Abstract
The biological effects of 1,25-dihydroxyvitamin D3 [1,25(OH)2D] are mediated through a soluble receptor protein termed the vitamin D receptor (VDR). The VDR binds 1,25(OH)2D with high affinity and high selectivity. In the target cell, the interaction of the 1,25(OH)2D hormone with VDR initiates a complex cascade of molecular events culminating in alterations in the rate of transcription of specific genes or gene networks. This chapter discusses the molecular biology of the VDR and focuses on various aspects of VDR function with an emphasis on the macromolecular interactions that are required for the transcriptional regulatory activity of the VDR. These macromolecular interactions include the association of VDR with the 1,25(OH)2D ligand, the mechanisms required for specific, high-affinity interaction of VDR with DNA, the heterodimeric interaction of VDR with retinoid X receptor (RXR), and protein–protein contacts that comprise the communication links between the VDR and the transcriptional machinery. This chapter also touches on some recent data that suggest that the VDR has transcriptional activity independent of the 1,25(OH)2D ligand in the hair follicle and in the skin. This last aspect demonstrates a novel role for the VDR and its implications in the transcriptional mechanism of the VDR are profound.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Key Words
- Vitamin D receptor (VDR)
- 1,25-dihydroxyvitamin D3
- vitamin D
- cholicalciferol
- transcription
- steroid hormone receptor
- nuclear receptor
1 Introduction
Vitamin D was discovered as a micronutrient that is essential for normal skeletal development and for maintaining bone integrity. However, vitamin D is more appropriately classified as a hormone and it is the vitamin D endocrine system that regulates skeletal homeostasis. Its predominant role is to preserve skeletal calcium by ensuring that adequate absorption of dietary calcium and phosphorous takes place. In addition to this calciotropic role, vitamin D functions in a plethora of cellular actions, perhaps the most fundamental of which is cellular differentiation (1). In skeletal tissue, the hormonal form of vitamin D, 1,25-dihydroxyvitamin D3 [1,25(OH)2D], increases osteoclast number (2) possibly by inducing the differentiation of preosteoclasts into mature bone-resorbing cells (3). Vitamin D also acts directly on the osteoblast wherein one well-established effect is stimulating the synthesis of several bone matrix proteins including osteocalcin and osteopontin. Thus, it is via an integrated series of diverse effects that vitamin D is thought to preserve and maintain the integrity of the bony tissues.
The biological effects of 1,25(OH)2D are mediated through a soluble receptor protein termed the vitamin D receptor (VDR). The VDR binds 1,25(OH)2D with high affinity and high selectivity. In the target cell, the interaction of the 1,25(OH)2D hormone with VDR initiates a complex cascade of molecular events culminating in alterations in the rate of transcription of specific genes or gene networks. Central to this mechanism is the requisite interaction of VDR with retinoid X receptor (RXR) to form a heterodimeric complex. This complex binds to specific DNA sequence elements (VDREs) in vitamin D-responsive genes and ultimately influences the rate of RNA polymerase II-mediated transcription. Thus, the VDR–RXR heterodimer serves as the functional transcriptional enhancer in vitamin D-activated transcription. Following VDR–RXR interaction with the VDRE, protein–protein interactions between the VDR–RXR heterodimer and the transcription machinery are essential for the mechanism of vitamin D-mediated gene expression.
This chapter discusses the molecular biology of the VDR and focuses on various aspects of VDR function with an emphasis on the macromolecular interactions that are required for the transcriptional regulatory activity of the VDR. These macromolecular interactions include the association of VDR with the 1,25(OH)2D ligand, the mechanisms required for specific, high-affinity interaction of VDR with DNA, the heterodimeric interaction of VDR with retinoid X receptor (RXR), and protein–protein contacts that comprise the communications links between VDR and the transcriptional machinery. This chapter also touches on some recent data that suggest that the VDR has transcriptional activity independent of the 1,25(OH)2D ligand in the hair follicle and in the skin. This last aspect demonstrates a novel role for the VDR and its implications in the transcriptional mechanism of the VDR are profound.
2 The Vitamin D Receptor Gene
The location of the human VDR (hVDR) gene on chromosome 12 was originally determined using Southern blot analysis of DNA from human-Chinese hamster cell hybrids (4). This was further refined to the 12q13-14 region using somatic cell hybrid mapping (5) and in situ hybridization and linkage analysis (6). Human chromosome 12q13.3 is also the location of the 1α-hydroxylase gene which is involved in pseudovitamin D-deficient rickets (PDDR) (6, 7). This autosomal recessive disorder, caused by impaired activity of the renal 1α-hydroxylase, results in insufficient levels of serum 1,25(OH)2D. It is intriguing that the two most crucial components of the vitamin D endocrine system, namely VDR and the 1α-hydroxylase, map close to each other on the same region of human chromosome 12.
The gene encoding the hVDR was originally isolated from a human liver genomic DNA library (8) and subsequent reports characterized the hVDR gene and its promoter (8)–(12) (Fig. 1). The hVDR gene spans over 70 kb of genomic DNA and is a complex structure consisting of 14 exon sequences interrupted by intronic sequences ranging in size from 0.2 to 13 kb. A GC-rich, TATA-less promoter directs the transcription of at least three VDR mRNA transcripts (11). The noncoding 5′-end of the gene includes exons 1a–f, while eight additional exons (exons 2–9) encode the VDR protein (11, 12). Exon 2 encodes the two known translation initiation codons. The first initiation codon encodes a protein of 427 amino acids. A T to C transition in the initiation codon creates a polymorphic FokI site and results in initiation of translation at an alternative ATG start codon beginning at the tenth nucleotide downstream, encoding a receptor protein of 424 amino acids. Exon 2 also encodes sequences for the first zinc finger of the DNA-binding domain (see below). Exon 3 contains the sequence for the second zinc finger. The observation that the two motifs of the zinc-finger DNA-binding domain are encoded by separate exons is a characteristic trait of the steroid receptor superfamily. The majority of the C-terminal ligand-binding domain is encoded by exons 8 and 9.
The human VDR cDNA or mRNA transcript consists of 4,605 bp containing a 115 bp noncoding leader sequence, a 1,281-bp open reading frame, and 3,209-bp of 3′-noncoding sequence (13). The functional significance of this rather long 3′-untranslated region is unknown, but it is a characteristic feature of the steroid receptor superfamily. The size of this cDNA agrees well with the predominant mRNA species observed in human extracts (approximately 4.6 kb). While cDNAs were identified which varied in their 5′-untranslated region, most use either the Met1 or polymorphic Met4 translation initiation codon, thereby producing a 48-kDa VDR protein. Recent studies have reported that a small proportion of transcripts produce a 54 kDa protein with a slightly longer N-terminus, suggesting that expression of the VDR is under the complex control of multiple promoters (12, 14). This larger protein, VDRB1, was co-expressed at a 1:3 ratio with the 48 kDa VDR protein (VDRA), had 60% of the transcriptional activity of VDRA, and was localized differently in the cell.
Sequence comparison of the VDR cDNAs showed striking similarity with the members of the superfamily of nuclear receptors for steroid and thyroid hormones. An area of high sequence relatedness is found in the DNA-binding domain (DBD) of these receptors. This 70 amino acid domain is rich in cysteine, lysine, and arginine residues and it is the region of the receptor that is responsible for high-affinity interaction with specific DNA sequence elements. An area of more limited homology is in the large C-terminal domain which is responsible for high-affinity interaction with the various hormones, termed the ligand-binding domain (LBD). Interestingly, distinct regions of high similarity are present in the LBDs of the various receptors despite the obvious structural nonrelatedness of their individual ligands. This may be due to the fact that this C-terminal domain functions in other related areas of receptor function (e.g., in protein–protein interactions; see below).
The deduced amino acid sequences of the VDR from several species are illustrated in Fig. 2. A comparison of the coding regions of the human, rat, mouse, bovine, and avian VDRs shows a high degree of sequence identity particularly within the N-terminal portion (aa 16–116 of hVDR) that constitutes the DBD (97% identity) and the C-terminal domain (aa 226–427 of hVDR) which comprises most of the 1,25(OH)2D-binding domain (85% identity). The sequences tend to diverge in the hinge region of the receptors which is the region located between the DBD and the LBD. The high degree of sequence conservation of VDR between species supports the fundamental roles that these domains serve in VDR function.
3 Molecular Analysis Of The Functional Domains Of The Vdr
The nuclear receptor superfamily is characterized by a modular structure consisting of regions required for specific functions. The amino terminus is of a variable length and contains a transactivation domain termed AF-1. In the VDR, this region is very short. The central DBD has two zinc-finger motifs that are responsible for protein–DNA interactions. The carboxy-terminal domain contains the ligand-binding domain (LBD) and the AF-2 domain. These regions are discussed in more detail below.
3.1 The DNA-Binding Domain (DBD)
The location of the DBD of VDR was originally mapped to the N-terminal 113 amino acids of the receptor (15, 16). Sone et al. showed that the N-terminal 21 amino acids immediately preceding the first cysteine residue could be removed without affecting DNA binding or the transcriptional activation potential of the VDR (17). Thus, the minimal domain of the VDR that mediates VDR–DNA interactions was determined to reside between amino acid residues 22 and 113 in the human sequence. There are nine cysteine residues within the DBD that are conserved throughout the members of the superfamily of receptor proteins. The first eight of these cysteines (counting from the N-terminus) tetrahedrally coordinate two zinc atoms to form two zinc-finger DNA-binding motifs (Fig. 3a). In mutagenesis studies, mutation of the first eight of the nine cysteine residues to serines eliminated VDR binding to both nonspecific and specific DNA sequences and eliminated VDR-mediated transactivation (17). A serine mutation at the ninth cysteine residue (C84S) had little effect on VDR function suggesting that this residue is not functionally analogous to the first eight cysteines. These data showed the essential nature of the first eight cysteines in the overall organization and structural integrity of the DBD zinc fingers. Later crystallization of the VDR DBD confirmed the structural predictions (18).
Binding of VDR to its response element. (a) The N-terminal, zinc-finger DNA-binding motifs of human vitamin D receptor. Residues circled are involved in the homodimers interface when bound to DNA. (b) Orientation of helix 1 and helix 2 when bound to DNA. (c) Orientation of the VDR–RXR heterodimers when bound to a VDRE.
While the VDR–RXR heterodimer is the active transcription factor, efforts to co-crystallize their DBDs with DNA have been unsuccessful. However, Shaffer and Gewirth successfully crystallized the VDR DBD homodimer bound to a DR3 response element (18). Thus, much of what is known of the nature of VDR–RXR–DNA interactions is modeled after functional and structural data of the homodimers bound to DNA as well as of other related nuclear receptors bound to their recognition sites. The common structural feature of the DBDs for all these receptors is the folding of two α-helices in the carboxyl terminal portion of the each zinc finger into a single DNA-binding domain. The first α-helix in the amino terminal finger (denoted helix 1 in Fig. 3b) lies across the major groove of DNA making specific contacts with the DNA-binding site and it is this region that contains the crucial amino acids that determine response element specificity for some receptors. The second α-helix (denoted helix 2 in Fig. 3b) folds across the first in a perpendicular arrangement. The DBD is rich in the positively charged amino acids, lysine and arginine, several of which form favorable electrostatic interactions with the negatively charged phosphate backbone of the DNA helix.
The VDR response element, VDRE, consists of two hexameric half-sites that are arranged as a direct repeat separated by a spacer of three nucleotides (DR3). Unlike some of the other nuclear receptors, VDR does not show any preference for particular sequences outside of the half-site to which it is bound (19). When bound to a DR3, the VDR and RXR each binds to half-site in a head-to-tail dimer with RXR occupying the 5′ or upstream half-site in each element (Fig. 3c) (20, 21). The VDR–VDR homodimer also binds to DR3 response elements in a head-to-tail arrangement (18). VDR dimerization contacts involve the side chains of Pro61, Phe62, and His75 of the upstream subunit and residues Asn37, Glu92, and Phe93 of the downstream subunit (18). These sequences are conserved among the nine known VDRs. Mutation of the core of this hydrophobic dimer interface abolished cooperative assembly on DR3 elements (22) and reporter gene activation (23).
Helix I of the VDR DBD is also referred to as the recognition helix, the proximal-box, or the P-box. The recognition helix confers target sequence selectivity for binding of the VDR homodimer to its element (see Fig. 3a). The key interactions occur with four conserved residues, Glu42, Lys45, Arg49, and Arg50, and each residue makes sequence-specific base contacts in the major groove of the half-site (18). Helix III, referred to as the T-box or C-terminal extension (CTE), resides just C-terminal to the second zinc finger and mediates homodimer interaction and interaction of nuclear receptors with DNA (24, 25). Mutations in the CTE of the VDR resulted in a dramatic reduction in VDR binding to DNA and in transactivation indicating an important role for this α-helical domain in VDR function. However, the crystal structure of VDR demonstrated that the CTE of VDR only contacted the DNA at two positions (18). This is in stark contrast to the thyroid hormone receptor CTE which makes 15 contacts with DNA. Shaffer and Gewirth’s studies further suggest that the primary role of the VDR CTE is to provide not additional protein–DNA contacts but the mechanism by which the protein discriminates the spacing between the half elements. Thus, the CTE in VDR most likely prevents dimerization on incorrectly spaced response elements.
3.2 The Multifunctional C-terminal Domain
Although the large C-terminal domain of VDR (aa 116 – 427 of hVDR) clearly functions as the binding motif for the 1,25(OH)2D ligand, it also fulfills several other critical roles in VDR function (reviewed in (26)). A prominent role for the LBD is that of a protein–protein interaction surface through which VDR contacts its heterodimeric partner RXR for high-order binding to DNA. It is through this C-terminal domain that VDR also interacts with other proteins such as TFIIB and coactivators that are important for the mechanism of VDR-mediated transcription. Finally, key serine residues in this domain serve as sites of phosphorylation that may be important in regulating the transcriptional activity of the VDR.
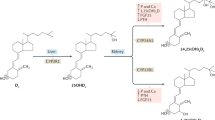
The LBD is responsible for high-affinity binding of 1,25(OH)2D, exhibiting equilibrium-binding constants on the order of 10–10–10–11 M (27)–(30). Both the 1-hydroxyl and 25-hydroxyl moieties are crucial for efficient recognition by VDR. For example, the monohydroxylated vitamin D3 compounds, 1(OH)D and 25(OH)D, bind with approximately 100-fold less affinity than 1,25(OH)2D. The nonhydroxylated parent vitamin D3 compound does not bind significantly to the VDR. Modifications in both the side chain structure and in the A ring of the secosteroid also dramatically reduce the affinity of the LBD for the analog.
The specific amino acid residues that are directly involved in 1,25(OH)2D interactions were identified when the LBD was crystallized bound to its natural ligand (31). While earlier attempts to crystallize the VDR failed, Rochel et al. achieved success upon removing a region that decreased protein solubility. The removal of this highly variable region did not affect hormone binding or transactivation and is unlikely to disrupt structural integrity. The structure of the VDR LBD is organized similar to other nuclear receptors into 13 α-helices and 3 β-sheets that together form a hydrophobic pocket that binds ligand. Helix 12, containing the ligand-dependent AF-2 region, contacts the ligand directly while creating a coactivator-binding surface. The VDR ligand-binding cavity is large compared to other receptors and the 1,25(OH)2D ligand occupies only about one half of the volume. Rochel et al. (31) conducted ligand-docking modeling of synthetic analogs to begin to understand the differences in activities of the various ligands. They observed variations in conformation of the ligand and receptor as well as different contacts in the binding pockets. These differences may result in changes in half-lives and transcriptional activities. The information obtained from the crystallization of the LBD and these docking studies will aid in the development of synthetic analogs that mimic the advantageous effects of 1,25(OH)2D without the hypercalcemic side effects.
In addition to hormone binding, the LBD is required for several other aspects of receptor function, including an importance in mediating protein–protein interactions. One important protein–protein contact is the heterodimerization of VDR with the retinoid X receptor (RXR) family of receptors. By drawing analogies to other related receptors, the crystal structure of the RXR–RXR homodimeric complex provides some insight into what may be the crucial heterodimerization surface of the VDR–RXR complex. The RXRα crystal structure revealed an α-helical-rich LBD (65%) consisting of 11 α-helices organized into what has been termed “a three-layer antiparallel sandwich” (32). The RXR dimer is symmetrically arranged with the interaction surface being formed mainly by helix 10 and, to a lesser extent, by helix 9. Helix 10 of RXR corresponds to the C-terminal region of VDR identified by Nakajima et al. (33) as being crucial for heterodimer formation. Like RXR, the VDR LBD also forms a sandwich in three layers of β-sheets (31). Thus, based on the structural relatedness between VDR and RXR, the C-terminal region of hVDR that includes residues 382–403 (helix 10) may directly contact helix 10 of RXR to comprise the major interaction surface which would generate a structurally symmetrical VDR–RXR heterodimeric complex.
The C-terminal domain also functions as a transactivation domain. Transcriptional activation domains are generally defined through mutations that selectively affect the transcriptional activity of the receptor without disrupting other receptor functions such as ligand binding, response element interaction, or nuclear localization. These domains are often transferable; that is, fusing the activation domain itself to a heterologous DNA-binding domain imparts high-order transcription to the heterologous fusion protein. The extreme C-terminus of the nuclear receptors has a highly conserved, constitutive, hormone-dependent activation domain known as the AF2 domain. Nakajima et al. showed that removing the 25 amino acid AF2 domain from the C-terminus of hVDR (Δ403-427) resulted in a complete loss of 1,25(OH)2D-/VDR-activated transcription (33). This loss of function was not due to altered binding of RXR, VDRE, or hormone, and the mutant receptor was appropriately targeted to the cell nucleus. Later studies demonstrated that the AF2 domain resides within helix 12 and the main structural feature is that of an amphipathic α-helix (31). Upon binding ligand, the AF-2 domain, located in helix 12, folds over the top of the LBD and creates a platform for the docking of nuclear receptor coactivators required for nuclear receptor-dependent transcription. Communication between the VDR and the transcriptional machinery is discussed below.
4 Molecular Mechanism Of Transcriptional Control By Vdr
4.1 VDR Interaction with Vitamin D-Responsive Elements
4.1.1 Vitamin D-Responsive Elements
Nuclear receptors modulate transcription by binding to specific DNA elements in the promoter regions of hormone-responsive genes. The specific VDR-interactive promoter sequences are termed vitamin D-responsive elements (VDREs). VDREs from a variety of vitamin D-responsive genes have been identified (Fig. 4) on the basis of several functional criteria: (1) deletion or mutation of the element resulted in the loss of promoter responsiveness to 1,25(OH)2D; (2) the sequence alone conferred vitamin D responsiveness to an otherwise unresponsive, heterologous promoter; and (3) the element served as a high-affinity binding site for VDR in vitro.
By comparing the limited number of natural response elements identified to date, the VDRE is generally described as an imperfect direct repeat of a core hexanucleotide sequence, G/A G G T G/C A, with a spacer region of three nucleotides separating each half element (also termed DR-3 for direct repeat with a three nucleotide spacer). This direct repeat motif is analogous to DNA elements that mediate retinoic acid and thyroid hormone responsiveness (RAREs and TREs) and it contrasts with responsive elements that mediate glucocorticoid- or estrogen-responsive genes (GREs and EREs) which are generally palindromic or inverted repeat sequences (Fig. 4, lower panel). It is apparent that some degree of plasticity exists in the sequence of each half element of the VDRE, suggesting that there is flexibility in the precise sequence that will mediate vitamin D responsiveness. However, there is little question that the nucleotide sequence of the element is critical for receptor-mediated transcription. Modest changes in the nucleotides of either half element in the rat osteocalcin VDRE disrupted VDR–VDRE interactions and compromised VDR-dependent transactivation (34).
An additional determinant of response specificity for this class of nuclear receptors is the length of the spacer region (35). In general, VDR, thyroid hormone receptor (TR), and retinoic acid receptor (RAR) all recognize similar direct repeat sequences, but with differing spacer regions of 3, 4, and 5 nucleotides, respectively. This phenomenon, termed the “3-4-5 rule” and later expanded to the “1-5 rule” (36, 37), illustrates that half-site spacing plays a major role in determining selective hormonal response. With regard to the VDRE, it is clear that the actual nucleotide sequence of the spacer region is also important since mutations in the spacer of the rat osteocalcin VDRE disrupt VDR binding and transactivation (34).
The natural VDREs identified thus far provide only a snapshot of the DNA sequences that mediate the transcriptional effects of the VDR and the elements in Fig. 4 are certainly a limited sample. Variations on the DR-3 motif for VDREs have been identified in the elements that mediate vitamin D responsiveness in the calbindin D9k and calbindin D28k genes (38, 39). Moreover, several synthetic elements with large spacer regions and inverted arrangements can mediate vitamin D responsiveness under certain conditions (40). It is likely that the affinity of VDR for these atypical elements may vary from that of the classic DR-3 motif adding yet another level of regulatory complexity to the process of VDR-mediated gene expression. Interestingly, VDREs and other NR regulatory elements may function over very long distances. For example, Nerenz et al. recently identified a 1,25(OH)2D-responsive enhancer in the human RANKL promoter that resides –20 kb upstream of the transcriptional start site (41). This region contains a VDRE to which the VDR/RXR heterodimer can bind and which is responsible for induction by 1,25(OH)2D.
4.1.2 A R ole for the 1,25(OH) 2 D L igand
Insight into a role for the 1,25(OH)2D ligand in the transactivation process has emerged from studies examining VDR, RXR, and VDRE interactions. Nanomolar amounts of the 1,25(OH)2D ligand dramatically enhance VDR–RXR heterodimerization, both the direct interaction of VDR with RXR in solution (17, 42) and the interaction of the VDR–RXR heterodimer with the VDRE (17, 43, 44). Surface plasmon resonance quantitated the binding constants for these interactions and showed a clear 1,25(OH)2D-dependent decrease in VDR interaction with itself (i.e., VDR homodimers) and a concomitant increase in VDR heterodimerization with RXR (45). These ligand-induced changes in VDR–RXR interactions are likely due to altered conformations of the VDR in the absence and presence of 1,25(OH)2D (46). Thus, one putative role for the 1,25(OH)2D in VDR-mediated transcription may be to induce a distinct conformational change in VDR that disrupts weak homodimers of unliganded VDR and promotes liganded VDR heterodimerization with RXR. The interaction of VDR and RXR generates a heterodimeric complex that is highly competent to bind DR-3 VDREs and subsequently affect the transcriptional process.
The possibility exists that other natural compounds may serve as activators of VDR-mediated responses. Makishima et al. (47) screened classical nuclear receptors to identify those that were activated by the bile acid lithocholic acid (LCA). They found that micromolar concentrations of LCA and its metabolites directly bind to VDR and activate transcription. Moreover, LCA- or 1,25(OH)2D-liganded VDR also stimulates the expression of endogenous CYP3A, the P450 enzyme responsible for degradation of LCA in the liver and the intestine. While LCA is implicated as a toxin that promotes colorectal carcinogenesis (48, 49), 1,25(OH)2D protects against colon cancer (50). Thus, induction of CYP3A by 1,25(OH)2D and LCA may represent a detoxification pathway for LCA and explain the potential preventative effects of 1,25(OH)2D in colon cancer.
4.1.3 G ene R egulation T hrough N egative R esponse E lements
The role of VDR as a transcriptional activator is well known, but numerous studies demonstrate that VDR also directly down-regulates the transcription of some genes. The genes encoding the parathyroid hormone (PTH), the parathyroid hormone-related peptide (PTHrP), and CYP27B1 are all down-regulated by VDR (51)–(53). Both the rat PTH promoter and the human PTHrP promoter contain negative VDREs (nVDREs) which closely resemble the consensus VDRE sequence and which are bound by either VDR homodimers or VDR/RXR heterodimers. However, negative regulation may not necessarily require both VDRE half-sites.
A second type of nVDRE, composed of E-box-type motifs (5′-CATCTG-3′), was identified in the promoters of the human CTP27B1 gene, the human PTH gene, and the human PTHrP gene (54, 55). These nVDREs are transcriptionally active in the absence of 1,25(OH)2D and are bound by a helix-loop-helix transcription factor known as VDR-interacting repressor (VDIR) (56). In the absence of 1,25(OH)2D, VDIR recruits CBP/p300 HAT activity to promote active transcription. However, in the presence of 1,25(OH)2D, ligand-induced coregulator switching occurs which is responsible for transcriptional repression (57). Basically, liganded VDR/RXR heterodimers associate with the VDRE-bound VDIR, not with the DNA itself. This results in the dissociation of the CBP/p300 HAT coactivator and the recruitment of histone deacetylases and NCoR/SMRT corepressors. Thus, the chromatin is subsequently remodeled and transcription inhibited.
4.1.4 A R ole for the U nliganded VDR in T ranscriptional R egulation
Studies of the VDR knockout (VDRKO) mice and the 25(OH)D-1α-hydroxylase knockout mice suggest a role for the unliganded receptor in controlling transcription (58, 59). The VDRKO mice exhibit an impairment of calcium absorption and progressive hypocalcemia, hypophosphatemia, and compensatory hyperparathyroidism. These metabolic imbalances result in growth retardation, severe skeletal defects including decreased bone mineral density, thinned bone cortex, and widened undermineralized growth plates. These bone defects are secondary to the malabsorption of calcium, since VDRKO mice fed a rescue diet rich in calcium and phosphorus develop normally without bone abnormalities. The VDRKO mice also develop alopecia which, in contrast to the skeletal phonotype, is not corrected by the rescue diet. This indicates a direct role for VDR in hair follicle cycling. Interestingly, mice that lack the 25(OH)D-1α(OH)ase enzyme and that are totally devoid of the 1,25(OH)2D ligand do not exhibit alopecia (59). Additional studies using VDR transgenes with mutations in the ligand-binding domain demonstrated that these VDR mutants were able to restore hair growth (60). VDR ablation also sensitizes mice to chemically and UV-induced tumorigenesis (61, 62), a response that was not displayed in the 25(OH)D-1α(OH)ase knockout. Thus, the VDR, but not its ligand, is required for protection against carcinogenesis (61). Taken together with the alopecia data, these studies suggest that in the epidermis, the VDR has functions that are independent of the 1,25(OH)2D ligand but require interactions with nuclear factors. There is precedent for this, as in vitro studies show that VDR can associate with various transcription factors and induce select genes in the absence in ligand (63)–(66). Indeed, VDR has been shown to activate target promoters in the absence of 1,25(OH)2D selectively in primary keratinocyte cell culture models (67). Alternatively, unliganded VDR may repress a subset of target genes in a manner analogous to other nuclear receptors through corepressor interactions.
4.2 Communication Between VDR and the Transcriptional Machinery
The past decade has witnessed significant progress in understanding the sequence of events that follow VDR–RXR heterodimer binding to the VDRE and lead to RNA polymerase II-directed transcription. Central to the process of activated transcription are the general transcription factors and the ordered assembly of the preinitiation complex (PIC), beginning with TATA-binding protein (TBP, a subunit of TFIID), binding to the TATA element of class II promoters in a process that is facilitated by TFIIA [reviewed in (68)]. Then, TFIIB enters the complex by direct interaction with TBP. RNA Pol II, in association with TFIIF, binds to this early complex by contacting TFIIB. The further association with TFIIE and other general factors results in a complex capable of accurately initiating RNA synthesis. Transcription initiated by these minimal components represents basal-level transcription which can be stimulated (or repressed) by sequence-specific, trans-acting factors such as the VDR.
The nuclear receptor superfamily contains two regions responsible for transcriptional activation: AF1, located in the extreme N-terminus, and AF2, located in the C-terminus. The N-terminus of the VDR is truncated compared to other nuclear receptors and thus it was thought to be unlikely that an analogous constitutive AF-1 domain exists N-terminal to the DBD in VDR. Indeed, as previously mentioned, removing the N-terminal 22 amino acids preceding the DBD had no affect on VDR-activated transcription. However, mutation of Arg-18 and Arg-22 compromised hVDR transcriptional activity presumably by compromising interaction with transcription factor TFIIB (69). Moreover, a polymorphic variant, lacking the first three amino acids at the N-terminus has increased transactivation potency due to better interaction with TFIIB (69). Thus, the short N-terminus of VDR may act as a docking site for TFIIB. VDR also interacts with TFIIB via its C-terminus (42, 70), an interaction that leads to a functional increase in vitamin D-mediated transcription (70). These studies clearly indicate that the VDR–TFIIB interaction is functionally significant and they suggest that the VDR–RXR heterodimer may communicate with the PIC, in part, through specific protein–protein contacts between VDR and TFIIB.
In addition to contacting components of the PIC, VDR and other nuclear receptors are known to interact with a variety of coregulators, i.e., coactivator (CoA) and corepressor (CoR) proteins that also function in the mechanism of steroid-regulated gene transcription (26). Coregulators can be classified into two main groups. The first group contains factors that are recruited by the nuclear receptor to the target promoter to modulate the chromatin architecture. This is done by covalently modifying histones by such processes as acetylation/deacetylation and methylation/demethylation. The second group of coregulators includes ATP-dependent chromatin remodeling factors that modulate promoter accessibility to transcription factors and to the basal transcriptional machinery.
Basically, in the absence of ligand, the nuclear receptor may interact with CoR proteins such as nuclear corepressor (NCoR), silencing mediator of retinoic acid and thyroid hormone receptor (SMART), hairless, and Alien, which in turn associate with histone deacetylases leading to a locally increased chromatin packaging and decrease in gene expression. The binding of ligand induces the dissociation of the CoR and the association of a CoA of the p160/SRC family. Ligand binding results in conformational changes that create a hydrophobic cleft composed of helices 3, 4, 5, and 12. The hydrophobic cleft serves as a docking surface for the p160/SRC family of proteins through their LXXLL domains, and this provides a scaffold for further recruitment of coactivators including histone acetytransferases (HATs) such as p300/CBP and histone methyltransferases (HMTs). By covalently modifying histones, these enzymes cause the relaxation in the chromatin architecture and provide a signal for recruitment of additional co-regulatory proteins. Ligand-activated nuclear receptors then change rapidly from interacting with the p160/SRC family to interacting with mediator/VDR-interacting protein (DRIP) complexes, such as Med1 (71). The mediator/DRIP complexes consist of approximately 15–20 proteins and build a bridge to the basal transcription machinery (71, 72). In fact, liganded VDR binds to DRIP/mediator and this ternary complex can then bind directly to the Pol II holoenzyme and recruit the transcriptional machinery to the promoter (73). In this way, ligand-activated nuclear receptors execute two tasks, modification of chromatin and regulation of transcription.
In addition, several other proteins that potentiate VDR-mediated transcription have been described. One example is NCoA-62/ski-interacting protein (SKIP), a coactivator which binds VDR simultaneously with SRC-1 to form a ternary complex that synergistically enhances VDR-stimulated transcription (63, 74). Subsequent studies identified NCoA62/SKIP in subcomplexes of the spliceosome (75, 76). This, combined with NCoA62/SKIP’s ability to contact varied transcription factors such as the VDR, suggests a potentially important role in coupling nuclear receptor-mediated transcription with mRNA splicing (77, 78).
A current model for VDR-mediated transcription is illustrated in Fig. 5. This model incorporates numerous properties of VDR that were discussed in this chapter. The initial event in this model is high-affinity binding of the 1,25(OH)2D ligand to the VDR. Ligand binding induces VDR/RXR heterodimerization and the heterodimer specifically binds VDREs in the promoter regions of vitamin D-responsive genes. The VDR–RXR heterodimer serves as a platform for the sequential binding of a wide variety of proteins needed for efficient transcriptional regulation. These include basal transcriptional components such as TFIIB and mediator/DRIP complexes. Coactivator proteins form additional contacts between the VDR and the PIC and it is the interaction with and the communication between VDR, RXR, TFIIB, and other ligand-dependent coactivator proteins such as SRC-1 that may determine the overall transcriptional activity of a vitamin D-responsive gene. It also incorporates proteins such as NCoA62/SKIP, which may enter later to act at more distal steps such as the processing of the nascent RNA transcript. Understanding the complex interplay that occurs between these various factors is crucial to unraveling the complexities of activated or repressed transcription mediated by vitamin D and the VDR.
References
Abe E, Miyaura C, Sakagami H et al (1981) Differentiation of mouse myeloid leukemia cells induced by 1α,25-dihydroxyvitamin D3. Proc Natl Acad Sci USA 78:4990–4994
Holtrop ME, Cox KA, Clark MB et al (1981) 1,25-dihydroxycholecalciferol stimulates osteoclasts in rat bones in the absence of parathyroid hormone. Endocrinology 108:2293–2301
Bar-Shavit Z, Teitelbaum SL, Reitsma P et al (1983) Induction of moncytic differentiation and bone resorption by 1,25(OH)2D3. Proc Natl Acad Sci USA 80:5908–5911
Faraco JH, Morrison NA, Baker A et al (1989) ApaI dimorphism at the human vitamin D receptor gene locus. Nucleic Acids Res 17:2150
Szpirer J, Szpirer C, Riviere M et al (1991) The Sp1 transcription factor gene (SP1) and the 1,25-dihydroxyvitamin D3 receptor gene (VDR) are colocalized on human chromosome arm 12q and rat chromosome 7. Genomics 11:168–173
Labuda M, Fujiwara TM, Ross MV et al (1992) Two hereditary defects related to vitamin D metabolism map to the same region of human chromosome 12q13-14. J Bone Miner Res 7:1447–1453
Kitanaka S, Takeyama K, Murayama A et al (1998) Inactivating mutations in the 25-hydroxyvitamin D3 1alpha-hydroxylase gene in patients with pseudovitamin D-deficiency rickets. N Engl J Med 338:653–661
Pike JW, Kesterson RA, Scott RA et al (1988) Vitamin D3 receptors: molecular structure of the protein and its chromosomal gene. In: Norman AW, Schaefer K et al (ed) Vitamin D: molecular, cellular and clinical endocrinology. Walter de Gruyter & Co., Berlin, New York
Sone T, Marx SJ, Liberman UA et al (1990) A unique point mutation in the human vitamin D receptor chromosomal gene confers hereditary resistance to 1,25-dihydroxyvitamin D3. Mol Endocrinol 4:623–631
Hughes MR, Malloy PJ, Kieback DG et al (1988) Point mutations in the human vitamin D receptor gene associated with hypocalcemic rickets. Science 242:1702–1705
Miyamoto K, Kesterson RA, Yamamoto H et al (1997) Structural organization of the human vitamin D receptor chromosomal gene and its promoter. Mol Endocrinol 11:1165–1179
Crofts L, Hancock M, Morrison N et al (1998) Multiple promoters direct the tissue-specific expression of novel N-terminal variant human vitamin D receptor gene transcripts. Proc Natl Acad Sci USA 95:10529–10534
Baker AR, McDonnell DP, Hughes M et al (1988) Cloning and expression of full-length cDNA encoding human vitamin D receptor. Proc Natl Acad Sci USA 85:3294–3298
Sunn K, Cock T, Crofts L et al (2001) Novel N-terminal variant of human VDR. Mol Endocrinol 15:1599–1609
Allegretto EA, Pike JW, Haussler MR (1987) Immunochemical detection of unique proteolytic fragments of the chick 1,25-dihydroxyvitamin D3 receptor. Distinct 20 kDa DNA-binding and 45 kDa hormone-binding species. J Biol Chem 262:1312–1319
McDonnell DP, Scott RA, Kerner SA et al (1989) Functional domains of the human vitamin D3 receptor regulate osteocalcin gene expression. Mol Endocrinol 3:635–644
Sone T, Kerner S, Pike JW (1991) Vitamin D receptor interaction with specific DNA: association as a 1,25-dihydroxyvitamin D3-modulated heterodimer. J Biol Chem 266:23296–23305
Shaffer PL, Gewirth DT (2002) Structural basis of VDR–DNA interactions on direct repeat response elements. EMBO J 21:2242–2252
Freedman LP, Arce V, Perez Fernandez R (1994) DNA sequences that act as high affinity targets for the vitamin D3 receptor in the absence of the retinoid X receptor. Mol Endocrinol 8:265–273
Jin CH, Pike JW (1996) Human vitamin D receptor-dependent transactivation in Saccharomyces cerevisiae requires retinoid X receptor. Mol Endocrinol 10:196–205
Schrader M, Nayeri S, Kahlen JP et al (1995) Natural vitamin D3 response elements formed by inverted palindromes: polarity-directed ligand sensitivity of vitamin D3 receptor-retinoid X receptor heterodimer-mediated transactivation. Mol Cell Biol 15:1154–1161
Towers TL, Luisi BF, Asianov A et al (1993) DNA target selectivity by the vitamin D3 receptor: mechanism of dimer binding to an asymmetric repeat element. Proc Natl Acad Sci USA 90:6310–6314
Quack M, Szafranski K, Rouvinen J et al (1998) The role of the T-box for the function of the vitamin D receptor on different types of response elements. Nucleic Acids Res 26:5372–5378
Lee MS, Kliewer SA, Provencal J et al (1993) Structure of the retinoid X receptor α DNA binding domain: a helix required for homodimeric DNA binding. Science 260:1117–1121
Wilson TE, Paulsen RE, Padgett KA et al (1992) Participation of non-zinc finger residues in DNA binding by two nuclear orphan receptors. Science 256:107–110
Sutton AL, MacDonald PN (2003) Vitamin D: more than a "bone-a-fide" hormone. Mol Endocrinol 17:777–791
Brumbaugh PF, Haussler MR (1975) Specific binding of 1α,25-dihydroxycholecalciferol to nuclear components of chick intestine. J Biol Chem 250:1588–1594
Brumbaugh PF, Haussler MR (1974) 1,25-dihydroxycholecalciferol receptors in the chick intestine. II. Temperature dependent transfer of the hormone to chromatin via a specific cytosol receptor. J Biol Chem 249:1258–1262
Wecksler WR, Norman AW (1980) A kinetic and equilibrium binding study of 1α,25-dihydroxyvitamin D3 with its cytosol receptor from chick intestinal mucosa. J Biol Chem 255:3571–3574
Mellon W, DeLuca HF (1979) An equilibrium and kinetic study of 1,25-dihydroxyvitamin D3 binding to chicken intestinal cytosol employing high specific activity 1,25-dihydroxy[3H-26,27]vitamin D3. Arch Biochem Biophys 197:90–95
Rochel N, Wurtz JM, Mitschler A et al (2000) The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol Cell 5:173–179
Bourguet W, Ruff M, Chambon P et al (1995) Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-α. Nature 375:377–382
Nakajima S, Hsieh J-C, MacDonald PN et al (1994) The C-terminal region of the vitamin D receptor is essential to form a complex with a receptor auxiliary factor required for high affinity binding to the vitamin D-responsive element. Mol Endocrinol 8:159–172
Demay MB, Kiernan MS, DeLuca HF et al (1992) Characterization of 1,25-dihydroxyvitamin D3 receptor interactions with target sequences in the rat osteocalcin gene. Mol Endocrinol 6:557–562
Umesono K, Evans RM (1989) Determinants of target gene specificity for steroid/thyroid hormone receptors. Cell 57:1139–1146
Leid M, Kastner P, Chambon P (1992) Multiplicity generates diversity in the retinoic acid signalling pathways. Trends Biochem Sci 17:427–433
Mangelsdorf DJ, Umesono K, Kliewer SA et al (1991) A direct repeat in the cellular retinol-binding protein type II gene confers differential regulation by RXR and RAR. Cell 66:555–561
Darwish HM, DeLuca HF (1992) Identification of a 1,25-dihydroxyvitamin D3-response element in the 5'-flanking region of the rat calbindin D-9 k gene. Proc Natl Acad Sci USA 89:603–607
Gill RK, Christakos S (1993) Identification of sequence elements in mouse calbindin-D28 k gene that confer 1,25-dihydroxyvitamin D3- and butyrate-inducible responses. Proc Natl Acad Sci USA 90:2984–2988
Carlberg C, Bendik I, Wyss A et al (1993) Two nuclear signalling pathways for vitamin D. Nature 361:657–660
Nerenz RD, Martowicz ML, Pike JW (2008) An enhancer 20 kilobases upstream of the human receptor activator of nuclear factor-κB ligand gene mediates dominant activation by 1,25-dihydroxyvitamin D3. Mol Endocrinol 22:1044–1056
MacDonald PN, Sherman DR, Dowd DR et al (1995) The vitamin D receptor interacts with general transcription factor IIB. J Biol Chem 270:4748–4752
Liao J, Ozono K, Sone T et al (1990) Vitamin D receptor interaction with specific DNA requires a nuclear protein and 1,25-dihydroxyvitamin D3. Proc Natl Acad Sci USA 87:9751–9755
MacDonald PN, Dowd DR, Nakajima S et al (1993) Retinoid X receptors stimulate and 9-cis retinoic acid inhibits 1,25-dihydroxyvitamin D3-activated expression of the rat osteocalcin gene. Mol Cell Biol 13:5907–5917
Cheskis B, Freedman LP (1996) Modulation of nuclear receptor interactions by ligands: kinetic analysis using surface plasmon resonance. Biochemistry 35:3309–3318
Peleg S, Sastry M, Collins ED et al (1995) Distinct conformational changes induced by the 20-epi analogues of 1α,25-dihydroxyvitamin D3 are associated with enhanced activation of the vitamin D receptor. J Biol Chem 270:10551–10558
Makishima M, Lu TT, Xie W et al (2002) Vitamin D receptor as an intestinal bile acid sensor. Science 296:1313–1316
Debruyne PR, Bruyneel EA, Karaguni IM et al (2002) Bile acids stimulate invasion and haptotaxis in human colorectal cancer cells through activation of multiple oncogenic signaling pathways. Oncogene 21:6740–6750
Kozoni V, Tsioulias G, Shiff S et al (2000) The effect of lithocholic acid on proliferation and apoptosis during the early stages of colon carcinogenesis: differential effect on apoptosis in the presence of a colon carcinogen. Carcinogenesis 21:999–1005
Lamprecht SA, Lipkin M (2001) Cellular mechanisms of calcium and vitamin D in the inhibition of colorectal carcinogenesis. Ann N Y Acad Sci 952:73–87
Demay MB, Kiernan MS, DeLuca HF et al (1992) Sequences in the human parathyroid hormone gene that bind the 1,25-dihydroxyvitamin D3 receptor and mediate transcriptional repression in response to 1,25-dihydroxyvitamin D3. Proc Natl Acad Sci USA 89:8097–8101
Falzon M (1996) DNA sequences in the rat parathyroid hormone-related peptide gene responsible for 1,25-dihydroxyvitamin D3-mediated transcriptional repression. Mol Endocrinol 10:672–681
Russell J, Ashok S, Koszewski NJ (1999) Vitamin D receptor interactions with the rat parathyroid hormone gene: synergistic effects between two negative vitamin D response elements. J Bone Miner Res 14:1828–1837
Kim MS, Fujiki R, Murayama A et al (2007) 1Alpha,25(OH)2D3-induced transrepression by vitamin D receptor through E-box-type elements in the human parathyroid hormone gene promoter. Mol Endocrinol 21:334–342
Murayama A, Takeyama K, Kitanaka S et al (1998) The promoter of the human 25-hydroxyvitamin D3 1 alpha-hydroxylase gene confers positive and negative responsiveness to PTH, calcitonin, and 1 alpha,25(OH)2D3. Biochem Biophys Res Commun 249:11–16
Murayama A, Kim MS, Yanagisawa J et al (2004) Transrepression by a liganded nuclear receptor via a bHLH activator through co-regulator switching. EMBO J 23:1598–1608
Fujiki R, Kim MS, Sasaki Y et al (2005) Ligand-induced transrepression by VDR through association of WSTF with acetylated histones. EMBO J 24:3881–3894
Sakai Y, Kishimoto J, Demay MB (2001) Metabolic and cellular analysis of alopecia in vitamin D receptor knockout mice. J Clin Invest 107:961–966
Panda DK, Miao D, Tremblay ML et al (2001) Targeted ablation of the 25-hydroxyvitamin D 1alpha -hydroxylase enzyme: evidence for skeletal, reproductive, and immune dysfunction. Proc Natl Acad Sci USA 98:7498–7503
Skorija K, Cox M, Sisk JM et al (2005) Ligand-independent actions of the vitamin D receptor maintain hair follicle homeostasis. Mol Endocrinol 19:855–862
Ellison TI, Smith MK, Gilliam AC et al (2008) Inactivation of the vitamin D receptor enhances susceptibility of murine skin to UV-induced tumorigenesis. J Invest Dermatol
Zinser GM, Sundberg JP, Welsh J (2002) Vitamin D(3) receptor ablation sensitizes skin to chemically induced tumorigenesis. Carcinogenesis 23:2103–2109
Baudino TA, Kraichely DM, Jefcoat SC Jr. (1998) Isolation and characterization of a novel coactivator protein, NCoA-62, involved in vitamin D-mediated transcription. J Biol Chem 273:16434–16441
Masuyama H, Jefcoat SC, MacDonald PN (1997) The N-terminal domain of transcription factor IIB is required for direct interaction with the vitamin D receptor and participates in vitamin D-mediated transcription. Mol Endocrinol 11:218–228
Lavigne AC, Mengus G, Gangloff YG et al (1999) Human TAF(II)55 interacts with the vitamin D(3) and thyroid hormone receptors and with derivatives of the retinoid X receptor that have altered transactivation properties [In Process Citation]. Mol Cell Biol 19:5486–5494
Tolon RM, Castillo AI, Jimenez-Lara AM et al (2000) Association with Ets-1 causes ligand- and AF2-independent activation of nuclear receptors. Mol Cell Biol 20:8793–8802
Ellison TI, Eckert RL, MacDonald PN (2007) Evidence for 1,25-dihydroxyvitamin D3-independent transactivation by the vitamin D receptor: uncoupling the receptor and ligand in keratinocytes. J Biol Chem 282:10953–10962
Zawel L, Reinberg D (1992) Advances in RNA polymerase II transcription. Curr Opin Cell Biol 4:488–495
Jurutka PW, Remus LS, Whitfield GK et al (2000) The polymorphic N terminus in human vitamin D receptor isoforms influences transcriptional activity by modulating interaction with transcription factor IIB. Mol Endocrinol 14:401–420
Blanco JCG, Wang I-M, Tsai SY et al (1995) Transcription factor TFIIB and the vitamin D receptor cooperatively activate ligand-dependent transcription. Proc Natl Acad Sci USA 92:1535–1539
Rachez C, Suldan Z, Ward J et al (1998) A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell- free system. Genes Dev 12:1787–1800
Rachez C, Lemon BD, Suldan Z et al (1999) Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature 398:824–828
Chiba N, Suldan Z, Freedman LP et al (2000) Binding of liganded vitamin D receptor to the vitamin D receptor interacting protein coactivator complex induces interaction with RNA polymerase II holoenzyme. J Biol Chem 275:10719–10722
Zhang C, Baudino TA, Dowd DR et al (2001) Ternary complexes and cooperative interplay between NCoA-62/Ski-interacting protein and steroid receptor coactivators in vitamin D receptor-mediated transcription. J Biol Chem 276:40614–40620
Makarov EM, Makarova OV, Urlaub H et al (2002) Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298:2205–2208
Zhou Z, Licklider LJ, Gygi SP et al (2002) Comprehensive proteomic analysis of the human spliceosome. Nature 419:182–185
Auboeuf D, Honig A, Berget SM et al (2002) Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science 298:416–419
Zhang C, Dowd DR, Staal A et al (2003) Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J Biol Chem 278:35325–35336
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2010 Springer Science+Business Media, LLC
About this chapter
Cite this chapter
Dowd, D.R., MacDonald, P.N. (2010). The Molecular Biology of the Vitamin D Receptor. In: Holick, M. (eds) Vitamin D. Nutrition and Health. Humana Press. https://doi.org/10.1007/978-1-60327-303-9_5
Download citation
DOI: https://doi.org/10.1007/978-1-60327-303-9_5
Published:
Publisher Name: Humana Press
Print ISBN: 978-1-60327-300-8
Online ISBN: 978-1-60327-303-9
eBook Packages: MedicineMedicine (R0)