Abstract
Iron and citrate are essential for the metabolism of most organisms, and regulation of iron and citrate biology at both the cellular and systemic levels is critical for normal physiology and survival. Mitochondrial and cytosolic aconitases catalyze the interconversion of citrate and isocitrate, and aconitase activities are affected by iron levels, oxidative stress and by the status of the Fe–S cluster biogenesis apparatus. Assembly and disassembly of Fe–S clusters is a key process not only in regulating the enzymatic activity of mitochondrial aconitase in the citric acid cycle, but also in controlling the iron sensing and RNA binding activities of cytosolic aconitase (also known as iron regulatory protein IRP1). This review discusses the central role of aconitases in intermediary metabolism and explores how iron homeostasis and Fe–S cluster biogenesis regulate the Fe–S cluster switch and modulate intracellular citrate flux.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
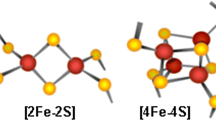
Iron–sulfur (Fe–S) proteins constitute one of the most ubiquitous and functionally versatile classes of metalloproteins (Beinert et al. 1997; Johnson et al. 2005). More than 120 distinct types of proteins contain Fe–S clusters and many Fe–S proteins are involved in fundamental processes such as respiration, photosynthesis, intermediary metabolism and nitrogen fixation. The citric acid cycle enzyme aconitase contains a [4Fe-4S] cluster and the enzymatic reaction catalyzed by aconitase involves substrate coordination to a specific iron atom in this cluster (Beinert et al. 1996). Two aconitase isozymes are present in mammalian cells: the mitochondrial enzyme (m-aconitase) that functions in the citric acid cycle, and the bifunctional cytosolic enzyme (c-aconitase/IRP1) which also plays a role in the regulation of iron metabolism. Moreover, recent studies in yeast suggested that mitochondrial aconitase is also essential for mtDNA maintenance and this activity is independent of its enzymatic activity (Chen et al. 2005). Thus, modulation of the aconitases can impact a wide spectrum of cellular activities under both normal and pathophysiological conditions. Here, we will present an overview of the physiological roles of mitochondrial aconitase and cytosolic aconitase/IRP1, and review recent findings related to the biogenesis/repair of Fe–S clusters that have shed new lights on the regulation of aconitase functions and emphasized the intricate links between iron and citrate metabolism.
Cellular and systemic citrate metabolism
Citrate in energy metabolism pathways
Aconitase is best known for its function in catalyzing the reversible isomerization of tricarboxylic acids-citrate, cis-aconitate, and isocitrate (Breusch 1937). Citrate is a key intermediate in several major pathways of energy and intermediary metabolism (Fig. 1). In the mitochondria, citrate is an intermediate in the citric acid cycle, which converts acetyl-CoA to two molecules of CO2 with concomitant generation of NADH and FADH2. Reoxidation of NADH and FADH2 via the electron transport chain yields ATP. Citric acid cycle flux is in part controlled by the activity of isocitrate dehydrogenase through allosteric inhibition by ATP and product inhibition by NADH (Lawlis and Roche 1980). Thus, when the need for ATP synthesis is low, citrate accumulates and can be transported across the inner mitochondrial membrane via the tricarboxylate carrier (Bisaccia et al. 1989). In the cytosol, citrate is the substrate for ATP-citrate lyase, which generates acetyl-coenzyme A (acetyl-CoA), the building block for cholesterol and fatty acid biosynthesis in liver and adipose tissue. Citrate is also metabolized via cytosolic aconitase and cytosolic NADP+-dependent isocitrate dehydrogenase to generate α-ketoglutarate. This process reduces NADP+ to NADPH, which is an essential cofactor for many enzymatic reactions involved in glutathione metabolism and lipid and cholesterol biosynthesis (Koh et al. 2004; Minard and McAlister-Henn 2005).
Overview of the roles of citrate and aconitases in the major energy metabolism pathways. Citrate is a key intermediate that interconnects the metabolic pathways of the citric acid cycle and oxidative phosphorylation in the mitochondria to glycolysis and fatty acid synthesis in the cytosol. When the need for ATP synthesis is high, citrate is metabolized through the citric acid cycle to generate NADH and FADH2, the reducing equivalents needed for ATP production. When the need for ATP synthesis is low, citrate is exported into the cytosol and channeled into fatty acid biosynthesis for energy storage. Inhibition of aconitases due to iron deficiency or oxidative damage to the Fe–S cluster could decrease ATP production, promote fat accumulation, decrease glycolysis, and decrease fatty acid oxidation. In addition, decreased citrate flux through c-aconitase may decrease production of cytosolic NADPH, a major source of reducing equivalents for fatty acid synthesis and an important defense against cytosolic oxidative stress
In addition to being an intermediate, citrate also has regulatory roles in glycolysis, fatty acid synthesis and oxidation. Citrate is a negative regulator of the glycolytic enzyme phosphofructokinase (Denton and Randle 1966; Randle 1998). Since citrate efflux from mitochondria occurs when ATP and NADH are high, glycolysis would be repressed. Citrate is also an allosteric activator of acetyl-CoA carboxylase, the enzyme that generates malonyl-CoA (Munday 2002). In turns, malony-CoA is a potent allosteric inhibitor of carnitine palmitoyltransferase-1 (McGarry et al. 1977), which controls the transfer of long chain acyl-CoA into mitochondria, the site of fatty acid oxidation (Saha and Ruderman 2003). An increase in malonyl-CoA concentration inhibits fatty acid β-oxidation and diverts acetyl-CoA into lipogenesis for energy storage or membrane assembly. Conversely, a decrease in malonyl-CoA level directs long chain acyl-CoA into the β-oxidation pathway in the mitochondria. Thus, through its complex biological effects on malonyl-CoA production, glucose utilization, fatty acid synthesis and oxidation, citrate biology can affect the pathophysiology of obesity, insulin resistance and diabetes (Belfiore and Iannello 1998; Wolfgang and Lane 2006).
In multicellular organisms, the complex network of processes involved in energy metabolism are distributed among different subcellular sites as well as in different organs of the body. Thus, the mediated membrane transport and systemic trafficking of citrate also plays an essential metabolic role. The mitochondrial tricarboxylate carrier activity that mediates citrate export is low in heart and high in liver, reflecting the high ATP demand in heart and the cytosolic location of fatty acid biogenesis in liver (Sluse et al. 1971; LaNoue and Schoolwerth 1979). Furthermore, the mitochondrial tricarboxylate carrier is repressed in type I diabetes (Kaplan et al. 1990) and during starvation (Siculella et al. 2002), and enhanced during hyperthyroidism (Paradies and Ruggiero 1990). Serum contains significant levels of citrate (∼0.1 mM), and the serum levels of citrate varies appreciably between feedings and fastings (Hodgkinson 1963). Recent studies in Drosophila showed that decreased expression of a plasma membrane citrate transporter Indy resulted in decreased lipid content and increased life span (Rogina et al. 2000; Knauf et al. 2006). Functional knockdown of ceNAC-2, the Indy orthologue in C. elegans, also led to decreased body size, decreased fat content, and a significant increase in life span (Fei et al. 2004). The presence of the mammalian orthologue NaC2 (formerly know as NaCT) in liver and in brain is consistent with the uptake of serum citrate for the synthesis of fatty acid acids and cholesterol in liver, and for energy synthesis in liver and brain (Inoue et al. 2002). Taken together, these studies indicated that systemic citrate homeostasis is of great importance for an organism’s health and survival.
Citrate in specialized cells
In cholinergic neurons, citrate can be used to generate acetyl-CoA for acetylcholine synthesis. In the central nervous system and the retina, aconitase is important for the metabolic pathway that generates glutamate, the major excitatory neurotransmitter (McGahan et al. 2005). It is also reported that, due to the absence of pyruvate carboxylase, neurons lack the capacity to perform de novo synthesis of glutamate from glucose and are therefore dependent on a supply of glutamate and citric acid cycle intermediates synthesized in astrocytes (Hertz and Zielke 2004). Studies in primary cultures indicated that citrate is synthesized and released from astrocytes (Sonnewald et al. 1991), and plasma membrane Na+-coupled citrate transporters have been shown to be expressed in the brain (Inoue et al. 2002; Wada et al. 2006).
In the kidney, citrate is an important inhibitor of urinary stone formation (Caudarella et al. 2003), and regulation of renal cortical m-aconitase has been shown to affect urinary citrate excretion (Melnick et al. 1998). Citrate secretion requires that the rate of citrate synthesis exceed the rate of citrate oxidation via the citric acid cycle. Thus, in normal citrate-secreting prostate epithelial cells, m-aconitase expression is hormonally suppressed (Costello and Franklin 2002). Elevated zinc levels can also inhibit m-aconitase (Costello et al. 1997) and may be responsible for the impaired citrate oxidation observed in normal prostate epithelial cells (Dakubo et al. 2006). Conversely, in prostate cancer, normal citrate-secreting epithelial cells are metabolically transformed to malignant citrate-oxidizing cells, and the depletion of zinc in malignant cells is thought to be an important factor in this metabolic transformation (Singh et al. 2006).
Citrate as a chelator of divalent metal ions
Citrate can chelate divalent cations such as Fe2+, Ca2+, and Zn2+, and various studies have indicated that citrate has complex functions in the homeostasis of these divalent metal ions. Serum citrate is thought to be one of the carriers of non-transferrin-bound iron (Grootveld et al. 1989) that can contribute to hepatic iron loading in hemochromatosis (Chua et al. 2004). Although it has been suggested that cytoplasmic citrate might be needed for transport of iron into mitochondria (Melefors and Hentze 1993), in vitro experiments and yeast genetic studies have shown that citrate–iron complexes (Martin 1986; Pierre and Gautier-Luneau 2000) can promote autooxidation of ferrous iron and may contribute to iron-dependent toxicity (Chen et al. 2002). Serum citrate can also effect anticoagulation by binding Ca2+ and rending Ca2+ unavailable to the clotting cascade (Weinstein 2001). Systemic hypocalcemia due to chelation of Ca2+ by citrate can depress cardiac function by causing hypotension, decreased cardiac output, cardiac arrest and muscle tetany (Bosakowski and Levin 1986; Dzik and Kirkley 1988). In addition, the relatively high concentration (0.4 mM) of citrate in the cerebrospinal fluid (Bell et al. 1987) and the ability of citrate to chelate Zn2+, Ca2+and Mg2+ are thought to play a role in modulating the activity of N-methyl-d-aspartate (NMDA) receptors and neuronal excitability (Westergaard et al. 1995).
Cytosolic aconitase in the regulation of iron metabolism
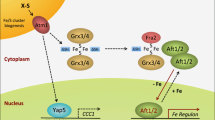
Biochemical and phylogenetic studies have suggested that during the evolution of the aconitase family, an early gene duplication allowed cytosolic aconitase to evolve independently from mitochondrial aconitase, and a second duplication of the cytosolic aconitase produced two cytosolic homologues in animals, which subsequently acquired the ability to bind to RNAs containing the iron responsive element (IRE) (Gruer et al. 1997). Hence, unicellular eukaryotes such as Saccharomyces cerevisiae (Regev-Rudzki et al. 2005) and protozoan parasites such as Trypanosoma brucei (Saas et al. 2000) contain a single aconitase gene which encodes isozymes that function in the cytosol and in the mitochondria, whereas multicellular eukaryotes have separate genes that encode m- and c-aconitases (Kennedy et al. 1992; Muckenthaler et al. 1998). In the case of C. elegans, the c-aconitase has no RNA-binding activities (Gourley et al. 2003), whereas in Drosphila, one of the two c-aconitases also functions as an RNA-binding, iron regulatory protein (Lind et al. 2006). In mammalia, IRP1 switches between aconitase and RNA-binding functions, whereas IRP2 has no aconitase activity and functions solely as an RNA-binding protein (Fig. 2) (Pantopoulos 2004).
Regulation of intermediary metabolism and intracellular iron metabolism by iron and Fe–S cluster biogenesis in the cytosol and in the mitochondria of mammalian cells. In the mitochondria, the citic acid cycle, respiration complexes, and heme biosynthesis are all dependent on iron availability and Fe–S cluster biogenesis. In addition, Fe–S cluster biogenesis is a key factor in the regulation of iron homeostasis in both the cytosol and the mitochondria. In most tissues, due to low oxygen concentration and efficient Fe–S cluster assembly in IRP1, IRP2 assumes a greater role in regulating intracellular iron homeostasis, although the substantial pool of latent IRE-binding activity of IRP1 may be activated by oxidative stress or decreased Fe–S cluster biogenesis. In the mitochondria, decreased Fe–S cluster biogenesis results in mitochondrial iron accumulation and cytosolic iron depletion, leading to activation of IRE-binding by IRP1 and IRP2, which can result in altered expressions of TfR, ferritin, DMT1 and iron exporter ferroportin. In addition to facilitating compartmentalization of iron homeostasis, the spatial separation of Fe–S cluster assembly in m- and c-aconitase may also allow cells to perform anabolic and catabolic processes in a finely coordinated and efficient fashion (see Fig. 1)
Iron is a vital micronutrient, but excess free iron is cytotoxic and misregulation of iron homeostasis can contribute to a number of hematological, metabolic and neurodegenerative diseases (Pietrangelo 2003; Hentze et al. 2004; Napier et al. 2005). In mammalian cells, intracellular iron homeostasis is largely achieved by the iron-dependent regulation of transferrin receptor (TfR) and and iron storage protein ferritin. The mRNAs of TfR and ferritin contain IREs in their untranslated regions. In iron-depleted cells, binding of the IRP1 and IRP2 to the IREs leads to stabilization of the TfR mRNA and translational inhibition of ferritin mRNAs, resulting in increased TfR-dependent iron uptake and decreased iron sequestration into ferritin. Conversely, decreased IRP/IRE interactions in iron-replete cells results in TfR mRNA degradation and ferritin translation. The list of IRE-containing transcripts also includes the iron transporter ferroportin (Abboud and Haile 2000; Donovan et al. 2000; McKie et al. 2000) and one of the spliced forms of the divalent metal tranporter DMT1 (Gunshin et al. 1997), although the exact role for IRPs in the regulation of these proteins is still under investigation. In addition to proteins involved in iron homeostasis, the identification and characterization of IREs in the transcripts of erythrocyte-specific aminolevulinate synthase (Cox et al. 1991; Zoller et al. 2002; Cooperman et al. 2005), mammalian mitochondrial aconitase (Kim et al. 1996; Ross and Eisenstein 2002) and insect succinate dehydrogenase (SDH) (Kohler et al. 1995) indicated that the function of IRP/IRE regulatory system extends to the control of heme biosynthesis and citric acid cycle.
Although IRP1 and IRP2 are highly homologous, they sense cytosolic iron levels by different mechanisms. IRP2 is regulated through protein degradation in the presence of iron and oxygen. Various studies have suggested that iron might be involved in targeting IRP2 for degradation by directly binding to IRP2 or indirectly through a mechanism that involves heme or Fe(II) α-ketoglutarate-dependent oxygenases (Hanson et al. 2003; Pantopoulos 2004; Ishikawa et al. 2005). In contrast, IRP1 registers cytosolic iron and oxidative stress through its labile Fe–S cluster (Pantopoulos 2004). In the absence of the Fe–S cluster, IRP1 loses aconitase activity and binds to IREs with high affinity. When iron is abundant, the Fe–S cluster is efficiently repaired/regenerated, and IRP1 functions mainly as an aconitase. In vitro experiments showed that IRP1 and IRP2 bind with similar affinities to IRE-containing transcripts (Kim et al. 1995), and studies in rats showed that dietary iron intake can modulate the activity of both IRP1 and IRP2 in rat liver (Chen et al. 1997). However, IRP1 -/-mice developed no overt phenotype, whereas IRP2-/-mice developed microcytic anemia, hyperferritinemia and erythropoietic protoporphyria (Cooperman et al. 2005; Galy et al. 2005) as well as adult-onset neurodegeneration (LaVaute et al. 2001). A major reason that IRP2 assumes a much greater regulatory role than IRP1 in the mouse models is that IRP2 is stable and active at normal tissue oxygen concentrations (3-6%) (Meyron-Holtz et al. 2004b). In contrast, under the same conditions, the Fe–S cluster in IRP1 is stable, and IRP1 functions mainly as an active aconitase. Furthermore, iron diet studies in mice showed that IRP2 is more responsive to dietary iron changes than IRP1 (Meyron-Holtz et al. 2004a). Although IRP1 is not required for the regulation of iron metabolism in wild type mice, IRP2-/-animals that also lack one copy of IRP1 developed more severe anemia and neurodegeneration (Smith et al. 2004), and IRP1-/-IRP2-/-embryos die at the blastocyst stage (Smith et al. 2006), indicating that, in the absence of IRP2, retention of at least one allele of IRP1 is critical for survival through embryonic development.
As mentioned above, the presence of an IRE in the mRNA of mammalian m-aconitase makes it a potential target for regulation by IRPs. Indeed, studies have shown that iron deficiency decreased the abundance of m-aconitase (Kim et al. 1996) and enhanced citrate utilization in rat liver, although no increase in liver lipogenesis was observed during the course of the study (Ross and Eisenstein 2002). Several interesting hypotheses have been proposed for the reason why the proteins involved in cellular energy production, such as m-aconitase and SDH, should be coupled to iron availability. First, it has been suggested that cytosolic citrate might be needed for transport of iron into mitochondria and down-regulation of aconitases in iron-depleted cells would maintain a certain level of citrate needed for this transport (Melefors and Hentze 1993). Second, some repression of these Fe–S proteins may be advantageous in iron-depleted cells, since lack of sufficient iron could lead to synthesis of non-functional apoproteins (Kim et al. 1996). The possibility that intermediary metabolism may be regulated by the reactive oxygen species (ROS) generated in mitochondrial respiration forms the basis of a third hypothesis, in which oxidative stress not only inactivates m-aconitase directly through chemical modification of the Fe–S cluster, but also activates the IRE-binding activity of IRP1, resulting in repression of m-aconitase synthesis (Gray et al. 1996). Decreased m-aconitase activity would decrease NADH production and electron flux through the respiratory complexes, thereby creating a feedback loop that decreases respiration-generated ROS (Armstrong et al. 2004). Last but not least, recent studies in yeast have demonstrated that, in response to iron deprivation, the RNA-binding protein Cth2 down-regulates several Fe-dependent pathways to limit iron expenditure (Puig et al. 2005). From this prospective, the IRP-dependent suppression of mammalian m-aconitase and insect SDH may serve to limit the utilization of iron in iron deficient cells. Interestingly, iron deprivation also decreased the expression of the mammalian Fe–S cluster assembly proteins ISCU (Tong and Rouault 2006) and ISCS (Liew and Shaw 2005), further suggesting that selective suppression of Fe-dependent pathways is a general cellular response to iron deficiency in mammalian cells.
Biochemical and physiological features of aconitase inhibition
Although m-aconitase is not generally considered to be the rate-limiting step (Williamson and Cooper 1980), in cardiac myocytes, decreased aconitase activity led to decreases in citric acid cycle flux (Janero and Hreniuk 1996). Mitochondrial aconitase is also a major target of oxidative demage during aging (Delaval et al. 2004; Yarian et al. 2006), and oxidative inactivation of aconitase has been associated with decreased life span in Drosophila (Yan et al. 1997). Cellular inactivation of aconitase is largely attributable to the enzyme’s sensitivity to oxidative stress and iron availability (Fig. 3) (Chen et al. 1997; Gardner 1997; Tong and Rouault 2006), although phosphorylation may also affect the stabilities of the Fe–S cluster and the protein (Fillebeen et al. 2005; Clarke et al. 2006). The Fe–S clusters in both m-aconitase and c-aconitase can be disassembled upon exposure to oxidants including O -2 , H2O2, NO, and ONOO- (Pantopoulos and Hentze 1995; Gardner 1997; Bouton and Drapiers 2003; Han et al. 2005), and the liberation of reactive iron may further induce oxidative damage of cellular constituents (Lipinski et al. 2005). Oxidative damage can also increase the proteolytic susceptibility of m-aconitase and IRP1, and severe oxidation can cause aggregation of the aconitase protein (Das et al. 2001; Bota and Davies 2002; Starzynski et al. 2005). Since citrate can be channeled into cholesterol and lipid biosynthesis if the citric acid cycle is inhibited, it has been suggested that the massive lipid accumulation observed in the livers of MnSOD-deficient mice results from oxidative damage to the Fe–S clusters in mitochondrial aconitase and SDH (Li et al. 1995; Armstrong et al. 2004). Consistent with this hypothesis, recent studies in Drosphila and C. elegans have shown that decreased expression of the plasma membrane citrate transporters resulted in decreased lipid content and increased life span (Rogina et al. 2000; Fei et al. 2004), further underscoring the pivotal role of citrate flux in energy homeostasis.
Aconitase activities are affected by iron levels, oxidative stress and Fe–S cluster assembly activity. (A) Mitochondrial and cytosolic aconitases can be selectively inactivated by different inducers of oxidative stress. HeLa S3 cells were treated for 1 h with H2O2 or DEA/NO, then washed and incubated in normal growth medium (chase). Treatment with H2O2 (lanes 1, 5, 9,13) led to dose-dependent inactivation of c-aconitase (Tong and Rouault 2006), whereas treatment with DEA/NO (lanes 1, 4, 8,12) resulted in inactivation of m-aconitase (Tong et al. unpublished data). Subsequent withdrawal of either oxidant resulted in reactivation of aconitase activity, indicating robust repair/regeneration of the [4Fe-4S] cluster in both the mitochondrial and the cytosolic compartments. (B) Iron deficiency, induced by treatment with the iron chelator Desferal (Dfo), lowers both m- and c-aconitase activities and activates the IRE-binding activities of both IRP1 and IRP2. Reactivation of aconitase activities and deactivation of IRE-binding activities depends on the presence of the Fe–S cluster assembly protein ISCU. (C) Isoform-specific depletion of cytosolic ISCU (c-ISCU) impairs Fe–S cluster assembly in c-aconitase only (compare lane 3 and 6). In comparison, Fe–S cluster assembly in m-aconitase were unaffected due to the presence of a distinct mitochondrial ISCU. Reprinted from Cell Metabolism, v.3, Tong and Rouault, “Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis” pp. 199–210, 2006, with permission from Elsevier
In addition to affecting citrate metabolism, it has been suggested that oxidative or nitrosative modulation of IRPs may cause changes in iron metabolism during inflammation and anaemia of chronic disease (Cairo et al. 2002a). The response of IRPs to oxidative stress is very complex. Various studies in different cell culture systems have shown that different oxidizing species can activate or inactivate IRP1 and IRP2 (Cairo et al. 2002b; Bouton and Drapiers 2003; Hanson et al. 2003; Kim and Ponka 2003; Pantopoulos 2004). Likewise, studies in tissue or animal models indicated that, while the IRE-binding activity of IRP1 increased in rat liver perfused with a H2O2-generating system that mimicked a physiological inflammatory response (Mueller et al. 2001), mice that lack the cytosolic superoxide dismutase (SOD) display a marked decrease in IRP1 aconitase activity and protein levels, but a normal iron metabolism phenotype (Starzynski et al. 2005).
Citric acid cycle genetic defects in humans are rare, and the most severe are likely embryonic lethal events (Rustin et al. 1997). Yeast with mutations in Aco1, the single gene that encodes both m- and c-aconitase, exhibit glutamate auxotrophy, defects in respiratory and cytosolic glyoxylate pathways, and are unable to grow on non-fermentable carbon source (Gangloff et al. 1990; Regev-Rudzki et al. 2005). While a genetic model for the ablation of mitochondrial aconitase in vertebrates has not been reported, IRP1-/-IRP2-/-mice are not viable past the blastocyst stage (Smith et al. 2006). Surprisingly, IRP1-/-mice have no obvious defects (Meyron-Holtz et al. 2004a). In situ hybridization and activity assays indicated that IRP1 levels are highest in the kidney, liver and brown fat (Mullner et al. 1992; Meyron-Holtz et al. 2004a), and aconitase activity assays indicated that c-aconitase accounts for ∼50% of the total aconitase activities in liver and kidney (Konstantinova and Russanov 1996; Huang et al. 2002). However, there is no evidence for compromised renal function, abnormal blood chemistry, tissue pathology, or difference in body weight or brown fat in IRP1-/-mice (Meyron-Holtz et al. 2004a), indicating that the presence of mitochondrial aconitase and IRP2 can substitute for the lack of IRP1 under basal metabolic conditions.
The complex roles of aconitases in normal physiology were also revealed by extensive toxicological studies of fluoroacetate (Goncharov et al. 2006). Fluoroacetate undergoes a series of metabolic conversions to yield the extremely toxic compound, fluorocitrate, a potent mechanism-based inhibitor of aconitase. Acute fluoroacetate poisoning in humans mainly affects the central nervous system, cardiovascular system and kidney, and the long list of biochemical effects include TCA cycle blockage, respiratory failure, excessive citrate accumulation in heart, kidney and spleen, deviation in carbohydrate metabolism regulation, metabolic acidosis, increased serum citrate, decreased plasma Ca2+, and lactate accumulation in cerebrospinal fluid.
Aconitase activities in mammalian cells are modulated by the mitochondrial and cytosolic Fe–S cluster assembly machineries
In recent years, Fe–S cluster biogenesis has received increasing attention, not only for its role in supporting the functions of many metabolic pathways, but also for its involvement in the sensing of cytosolic and mitochondrial iron status, and the communication of iron needs in the different subcellular compartments (Fig. 2). In the cytosol, Fe–S cluster assembly and disassembly controls the function of one of the iron regulatory proteins IRP1. In the mitochondria, disruption in Fe–S cluster biogenesis results in mitochondrial iron overload, cytosolic iron depletion, activation of cellular iron uptake and increased oxidative stress (Babcock et al. 1997; Knight et al. 1998; Garland et al. 1999; Foury and Talibi 2001; Yang et al. 2006). In patients with the neurodegenerative disease, Friedreich ataxia, decreased frataxin levels results in reduced heme and Fe–S cluster biogenesis, defective mitochondrial respiration, mitochondrial iron overload, and increased oxidative damage (Rotig et al. 1997; Pandolfo 2003; Seznec et al. 2004; Napoli et al. 2006).
The complexity and importance of the process of Fe–S cluster biogenesis is underscored by the number of genes involved and the lethal phenotypes associated with disruption of many such genes (Fig. 4) (Zheng et al. 1998; Li et al. 1999; Schilke et al. 1999; Cossee et al. 2000; Seidler et al. 2001; Pondarre et al. 2006). Excellent summaries of the current understanding of Fe–S cluster biogenesis in bacteria, yeast, plant and animal are available in several recent reviews (Johnson et al. 2005; Lill and Muhlenhoff 2005; Rouault and Tong 2005; Pilon et al. 2006). E. coli and S. cerevisiae have the most extensively characterized Fe–S cluster assembly systems. More than a dozen genes have been implicated in Fe–S cluster biogenesis, and homologues of some of them have been characterized in mammalian systems. Briefly, cysteine desulphurases cleave cysteine to yield sulfur in the form of an enzyme-bound persulfide (Zheng et al. 1993), whereas frataxin may function as the iron donor (Gerber et al. 2003; Yoon and Cowan 2003; Bulteau et al. 2004) (Fig. 4A). Sulfur transfer (Smith et al. 2001; Urbina et al. 2001) and iron acquisition leads to assembly of [2Fe-4S] and [4Fe-4S] clusters on the scaffold proteins (Agar et al. 2000). Chaperone proteins are thought to facilitate cluster transfer to target proteins (Silberg et al. 2004; Dutkiewicz et al. 2006), and redox proteins (Lange et al. 2000; Wingert et al. 2005) provide the reducing equivalents that are necessary for cluster assembly and cluster transfer. Genetic and biochemical studies have indicated that many other proteins are also involved in Fe–S cluster biogenesis, although their exact functions are still under active investigation (Fig. 4B) (Balk et al. 2005; Johnson et al. 2005; Lill and Muhlenhoff 2005; Tury et al. 2005; Adam et al. 2006; Balasubramanian et al. 2006; Molina-Navarro et al. 2006).
Fe–S cluster assembly proteins and their sub-cellular localization in S. cerevisiae and in mammalian cells. (A) Formation of intracellular Fe–S cluster involves a complex biosynthetic machinery that assembles rudimentary Fe–S clusters in scaffold proteins, and subsequently transfers these clusters to target proteins. (B) Up to 19 different proteins have been proposed to be involved in Fe–S cluster biogenesis in S. cerevisiae. A number of the human homologues that have been characterized biochemically and cytologically are shown here, and putative homologues of many others were revealed by sequence comparison. In yeast, many components of the Fe–S cluster assembly machinery, including the yeast homolog of frataxin, Yfh1, the scaffold proteins Isu1, Isu2, and Nfu, the chaperones Ssq1 and Jac1, the redox proteins Yah1, Arh1, and Grx5, and other proteins such as Isa1, Isa2 and Isd11 have been detected in the mitochondria. Isa2 is also in the intermembrane space and the cysteine desulfurase Nfs1 is present in both the mitochondria and the nucleus. The ABC type transporter Atm1 is in the mitochondrial inner membrane, and the sulfhydryl oxidase Erv1 is in the intermembrane space. In yeast cytosol, Cfd1, Nar1, Cia1 and Nbp35 were proposed to assist in maturation of Fe–S protein. In contrast to yeast, small but significant amounts of the cysteine desulfurase ISCS, scaffold proteins ISCU and NFU, and frataxin have been identified in the cytosol of mammalian cells, and recent functional studies have confirmed that Fe–S cluster biogenesis occurs in the cytosol as well as in the mitochondria of mammalian cells
In yeast and animal cells, most of the known Fe–S proteins are found in the mitochondrial matrix and the respiratory chain of the mitochondrial inner membrane, and mitochondria constitute the major subcellular site of Fe–S -cluster assembly (Lill and Muhlenhoff 2005; Rouault and Tong 2005). In human, multiple tissue Northern blot analyses have indicated that mitochondrial-rich tissues such as heart and skeletal muscle have the highest levels of ABC7, ISCS, ISCU and ISCA transcripts, reflecting the high demand for Fe–S cluster biogenesis in these tissues (Allikmets et al. 1999; Tong and Rouault 2000). Frataxin is also abundant in the adult heart and the developing brain, which correlates with the hypertrophic cardiomyopathy and neuropathy observed in Friedreich ataxia patients (Campuzano et al. 1996; Koutnikova et al. 1997).
In yeast, many of the Fe–S cluster assembly proteins have thus far been detected only in the mitochondria (Fig. 4B), leading to the proposal that Fe–S cluster assembly occurs exclusively in the mitochondria, and that preformed Fe–S clusters are exported through the ABC transporter Atm1 into the cytosol, where several auxiliary factors facilitate the incorporation of these clusters into cytosolic proteins (Lill and Muhlenhoff 2005). However, Fe–S cluster biogenesis is not confined to the mitochondrial matrix in plants and animals. In plants, genomic and biochemical analysis have revealed that isoforms of several Fe–S cluster assembly proteins exist in both the chloroplast and the mitochondria (Pilon et al. 2006). In mammalian cells, a small but significant extramitochondrial fraction of Fe–S cluster assembly proteins including ISCS (Land and Rouault 1998), ISCU (Tong and Rouault 2000; Acquaviva et al. 2005), NFU (also known as HIRIP5) (Lorain et al. 2001; Ganesh et al. 2003; Tong et al. 2003) and frataxin (Acquaviva et al. 2005; Condo et al. 2006) have been identified (Fig. 4B). More recently, three studies have provided further evidence for extramitochondrial Fe–S cluster biogenesis. First, the mitochondrial and cytosolic ISCU proteins can be selectively suppressed by isoform-specific siRNA, and silencing of both m-and c-ISCU impaired Fe–S cluster assembly in both m- and c-aconitase (Fig. 3B) (Tong and Rouault 2006). Interestingly, Fe–S cluster assembly in c-aconitase was specifically affected by silencing of cytosolic ISCU (Fig. 3C) whereas m-aconitase was more sensitive to the depletion of m-ISCU, in keeping with the presence of a functional Fe–S cluster assembly machinery in the cytosol (Tong and Rouault 2006). Second, in vitro studies confirmed that recombinant human cytosolic ISCS and cytosolic ISCU can promote Fe–S cluster assembly in c-aconitase/IRP1 (Li et al. 2006). Third, expression of extramitochondrial frataxin can promote survival of frataxin-deficient cells from a Friedreich ataxia patient (Condo et al. 2006).
Compartmentalization is a valuable tool for creating functional diversity, regulatory flexibility, and specificity in signaling. Spatial separation of the Fe–S cluster assembly apparatus into different subcellular compartments provides the cell increased capacity to respond to changing metabolic needs (Figs. 1 and 2). The redox environment also varies markedly in the different subcellular compartments (Hansen et al. 2006), and mitochondrial and cytosolic aconitases can be selectively inactivated by different inducers of oxidative stress (Fig. 3A). Studies in Drosophila and S. cerevisiae have shown that the mitochondrial and cytosolic SOD provide independent protection to Fe–S proteins against damage by O -2 generated within those compartments (Missirlis et al. 2003; Wallace et al. 2004), whereas compartment-specific Fe–S cluster assembly would facilitate robust repair/regeneration of Fe–S clusters in the different locations (Tong and Rouault 2006). Furthermore, separate Fe–S cluster assembly machineries allow independent sensing of iron status in the cytosol and in the mitochondria in mammalian cells (Fig. 2). In the cytosol, Fe–S cluster biogenesis is important for the switch between cytosolic aconitase and IRE-binding activities of IRP1. In the mitochondria, Fe–S cluster biogenesis is needed not only to support respiratory functions and heme biosynthesis, but also for sensing and regulation of mitochondrial iron homeostasis (Rouault and Tong 2005). When the need for Fe–S cluster-dependent activities in the mitochondria is high, the cell may respond by increasing delivery of iron to the mitochondria and decreasing mitochondrial iron export. Activation of IRE-binding activities as a result of cytosolic iron depletion and inadequate Fe–S cluster biogenesis (Fosset et al. 2006; Tong and Rouault 2006) would lead to increased TfR-dependent iron uptake and decreased iron sequestration by ferritin (Anderson et al. 2005; Fosset et al. 2006). Since expression of m-ISCU and m-ISCS are down-regulated by iron depletion (Liew and Shaw 2005; Tong and Rouault 2006), iron deficiency would suppress Fe–S proteins both by decreasing availability of a basic component of the Fe–S cluster, and by decreasing the Fe–S cluster assembly protein levels. Taken together, these results place Fe–S cluster biogenesis at the forefront of iron sensing and reprogramming of intracellular iron trafficking in response to changing iron needs.
Concluding remarks
Iron deficiency is the most common and devastating nutritional disorder in the world (Lewis 2005), and much effort has been directed to understanding the metabolic consequences of iron deficiency (Beard and Connor 2003; Umbreit 2005). We provided here one example of how iron deficiency can affect many aspects of cellular metabolism through its effects on aconitase activities. Iron deficiency can decrease aconitase protein levels and limit the assembly of the Fe–S clusters required for their enzymatic activities. As a result, iron deficiency can potentially affect the citric acid cycle, lipid biosynthesis, carbohydrate metabolism and many other biological pathways that involve citrate. Similarly, iron deficiency and suppression of Fe–S cluster biogenesis may hamper the functions of many other Fe–S proteins in intermediary metabolism, respiratory complexes, heme biosynthesis and DNA repair. The selective suppression of iron-containing proteins in the mitochondria can also lead to decoupling of the electron transport chain and increased oxidative stress. Although tremendous progress has been made recently in understanding the impact of Fe–S cluster biogenesis on cellular iron metabolism, the mechanism by which this system communicates information about mitochondrial iron status to modulate cellular iron trafficking remains to be elucidated. Lessons from this new and exciting area of research should provide new insights into the mechanisms of mitochondrial signaling and the interrelationship between iron and energy metabolism.
References
Abboud S, Haile DJ (2000) A novel mammalian iron-regulated protein involved in intracellular iron metabolism. J Biol Chem 275:19906–19912
Acquaviva F, De Biase I, Nezi L et al (2005) Extra-mitochondrial localisation of frataxin and its association with IscU1 during enterocyte-like differentiation of the human colon adenocarcinoma cell line Caco-2. J Cell Sci 118:3917–3924
Adam AC, Bornhovd C, Prokisch H, Neupert W, Hell K (2006) The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. EMBO J 25:174–183
Agar JN, Krebs C, Frazzon J, Huynh BH, Dean DR, Johnson MK (2000) IscU as a scaffold for iron–sulfur cluster biosynthesis: sequential assembly of [2Fe-4S] and [4Fe-4S] clusters in IscU. Biochemistry 39:7856–7862
Allikmets R, Raskind WH, Hutchinson A, Schueck ND, Dean M, Koeller DM (1999) Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A). Hum Mol Genet 8:743–749
Anderson PR, Kirby K, Hilliker AJ, Phillips JP (2005) RNAi-mediated suppression of the mitochondrial iron chaperone, frataxin, in Drosophila. Hum Mol Genet 14:3397–3405
Armstrong JS, Whiteman M, Yang H, Jones DP (2004) The redox regulation of intermediary metabolism by a superoxide-aconitase rheostat. Bioessays 26:894–900
Babcock M, de Silva D, Oaks R et al (1997) Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science 276:1709–1712
Balasubramanian R, Shen G, Bryant DA, Golbeck JH (2006) Regulatory roles for IscA and SufA in iron homeostasis and redox stress responses in the cyanobacterium Synechococcus sp. strain PCC 7002. J Bacteriol 188:3182–3191
Balk J, Aguilar Netz DJ, Tepper K, Pierik AJ, Lill R (2005) The essential WD40 protein Cia1 is involved in a late step of cytosolic and nuclear iron–sulfur protein assembly. Mol Cell Biol 25:10833–10841
Beard JL, Connor JR (2003) Iron status and neural functioning. Annu Rev Nutr 23:41–58
Beinert H, Holm RH, Munck E (1997) Iron–sulfur clusters: nature’s modular, multipurpose structures. Science 277:653–659
Beinert H, Kennedy MC, Stout CD (1996) Aconitase as iron–protein, enzyme, and iron-regulatory protein. Chem Rev 96:2335–2374
Belfiore F, Iannello S (1998) Insulin resistance in obesity: metabolic mechanisms and measurement methods. Mol Gen Metab 65:121–128
Bell JD, Brown JC, Sadler PJ et al (1987) High resolution proton nuclear magnetic resonance studies of human cerebrospinal fluid. Clin Sci (Lond). 72:563–570
Bisaccia F, De Palma A, Palmieri F (1989) Identification and purification of the tricarboxylate carrier from rat liver mitochondria. Biochim Biophys Acta 977:171–176
Bosakowski T, Levin AA (1986) Serum citrate as a peripheral indicator of fluoroacetate and fluorocitrate toxicity in rats and dogs. Toxicol Appl Pharmacol 85:428–436
Bota DA, Davies KJ (2002) Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol 4:674–680
Bouton C, Drapiers JC (2003) Iron regulatory proteins as NO signal transducers. Sci STKE 182:pe17
Breusch FL (1937) Citric acid in tissue metabolism. Physiol Chem 250:262–280
Bulteau AL, O’Neill HA, Kennedy MC, Ikeda-Saito M, Isaya G, Szweda LI (2004) Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science 305:242–245
Cairo G, Recalcati S, Pietrangelo A, Minotti G (2002a) The iron regulatory proteins: targets and modulators of free radical reactions and oxidative damage. Free Radic Biol Med 32:1237–1243
Cairo G, Ronchi R, Recalcati S, Campanella A, Minotti G (2002b) Nitric oxide and peroxynitrite activate the iron regulatory protein-1 of J774A.1 macrophages by direct disassembly of the Fe–S cluster of cytoplasmic aconitase. Biochemistry 41:7435–7442
Campuzano V, Montermini L, Molto M et al (1996) Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271:1423–1427
Caudarella R, Vescini F, Buffa A, Stefoni S (2003) Citrate and mineral metabolism: kidney stones and bone disease. Front Biosci 8:s1084–1106
Chen OS, Hemenway S, Kaplan J (2002) Genetic analysis of iron citrate toxicity in yeast: implications for mammalian iron homeostasis. Proc Natl Acad Sci USA 99:16922–16927
Chen OS, Schalinske KL, Eisenstein RS (1997) Dietary iron intake modulates the activity of iron regulatory proteins and the abundance of ferritin and mitochondrial aconitase in rat liver. J Nutr 127:238–248
Chen XJ, Wang X, Kaufman BA, Butow RA (2005) Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science 307:714–717
Chua AC, Olynyk JK, Leedman PJ, Trinder D (2004) Non-transferrin-bound iron uptake by hepatocytes is increased in the Hfe knockout mouse model of hereditary hemochromatosis. Blood 104:1519–1525
Clarke SL, Vasanthakumar A, Anderson SA et al (2006) Iron-responsive degradation of iron-regulatory protein 1 does not require the Fe–S cluster. EMBO J 25:544–553
Condo I, Ventura N, Malisan F, Tomassini B, Testi R (2006) A pool of extramitochondrial frataxin that promotes cell survival. J Biol Chem 281:16750–16756
Cooperman SS, Meyron-Holtz EG, Olivierre-Wilson H, Ghosh MC, McConnell JP, Rouault TA (2005) Microcytic anemia, erythropoietic protoporphyria and neurodegeneration in mice with targeted deletion of iron regulatory protein 2. Blood 106:1084–1091
Cossee M, Puccio H, Gansmuller A et al (2000) Inactivation of the Friedreich ataxia mouse gene leads to early embryonic lethality without iron accumulation. Hum Mol Genet 9:1219–1226
Costello LC, Franklin RB (2002) Testosterone and prolactin regulation of metabolic genes and citrate metabolism of prostate epithelial cells. Horm Metab Res 34:417–424
Costello LC, Liu Y, Franklin RB, Kennedy MC (1997) Zinc inhibition of mitochondrial aconitase and its importance in citrate metabolism of prostate epithelial cells. J Biol Chem 272:28875–28881
Cox TC, Bawden MJ, Martin A, May BK (1991) Human erythroid 5-aminolevulinate synthase: promoter analysis and identification of an iron-responsive element in the mRNA. EMBO J 10:1891–1902
Dakubo GD, Parr RL, Costello LC, Franklin RB, Thayer RE (2006) Altered metabolism and mitochondrial genome in prostate cancer. J Clin Pathol 59:10–16
Das N, Levine RL, Orr WC, Sohal R (2001) Selectivity of protein oxidative damage during aging in Drosophila melanogaster. Biochem J 360:206–216
Delaval E, Perichon M, Friguet B (2004) Age-related impairment of mitochondrial matrix aconitase and ATP-stimulated protease in rat liver and heart. Eur J Biochem 271:4559–4564
Denton RM, Randle PJ (1966) Citrate and the regulation of adipose-tissue phosphofructokinase. Biochem J 100:420–423
Donovan A, Brownlie A, Zhou Y et al (2000) Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature 403:776–781
Dutkiewicz R, Marszalek J, Schilke B, Craig EA, Lill R, Muhlenhoff U (2006) The Hsp70 chaperone Ssq1p is dispensable for iron–sulfur cluster formation on the scaffold protein Isu1p. J Biol Chem 281:7801–7808
Dzik WH, Kirkley SA (1988) Citrate toxicity during massive blood transfusion. Transfus Med Rev 2:76–94
Fei YJ, Liu JC, Inoue K et al (2004) Relevance of NAC-2, an Na+-coupled citrate transporter, to life span, body size and fat content in Caenorhabditis elegans. Biochem J 379:191–198
Fillebeen C, Caltagirone A, Martelli A, Moulis JM, Pantopoulos K (2005) IRP1 Ser-711 is a phosphorylation site, critical for regulation of RNA-binding and aconitase activities. Biochem J 388:143–150
Fosset C, Chauveau MJ, Guillon B, Canal F, Drapier JC, Bouton C (2006) RNA silencing of mitochondrial m-Nfs1 reduces Fe–S enzyme activity both in mitochondria and cytosol of mammalian cells. J. Biol. Chem. Epub ahead of print
Foury F, Talibi D (2001) Mitochondrial control of iron homeostasis. A genome wide analysis of gene expression in a yeast frataxin-deficient strain. J Biol Chem 276:7762–7768
Galy B, Ferring D, Minana B et al (2005) Altered body iron distribution and microcytosis in mice deficient in iron regulatory protein 2 (IRP2). Blood 106:2580–2589
Ganesh S, Tsurutani N, Suzuki T et al (2003) The Lafora disease gene product laforin interacts with HIRIP5, a phylogenetically conserved protein containing a NifU-like domain. Hum Mol Genet 12:2359–2368
Gangloff SP, Marguet D, Lauquin GJ (1990) Molecular cloning of the yeast mitochondrial aconitase gene (ACO1) and evidence of a synergistic regulation of expression by glucose plus glutamate. Mol Cell Biol 10:3551–3561
Gardner PR (1997) Superoxide-driven aconitase FE–S center cycling. Biosci Rep 17:33–42
Garland SA, Hoff K, Vickery LE, Culotta VC (1999) Saccharomyces cerevisiae ISU1 and ISU2: members of a well-conserved gene family for iron–sulfur cluster assembly. J Mol Biol 294:897–907
Gerber J, Muhlenhoff U, Lill R (2003) An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep 4:906–911
Goncharov NV, Jenkins RO, Radilov AS (2006) Toxicology of fluoroacetate: a review, with possible directions for therapy research. J Appl Toxicol 26:148–161
Gourley BL, Parker SB, Jones BJ, Zumbrennen KB, Leibold EA (2003) Cytosolic aconitase and ferritin are regulated by iron in Caenorhabditis elegans. J Biol Chem 278:3227–3234
Gray NK, Pantopoulos K, Dandekar T, Ackrell BA, Hentze MW (1996) Translational regulation of mammalian and Drosophila citric acid cycle enzymes via iron-responsive elements. Proc Natl Acad Sci USA 93:4925–4930
Grootveld M, Bell JD, Halliwell B, Aruoma OI, Bomford A, Sadler PJ (1989) Non-transferrin bound iron in plasma or serum from patients with idiopathic hemochromatosis. J Biol Chem 264:4417–4422
Gruer MJ, Artymiuk PJ, Guest JR (1997) The aconitase family: three structural variations on a common theme. Trends Biochem Sci 22:3–6
Gunshin H, Mackenzie B, Berger UV et al (1997) Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature 388:482–488
Han D, Canali R, Garcia J, Aguilera R, Gallaher TK, Cadenas E (2005) Sites and mechanisms of aconitase inactivation by peroxynitrite: modulation by citrate and glutathione. Biochemistry 44:11986–11996
Hansen JM, Go YM, Jones DP (2006) Nuclear and mitochondrial compartmentation of oxidative stress and redox signaling. Annu Rev Pharmacol Toxicol 46:215–234
Hanson ES, Rawlins ML, Leibold EA (2003) Oxygen and iron regulation of iron regulatory protein 2. J Biol Chem 278:40337–40342
Hentze MW, Muckenthaler MU, Andrews NC (2004) Balancing acts: molecular control of mammalian iron metabolism. Cell 117:285–297
Hertz L, Zielke HR (2004) Astrocytic control of glutamatergic activity: astrocytes as stars of the show. Trends Neurosci 27:735–743
Hodgkinson A (1963) The relation between citric acid and calcium metabolism with particular reference to primary hyper-parathyroidism and idiopathic hypercalciuria. Clin Sci (Lond) 24:167–178
Huang TT, Raineri I, Eggerding F, Epstein CJ (2002) Transgenic and mutant mice for oxygen free radical studies. Methods Enzymol 349:191–213
Inoue K, Zhuang L, Maddox DM, Smith SB, Ganapathy V (2002) Structure, function, and expression pattern of a novel sodium-coupled citrate transporter (NaCT) cloned from mammalian brain. J Biol Chem 277:39469–39476
Ishikawa H, Kato M, Hori H et al (2005) Involvement of heme regulatory motif in heme-mediated ubiquitination and degradation of IRP2. Mol Cell 19:171–181
Janero DR, Hreniuk D (1996) Suppression of TCA cycle activity in the cardiac muscle cell by hydroperoxide-induced oxidative stress. Am J Physiol 270:C1735–C1742
Johnson DC, Dean DR, Smith AD, Johnson MK (2005) Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem 74:247–281
Kaplan RS, Oliveira DL, Wilson GL (1990) Streptozotocin-induced alterations in the levels of functional mitochondrial anion transport proteins. Arch Biochem Biophys 280:181–191
Kennedy MC, Mende-Mueller L, Blondin GA, Beinert H (1992) Purification and characterization of cytosolic aconitase from beef liver and its relationship to the iron-responsive element binding protein. Proc Natl Acad Sci USA 89:11730–11734
Kim HY, Klausner RD, Rouault TA (1995) Translational repressor activity is equivalent and is quantitatively predicted by in vitro RNA binding for two iron-responsive element-binding proteins, IRP1 and IRP2. J Biol Chem 270:4983–4986
Kim HY, LaVaute T, Iwai K, Klausner RD, Rouault TA (1996) Identification of a conserved and functional iron-responsive element in the 5’-untranslated region of mammalian mitochondrial aconitase. J Biol Chem 271:24226–24230
Kim S, Ponka P (2003) Role of nitric oxide in cellular iron metabolism. Biometals 16:125–135
Knauf F, Mohebbi N, Teichert C et al (2006) The life-extending gene Indy encodes an exchanger for Krebs-cycle intermediates. Biochem J 397:25–29
Knight SA, Sepuri NB, Pain D, Dancis A (1998) Mt-Hsp70 homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. J Biol Chem 273:18389–18393
Koh HJ, Lee SM, Son BG et al (2004) Cytosolic NADP+-dependent isocitrate dehydrogenase plays a key role in lipid metabolism. J Biol Chem 279:39968–39974
Kohler SA, Henderson BR, Kuhn LC (1995) Succinate dehydrogenase b mRNA of Drosophila melanogaster has a functional iron-responsive element in its 5’-untranslated region. J Biol Chem 270:30781–30786
Konstantinova SG, Russanov EM (1996) Aconitase activity in rat liver. Comp Biochem Physiol B Biochem Mol Biol 113:125–130
Koutnikova H, Campuzano V, Foury F, Dolle P, Cazzalini O, Koenig M (1997) Studies of human, mouse and yeast homologues indicate a mitochondrial function for frataxin. Nat Genet 16:345–351
Land T, Rouault TA (1998) Targeting of a human iron–sulfur cluster assembly enzyme, nifs, to different subcellular compartments is regulated through alternative AUG utilization. Mol Cell 2:807–815
Lange H, Kaut A, Kispal G, Lill R (2000) A mitochondrial ferredoxin is essential for biogenesis of cellular iron–sulfur proteins. Proc Natl Acad Sci USA 97:1050–1055
LaNoue KF, Schoolwerth AC (1979) Metabolite transport in mitochondria. Annu Rev Biochem 48:871–922
LaVaute T, Smith S, Cooperman S et al (2001) Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat Genet 27:209–214
Lawlis VB, Roche TE (1980) Effect of micromolar Ca2+ on NADH inhibition of bovine kidney alpha-ketoglutarate dehydrogenase complex and possible role of Ca2+ in signal amplification. Mol Cell Biochem 32:147–152
Lewis SM (2005) Introduction—the global problem of nutritional anemias. Hematology 10:224–226
Li Y, Huang TT, Carlson EJ et al (1995) Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet 11:376–381
Li J, Kogan M, Knight SAB, Pain D, Dancis A (1999) Yeast mitochondrial protein, Nfs1p, coordinately regulates iron–sulfur cluster proteins, cellular iron uptake, and iron distribution. J Biol Chem 274:33025–33034
Li K, Tong WH, Hughes RM, Rouault TA (2006) Roles of the mammalian cytosolic cysteine desulfurase, ISCS, and scaffold protein, ISCU, in iron–sulfur cluster assembly. J Biol Chem 281:12344–12351
Liew YF, Shaw NS (2005) Mitochondrial cysteine desulfurase iron–sulfur cluster S and aconitase are post-transcriptionally regulated by dietary iron in skeletal muscle of rats. J Nutr 135:2151–2158
Lill R, Muhlenhoff U (2005) Iron–sulfur–protein biogenesis in eukaryotes. Trends Biochem Sci 30:133–141
Lind MI, Missirlis F, Melefors O et al (2006) Of two cytosolic aconitases expressed in Drosophila, only one functions as an iron regulatory protein. J Biol Chem 281:18707–18714
Lipinski P, Starzynski RR, Drapier JC et al (2005) Induction of iron regulatory protein 1 RNA-binding activity by nitric oxide is associated with a concomitant increase in the labile iron pool: implications for DNA damage. Biochem Biophys Res Commun 327:349–355
Lorain S, Lecluse Y, Scamps C, Mattei MG, Lipinski M (2001) Identification of human and mouse HIRA-interacting protein-5 (HIRIP5), two mammalian representatives in a family of phylogenetically conserved proteins with a role in the biogenesis of Fe/S proteins. Biochim Biophys Acta 1517:376–383
Martin RB (1986) Citrate binding of Al3+ and Fe3+. J Inorg Biochem 28:181–187
McGahan MC, Harned J, Mukunnemkeril M, Goralska M, Fleisher L, Ferrell JB (2005) Iron alters glutamate secretion by regulating cytosolic aconitase activity. Am J Physiol Cell Physiol 288:C1117–C1124
McGarry JD, Mannaerts GP, Foster DW (1977) A possible role for malonyl-CoA in the regulation of hepatic fatty acid oxidation and ketogenesis. J Clin Invest 60:265–270
McKie AT, Marciani P, Rolfs A et al (2000) A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol Cell 5:299–309
Melefors O Hentze MW (1993) Translational regulation by mRNA/protein interactions in eukaryotic cells: ferritin and beyond. Bioessays 15:85–90
Melnick JZ, Preisig PA, Moe OW, Srere P, Alpern RJ (1998) Renal cortical mitochondrial aconitase is regulated in hypo- and hypercitraturia. Kidney Int 54:160–165
Meyron-Holtz EG, Ghosh MC, Iwai K et al (2004a) Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. EMBO J 23:386–395
Meyron-Holtz EG, Ghosh MC, Rouault TA (2004b) Mammalian tissue oxygen levels modulate iron-regulatory protein activities in vivo. Science 306:2087–2090
Minard KI, McAlister-Henn L (2005) Sources of NADPH in yeast vary with carbon source. J Biol Chem 280:39890–39896
Missirlis F, Hu J, Kirby K, Hilliker AJ, Rouault TA, Phillips JP (2003) Compartment-specific protection of iron–sulfur proteins by superoxide dismutase. J Biol Chem 278:47365–47369
Molina-Navarro MM, Casas C, Piedrafita L, Belli G, Herrero E (2006) Prokaryotic and eukaryotic monothiol glutaredoxins are able to perform the functions of Grx5 in the biogenesis of Fe/S clusters in yeast mitochondria. FEBS Lett 580:2273–2280
Muckenthaler M, Gunkel N, Frishman D, Cyrklaff A, Tomancak P, Hentze MW (1998) Iron-regulatory protein-1 (IRP-1) is highly conserved in two invertebrate species—characterization of IRP-1 homologues in Drosophila melanogaster and Caenorhabditis elegans. Eur J Biochem 254:230–237
Mueller S, Pantopoulos K, Hubner CA, Stremmel W, Hentze MW (2001) IRP1 activation by extracellular oxidative stress in the perfused rat liver. J Biol Chem 276:23192–23196
Mullner EW, Rothenberger S, Muller AM, Kuhn LC (1992) In vivo and in vitro modulation of the mRNA-binding activity of iron-regulatory factor. Tissue distribution and effects of cell proliferation, iron levels and redox state. Eur J Biochem 208:597–605
Munday MR (2002) Regulation of mammalian acetyl-CoA carboxylase. Biochem Soc Trans 30:1059–1064
Napier I, Ponka P, Richardson DR (2005) Iron trafficking in the mitochondrion: novel pathways revealed by disease. Blood 105:1867–1874
Napoli E, Taroni F, Cortopassi GA (2006) Frataxin, iron–sulfur clusters, heme, ROS, and aging. Antioxid Redox Signal 8:506–516
Pandolfo M (2003) Friedreich ataxia. Semin Pediatr Neurol 10:163–172
Pantopoulos K (2004) Iron metabolism and the IRE/IRP regulatory system: an update. Ann N Y Acad Sci 1012:1–13
Pantopoulos K, Hentze MW (1995) Rapid responses to oxidative stress mediated by iron regulatory protein. EMBO J 14:2917–2924
Paradies G, Ruggiero FM (1990) Enhanced activity of the tricarboxylate carrier and modification of lipids in hepatic mitochondria from hyperthyroid rats. Arch Biochem Biophys 278:425–430
Pierre JL, Gautier-Luneau I (2000) Iron and citric acid: a fuzzy chemistry of ubiquitous biological relevance. Biometals 13:91–96
Pietrangelo A (2003) Iron-induced oxidant stress in alcoholic liver fibrogenesis. Alcohol 30:121–129
Pilon M, Abdel-Ghany SE, Van Hoewyk D, Ye H, Pilon-Smits EA (2006) Biogenesis of iron–sulfur cluster proteins in plastids. Genet Eng (N Y). 27:101–117
Pondarre C, Antiochos BB, Campagna DR et al (2006) The mitochondrial ATP-binding cassette transporter Abcb7 is essential in mice and participates in cytosolic iron–sulfur cluster biogenesis. Hum Mol Genet 15:953–964
Puig S, Askeland E, Thiele DJ (2005) Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120:99–110
Randle PJ (1998) Regulatory interactions between lipids and carbohydrates: the glucose fatty acid cycle after 35 years. Diabetes Metab Rev 14:263–283
Regev-Rudzki N, Karniely S, Ben-Haim NN, Pines O (2005) Yeast aconitase in two locations and two metabolic pathways: seeing small amounts is believing. Mol Biol Cell 16:4163–4171
Rogina B, Reenan RA, Nilsen SP, Helfand SL (2000) Extended life-span conferred by cotransporter gene mutations in Drosophila. Science 290:2137–2140
Ross KL, Eisenstein RS (2002) Iron deficiency decreases mitochondrial aconitase abundance and citrate concentration without affecting tricarboxylic acid cycle capacity in rat liver. J Nutr 132(4):643–651
Rotig A, de Lonlay P, Chretien D et al (1997) Aconitase and mitochondrial iron–sulphur protein deficiency in Friedreich ataxia. Nat Genet 17:215–217
Rouault TA, Tong WH (2005) Iron–sulphur cluster biogenesis and mitochondrial iron homeostasis. Nat Rev Mol Cell Biol 6:345–351
Rustin P, Bourgeron T, Parfait B, Chretien D, Munnich A, Rotig A (1997) Inborn errors of the Krebs cycle: a group of unusual mitochondrial diseases in human. Biochim Biophys Acta 1361:185–197
Saas J, Ziegelbauer K, von Haeseler A, Fast B, Boshart M (2000) A developmentally regulated aconitase related to iron-regulatory protein-1 is localized in the cytoplasm and in the mitochondrion of Trypanosoma brucei. J Biol Chem 275:2745–2755
Saha AK, Ruderman NB (2003) Malonyl-CoA and AMP-activated protein kinase: an expanding partnership. Mol Cell Biochem 253:65–70
Schilke B, Voisine C, Beinert H, Craig E (1999) Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 96:10206–10211
Seidler A, Jaschkowitz K, Wollenberg M (2001) Incorporation of iron–sulphur clusters in membrane-bound proteins. Biochem Soc Trans 29:418–421
Seznec H, Simon D, Monassier L et al (2004) Idebenone delays the onset of cardiac functional alteration without correction of Fe–S enzymes deficit in a mouse model for Friedreich ataxia. Hum Mol Genet 13:1017–1024
Siculella L, Sabetta S, di Summa R et al (2002) Starvation-induced posttranscriptional control of rat liver mitochondrial citrate carrier expression. Biochem Biophys Res Commun 299:418–423
Silberg JJ, Tapley TL, Hoff KG, Vickery LE (2004) Regulation of the HscA ATPase reaction cycle by the co-chaperone HscB and the iron–sulfur cluster assembly protein IscU. J Biol Chem 279:53924–53931
Singh KK, Desouki MM, Franklin RB, Costello LC (2006) Mitochondrial aconitase and citrate metabolism in malignant and non-malignant human prostate tissues. Mol Cancer 5:14
Sluse FE, Meijer AJ, Tager JM (1971) Anion translocators in rat-heart mitochondria. FEBS Lett 18:149–153
Smith AD, Agar JN, Johnson KA et al (2001) Sulfur transfer from IscS to IscU: the first step in iron–sulfur cluster biosynthesis. J Am Chem Soc 123:11103–11104
Smith SR, Cooperman S, Lavaute T et al (2004) Severity of neurodegeneration correlates with compromise of iron metabolism in mice with iron regulatory protein deficiencies. Ann N Y Acad Sci 1012:65–83
Smith SR, Ghosh MC, Ollivierre-Wilson H, Tong W-H, Rouault TA (2006) Complete loss of iron regulatory proteins 1 and 2 prevents viability of murine zygotes beyond the blastocyst stage of embryonic development. Blood Cells Mol Dis 36:283–287
Sonnewald U, Westergaard N, Krane J, Unsgard G, Petersen SB, Schousboe A (1991) First direct demonstration of preferential release of citrate from astrocytes using [13C]NMR spectroscopy of cultured neurons and astrocytes. Neurosci Lett 128:235–239
Starzynski RR, Lipinski P, Drapier J-C et al (2005) Down-regulation of iron regulatory protein 1 activities and expression in superoxide dismutase 1 knock-out mice is not associated with alterations in iron metabolism. J Biol Chem 280:4207–4212
Tong WH, Jameson GN, Huynh BH, Rouault TA (2003) Subcellular compartmentalization of human Nfu, an iron–sulfur cluster scaffold protein, and its ability to assemble a [4Fe-4S] cluster. Proc Natl Acad Sci USA 100:9762–9767
Tong W-H, Rouault T (2000) Distinct iron–sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells. EMBO J 19:5692–5700
Tong WH, Rouault TA (2006) Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron–sulfur cluster biogenesis and iron homeostasis. Cell Metab 3:199–210
Tury A, Mairet-Coello G, Lisowsky T, Griffond B, Fellmann D (2005) Expression of the sulfhydryl oxidase ALR (Augmenter of Liver Regeneration) in adult rat brain. Brain Res 1048:87–97
Umbreit J (2005) Iron deficiency: a concise review. Am J Hematol 78:225–231
Urbina HD, Silberg JJ, Hoff KG, Vickery LE 2001 Transfer of sulfur from IscS to IscU during Fe/S cluster assembly. J Biol Chem 276:44521–44526
Wada M, Shimada A, Fujita T (2006) Functional characterization of Na+ -coupled citrate transporter NaC2/NaCT expressed in primary cultures of neurons from mouse cerebral cortex. Brain Res 1081:92–100
Wallace MA, Liou LL, Martins J et al (2004) Superoxide inhibits 4Fe-4S cluster enzymes involved in amino acid biosynthesis. Cross-compartment protection by CuZn-superoxide dismutase. J Biol Chem 279:32055–32062
Weinstein R (2001) Hypocalcemic toxicity and atypical reactions in therapeutic plasma exchange. J Clin Apher 16:210–211
Westergaard N, Banke T, Wahl P, Sonnewald U, Schousboe A (1995) Citrate modulates the regulation by Zn2+ of N-methyl-d-aspartate receptor-mediated channel current and neurotransmitter release. Proc Natl Acad Sci USA 92:3367–3370
Williamson JR, Cooper RH (1980) Regulation of the citric acid cycle in mammalian systems. FEBS Lett 117(Suppl):K73–K85
Wingert RA, Galloway JL, Barut B et al (2005) Deficiency of glutaredoxin 5 reveals Fe–S clusters are required for vertebrate haem synthesis. Nature 436:1035–1039
Wolfgang MJ, Lane MD (2006) Control of energy homeostasis: role of enzymes and intermediates of fatty acid metabolism in the central nervous system. Annu Rev Nutr 26:23–44 (Epub ahead of print)
Yan L-J, Levine RL, Sohal RS (1997) Oxidative damage during aging targets mitochondrial aconitase. Proc Natl Acad Sci USA 94:11168–11172
Yang M, Cobine PA, Molik S et al (2006) The effects of mitochondrial iron homeostasis on cofactor specificity of superoxide dismutase 2. EMBO J 25:1775–1783
Yarian CS, Toroser D, Sohal RS (2006) Aconitase is the main functional target of aging in the citric acid cycle of kidney mitochondria from mice. Mech Aging Dev 127:79–84
Yoon T, Cowan JA (2003) Iron–sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe–4S] clusters in ISU-type proteins. J Am Chem Soc 125:6078–6084
Zheng L, Cash VL, Flint DH, Dean DR (1998) Assembly of iron–sulfur clusters: identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J Biol Chem 273:13264–13272
Zheng L, White RH, Cash VL, Jack RF, Dean DR (1993) Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc Natl Acad Sci USA 90:2754–2758
Zoller H, Decristoforo C, Weiss G (2002) Erythroid 5-aminolevulinate synthase, ferrochelatase and DMT1 expression in erythroid progenitors: differential pathways for erythropoietin and iron-dependent regulation. Br J Haematol 118:619–626
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tong, WH., Rouault, T.A. Metabolic regulation of citrate and iron by aconitases: role of iron–sulfur cluster biogenesis. Biometals 20, 549–564 (2007). https://doi.org/10.1007/s10534-006-9047-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10534-006-9047-6