocitrate [19]. IRP2 also undergoes phosphorylation in cells treated with phorbol esters [73]. This phosphorylation occurs at serine 157 and is coordinated with cell cycle progression, with phosphorylation mediated by Cdk1/cyclin B and dephosphorylation mediated by Cdc14a. The phosphorylation reduces the IRE-binding activity of IRP2 during mitosis, which increases ferritin synthesis and reduces TfR1 synthesis, perhaps to diminish free iron and iron-dependent oxidation events during a time when the DNA of the cell is exposed and vulnerable to damage [74].
8 Summary
The IRE–IRP regulatory system is found in rudimentary form in flies, but its breadth and importance in regulation of cytosolic iron status is most extensive in mammalian cells. The regulatory system is based mainly on the ability of IRP1 and IRP2 to change their ability to bind to IREs in various transcripts, depending on cellular iron status. Binding to an IRE in the 5′UTR of transcripts generally represses translation, whereas binding to IREs in the 3′UTR likely generally increases mRNA stability. Functional IREs are found in numerous transcripts of iron metabolism genes and it is likely that the list of IRE-containing target transcripts will continue to grow as new genes are found and 5′ and 3′UTRs are carefully sequenced and analyzed.
References
Aisen P. Transferrin receptor 1. Int J Biochem Cell Biol. 2004;36:2137–43.
Mims MP, Prchal JT. Divalent metal transporter 1. Hematology. 2005;10:339–45.
Torti FM, Torti SV. Regulation of ferritin genes and protein. Blood. 2002;99:3505–16.
Theil EC, Matzapetakis M, Liu X. Ferritins: iron/oxygen biominerals in protein nanocages. J Biol Inorg Chem. 2006;11:803–10.
McKie AT, Barlow DJ. The SLC40 basolateral iron transporter family (IREG1/ferroportin/MTP1). Pflugers Arch. 2004;447:801–6.
Donovan A, Lima CA, Pinkus JL, et al. The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metab. 2005;1:191–200.
Ohgami RS, Campagna DR, Greer EL, et al. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat Genet. 2005;37:1264–9.
Wallander ML, Leibold EA, Eisenstein RS. Molecular control of vertebrate iron homeostasis by iron regulatory proteins. Biochim Biophys Acta. 2006;1763:668–89.
Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat Chem Biol. 2006;2:406–14.
Muckenthaler MU, Galy B, Hentze MW. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu Rev Nutr. 2008;28:197–213.
Addess KJ, Basilion JP, Klausner RD, Rouault TA, Pardi AJ. Structure and dynamics of the iron responsive element RNA: implications for binding of the RNA by iron regulatory proteins. J Mol Biol. 1997;274:72–83.
Ke Y, Wu J, Leibold EA, Walden WE, Theil EC. Loops and bulge/loops in iron-responsive element isoforms influence iron regulatory protein binding. Fine-tuning of mRNA regulation? J Biol Chem. 1998;273:23637–40.
Volz K. The functional duality of iron regulatory protein 1. Curr Opin Struct Biol. 2008;18:106–11.
Walden WE, Selezneva AI, Dupuy J, et al. Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science. 2006;314:1903–8.
Tong WH, Rouault TA. Metabolic regulation of citrate and iron by aconitases: role of iron-sulfur cluster biogenesis. Biometals. 2007;20:549–64.
Beinert H, Kennedy MC, Stout DC. Aconitase as iron–sulfur protein, enzyme, and iron-regulatory protein. Chem Rev. 1996;96:2335–73.
Dupuy J, Volbeda A, Carpentier P, Darnault C, Moulis JM, Fontecilla-Camps JC. Crystal structure of human iron regulatory protein 1 as cytosolic aconitase. Structure. 2006;14:129–39.
Rouault TA, Klausner RD. Iron–sulfur clusters as biosensors of oxidants and iron. Trends Biochem Sci. 1996;21:174–7.
Brown NM, Anderson SA, Steffen DW, et al. Novel role of phosphorylation in Fe–S cluster stability revealed by phosphomimetic mutations at Ser-138 of iron regulatory protein 1. Proc Natl Acad Sci USA. 1998;95:15235–40.
Clarke SL, Vasanthakumar A, Anderson SA, et al. Iron-responsive degradation of iron-regulatory protein 1 does not require the Fe–S cluster. EMBO J. 2006;25:544–53.
Rouault TA, Tong WH. Opinion: iron–sulphur cluster biogenesis and mitochondrial iron homeostasis. Nat Rev Mol Cell Biol. 2005;6:345–51.
Rouault TA, Tong WH. Iron–sulfur cluster biogenesis and human disease. Trends Genet. 2008;24:398–407.
Lill R, Muhlenhoff U. Maturation of iron–sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu Rev Biochem. 2008;77:669–700.
Samaniego F, Chin J, Iwai K, Rouault TA, Klausner RD. Molecular characterization of a second iron responsive element binding protein, iron regulatory protein 2 (IRP2): structure, function and post-translational regulation. J Biol Chem. 1994;269:30904–10.
Guo B, Yu Y, Leibold EA. Iron regulates cytoplasmic levels of a novel iron-responsive element-binding protein without aconitase activity. J Biol Chem. 1994;269:24252–60.
Iwai K, Klausner RD, Rouault TA. Requirements for iron-regulated degradation of the RNA binding protein, iron regulatory protein 2. EMBO J. 1995;14:5350–7.
Guo B, Phillips JD, Yu Y, Leibold EA. Iron regulates the intracellular degradation of iron regulatory protein 2 by the proteasome. J Biol Chem. 1995;270:21645–51.
Meyron-Holtz EG, Ghosh MC, Iwai K, et al. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J. 2004;23:386–95.
Iwai K, Drake SK, Wehr NB, et al. Iron-dependent oxidation, ubiquitination, and degradation of iron regulatory protein 2: implications for degradation of oxidized proteins. Proc Natl Acad Sci USA. 1998;95:4924–8.
Yamanaka K, Ishikawa H, Megumi Y, et al. Identification of the ubiquitin-protein ligase that recognizes oxidized IRP2. Nat Cell Biol. 2003;5:336–40.
Zumbrennen KB, Hanson ES, Leibold EA. HOIL-1 is not required for iron-mediated IRP2 degradation in HEK293 cells. Biochim Biophys Acta. 2008;1783:246–52.
Wang J, Chen G, Muckenthaler M, Galy B, Hentze MW, Pantopoulos K. Iron-mediated degradation of IRP2, an unexpected pathway involving a 2-oxoglutarate-dependent oxygenase activity. Mol Cell Biol. 2004;24:954–65.
Hanson ES, Rawlins ML, Leibold EA. Oxygen and iron regulation of iron regulatory protein 2. J Biol Chem. 2003;278:40337–42.
Liu XB, Hill P, Haile DJ. Role of the ferroportin iron-responsive element in iron and nitric oxide dependent gene regulation. Blood Cells Mol Dis. 2002;29:315–26.
Ke Y, Sierzputowska-Gracz H, Gdaniec Z, Theil EC. Internal loop/bulge and hairpin loop of the iron-responsive element of ferritin mRNA contribute to maximal iron regulatory protein 2 binding and translational regulation in the iso-iron-responsive element/iso-iron regulatory protein family. Biochemistry. 2000;39:6235–42.
Kim HY, Klausner RD, Rouault TA. Translational repressor activity is equivalent and is quantitatively predicted by in vitro RNA binding for two iron-responsive element binding proteins, IRP1 and IRP2. J Biol Chem. 1995;270:4983–6.
LaVaute T, Smith S, Cooperman S, et al. Targeted deletion of iron regulatory protein 2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat Genet. 2001;27:209–14.
Cooperman SS, Meyron-Holtz EG, Olivierre-Wilson H, Ghosh MC, McConnell JP, Rouault TA. Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood. 2005;106:1084–91.
Galy B, Ferring D, Minana B, et al. Altered body iron distribution and microcytosis in mice deficient in iron regulatory protein 2 (IRP2). Blood. 2005;106:2580–9.
Galy B, Holter SM, Klopstock T, et al. Iron homeostasis in the brain: complete iron regulatory protein 2 deficiency without symptomatic neurodegeneration in the mouse. Nat Genet. 2006;38:967–9.
Meyron-Holtz EG, Ghosh MC, Rouault TA. Mammalian tissue oxygen levels modulate iron-regulatory protein activities in vivo. Science. 2004;306:2087–90.
Smith SR, Cooperman S, Lavaute T, et al. Severity of neurodegeneration correlates with compromise of iron metabolism in mice with iron regulatory protein deficiencies. Ann N Y Acad Sci. 2004;1012:65–83.
Smith SR, Ghosh MC, Ollivierre-Wilson H, Tong WH, Rouault TA. Complete loss of iron regulatory proteins 1 and 2 prevents viability of murine zygotes beyond the blastocyst stage of embryonic development. Blood Cells Mol Dis. 2006;36:283–7.
Galy B, Ferring-Appel D, Kaden S, Grone HJ, Hentze MW. Iron regulatory proteins are essential for intestinal function and control key iron absorption molecules in the duodenum. Cell Metab. 2008;7:79–85.
Cozzi A, Corsi B, Levi S, Santambrogio P, Albertini A, Arosio P. Overexpression of wild type and mutated human ferritin H-chain in HeLa cells: in vivo role of ferritin ferroxidase activity. J Biol Chem. 2000;275:25122–9.
Gourley BL, Parker SB, Jones BJ, Zumbrennen KB, Leibold EA. Cytosolic aconitase and ferritin are regulated by iron in Caenorhabditis elegans. J Biol Chem. 2003;278:3227–34.
Piccinelli P, Samuelsson T. Evolution of the iron-responsive element. RNA. 2007;13:952–66.
Lind MI, Missirlis F, Melefors O, et al. Of two cytosolic aconitases expressed in Drosophila, only one functions as an iron-regulatory protein. J Biol Chem. 2006;281:18707–14.
Kohler SA, Henderson BR, Kuhn LC. Succinate dehydrogenase b mRNA of Drosophila melanogaster has a functional iron-responsive element in its 5′-untranslated region. J Biol Chem. 1995;270:30781–6.
Rouault TA. Biochemistry. If the RNA fits, use it. Science. 2006;314:1886–7.
Kim HY, LaVaute T, Iwai K, Klausner RD, Rouault TA. Identification of a conserved and functional iron-responsive element in the 5′UTR of mammalian mitochondrial aconitase. J Biol Chem. 1996;271:24226–30.
Schalinske KL, Chen OS, Eisenstein RS. Iron differentially stimulates translation of mitochondrial aconitase and ferritin mRNAs in mammalian cells. Implications for iron regulatory proteins as regulators of mitochondrial citrate utilization. J Biol Chem. 1998;273:3740–6.
Gray NK, Pantopoulos K, Dandekar T, Ackrell BAC, Hentze MW. Translational regulation of mammalian and drosophila citric-acid cycle enzymes via iron-responsive elements. Proc Natl Acad Sci USA. 1996;93:4925–30.
Gunshin H, Allerson CR, Polycarpou-Schwarz M, et al. Iron-dependent regulation of the divalent metal ion transporter. FEBS Lett. 2001;509:309–16.
Sanchez M, Galy B, Dandekar T, et al. Iron regulation and the cell cycle: identification of an iron-responsive element in the 3′-untranslated region of human cell division cycle 14A mRNA by a refined microarray-based screening strategy. J Biol Chem. 2006;281:22865–74.
Sanchez M, Galy B, Muckenthaler MU, Hentze MW. Iron-regulatory proteins limit hypoxia-inducible factor-2alpha expression in iron deficiency. Nat Struct Mol Biol. 2007;14:420–6.
Lofstedt T, Fredlund E, Holmquist-Mengelbier L, et al. Hypoxia inducible factor-2alpha in cancer. Cell Cycle. 2007;6:919–26.
Hetet G, Devaux I, Soufir N, Grandchamp B, Beaumont C. Molecular analyses of patients with hyperferritinemia and normal serum iron values reveal both L ferritin IRE and 3 new ferroportin (slc11A3) mutations. Blood. 2003;102:1904–10.
Ghosh MC, Tong WH, Zhang D, et al. Tempol-mediated activation of latent iron regulatory protein activity prevents symptoms of neurodegenerative disease in IRP2 knockout mice. Proc Natl Acad Sci USA. 2008;105:12028–33.
Kaelin WGJ. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. 2008;8:865–73.
Tacchini L, Bianchi L, Bernelli-Zazzera A, Cairo G. Transferrin receptor induction by hypoxia. HIF-1-mediated transcriptional activation and cell-specific post-transcriptional regulation. J Biol Chem. 1999;274:24142–6.
Lok CN, Ponka P. Identification of a hypoxia response element in the transferrin receptor gene. J Biol Chem. 1999;274:24147–52.
Alberghini A, Recalcati S, Tacchini L, Santambrogio P, Campanella A, Cairo G. Loss of the von Hippel Lindau tumor suppressor disrupts iron homeostasis in renal carcinoma cells. J Biol Chem. 2005;280:30120–8.
Shah YM, Matsubara T, Ito S, Yim SH, Gonzalez FJ. Intestinal hypoxia-inducible transcription factors are essential for iron absorption following iron deficiency. Cell Metab. 2009;9:152–64.
Peyssonnaux C, Zinkernagel AS, Schuepbach RA, et al. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs). J Clin Invest. 2007;117:1926–32.
Kerenyi MA, Grebien F, Gehart H, et al. Stat5 regulates cellular iron uptake of erythroid cells via IRP-2 and TfR-1. Blood. 2008;112:3878–88.
Pietsch EC, Chan JY, Torti FM, Torti SV. Nrf2 mediates the induction of ferritin H in response to xenobiotics and cancer chemopreventive dithiolethiones. J Biol Chem. 2003;278:2361–9.
Tsuji Y, Ayaki H, Whitman SP, Morrow CS, Torti SV, Torti FM. Coordinate transcriptional and translational regulation of ferritin in response to oxidative stress. Mol Cell Biol. 2000;20:5818–27.
McKie AT, Marciani P, Rolfs A, et al. A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol Cell. 2000;5:299–309.
Falzacappa MVV, Spasic MV, Kessler R, Stolte J, Hentze MW, Muckenthaler MU. STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood. 2007;109:353–8.
Wrighting DM, Andrews NC. Interleukin-6 induces hepcidin expression through STAT3. Blood. 2006;108:3204–9.
Wang RH, Li C, Xu X, et al. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2:399–409.
Kl S, Rs E. Phosphorylation and activation of both iron regulatory proteins 1 and 2 in HL60 cells. J Biol Chem. 1996;271:7168–76.
Wallander ML, Zumbrennen KB, Rodansky ES, Romney SJ, Leibold EA. Iron-independent phosphorylation of iron regulatory protein 2 regulates ferritin during the cell cycle. J Biol Chem. 2008;283:23589–98.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2012 Springer Science+Business Media, LLC
About this chapter
Cite this chapter
Rouault, T.A. (2012). Regulation of Iron Metabolism in Mammalian Cells. In: Anderson, G., McLaren, G. (eds) Iron Physiology and Pathophysiology in Humans. Nutrition and Health. Humana Press. https://doi.org/10.1007/978-1-60327-485-2_3
Download citation
DOI: https://doi.org/10.1007/978-1-60327-485-2_3
Published:
Publisher Name: Humana Press
Print ISBN: 978-1-60327-484-5
Online ISBN: 978-1-60327-485-2
eBook Packages: MedicineMedicine (R0)