Abstract
Agrobacterium vitis is the primary causal agent of grapevine crown gall worldwide. Symptoms of grapevine crown gall disease include tumor formation on the aerial plant parts, whereas both tumorigenic and nontumorigenic strains of A. vitis cause root necrosis. Genetic and genomic analyses indicated that A. vitis is distinguishable from the members of the Agrobacterium genus and its transfer to the genus Allorhizobium was suggested. A. vitis is genetically diverse, with respect to both chromosomal and plasmid DNA. Its pathogenicity is mainly determined by a large conjugal tumor-inducing (Ti) plasmid characterized by a mosaic structure with conserved and variable regions. Traditionally, A. vitis Ti plasmids and host strains were differentiated into octopine/cucumopine, nopaline, and vitopine groups, based on opine markers. However, tumorigenic and nontumorigenic strains of A. vitis may carry other ecologically important plasmids, such as tartrate- and opine-catabolic plasmids. A. vitis colonizes vines endophytically. It is also able to survive epiphytically on grapevine plants and is detected in soil exclusively in association with grapevine plants. Because A. vitis persists systemically in symptomless grapevine plants, it can be efficiently disseminated to distant geographical areas via international trade of propagation material. The use of healthy planting material in areas with no history of the crown gall represents the crucial measure of disease management. Moreover, biological control and production of resistant grape varieties are encouraging as future control measures.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
1 Introduction
Agrobacterium vitis (i.e., Agrobacterium biovar 3) is considered one of the most significant and destructive bacterial pathogens of grapevine worldwide. This bacterium is the primary causal agent of crown gall of grapevine (Vitis vinifera) (Burr et al. 1998; Burr and Otten 1999). However, tumorigenic strains belonging to the Agrobacterium tumefaciens complex (i.e., Agrobacterium biovar 1/Agrobacterium tumefaciens) and Rhizobium rhizogenes (i.e., Agrobacterium biovar 2/Agrobacterium rhizogenes) are occasionally associated with disease (see below Sect. 6.3).
Typical symptoms of grapevine crown gall disease include tumor formation on the aerial plant parts, unlike symptoms on most other hosts of tumorigenic agrobacteria. A. vitis also causes root necrosis on grapevines (Burr et al. 1987a).
To our knowledge, the first report on grapevine crown gall was made by Fabre and Dunal (1853) describing the disease in France. The infectious nature of this bacterium as a causal agent of grapevine crown gall was demonstrated by Cavara (1897) in Italy. However, at that time A. vitis was not established as a separate species. The evolution of classification and nomenclature of A. vitis will be reviewed in the next section referring to the taxonomy of A. vitis (see below Sect. 2).
Crown gall of grapevine is an economically important plant disease and is particularly serious in regions prone to temperatures that cause freeze injuries to dormant trunks and canes. Potential economic losses in replant and wine sales over a six-year period are calculated to be notably high (Stewart et al. 2013). The disease reduces the vigor and yield of infected plants by up to 40% depending on the extent of infection (Schroth et al. 1988). Severe infections can lead to dieback of canes or whole plants. Crown gall is especially destructive when the graft union is affected. Serious economic losses occur especially in nurseries because grafted grapevines with visible symptoms are unmarketable and are generally discarded. Moreover, both tumorigenic and nontumorigenic strains of A. vitis may negatively affect graft strength and subsequent nursery production and vineyard establishment (Hao et al. 2017).
2 Taxonomy of A. vitis
A. vitis belongs to the family Rhizobiaceae, order Rhizobiales, class Alphaproteobacteria. As with related species of the Agrobacterium and Rhizobium genera, A. vitis is an aerobic, nonsporeforming, Gram-negative, rod-shaped bacterium with peritrichous flagella (Young et al. 2005). In general, A. vitis grows optimally at 25–28 °C and produces copious amounts of extracellular polysaccharide slime on carbohydrate-rich media.
Initially, tumorigenic strains exclusively isolated from grapevine were defined as an atypical group that could not be classified to Agrobacterium biovar 1 or biovar 2 (Panagopoulos and Psallidas 1973). These atypical strains were subsequently classified to Agrobacterium biovar 3 (biotype 3) based on biochemical and physiological properties (Kerr and Panagopoulos 1977; Panagopoulos et al. 1978; Süle 1978). Agrobacterium biovar 3 could also be differentiated by serological analysis using monoclonal antibodies (Bishop et al. 1989). Finally, Ophel and Kerr (1990) formally described a new species A. vitis (vi’tis. L. n. Vitis, generic name of grapevines) based on polyphasic characterization of biovar 3 strains. However, it was recently shown that A. vitis is phylogenetically distinguishable from Agrobacterium spp. by using multilocus sequence analysis (MLSA), and its transfer to the revived genus Allorhizobium was suggested (Mousavi et al. 2014, 2015). Moreover, A. vitis has a different organization of genetic material compared to that of the genus Agrobacterium (see below Sect. 4.1), which also supports its distinctiveness (Ramírez-Bahena et al. 2014).
In recent years, genomics has significantly impacted the taxonomic status of bacteria. In particular, by using quantitative whole-genome comparison methods which include calculation of average nucleotide identity (ANI; Richter and Rossello-Mora 2009) and in silico DNA–DNA hybridization (isDDH) values (Meier-Kolthoff et al. 2013), it is possible to delineate prokaryotic species boundaries. Moreover, whole-genome-based phylogeny allows generation of highly robust phylogenetic trees based on large nucleotide or amino acid datasets. In this regard, whole-genome-based phylogeny based on 384 protein sequences conserved in the chromosomes also suggested that A. vitis represents a species distinct from Agrobacterium spp. and is more similar to Allorhizobium undicola (Ormeño-Orrillo et al. 2015). Moreover, our results based on genome comparisons (ANI and isDDH) of different A. vitis strains indicate that A. vitis is not a homogenous species but a species complex comprising at least three genomic species (unpublished data). Type strain K309T represents genomic species G1, whereas the well-known strains AB4 and S4 are representatives of genomic species G2 and G3, respectively. They all have similar average GC contents, ranging from 57.5 to 57.6%. Therefore, the taxonomy of A. vitis requires further elucidation.
3 Geographic Distribution of A. vitis
The geographic distribution of A. vitis generally reflects that of its host grapevine. So far, the presence of the pathogen has been reported in Australia (Ophel et al. 1988), Brazil (De Oliveira et al. 1994), Bulgaria (Genov et al. 2006a), Canada (Dhanvantari 1983), China (Ma et al. 1987), Egypt (Tolba and Zaki 2011), France (Ridé et al. 2000), Germany (Bien et al. 1990), Greece (Panagopoulos and Psallidas 1973), Hungary (Süle 1978), Italy (Bini et al. 2008b), Japan (Sawada et al. 1990), Jordan (Al-Momani et al. 2006), Iran (Mohammadi and Fatehi-Paykani 1999), Israel (Haas et al. 1991), Portugal (Nascimento et al. 1999), Russia (Ignatov et al. 2016), Serbia (Kuzmanović et al. 2014), Slovenia (Zidarič 2009), South Africa (Loubser 1978), South Korea (Lim et al. 2009), Spain (López et al. 1988), Tunisia (Chebil et al. 2013a), Turkey (Argun et al. 2002), and the USA (Burr and Hurwitz 1981) (Fig. 1). Although to the best of our knowledge, official reports have not been made, tumorigenic A. vitis strains originating from Afghanistan, Croatia, Moldova, Montenegro, Morocco, and Poland were included in some studies, confirming the presence of the pathogen in these countries (Kerr and Panagopoulos 1977; McGuire et al. 1991; Bini et al. 2008b; Kuzmanović et al. 2015). The pathogen is most likely present in more countries although, to the extent of our knowledge, this is not yet documented in the literature.
4 Genetic Characteristics and Diversity of A. vitis
4.1 Genome Organization of A. vitis
A. vitis has genome architecture that includes two circular chromosomes and a variable number of plasmids (Jumas-Bilak et al. 1998; Tanaka et al. 2006; Slater et al. 2009). On the other hand, members of the genus Agrobacterium are characterized by the presence of a circular chromosome and a linear chromosome (Allardet-Servent et al. 1993; Jumas-Bilak et al. 1998; Slater et al. 2009; Slater et al. 2013; Ramírez-Bahena et al. 2014). Strain S4 was the first A. vitis strain with a fully sequenced genome and so far the only one with high-quality, complete, and published chromosome and plasmid sequences (Slater et al. 2009). The genome of strain S4 contains two circular chromosomes and five plasmids. Plasmids play a substantial role in the pathogenicity and ecology of agrobacteria and rhizobia. The number of plasmids in tumorigenic strains of A. vitis may range from 2 to 5 (Perry and Kado 1982; Albiach and Lopez 1992). Their pathogenicity is primarily determined by a large conjugal tumor-inducing (Ti) plasmid. Besides the Ti plasmid, A. vitis may also carry other ecologically important plasmids, such as tartrate-catabolic plasmids and opine-catabolic plasmids (see below Sects. 4.3, 4.4 and 4.5).
4.2 Genetic Diversity of A. vitis
Knowledge of genetic variation and the relatedness between strains are of crucial importance, particularly in epidemiological studies and for a better understanding of the ecology and evolution of the pathogen. For this reason, the diversity of A. vitis was investigated both in terms of chromosomal and plasmid DNA. Here, we will focus on the genetic variation of chromosomal DNA, whereas the diversity of A. vitis plasmids will be discussed separately in the next section.
Overall, numerous studies demonstrated that A. vitis is genetically very diverse. A number of genetic groups were differentiated by analyzing populations of A. vitis in Australia (Gillings and Ophel-Keller 1995), Bulgaria (Genov et al. 2006b), Germany (Schulz et al. 1993), Iran (Rouhrazi and Rahimian 2012b), Japan (Kawaguchi et al. 2008b), Serbia (Kuzmanović et al. 2014, 2015), Spain (Palacio-Bielsa et al. 2009b), USA (Irelan and Meredith 1996; Otten et al. 1996b; Momol et al. 1998; Burr et al. 1999), and Turkey (Argun et al. 2002; Canik Orel et al. 2016). Moreover, strains isolated from the same locality and grapevine cultivar may belong to different genetic clusters (Kuzmanović et al. 2015).
Methods targeting sequences distributed throughout the whole genome, such as restriction analysis of total genomic DNA, as well as by PCR-based methods such as repetitive sequence-based PCR (rep-PCR) and randomly amplified polymorphic DNA (RAPD), allow comparison of closely related strains and assessment of clonality among them, which is particularly valuable in epidemiological studies. Additionally, the 16S-23S rRNA internal transcribed spacer (ITS) region is highly variable and therefore suitable for estimating epidemiological relationships among strains. Interestingly, Bautista-Zapanta et al. (2009) reported the existence of intercistronic heterogeneity of the 16S–23S rRNA ITS region among some A. vitis strains. However, the frequency of this phenomenon among a broader collection of A. vitis strains is unknown.
By applying the methodologies mentioned above, grouping of A. vitis strains isolated worldwide did not reflect the geographic origin of the strains. Some homogenous genetic groups were comprised of strains isolated in different countries and even different continents (Gillings and Ophel-Keller 1995; Otten et al. 1996b; Momol et al. 1998; Kuzmanović et al. 2014, 2015), which is consistent with the main means of spread of A. vitis via grapevine propagation material.
The 16S rDNA is a widely used phylogenetic marker for classification and discrimination of bacteria. However, it generally lacks the resolution power to discriminate among strains at the intraspecies or even intragenus level. Indeed, 16S rDNA sequences of A. vitis-type strain K309 and strain S4 differ by only one nucleotide (Otten et al. 1996a), although whole-genome comparisons suggest that they belong to different genomic species (Kuzmanović, unpublished). The 16S rDNA sequences of various A. vitis strains isolated in Spain also differed by one or two nucleotides at the same position (Palacio-Bielsa et al. 2009b). On the other hand, housekeeping genes, which are responsible for basic cellular functions and are relatively conserved on chromosomes, are particularly suitable markers for the assessment of phylogenetic relatedness among bacteria (Gevers et al. 2005). They were therefore exploited for analyzing A. vitis strains originating from various geographic locations (Kawaguchi et al. 2008b; Kuzmanović et al. 2015). Sequence analysis of dnaK, gyrB, and recA housekeeping genes was employed to characterize a representative collection of A. vitis strains originating from several European countries, Africa, North America, and Australia (Kuzmanović et al. 2015). Nucleotide sequence analysis indicated high genetic diversity among the strains studied and suggested the presence of recombination events in A. vitis, particularly affecting the dnaK locus. Phylogeny based on recA gene sequences revealed four main phylogenetic groups among the A. vitis strains studied. Strains K309, AB4, and S4 were clustered in separate phylogenetic groups (Kuzmanović et al. 2015), which was in accordance with whole-genome comparisons mentioned above and suggested that they belong to three different genomic species within A. vitis.
4.3 Ti Plasmids
As for other agrobacteria, the Ti plasmid of A. vitis carries the primary genes required for pathogenicity and can determine the host range of the pathogen (host range is further discussed in Sect. 5.3). The size of the Ti plasmid is approximately 200 kbp (Table 1). The Ti plasmid consists of the following functional elements: transferred DNA (T-DNA), virulence (vir) region, opine catabolism genes, replication (rep) region, and conjugative transfer genes (tra and trb loci). T-DNA carries genes responsible for production of plant hormones and tumor induction (oncogenes), and genes encoding biosynthesis of low molecular weight molecules termed opines (more details are given in Sect. 5.1).
Ti plasmids belong to the repABC family of megaplasmids and encode two independent type IV secretion systems (T4SSs; Suzuki et al. 2009; Pappas and Cevallos 2011; Christie and Gordon 2014). The first T4SS system (vir) is responsible for processing and transfer of T-DNA from the bacterium to host plant cells, whereas the second (tra/trb) mediates conjugative transfer of the Ti plasmid (Christie and Gordon 2014). Conjugative transfer of the Ti plasmid is regulated by a quorum-sensing (QS) mechanism and is induced by opines produced in crown gall tumors (Dessaux et al. 1998; Farrand 1998; White and Winans 2007; Faure and Lang 2014).
Ti plasmids have mosaic structures composed of conserved and variable regions, suggesting that they most likely evolved through horizontal gene transfer and recombination (Otten et al. 1992, 1993). This fact hinders their employment in the development of a satisfactory classification scheme. However, Ti plasmids can be grouped based on their backbone genes, such as those controlling plasmid replication and partitioning (repABC operon), as well as conjugative transfer. Thus, Ti plasmids and related root-inducing (Ri) plasmids have been classified into four incompatibility groups (Otten et al. 2008). A. vitis Ti plasmids pTiB10/7 and pTiAT66 were classified into the IncRh1 group, whereas pTiS4 belongs to the IncRh4 group (Table 1) (Szegedi et al. 1996). This classification scheme is based on their incompatibility (Inc) characteristics, referring to the inability of two related plasmids to be propagated stably in the same host cell line (Couturier et al. 1988). Incompatibility is mainly determined by functional elements located within the repABC cassette or in its proximity (Pappas and Cevallos 2011; Pinto et al. 2012). Therefore, the phylogeny of RepA, RepB, and RepC proteins has been used for grouping plasmids of agrobacteria (Wetzel et al. 2015). The Ti plasmid of A. vitis S4 was included in this study and clustered together with the OC plasmid pAoF64/95 of R. rhizogenes F64/95, the symbiotic plasmid pCB782 of Rhizobium leguminosaurum bv. trifolii CB782, and the cryptic plasmid pOV14c harbored by Ensifer adhaerens OV14, unlike other Ti plasmids investigated.
Classification of plasmids based on the amino acid sequence of conjugative relaxase and T4SS proteins has also been proposed (Garcillan-Barcia et al. 2009; Smillie et al. 2010; Garcillan-Barcia et al. 2011). In this respect, Ti plasmids are classified into the mobilization (MOB) families MOBQ and MOBP, based on their TraA and VirD2 relaxase protein sequences, respectively (Table 1) (Christie and Gordon 2014). Both T4SSs of Ti plasmids are classified into the MPFT group based on TrbE and VirB4 mating pair formation (MPF) protein sequences (Table 1). The relaxase and T4SS proteins of the Ti plasmid harbored by A. vitis S4 are generally phylogenetically related to those of other Ti plasmids (Wetzel et al. 2015).
Plasmid backbone markers used for the classifications discussed above can be associated with different gene clusters having important roles in the ecology of the pathogen and epidemiology of crown gall. For example, Ti plasmids possess genes and gene clusters responsible for the synthesis and catabolism of opines, which are a specific class of compounds produced in crown gall tumors following genetic transformation by the pathogen. Opine markers were widely used for grouping of Ti plasmids and their host bacteria, and classification based on opines reflects different aspects involved in plant–pathogen and pathogen–pathogen interactions (see Sect. 5.1). Based on opine types, A. vitis Ti plasmids and host strains were differentiated into octopine/cucumopine (O/C), nopaline (N), and vitopine (V) groups (Table 1) (Szegedi et al. 1988; Paulus et al. 1989a). Additionally, an octopine-type Ti plasmid carried by atypical strain CG474 has been reported (Burr et al. 1998; Burr and Otten 1999). Interestingly, strains carrying both octopine and vitopine synthase genes were detected in Italy and Tunisia (Bini et al. 2008b; Chebil et al. 2013b). These strains might harbor both O/C- and V-type Ti plasmids, or a new Ti plasmid type with another arrangement of opine-related genes. In this regard, the existence of Ti plasmids having different combinations of genes encoding synthesis and catabolism of opines cannot be excluded, which complicates classification based on the presence of opine genes. Tumorigenic Agrobacterium biovar 1 strains isolated from grapevine harbor Ti plasmids typical for this species, although they may also carry O/C-type Ti plasmids characteristic of A. vitis (Szegedi et al. 2005).
The size of the O/C-type Ti plasmid of A. vitis AB3 (pTiAB3) is approximately 234 kb (Otten et al. 1995). On the other hand, the nopaline-type Ti plasmid of A. vitis AB4 (pTiAB4) is relatively small (~157 kb) (Otten and De Ruffray 1994). The vitopine-type Ti plasmid of A. vitis S4 (pTiS4) is particularly large (258,824 bp). Its GC content is 56.7%, which is similar to that of other Ti plasmids (Suzuki et al. 2009).
O/C-type Ti plasmids are predominant within the population of A. vitis (60%), followed by N Ti plasmids (30%), whereas the V type is less abundant (10%) (Burr et al. 1998). Studies performed in Hungary (Szegedi 2003), France (Ridé et al. 2000), and Turkey (Canik Orel et al. 2016) were more or less in accordance with this proportion. Strains carrying an O/C-type Ti plasmid were also predominant in Bulgaria (Genov et al. 2006a, 2015), China (Ma et al. 1987), Spain (Palacio-Bielsa et al. 2009b), and Serbia (Kuzmanović et al. 2014). In Germany, more or less equal numbers of O/C and N strains of A. vitis were isolated from grapevine tumors, whereas the vitopine strains were not detected (Bien et al. 1990). On the contrary, vitopine strains were more abundant in Italy (Bini et al. 2008b) and Iran (Rouhrazi and Rahimian 2012b). The N strains were not identified within A. vitis populations in Bulgaria, Italy, and Serbia. It is unclear whether these differences in distribution of particular opine types are influenced by ecological factors or if they are a consequence of distribution of specific grape cultivars and/or rootstocks.
The O/C, N, and V types of Ti plasmids are characterized by a complex structure and gene arrangement of their T-DNA. Different variants of T-DNA structures have been described and thoroughly reviewed previously (Paulus et al. 1989a; Huss et al. 1990; Burr et al. 1998; Burr and Otten 1999). The O/C-type Ti plasmids possess two independent T-DNA fragments, TA-DNA and TB-DNA. Although at least six different O/C Ti plasmid structures have been characterized, based on the structure of the TA-DNA, they are divided into two main sub-groups OS and OL, having a small or large TA-DNA region, respectively. T-DNA of the atypical octopine strain CG474 has a unique T-DNA structure lacking TB-DNA, although it has some similarities with classical octopine TL-DNA and O/C TA-DNA (Burr and Otten 1999). The N-type Ti plasmids comprise a single T-DNA (Otten and De Ruffray 1994), whereas V types possess three independent T-DNAs (Canaday et al. 1992). The N- and V-type Ti plasmids are less variable than are the O/C plasmids. Reconstruction of evolutionary relationships among different T-DNA variants is hindered because it has been shown that Ti plasmids mainly evolved through horizontal gene transfer, insertions, and deletions (Otten et al. 1992; van Nuenen et al. 1993; Otten and De Ruffray 1994). However, little is known about the ecological dynamics of different Ti plasmid variants.
In a study on distribution and localization of insertion elements in A. vitis strains, Paulus et al. (1989b) found a correlation between Ti plasmid genotype and the particular chromosomal background within an O/C group of strains. Later, Otten et al. (1996b) showed that most Ti plasmids in A. vitis are associated with a particular chromosomal background determined by restriction fragment length polymorphism (RFLP) analysis of the 16S-23S rRNA ITS region, which was further supported by Genov et al. (2006b). Similar studies reported a general coherence between 16S rRNA gene sequences, RFLP profiles of the 5′-end of the 23S rRNA gene, and RAPD fingerprints with a particular type of Ti plasmid (Momol et al. 1998; Palacio-Bielsa et al. 2009b). However, Turkish A. vitis strains were exceptions (Argun et al. 2002). Based on analysis of A. vitis strains isolated in Serbia, no strong correlation between 16S-23S rRNA ITS genotype of the strains and their type of Ti plasmid was found, although some 16S-23S rRNA ITS groups and clusters were composed of strains harboring a particular type of Ti plasmid (Kuzmanović et al. 2014). This plasmid–chromosome correlation found in A. vitis is unique among tumor-inducing Agrobacterium and Rhizobium species. For other agrobacteria and rhizobia causing crown gall, no correlation between chromosome and Ti plasmid type was found. Although it is assumed that Ti plasmid exchange or transfer occurs more or less frequently between A. vitis strains, certain Ti plasmids appear to be strongly and stably associated with a particular bacterial host. This may be influenced by the specific pathosystem of A. vitis, with the highly specialized pathogen persisting systemically in grapevines. This may limit the chance of its contact with diverse and large pools of tumorigenic strains in contrast to predominantly soil-inhabiting agrobacteria and rhizobia.
The diversity of vir region and opine catabolism gene clusters among A. vitis strains has not been studied extensively. However, O/C and N Ti plasmids have very similar or identical virulence regions (Otten and De Ruffray 1994). On the contrary, the vitopine Ti plasmid of A. vitis has a different organization of vir region (Gerard et al. 1992). Kawaguchi and Inoue (2009) analyzed the phylogeny of strains belonging to the A. vitis and A. tumefaciens complex, including Agrobacterium strains from other hosts, using the partial nucleotide sequences of the virC operon. The majority of A. vitis strains formed a separate cluster, whereas the remaining A. vitis strains formed additional monophyletic groups with strains of the A. tumefaciens complex isolated from grapevine. Strains from other hosts clustered separately on the phylogenetic tree.
4.4 Opine-Catabolic Plasmids
Opine-catabolic plasmids (pOC) are a specific group of replicons that, like Ti plasmids, carry genes encoding the uptake and catabolism of opines but do not contain vir genes and T-DNA required for pathogenicity. pOCs have been identified both in tumorigenic and in nontumorigenic Rhizobiaceae strains isolated from tumors or soil around diseased plants (Merlo and Nester 1977; Sciaky et al. 1978; Wabiko et al. 1990; Szegedi et al. 1999; Wetzel et al. 2014; Puławska et al. 2016). Therefore, nonpathogenic strains carrying this plasmid are provided with the ability to metabolize opines and to proliferate inside or near tumors and diseased plants. Plasmids pAtK84b and pAoF64/95 carried by nonpathogenic R. rhizogenes strains K84 and F64/95, respectively, are the only pOCs that have been studied in detail (Clare et al. 1990; Wetzel et al. 2014). As with Ti plasmids, these two plasmids carry all genes required for conjugative transfer that is induced by opines and regulated by a QS system (Oger and Farrand 2002; Wetzel et al. 2014).
Data regarding the distribution of pOCs in strains associated with grapevine tumors are limited. Sciaky et al. (1978) characterized plasmids of various agrobacteria, including one avirulent Agrobacterium biovar 1 strain AG19 isolated from tumor tissue on grapevine in Greece. Although this strain did not confer tumorigenicity, it could utilize octopine and harbored a plasmid encoding this ability. Additional Agrobacterium biovar 1 strains from grapevine carrying large plasmids encoding catabolism of octopine were also described (Knauf et al. 1983). Later, Szegedi et al. (1999) reported the existence of pOC in nonpathogenic A. vitis strain F2/5 encoding the catabolism of octopine. Strain F2/5 is a potential biocontrol agent able to inhibit crown gall disease on grapevines. Similarly, pAtK84b conferring utilization of nopaline is harbored by the nonpathogenic biocontrol strain R. rhizogenes K84 (Clare et al. 1990).
4.5 Tartrate Utilization Plasmids
Tartrate utilization plasmids (pTrs) are a group of plasmids described in strains of A. vitis and related Agrobacterium species associated with grapevine. Gallie et al. (1984) first identified a plasmid encoding utilization of tartrate (named pTAR) in an atypical nonpathogenic Agrobacterium biovar 1 strain isolated from grapevine. The amino acid sequence of the replication protein RepA encoded by pTAR showed close homology to RepA protein of pAgK84, a plasmid that encodes synthesis of agrocin 84 in the biocontrol strain R. rhizogenes K84 (Gallie and Kado 1988; Kim et al. 2006). These two plasmids may belong to the same family and have a common evolutionary origin.
Szegedi et al. (1992) subsequently demonstrated that various tumorigenic A. vitis strains also carry large pTrs, some of which were self-conjugal. The pTr can be also carried by nonpathogenic A. vitis strains, for instance by the biocontrol strain F2/5 (Szegedi et al. 1999). The pTrs identified in tumorigenic A. vitis strains represent a diverse group of plasmids differing in size, transfer frequency, and stability in recipient Agrobacterium biovar 1 strains (Szegedi et al. 1992). They were also clearly different from the pTAR plasmid described by Gallie et al. (1984). pTrs also showed a high diversity in their incompatibility properties (Szegedi and Otten 1998). Therefore, they could coexist with pTis from different incompatibility groups.
The tartrate utilization system of the nopaline strain AB4 has been characterized and the corresponding genes identified as an operon (Crouzet and Otten 1995). Tartrate utilization in this strain is encoded by the 170-kb conjugative plasmid pTrAB4. On the other hand, the O/C strain AB3 carries two independent tartrate utilization systems, one encoded by the Ti plasmid (pTiAB3) and another by pTrAB3 (245 kb; Otten et al. 1995). Regions encoding tartrate utilization genes are found in most of A. vitis strains, and they have been grouped into three different types based on analysis of their sequences (Salomone et al. 1996; Salomone and Otten 1999). Vitopine strains of A. vitis are also able to degrade tartrate, although their tartrate utilization system is different and has not yet been characterized (Salomone et al. 1996). In any case, tartrate degradation by vitopine strain S4 is encoded by a large plasmid pAtS4c (211,620 bp), which was initially named pTrS4 (Szegedi and Otten 1998). Interestingly, different genetic regions encoding tartrate utilization showed complex distribution patterns among various A. vitis strains that correlated with their chromosomal backgrounds (Salomone et al. 1996). Some pTrs encode production of putative signal molecules used in QS regulation; however, their regulatory role remains unknown (Lowe et al. 2009).
Because tartrate is an abundant compound in grapevine, pTrs may enhance host strain competitiveness on this plant species (Kado 1998; Salomone et al. 1998). Both A. vitis and R. rhizogenes use tartrate as a sole carbon source (Kerr and Panagopoulos 1977; Moore et al. 2001). However, the latter species prefers glucose to tartrate, unlike A. vitis, that utilizes tartrate more intensively than glucose (Szegedi 1985). Although it was shown that Agrobacterium biovar 1 strains generally do not utilize tartrate (Kerr and Panagopoulos 1977; Moore et al. 2001), some biovar 1 strains isolated from grapevine were able to catabolize this compound (Gallie et al. 1984; Szegedi et al. 2005). Interestingly, most of the nontumorigenic A. vitis strains isolated from roots of asymptomatic feral Vitis riparia vines in the USA were not able to utilize tartrate (Burr et al. 1999).
5 Plant–Host Interactions
5.1 Tumor Induction and Opine Production
A. vitis can be present in symptomless propagation material and thereby cause infections on young plants in newly established vineyards. In such cases, infection mostly occurs on aerial plant parts through wounds caused by abiotic and biotic factors, especially by freezing temperatures and cultural practices (Burr et al. 1998; Burr and Otten 1999). However, in warmer climates, such as that of Israel and South Africa, high temperatures and humidity can cause injuries and initiate infection (Burr et al. 1998). On the other hand, wounds made by disbudding and grafting are particularly important for triggering infection in nurseries. For soil-borne infections, injuries made by cultural practices or nematode wounds may also be conducive for infection by the pathogen (Süle et al. 1995; Burr et al. 1998). Interestingly, unlike tumorigenic strains belonging to the A. tumefaciens complex, A. vitis was unable to induce tumors on in vitro grown grapevine stem segments (Szegedi et al. 2014). However, factors determining differences in the susceptibility of intact grapevines and explants to A. vitis remain to be elucidated.
Infection of plants by tumorigenic agrobacteria is a complex multistage process of natural genetic transformation of plants that includes DNA transfer from bacteria to plants (Hooykaas 2000; Zhu et al. 2000; Zupan et al. 2000; McCullen and Binns 2006; Pitzschke and Hirt 2010; Gelvin 2012). In brief, wounded plant tissue releases signal molecules that trigger infection through induction of vir genes of the bacteria. For a number of grapevine cultivars, syringic acid methyl ester was identified as a vir-inducing phenolic compound (Spencer et al. 1990). Vir proteins are involved in the processing and transfer of the T-DNA of the Ti plasmid and its stable integration into the plant host genome.
T-DNA genes are expressed in the host plant and encode biosynthesis of the phytohormones auxin and cytokinin (oncogenes) that lead to uncontrolled proliferation of plant cells, resulting in tumor formation. Tumors develop mainly on the lower trunks, graft unions, cordons, and canes (Fig. 2). Initial symptoms may be inconspicuous and remain unnoticed. However, as disease develops, tumor tissue can enlarge rapidly. Tumors can be localized or in the form of continuous proliferations that completely girdle the trunk (Fig. 2). Tumors are rarely observed on grapevine roots.
However, T-DNA genes also encode production of opines, as previously described in this chapter, which play important roles in the epidemiology of crown gall and the ecology of tumorigenic bacteria. Opines are typically conjugates of amino acids and α-ketoacids or sugars, and less frequently, they are sugar phosphodiesters (Dessaux et al. 1993, 1998; Chilton et al. 2001). So far, more than 20 different opine types belonging to different structural families have been described and characterized (Dessaux et al. 1993, 1998; Chilton et al. 2001).
In general, multiple opines belonging to different families may be produced in tumors as a consequence of plant genetic transformation. For example, O/C strains of A. vitis are responsible for production of octopine and cucumopine in tumor tissue (Szegedi et al. 1988; Paulus et al. 1989a). Although additional opines are produced in tumors induced by various tumorigenic strains carrying well-studied octopine-type Ti plasmids (Dessaux et al. 1998; Zhu et al. 2000), their presence has not been investigated in grapevine tumors caused by A. vitis carrying O/C-type Ti plasmids. Nopaline was the only opine detected in tumors caused by A. vitis strains carrying an N type of Ti plasmid. Finally, V strains of A. vitis induced tumors in which vitopine and ridéopine were produced (Szegedi et al. 1988; Paulus et al. 1989a; Chilton et al. 2001). Chilton et al. (2001) suggested that vitopine is identical to heliopine. Heliopine is one of the opines detected in tumors caused by Agrobacterium strains carrying a classical octopine-type plasmid (e.g., Agrobacterium biovar 1 strain 15955), and its structure has been published (Chang et al. 1989).
Primarily, opines serve as selective nutrient sources for the pathogen because, as indicated above, genes responsible for uptake and catabolism of opines are located on the Ti plasmid, outside of the T-DNA region. The presence of opines is not limited to tumors, and opines can translocate to other plant parts and can also be secreted from roots as a component of root exudates (Savka et al. 1996).
Some opines can also induce conjugative transfer of Ti plasmids among agrobacteria (Dessaux et al. 1998; Farrand 1998). Opines therefore contribute to dissemination of Ti plasmids. Although transferred Ti plasmid genes may encode production of more than one opine type in a particular tumor, thus far it appears that only some opines serve as conjugal inducers (Farrand 1998). The conjugal opines induce the QS system that directly regulates conjugative transfer of the Ti plasmid. Taken together, the conjugative transfer of the Ti plasmid is dependent on pathogen population density and requires the presence of a conjugal opine. Conjugation mechanisms of plasmids harbored by A. vitis strains likely behave similarly, but this has thus far not been studied.
Unwounded tobacco seedlings can elicit Agrobacterium vir gene induction and T-DNA transfer (Brencic et al. 2005). Intriguingly, transformation did not lead to tumor formation, although plants produced opines. These results suggested that genetic transformation of plants by tumorigenic bacteria does not require wounding and that cell division during wound healing may play a role in tumor formation. However, such interaction between A. vitis and grapevine has not yet been studied.
5.2 Root Necrosis and Associated Mechanisms
Unlike other tumorigenic agrobacteria, A. vitis causes necrosis on roots of grapevine plants (Fig. 3; Burr et al. 1987a). Necrosis may provide a niche for the bacterium to persist in the soil, as A. vitis can persist in grapevine root debris for at least two years (Burr et al. 1995). Necrosis develops within 24 to 48 h after inoculation and is generally restricted to localized lesions from which A. vitis can be consistently isolated.
Interestingly, both tumorigenic and nontumorigenic strains of A. vitis cause root necrosis. Later studies indicated that the enzyme polygalacturonase encoded by the chromosomal pehA gene represents a virulence factor associated with grape root decay (McGuire et al. 1991; Rodriguez-Palenzuela et al. 1991). Nonetheless, a mutant strain lacking the polygalacturonase gene could still induce grape root necrosis when higher concentrations of bacteria were used, suggesting that additional factors are associated with this process (Herlache 1999).
Polygalacturonase may also play a role in tumorigenesis on grapevine, because pehA mutants were less pathogenic on this host (Rodriguez-Palenzuela et al. 1991). However, both wild-type and pehA mutants were equally tumorigenic on potato disks. Synthesis of polygalacturonase appears to be associated with host specialization of A. vitis, because it may affect attachment of bacteria to grape roots and their multiplication at wound sites (Brisset et al. 1991).
Polygalacturonase of A. vitis showed significant similarity to those of two other plant pathogens, Pectobacterium carotovorum and Ralstonia solanacearum, although differences in their soft rotting effects on potato tuber tissue were reported (Herlache et al. 1997).
Interestingly, A. vitis also causes a hypersensitive-like response (HR) on nonhost plants such as tobacco (Herlache et al. 2001). The underlying mechanism of grape necrosis and HR response may be related. In this respect, both grape necrosis and the HR are regulated by a complex QS regulatory system (Hao et al. 2005; Li et al. 2005; Hao and Burr 2006). More recently, the ability of A. vitis to cause the HR and necrosis was shown to be associated with a phosphopantetheinyl transferase (PPTase), and bacterial polyketide and nonribosomal peptide synthase-associated genes (Zheng and Burr 2013).
The QS system associated with induction of necrosis on grape and a HR response on tobacco also regulates characteristic surface motility (swarming) of A. vitis, which is associated with surfactant secretion (Süle et al. 2009). A. vitis was the only of the tested agrobacteria expressing swarming activity, and such behavior may facilitate colonization of grapevine by this pathogen.
5.3 Host Range
A. vitis has been detected in nature almost exclusively in association with grapevine, suggesting a high degree of natural host specialization. Nevertheless, in one exceptional case, A. vitis was isolated from galls on the roots of kiwi in Japan (Sawada and Ieki 1992). Interestingly, it was recently reported that A. vitis can cause banana leaf blight in China (Huang et al. 2015). Although strains isolated from banana were related to A. vitis based on 16S rDNA analysis, further study is required for reliable identification.
Inoculation of various test plants under greenhouse environment conditions revealed differences in host range among A. vitis strains and generally among tumorigenic strains isolated from grapevine (Panagopoulos et al. 1978; Knauf et al. 1982; Ma et al. 1987; Bien et al. 1990). Although initial studies suggested that A. vitis has a narrow host range limited to grapevine and a few other test plants, it later became evident that there are limited and wide host range strains. In particular, A. vitis strains harboring an O/C Ti plasmid (previously named octopine Ti plasmid) showed different host range patterns and were divided into limited and wide host range strains (Thomashow et al. 1980, 1981; Knauf et al. 1982, 1983). In this respect, the Ti plasmid largely determines host range of the bacterium (Thomashow et al. 1980; Knauf et al. 1982). Knauf et al. (1982) showed that different grapevine cultivars can respond differently, depending on the Ti plasmid carried by the inoculant strain. Further studies identified particular Ti plasmid genes located either in T-DNA or in the vir region that are associated with A. vitis host range (Buchholz and Thomashow 1984a, b; Hoekema et al. 1984; Yanofsky et al. 1985a, b; Bonnard et al. 1989). Moreover, Ti plasmids of limited host range strains may have evolved from ones carried by wide host range strains (Paulus et al. 1991a, b). Although previous studies clearly showed differences in host range of A. vitis, they do not answer the question as to why this pathogen is largely restricted to grapevines in natural environments.
6 Epidemiology of Grapevine Crown Gall
6.1 Survival in and on Grapevine—The Role in Pathogen Dissemination
A. vitis can systemically infect grapevine and spread through xylem sap (Fig. 4). The first direct evidence of this phenomenon was provided by Lehoczky (1968), who isolated the pathogen from the xylem bleeding sap of symptomatic vines. Moreover, development of secondary tumors was observed on experimentally injured canes, which provided further evidence of bacteria internally present in the vascular system. Systemic colonization of grapevine by A. vitis was confirmed in subsequent studies (Lehoczky 1971; Burr and Katz 1983; Tarbah and Goodman 1987). A. vitis was also isolated from latently infected symptomless grapevine material (Lehoczky 1971; Burr and Katz 1984).
Although A. vitis rarely induces tumors on grapevine roots, the bacterium is able to persist in root tissues as it was consistently isolated from tumor-free grapevine roots (Lehoczky 1971, 1978; Süle 1986; Burr et al. 1987b, 1995; Thies et al. 1991). Lehoczky (1978) suggested that grapevine root systems represent a reservoir for A. vitis in which the pathogen can multiply and survive extreme environmental conditions. Additionally, A. vitis was frequently isolated from necrotic lesions on roots of infected vines (Fig. 4) (Burr et al. 1987b).
Tarbah and Goodman (1987) showed that A. vitis can move rapidly through xylem vessels of grapevine shoots. Within a period of 24 h, bacteria translocated 30 cm through the vascular system of shoots inoculated by dipping their freshly cut basal ends in bacterial suspension. Bauer et al. (1994) inoculated shoots of actively growing plants and demonstrated that migration of A. vitis from inoculation sites to the roots requires at least 15 weeks. It was later found that population dynamics of A. vitis in vines may vary with the cultivar and vegetation period (Burr et al. 1988, 1994; Stover et al. 1997b). Moreover, poor systemic movement of A. vitis and its presence in higher numbers mainly at inoculated sites were reported (Bauer et al. 1994; Stover et al. 1997b). Freezing may facilitate systemic movement of A. vitis in naturally infected and inoculated vines (Stover et al. 1997b).
Regarding seasonal fluctuations in pathogen population in grapevine, A. vitis was most abundant during the spring, decreased during the summer, and then in autumn returned to nearly the same levels as in spring (Bauer et al. 1994). In a recent study, Faist et al. (2016) compared the endophytic microbiota of organs from grapevine plants with or without crown gall disease symptoms using 16S rDNA amplicon sequencing. Their results also suggested that populations of A. vitis decreased in summer in comparison with spring and autumn, confirming the results of Bauer et al. (1994).
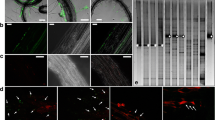
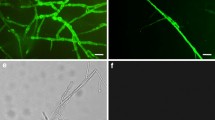
By testing plant material collected from a crown gall-infested vineyard, A. vitis could not be isolated from young green shoots until after the shoots became lignified (Burr et al. 1988). In addition, the pathogen was also absent from shoot tips. Similar results were reported by Bauer et al. (1994), who assayed experimentally inoculated grapevine plants. Nevertheless, Lehoczky (1989) infected healthy vines by grafting them with green shoots from infected plants, indirectly showing that A. vitis actually can be present in green shoots. Therefore, the detection methods used in previous studies may not have been sensitive enough for reliable detection of the pathogen. Indeed, using a PCR-based method, Poppenberger et al. (2002) showed that A. vitis can be translocated to shoot tips. Recently, by using a highly sensitive magnetic capture hybridization procedure followed by real-time polymerase chain reaction (real-time PCR), A. vitis was detected in shoot tips and meristem tissue of grapevine plants that were derived from symptomless cuttings grown in the greenhouse but latently infected with the pathogen (Johnson et al. 2016). The authors also revealed the frequent presence of tumorigenic A. vitis in dormant canes and green shoots. In addition, A. vitis is irregularly present in tested tissue. Intriguingly, Johnson et al. (2016) suggested that tumorigenic A. vitis is able to survive epiphytically on shoot tips. In a related study, Canik Orel et al. (2017) detected A. vitis in dormant grape buds and on surfaces of grapevine leaves collected from commercial vineyards (Fig. 4).
Considering its ability to survive in and on grapevine, A. vitis can be efficiently disseminated from nurseries to distant geographical areas via symptomless propagation material, which is regarded as the most important means of pathogen spread. Hence, A. vitis can be latently present in vines until favorable conditions for disease development arise. In this respect, some studies attempted to resolve the possible introduction pathway and evaluate epidemiological relationships among A. vitis strains. Gillings and Ophel-Keller (1995) suggested that A. vitis was introduced to South Australia in grapevine cuttings imported from California. Furthermore, a majority of A. vitis strains isolated predominantly from young commercial vineyards over the period of eight years (from 2003 to 2011) in six European countries were genotypically related, suggesting that they most likely had a common origin and were distributed following the movement of infected grapevine planting material (Kuzmanović et al. 2015).
The presence of A. vitis was also monitored in feral and wild grapevines. In this respect, nontumorigenic A. vitis strains were isolated from roots of asymptomatic feral V. riparia vines collected in different locations in the USA (Burr et al. 1999). In contrast, Genov et al. (2006a) isolated tumorigenic A. vitis strains from stem samples of wild V. vinifera ssp. sylvestris collected in natural forests in Bulgaria. Tumorigenic A. vitis was also detected in dormant canes collected from symptomless wild grapevines (V. riparia) in New York and feral grapevines (including Vitis californica) in California, which were either proximal to or distant from commercial vineyards (Johnson et al. 2016; Canik Orel et al. 2017). Therefore, wild grapevines may serve as a significant reservoir of inoculum.
6.2 Survival in Soil and Importance of Soil-Borne Infections
Members of the family Rhizobiaceae are generally soil-inhabiting and plant-associated bacteria. Accordingly, A. vitis strains are able to survive in soil. However, A. vitis has been detected in soil exclusively in association with grapevine plants and, to the best of our knowledge, has never been isolated from non vineyard soil (Fig. 4). Tumorigenic A. vitis, including tumorigenic Agrobacterium biovar 1 strains, are thus isolated from soil samples taken from the root zone of vines, only from samples collected during the period when fresh galls were present on trunks (Burr and Katz 1983). However, the ratio of pathogenic to nonpathogenic strains was remarkably low. In this respect, Bien et al. (1990) could not isolate pathogenic A. vitis strains from soil samples taken from the root zone of infected vines; only nonpathogenic Agrobacterium biovar 1 strains were isolated from this source. As determined by isolation of bacteria, A. vitis was also detected in nonrhizosphere soil of infected vineyards, although the pathogen may survive to a lesser extent in this environment and may preferentially inhabit the grapevine rhizosphere (Burr et al. 1987b). In their greenhouse study, Bishop et al. (1988) compared the population dynamics of A. vitis in grapevine and oat rhizosphere to that of fallow soil. Unlike fallow soil, both host and nonhost rhizospheres enhanced the survival of A. vitis. However, the pathogen population was greater in the grapevine rhizosphere without a population decline over the course of the study. As shown before for some other plant species (Guyon et al. 1993; Oger et al. 1997; Savka and Farrand 1997; Mansouri et al. 2002), opines exuded from roots of transformed grapevine might affect the composition of bacterial populations in the rhizosphere and promote the growth of opine-degrading bacteria, primarily A. vitis.
Although crown gall outbreaks are primarily associated with diseased or systemically infected propagation material, soil-borne A. vitis can be responsible for infections of grapevine plants grown in greenhouse conditions and vineyards (Bishop et al. 1988; Pu and Goodman 1993). However, planting of grapevine in soil containing lower levels of A. vitis (≤104 cfu/g) did not result in systemic infection within 10 weeks (Bishop et al. 1988). Soil-borne populations of A. vitis represent an important source of inoculum at higher levels of soil infestation (106 cfu/g). On the other hand, nematodes can enhance infection from soil. Combined inoculation of grapevine roots with Meloidogyne hapla and A. vitis at the level of 102−3 cfu/g of soil resulted in root infestation by A. vitis and subsequent systemic colonization of grapevine by the pathogen (Süle et al. 1995). It is still unclear if nematodes could be vectors of A. vitis, although nematodes have been used as vectors to transfer A. tumefaciens into plant roots in order to transform Arabidopsis plants (Karimi et al. 2000). A. vitis can also be spread over short or long distances in rhizosphere soil or on the rhizoplane of apparently healthy propagation material (Burr et al. 1987b).
A. vitis can survive in association with decaying grape roots and canes in soil for at least two years after plants were artificially inoculated with the pathogen (Fig. 4) (Burr et al. 1995). However, A. vitis can most likely survive longer in decaying grape and cane tissue as long as grapevine residues are present in soil, which could serve as significant reservoirs of inoculum.
Overall, the methods used in the studies described above may not be sensitive enough to detect A. vitis in soil at low population densities. Further investigations on the survival of A. vitis in soil are therefore needed. In this respect, a magnetic capture hybridization procedure followed by real-time PCR might be the method of choice for such studies.
6.3 Microbial Community Associated with Grapevine Crown Gall
Infected grapevine plants and especially crown gall tumors are dynamic ecological niches inhabited by diverse microorganisms, both pathogenic and nonpathogenic, and their genetic diversity remains largely unexplored. The possible roles of different members of the microbial community associated with grapevine tumors in relation to the ecology and epidemiology of crown gall disease are yet to be explored.
A. vitis is the predominant species identified as a causal agent of grapevine crown gall worldwide. Tumorigenic strains belonging to Agrobacterium biovar 1 and R. rhizogenes were, however, sporadically isolated from infected grapevine plants (Panagopoulos and Psallidas 1973; Panagopoulos et al. 1978; Süle 1978; Burr and Katz 1984; Ma et al. 1987; Thies et al. 1991; Ridé et al. 2000; Argun et al. 2002; Bini et al. 2008b; Kawaguchi and Inoue 2009; Palacio-Bielsa et al. 2009b; Rouhrazi and Rahimian 2012a; Abdellatif et al. 2013; Genov et al. 2015; Perović et al. 2015). In addition, tumorigenic strain 0 belonging to the newly described species Agrobacterium nepotum was isolated from a crown gall tumor on grapevine in Hungary (Süle and Kado 1980; Puławska et al. 2012; Mousavi et al. 2015).
Nontumorigenic A. vitis strains were isolated as cohabitants with tumorigenic A. vitis strains from grapevine tumors, roots, and sap (Panagopoulos and Psallidas 1973; Burr and Katz 1983; Staphorst et al. 1985; Burr et al. 1987a; Bien et al. 1990; Genov et al. 2006a; Bini et al. 2008b; Kawaguchi et al. 2008b; Rouhrazi and Rahimian 2012a; Canik Orel et al. 2017; Kuzmanović et al., unpublished). Their occurrence is most likely more frequent in diseased plants; however, they have not been studied in more detail. Nevertheless, it was determined that some nonpathogenic A. vitis strains isolated from callus tissue on dormant scion cuttings can catabolize octopine and nopaline, although pathogenic strains were not detected in the same plant material (Ophel et al. 1988). Most likely, these strains carry pOC encoding catabolism of opines.
Grapevine tumors were also inhabited by nontumorigenic strains belonging to novel phylogenetic groups within the genus Agrobacterium. In this respect, strains related to Agrobacterium rubi, but phylogenetically clearly different, were isolated from grapevine tumors (Kuzmanović, unpublished). As determined by PCR, these strains carried an ooxA gene-encoding oxidoreductase for conversion of octopine-type opines to pyruvate and corresponding basic amino acid, suggesting that they are able to catabolize octopine and likely harbor pOC. One of these strains (strain 384) forms a novel Agrobacterium species, Agrobacterium rosae, together with the atypical tumorigenic strain NCPPB 1650 isolated from hybrid tea rose (Rosa x hybrida) in South Africa, and three nonpathogenic strains isolated from tumors on raspberry and blueberry (Kuzmanović et al. 2018).
Opine utilization is not restricted to Agrobacterium spp. and related organisms. Several reports described that other microorganisms isolated from tumors, soil, and the rhizosphere, including fluorescent and nonfluorescent Pseudomonas spp., Arthrobacter spp., coryneforms, and several fungal species, are able to utilize opines (Beaulieu et al. 1983; Dahl et al. 1983; Bouzar and Moore 1987; Tremblay et al. 1987; Beauchamp et al. 1990; Bergeron et al. 1990; Nautiyal and Dion 1990; Beauchamp et al. 1991; Canfield and Moore 1991; Nautiyal et al. 1991; Moore et al. 1997). Moore et al. (1997) analyzed opine-catabolizing bacteria in tumors on several hosts, including grapevine. Interestingly, besides one nonpathogenic Agrobacterium strain utilizing octopine, they identified various fluorescent Pseudomonas strains having the ability to catabolize either octopine or nopaline, or both opines. Genes encoding opine catabolism in some non-Agrobacterium species were found on the chromosome, but not on plasmids (Watanabe et al. 2015).
Faist et al. (2016) investigated the bacterial endophytic community associated with grapevine plants with and without crown gall disease, including the surrounding vineyard soil over one year. The authors used cultivation-independent 16S rDNA-based analysis in preference to traditional isolation techniques. Taken together, they found the highest diversity of bacterial taxa in soil; the diversity decreased with the distance the soil was from roots and the graft union, and the cane. Crown gall disease affected the makeup of the bacterial community only on graft unions with visible tumors. Compared to graft unions on healthy plants, galls possessed higher species richness with a more stable bacterial population structure over time and shared more bacterial species with the soil microbial population. Besides A. vitis, the most abundant bacteria in graft unions of diseased plants were Pseudomonas sp. and Enterobacteriaceae sp. However, the reasons for the higher abundance of bacteria belonging to these genera are unknown.
7 Disease Management
7.1 Early Diagnosis—Detection of the Pathogen
The use of healthy planting material in areas with no history of the crown gall is crucial because, once established in a vineyard, A. vitis may be impossible to eliminate. Therefore, analysis of the grapevine propagation material for the presence of the pathogen is important for disease control. Additionally, it is also important to test the soil prior to planting.
Despite its destructiveness, A. vitis and other species causing grapevine crown gall are not considered quarantine pathogens in many countries. They are commonly regarded as harmful, widespread pathogens that can reduce the value of propagation material (quality pathogens), and grapevine material exchanges are not subject to strict phytosanitary control. Moreover, there is a lack of standardized protocols for pathogen detection and identification.
Testing of infected plants is mainly based on pathogen isolation on semiselective and/or differential media, followed by analysis of isolated strains using biochemical tests and pathogenicity assays (Moore et al. 2001). Although isolation of A. vitis from tumor tissue of infected vines is relatively straightforward, low numbers of bacteria in asymptomatic samples and their irregular distribution limit the efficiency of detection methods. A method involving callusing of dormant cuttings and isolation of bacteria from callus tissue on semiselective medium was initially established and used by some laboratories (Lehoczky 1971). Because of its systemic nature, the pathogen can be isolated from grapevine vascular sap by flushing water or buffer through dormant grapevine cuttings using a vacuum pump (Tarbah and Goodman 1986; Bazzi et al. 1987). In this respect, a greater number of A. vitis cells has been recovered when dormant cuttings were frozen prior to vacuum flushing (Stover et al. 1997b). However, these procedures are time-consuming and laborious and are therefore not suitable for routine analysis of large numbers of samples.
PCR-based techniques are the method of choice for rapid pathogen detection in plant material and soil samples (Burr et al. 2017). Although a number of different PCR primers have been reported (Palacio-Bielsa et al. 2009a), remarkable genetic variations in A. vitis and its Ti plasmids may limit the specificity of described protocols (Kuzmanović et al. 2016). Moreover, most PCR protocols developed so far are generally suitable for testing bacteria from pure culture and may not be sensitive enough for pathogen detection in plant material.
Efficient DNA extraction followed by highly sensitive real-time PCR currently represents the most promising tool for early pathogen detection in plant and soil samples. So far, two real-time PCR protocols for detection of A. vitis in grapevine samples have been reported, both based on SYBR Green I dye chemistry (Bini et al. 2008a; Johnson et al. 2013). The protocol developed by Johnson et al. (2013) involves sample enrichment followed by efficient DNA extraction via magnetic capture hybridization (MCH) and detection of tumorigenic strains by using virD2 gene-specific primers. This assay allowed detection of 101 CFU/ml and was able to detect A. vitis in asymptomatic grapevine material (Johnson et al. 2013; Johnson et al. 2016; Canik Orel et al. 2017).
7.2 Management Practices
Because A. vitis persists systemically in grapevines and there are no effective chemical controls for crown gall on grapes, the disease is especially challenging to manage. As covered in Sect. 7.4 in this chapter, biological control is encouraging as a future commercial control. The development of A. vitis-free propagation material is also a viable consideration which is covered in this chapter (see Sect. 7.3). However, the uncertainty of material being completely free of the pathogen and the potential sources of A. vitis in the environment that may infect “clean” vines will ultimately affect the effectiveness of this management strategy.
Currently in commercial vineyards, crown gall is managed primarily through the use of cultural practices that aim to reduce injuries to vines which may serve as infection sites for A. vitis (Moyer 2013). These practices include selecting vineyard sites that have well-drained soils and which are geographically located to have good air drainage and thus are not as prone to freezing temperatures. Crown gall is often most prevalent in low-lying regions of vineyards where standing water may accumulate and cold air may settle in frost pockets. Such wet soil conditions can affect late-season acclimation of vines, making them more prone to injury from sudden freezing temperatures. Where possible, management of irrigation water is another practice employed for slowing vine growth to facilitate hardening-off and making them more tolerant to winter freezes. Other factors that affect vine sensitivity to freezing temperatures and injury include excessive fertilization and over-cropping which can stimulate late growth of vines and affect the onset of dormancy.
In cold regions, the practice of hilling soil around grafts of vines in the fall is employed as a means to preserve vines and as a crown gall management tool. In this case, should winter freezes severely damage or kill vine trunks it becomes possible to train a new trunk the following year that is generated from the remaining living scion wood that was buried by soil and protected from the freeze. The training of multiple trunks per vine is often implemented as well. In this case, once a trunk becomes severely injured and diseased with crown gall, it can be removed and the remaining trunk or trunks will allow crop production as new trunks are trained.
There are no effective chemical controls for grape crown gall. Although antibacterial compounds, such as copper products, are lethal to A. vitis, topical treatments to vines have limited value considering the bacterium is systemic in the vine. Moreover, the plants affected by crown gall are genetically transformed and stay permanently infected.
Several papers have been published to demonstrate that grape species and varieties (scion and rootstock) differ in their susceptibility to infection by A. vitis (Ferreira and van Zyl 1986; Szegedi et al. 1989; Goodman et al. 1993; Stover et al. 1997a; Roh et al. 2003; Mahmoodzadeh et al. 2004; Jung et al. 2016). In general, V. vinifera is most susceptible, whereas V. riparia and hybrids of Vitis species used for scion and rootstock varieties are most resistant to infection, producing fewer and smaller galls. Among grape rootstocks, Courderc 3309 and Riparia Gloire are generally viewed as resistant. Differences within specific accessions have been noted among genotypes of these and other Vitis spp. Regardless, even those considered “resistant,” such as V. riparia, may carry internal populations of A. vitis with the possibility of spreading it to more susceptible grapevines.
One study compared the relative levels of crown gall susceptibility of 43 Vitis genotypes following inoculation with a diverse set of A. vitis strains, followed by measuring gall size and the proportion of inoculation sites with galls (Stover et al. 1997a). None of the genotypes were immune and, depending on genotype, galls formed at 10–100% of the inoculated sites; the mean gall size ranged from 1.0 to 12 mm when averaged across A. vitis strains. Significant strain by genotype interactions was observed. For example, Vitis amurensis was most susceptible to a limited host range A. vitis strain, AG57. Commonly used rootstocks, 3309C, T5C, Riparia Gloire, and 101–14 Mgt, were among the most crown gall-resistant genotypes (Stover et al. 1997a). Vitis flexuosa, Vitis piasezkii, and V. amurensis that had been reported as resistant previously developed some galls. Rootstocks 110R, 420A, and Dogridge were categorized as highly susceptible. In another study, Szegedi et al. (1989) inoculated various grapevine cultivars with A. vitis strains belonging to different opine groups. Grapevine varieties were separated into four groups based on their susceptibility or resistance. Both host and bacterial factors likely contribute to the susceptibility/resistance of grapevine to A. vitis.
A limited amount of research has been done on the genetics of crown gall resistance in grape. Szegedi and Kozma (1984) tested seedlings from 27 hybrid families by inoculating with A. vitis At-1. Crown gall resistance in these crosses originated from V. amurensis. Their results showed a segregation of 1:1 following crosses of resistant and susceptible phenotypes and 3:1 (resistant to susceptible) following selfing of resistant parents. Therefore, a Mendelian-dominant inheritance of crown gall resistance to strain AT-1 was proposed. Subsequent research (Kuczmog et al. 2012) developed molecular markers linked to the Rcg1 crown gall resistance gene from V. amurensis. This technology holds promise for future marker-assisted breeding of high-quality crown gall-resistant grape varieties.
Attempts to control grapevine crown gall by developing transgenic disease-resistant plants have been also made (Vidal et al. 2006; Krastanova et al. 2010; Galambos et al. 2013). In one such study, grapevine rootstock (Vitis berlandieri × V. rupestris cv. “Richter 110”) plants were transformed with an oncogene-silencing transgene based on iaaM and ipt oncogene sequences (Galambos et al. 2013). However, oncogene silencing in grapevine is highly strain-specific and thus has limited effectiveness in disease control.
7.3 Evaluation of Strategies for Producing A. vitis-Free Grapevines
The discovery that A. vitis survives systemically and distributes randomly in grapevine propagation material stimulated research to determine if and how vines free of the pathogen could be produced. One approach used hot water treatments of dormant cuttings (Burr et al. 1989; Bazzi et al. 1991). In this case, dormant cuttings were submersed in water at 50 °C for 30 min. Although populations of the pathogen could be greatly reduced with this treatment, the bacteria were not eliminated (Burr et al. 1996). When temperatures higher than 50 °C were tested, the potential for increased bud mortality became apparent. Subsequently, Wample et al. (1991) showed that dormant cuttings collected in Washington State could withstand higher temperatures without enduring bud kill. Therefore, factors (temperatures, cutting hardiness, etc.) prior to the treatment appear to have a significant effect on grape bud tolerance to heat and should be explored further to determine the effectiveness of such treatments on internal A. vitis populations. Despite the fact that hot water treatment may result in a certain amount of bud kill, the practice (50 °C for 30 min) is still used in some regions of the world and felt to be of benefit for disease management.
Plant tissue culture has also been used to eliminate viral and bacterial pathogens from plants, including grapes (Dula et al. 2007; Cassells 2012). Explants from shoot tips or meristems are cultured in specific media to facilitate plant development (Sim and Golino 2010). Shoot tip culture was previously tested and shown to be effective for elimination of A. vitis. However, the detection method for evaluating its effectiveness was much less sensitive than is magnetic capture hybridization in conjunction with real-time PCR (MCH real-time PCR) technology currently in use (Johnson et al. 2013).
More recently, the MCH real-time PCR method was used in multiple experiments to assay tissue culture plants that were propagated from vines collected from a commercial vineyard that had severe crown gall. Shoot tips and meristems from the plants were assayed in 2013. Eighteen of the first 29 plants propagated tested positive for A. vitis. These included meristems, shoot tips with meristems extracted, and shoot tips with meristems (Johnson et al. 2016). When the same plants were cut back and regrown, only 4 of the 29 were positive. These results indicate the irregular distribution of A. vitis in the tissues as well as the uncertainty of a negative result.
Similar experiments were done in 2014 using 31 plants that were grown from cuttings taken from Riesling vines that were heavily infected with crown gall. In this case, A. vitis was not detected in any of the meristems for the two repetitions of the experiment (second repetition involved evaluating the regrowth of the plants after they were cut back following the first set of assays). For shoot tips with the meristems removed, four tested positive in the first repetition, but none were positive in the second (Johnson et al. 2016). Detecting A. vitis associated with grape tissue culture plants is not totally unexpected and has been reported previously (Poppenberger et al. 2002). A. vitis can persist on surfaces of grape leaves, thus epiphytically on grapevines (Canik Orel et al. 2017). Additional research on the use of tissue culture is underway to determine if in fact vines free of the pathogen can be generated. However, from work concluded thus far it is apparent that shoot tips do not necessarily comprise tissue that is free of the pathogen. Another consideration is the sensitivity of the MCH real-time PCR assay, which was found to be about 10 bacterial cells per sample. Therefore, if populations lower than 10 cells are present in a sample they may not be detected. From this research, we conclude that tissue culture alone may not eliminate A. vitis from grape explants and that additional practices such as incorporation of effective antibiotics that do not inhibit plant growth and/or the use of heat therapy should be evaluated as a component of the tissue culture propagation.
7.4 Biological Control
Biological control of crown gall disease caused by tumorigenic agrobacteria represents a major success story in the field of plant pathology, resulting from the discovery of the nontumorigenic R. rhizogenes (former name Agrobacterium radiobacter) strain K84 by Kerr (1972). Control of crown gall by K84 has been implemented on different plant hosts in many regions worldwide. The primary mode of action of K84 is by antibiosis through the production of agrocin 84, which is encoded by the conjugative plasmid pAgK84 (Kim et al. 2006). Subsequently, a genetically modified form of K84 (strain K1026) was developed having a deleted fragment of a tra gene responsible for plasmid transfer, thereby preventing its transfer to pathogenic strains (Jones et al. 1988). Such transfer can result in strains becoming resistant to agrocin 84. Nevertheless, K84 and K1026 are not effective in preventing crown gall caused by some tumorigenic agrobacteria including A. vitis, the primary cause of the grapevine crown gall disease.
The impressive success of K84 and K1026 together with their ineffectiveness against A. vitis on grape led several researchers to search for bacterial strains that may control crown gall on grape. A number of strains have shown an ability to inhibit growth of A vitis and tumor formation, including A. vitis strains E26 (Yang et al. 2009) and VAR03-1 and ARK-1 (Kawaguchi et al. 2005, 2007, 2008a, 2014, 2017; Kawaguchi and Inoue 2012; Kawaguchi 2013, 2014, 2015; Saito et al. 2018), the Agrobacterium biovar 1 strain HLB2 (Pu and Goodman 1993), and the R. rhizogenes strain J73 (Webster et al. 1986). The potential of strains belonging to other bacterial genera to control grapevine crown gall has been also tested. These investigations included various endophytic strains isolated from grapevine (Bell et al. 1995; Ferrigo et al. 2017), various Pseudomonas spp. strains (Khmel et al. 1998; Eastwell et al. 2006; Biondi et al. 2009), and Rahnella aquatilis strain HX2 (Chen et al. 2007). These strains have shown a range of effects on pathogen growth as well as on various levels of disease suppression in experiments done under laboratory and greenhouse conditions. In addition, preliminary screens of bacteria in vitro and on different indicator plants in the greenhouse have revealed bacterial strains belonging to different genera that have activity against A. vitis (Habbadi et al. 2017a). Interestingly, one fungal isolate belonging to the genus Acremonium and commercial biological control agents Bacillus subtilis SR63 and Trichoderma asperellum T1 were also effective against A. vitis (Ferrigo et al. 2017). A thorough review of bacterial strains that have been tested for activity against A. vitis has been published (Filo et al. 2013).
The nontumorigenic A. vitis strain F2/5, which was originally isolated from grape and shown to inhibit grape crown gall in South Africa (Staphorst et al. 1985), has been further studied by several laboratories (Burr and Reid 1994; Bazzi et al. 1999; Zäuner et al. 2006). Strain F2/5 inhibits tumor formation by diverse A. vitis strains on different grape varieties (Burr and Reid 1994). Although tumor formation by most strains of A. vitis is greatly inhibited by F2/5, a few strains appear to be unaffected (Staphorst et al. 1985; Burr and Reid 1994). As with other A. vitis strains, F2/5 also causes necrosis on grape roots (Burr et al. 1987a) which was recently shown to have a negative impact on graft wound healing and on plant growth but was not required for grape tumor inhibition (GTI; Hao et al. 2017). The molecular mechanism involved in necrosis is not fully understood but includes QS regulation and the involvement of specific polyketide and nonribosomal peptide synthases (Zheng et al. 2003; Hao et al. 2005; Zheng et al. 2012; Zheng and Burr 2013). Our current understanding of the biochemical pathways associated with necrosis and tumor inhibition by F2/5 shows overlap, but also that they are distinct processes (Zheng and Burr 2016).
Additional research has focused on characterizing GTI and identifying factors associated with the ability of F2/5 to inhibit crown gall. For example, although F2/5 inhibits A. vitis from causing tumors on grapevine, it does not block tumor formation on most other plants such as tobacco. An exception to this rule was reported to be inhibition of tumors on Ricinus (Zäuner et al. 2006). In addition, for F2/5 to inhibit tumor formation it must be applied to wounds at the same time or prior to the tumorigenic strain, and usually at cell numbers equal to or greater (Burr and Reid 1994). Through mutational analyses, it was shown that tumor inhibition was not associated with antibiosis even though an antibiosis phenotype could be demonstrated in vitro (Burr et al. 1997). Additionally, this study also demonstrated that tumor inhibition was not the result of competition for attachment sites on plant wounds. More recently, it was shown that F2/5 does not reduce populations of the tumorigenic strain at grape wound surfaces but, by an unknown mechanism, inhibits A. vitis from causing crown gall on grape (Kaewnum et al. 2013). This study also demonstrated that the genetic mechanism of gall inhibition is associated with at least two regulatory mechanisms that include QS and the involvement of clp protease genes.
Essential oils of Origanum compactum and Thymus vulgaris showed in vitro and in planta antibacterial activity against A. vitis (Habbadi et al. 2017b). Therefore, the use of essential oils of medicinal and aromatic plants could be a valuable alternative strategy in the control of grapevine crown gall. Moreover, in one recent study, a specific phage display-selected peptide displayed inhibitory effect toward A. vitis polygalacturonase, which could be a promising approach in disease control strategy (Warren et al. 2016).
References
Abdellatif E, Valentini F, Janse JD et al (2013) Occurrence of crown gall of the grapevine in Tunisia and characterization of Tunisian Agrobacterium vitis and A. tumefaciens strains. J Plant Pathol 95:115–126
Albiach MR, Lopez MM (1992) Plasmid heterogeneity in Spanish isolates of Agrobacterium tumefaciens from thirteen different hosts. Appl Environ Microbiol 58:2683–2687
Allardet-Servent A, Michaux-Charachon S, Jumas-Bilak E et al (1993) Presence of one linear and one circular chromosome in the Agrobacterium tumefaciens C58 genome. J Bacteriol 175:7869–7874
Al-Momani F, Albasheer S, Saadoun I (2006) Distribution of Agrobacterium tumefaciens biovars in Jordan and variation of virulence. Plant Pathol J 22:318–322
Argun N, Momol MT, Maden S et al (2002) Characterization of Agrobacterium vitis strains isolated from Turkish grape cultivars in the Central Anatolia region. Plant Dis 86:162–166
Bauer C, Schulz TF, Lorenz D et al (1994) Population dynamics of Agrobacterium vitis in two grapevine varieties during the vegetation period. Vitis 33:25–29
Bautista-Zapanta JN, Arafat HH, Tanaka K et al (2009) Variation of 16S-23S internally transcribed spacer sequence and intervening sequence in rDNA among the three major Agrobacterium species. Microbiol Res 164:604–612
Bazzi C, Piazza C, Burr TJ (1987) Detection of Agrobacterium tumefaciens in grapevine cuttings. EPPO Bulletin 17:105–112
Bazzi C, Stefani E, Gozzi R et al (1991) Hot-water treatment of dormant grape cuttings: its effects on Agrobacterium tumefaciens and on grafting and growth of vine. Vitis 30:177–187
Bazzi C, Alexandrova M, Stefani E et al (1999) Biological control of Agrobacterium vitis using non-tumorigenic agrobacteria. Vitis 38:31–35
Beauchamp CJ, Chilton WS, Dion P et al (1990) Fungal catabolism of crown gall opines. Appl Environ Microbiol 56:150–155
Beauchamp CJ, Kloepper JW, Lifshitz R et al (1991) Frequent occurrence of the ability to utilize octopine in rhizobacteria. Can J Microbiol 37:158–164
Beaulieu C, Coulombe LJ, Granger RL et al (1983) Characterization of opine-utilizing bacteria isolated from Quebec. Phytoprotect 64:61–68
Bell CR, Dickie GA, Chan JWYF (1995) Variable response of bacteria isolated from grapevine xylem to control grape crown gall disease in planta. Am J Enol Vitic 46:499–508
Bergeron J, Macleod RA, Dion P (1990) Specificity of octopine uptake by Rhizobium and Pseudomonas strains. Appl Environ Microbiol 56:1453–1458
Bien E, Lorenz D, Eichhorn K et al (1990) Isolation and characterization of Agrobacterium tumefaciens from the German vineregion Rheinpfalz. J Plant Dis Protect 97:313–322
Bini F, Geider K, Bazzi C (2008a) Detection of Agrobacterium vitis by PCR using novel virD2 gene-specific primers that discriminate two subgroups. Eur J Plant Pathol 122:403–411
Bini F, Kuczmog A, Putnoky P et al (2008b) Novel pathogen-specific primers for the detection of Agrobacterium vitis and Agrobacterium tumefaciens. Vitis 47:181–189
Biondi E, Bini F, Anaclerio F et al (2009) Effect of bioagents and resistance inducers on grapevine crown gall. Phytopathol Mediter 48:379–384
Bishop AL, Katz BH, Burr TJ (1988) Infection of grapevines by soilborne Agrobacterium tumefaciens biovar 3 and population dynamics in host and nonhost rhizospheres. Phytopathol 78:945–948
Bishop AL, Burr TJ, Mittak VL et al (1989) A monoclonal antibody specific to Agrobacterium tumefaciens biovar 3 and its utilization for indexing grapevine propagation material. Phytopathol 79:995–998
Bonnard G, Tinland B, Paulus F et al (1989) Nucleotide sequence, evolutionary origin and biological role of a rearranged cytokinin gene isolated from a wide host range biotype III Agrobacterium strain. Mol Gen Genet 216:428–438
Bouzar H, Moore LW (1987) Isolation of different Agrobacterium biovars from a natural oak savanna and tallgrass prairie. Appl Environ Microbiol 53:717–721
Brencic A, Angert ER, Winans SC (2005) Unwounded plants elicit Agrobacterium vir gene induction and T-DNA transfer: transformed plant cells produce opines yet are tumour free. Mol Microbiol 57:1522–1531
Brisset M-N, Rodriguez-Palenzuela P, Burr TJ et al (1991) Attachment, chemotaxis, and multiplication of Agrobacterium tumefaciens biovar 1 and biovar 3 on grapevine and pea. Appl Environ Microbiol 57:3178–3182
Buchholz WG, Thomashow MF (1984a) Comparison of T-DNA oncogene complements of Agrobacterium tumefaciens tumor-inducing plasmids with limited and wide host ranges. J Bacteriol 160:319–326
Buchholz WG, Thomashow MF (1984b) Host range encoded by the Agrobacterium tumefaciens tumor-inducing plasmid pTiAg63 can be expanded by modification of its T-DNA oncogene complement. J Bacteriol 160:327–332
Burr TJ, Hurwitz B (1981) Occurrence of Agrobacterium radiobacter pv. tumefaciens (Smith & Townsend) biotype 3 on grapevine in New York State (Abstr.). Phytopathol 71:26
Burr TJ, Katz BH (1983) Isolation of Agrobacterium tumefaciens biovar 3 from grapevine galls and sap, and from vineyard soil. Phytopathol 73:163–165
Burr TJ, Katz BH (1984) Grapevine cuttings as potential sites of survival and means of dissemination of Agrobacterium tumefaciens. Plant Dis 68:976–978
Burr TJ, Otten L (1999) Crown gall of grape: Biology and disease management. Annu Rev Phytopathol 37:53–80
Burr TJ, Reid CL (1994) Biological control of grape crown gall with non-tumorigenic Agrobacterium vitis strain F2/5. Am J Enol Vitic 45:213–219
Burr TJ, Bishop AL, Katz BH et al (1987a) A root-specific decay of grapevine caused by Agrobacterium tumefaciens and A. radiobacter biovar 3. Phytopathol 77:1424–1427
Burr TJ, Katz BH, Bishop AL (1987b) Populations of Agrobacterium in vineyard and non vineyard soils and grape roots in vineyards and nurseries. Plant Dis 71:617–620
Burr TJ, Katz BH, Bishop AL et al (1988) Effect of shoot age and tip culture propagation of grapes on systemic infestations by Agrobacterium tumefaciens biovar 3. Am J Enol Vitic 39:67–70
Burr TJ, Ophel K, Kerr A (1989) Effect of hot water treatment on systemic Agrobacterium tumefaciens biovar 3 in dormant grape cuttings. Plant Dis 73:242–245
Burr TJ, Reid CL, Tagliatti E et al (1995) Survival and tumorigenicity of Agrobacterium vitis in living and decaying grape roots and canes in soil. Plant Dis 79:677–682
Burr TJ, Reid CL, Splittstoesser DF et al (1996) Effect of heat treatments on grape bud mortality and survival of Agrobacterium vitis in vitro and in dormant grape cuttings. Am J Enol Vitic 47:119–123
Burr TJ, Reid CL, Tagliati E et al (1997) Biological control of grape crown gall by strain F2/5 is not associated with agrocin production or competition for attachment sites on grape cells. Phytopathol 87:706–711
Burr TJ, Bazzi C, Süle S et al (1998) Crown gall of grape: Biology of Agrobacterium vitis and the development of disease control strategies. Plant Dis 82:1288–1297
Burr TJ, Reid CL, Adams CE et al (1999) Characterization of Agrobacterium vitis strains isolated from feral Vitis riparia. Plant Dis 83:102–107
Burr TJ, Johnson KL, Kaewnum S et al (2017) Detection of Agrobacterium spp. in grapevines (Chapter 43). In: Fatmi MB, Walcott RR, Schaad NW (eds) Detection of plant-pathogenic bacteria in seed and other planting material. Second edn. APS Press, Minneapolis, MN, USA
Canaday J, Gerad JC, Crouzet P et al (1992) Organization and functional analysis of three T-DNAs from the vitopine Ti plasmid pTiS4. Mol Gen Genet 235:292–303
Canfield ML, Moore LW (1991) Isolation and characterization of opine-utilizing strains of Agrobacterium tumefaciens and fluorescent strains of Pseudomonas spp. from rootstocks of Malus. Phytopathol 81:440–443
Canik Orel D, Karagoz A, Durmaz R et al (2016) Phenotypic and molecular characterization of Rhizobium vitis strains from vineyards in Turkey. Phytopathol Mediter 55
Canik Orel D, Reid CL, Fuchs M et al (2017) Identifying environmental sources of Agrobacterium vitis in vineyards and wild grapevines. Am J Enol Vitic 68:213–217
Cassells AC (2012) Pathogen and biological contamination management in plant tissue culture: phytopathogens, vitro pathogens, and vitro pests. Methods in molecular biology (Clifton, NJ) 877:57–80
Cavara F (1897) Tubercolosi della vite. Intorno alla eziologia de alcune malattie di piante coltivate. Stazoni Sperimentali Agrarie Italiane 30:483–487
Chang CC, Jayaswal RK, Chen CM et al (1989) Altered imino diacid synthesis and transcription in crown gall tumors with transposon Tn5 insertions in the 3′ end of the octopine synthase gene. J Bacteriol 171:5922–5927
Chebil S, Fersi R, Chenenaoui S et al (2013a) First report of Agrobacterium vitis as causal agent of crown gall disease of grapevine in Tunisia. Plant Dis 97:836
Chebil S, Fersi R, Chenenaoui S et al (2013b) Occurrence of Agrobacterium vitis carrying two opine-type plasmids in Tunisian vineyards. J Plant Pathol Microbiol 4:175
Chen F, Guo YB, Wang JH et al (2007) Biological control of grape crown gall by Rahnella aquatilis HX2. Plant Dis 91:957–963
Chilton WS, Petit A, Chilton MD et al (2001) Structure and characterization of the crown gall opines heliopine, vitopine and rideopine. Phytochem 58:137–142
Christie PJ, Gordon JE (2014) The Agrobacterium Ti Plasmids. Microbiology spectrum 2. https://doi.org/10.1128/microbiolspec.plas-0010-2013
Clare BG, Kerr A, Jones DA (1990) Characteristics of the nopaline catabolic plasmid in Agrobacterium strains K84 and K1026 used for biological control of crown gall disease. Plasmid 23:126–137
Couturier MA, Bex F, Bergquist PL, Maas WK (1988) Identification and classification of bacterial plasmids. Microbiol rev 52(3):375–395 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC373151/
Crouzet P, Otten L (1995) Sequence and mutational analysis of a tartrate utilization operon from Agrobacterium vitis. J Bacteriol 177:6518–6526
Dahl GA, Guyon P, Petit A et al (1983) Characterization of opine utilizing bacteria isolated from Quebec Canada. Phytoprotect 64:61–68
De Oliveira JR, Da Silva Romeiro R, De Souza Leäo Lacerda B (1994) Occurrence of Agrobacterium tumefaciens biovar 3 on grapevine in Brazil. J Phytopathol 140:363–366
Dessaux Y, Petit A, Tempe J (1993) Chemistry and biochemistry of opines, chemical mediators of parasitism. Phytochem 34:31–38
Dessaux Y, Petit A, Farrand SK et al (1998) Opines and opine-like molecules involved in plant-Rhizobiaceae interactions. In: Spaink HP, Kondorosi A, Hooykaas PJJ (eds) The Rhizobiaceae, molecular biology of model plant-associated bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 173–197
Dhanvantari BN (1983) Etiology of grape crown gall in Ontario. Can J Bot 61:2641–2646
Dula T, Kolber M, Lazar J et al (2007) Production of healthy grapevine propagating material: Pathogens and methods. Online publication http://oiv2007hu/documents/viticulture/Szegedi_OIV_2007_textpdf
Eastwell KC, Sholberg PL, Sayler RJ (2006) Characterizing potential bacterial biocontrol agents for suppression of Rhizobium vitis, causal agent of crown gall disease in grapevines. Crop Protect 25:1191–1200
Fabre E, Dunal MF (1853) Observations sur les maladies régnantes de la vigne. Bulletin de la Société d’Agriculture du Département de l’Hérault 40:46
Faist H, Keller A, Hentschel U et al (2016) Grapevine (Vitis vinifera) crown galls host distinct microbiota. Appl Environ Microbiol 82:5542–5552
Farrand SK (1998) Conjugal plasmids and their transfer. In: Spaink HP, Kondorosi A, Hooykaas PJJ (eds) The rhizobiaceae: molecular biology of model plant-associated bacteria. Springer, Netherlands, Dordrecht, pp 199–233
Faure D, Lang J (2014) Functions and regulation of quorum-sensing in Agrobacterium tumefaciens. Front Plant Sci 5
Ferreira JHS, van Zyl FGH (1986) Susceptibility of grapevine rootstocks to strains of Agrobacterium tumefaciens biovar 3. S Afr J Enol Vitic 7:101–104
Ferrigo D, Causin R, Raiola A (2017) Effect of potential biocontrol agents selected among grapevine endophytes and commercial products on crown gall disease. Biocontrol 62:821–833
Filo A, Sabbatini P, Sundin GW et al (2013) Grapevine crown gall suppression using biological control and genetic engineering: a review of recent research. Am J Enol Vitic 64:1–14
Galambos A, Zok A, Kuczmog A et al (2013) Silencing Agrobacterium oncogenes in transgenic grapevine results in strain-specific crown gall resistance. Plant Cell Rep 32:1751–1757
Gallie DR, Kado CI (1988) Minimal region necessary for autonomous replication of pTAR. J Bacteriol 170:3170–3176
Gallie DR, Zaitlin D, Perry KL et al (1984) Characterization of the replication and stability regions of Agrobacterium tumefaciens plasmid pTAR. J Bacteriol 157:739–745
Garcillan-Barcia MP, Francia MV, de la Cruz F (2009) The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev 33:657–687
Garcillan-Barcia MP, Alvarado A, de la Cruz F (2011) Identification of bacterial plasmids based on mobility and plasmid population biology. FEMS Microbiol Rev 35:936–956
Gelvin SB (2012) Traversing the cell: Agrobacterium T-DNA’s journey to the host genome. Front Plant Sci 3:52
Genov I, Atanassov I, Tsvetkov I et al (2006a) Isolation and characterization of Agrobacterium strains from grapevines in Bulgarian vineyards and wild grapes, V. vinifera ssp. silvestris. Vitis 45:97–101
Genov I, Atanassov I, Yordanov Y et al (2006b) Genetic diversity of Agrobacterium vitis strains, isolated from grapevines and wild grapes in Bulgaria, assessed by cleaved amplified polymorphic sequences analysis of 16S-23S rDNA. Vitis 45:125–130
Genov N, Llop P, Lopez MM et al (2015) Molecular and phenotypic characterization of Agrobacterium species from vineyards allows identification of typical Agrobacterium vitis and atypical biovar 1 strains. J Appl Microbiol 118:1465–1477
Gerard JC, Canaday J, Szegedi E et al (1992) Physical map of the vitopine Ti plasmid pTiS4. Plasmid 28:146–156
Gevers D, Cohan FM, Lawrence JG et al (2005) Re-evaluating prokaryotic species. Nat Rev. Micro 3:733–739
Gillings M, Ophel-Keller K (1995) Comparison of strains of Agrobacterium vitis from grapevine source areas in Australia. Austral Plant Pathol 24:29–37
Goodman RN, Grimm R, Frank M (1993) The influence of grape rootstocks on the crown gall infection process and on tumor development. Am J Enol Vitic 44:22–26
Guyon P, Petit A, Tempe J et al (1993) Transformed plants producing opines specifically promote growth of opine-degrading agrobacteria. Mol Plant-Microbe Interact 6:92–98
Haas JH, Zveibil A, Zutra D et al (1991) The presence of crown gall of grape incited by Agrobacterium tumefaciens biovar 3 in Israel. Phytoparasitica 19:311–318
Habbadi K, Benkirane R, Benbouazza A et al (2017a) Biological control of grapevine crown gall caused by Allorhizobium vitis using bacterial antagonists. Int J Science Res 6:1390–1397
Habbadi K, Meyer T, Vial L et al (2017b) Essential oils of Origanum compactum and Thymus vulgaris exert a protective effect against the phytopathogen Allorhizobium vitis. Environ Sci Pollut Res Int
Hao G, Burr TJ (2006) Regulation of long-chain N-acyl-homoserine lactones in Agrobacterium vitis. J Bacteriol 188:2173–2183
Hao G, Zhang H, Zheng D et al (2005) luxR homolog avhR in Agrobacterium vitis affects the development of a grape-specific necrosis and a tobacco hypersensitive response. J Bacteriol 187:185–192
Hao L, Kemmenoe DJ, Orel DC et al (2017) The impacts of tumorigenic and non-tumorigenic Agrobacterium vitis strains on graft strength and growth of grapevines. Plant Dis
Herlache TC (1999) Biochemical and molecular genetic investigations of the Agrobacterium vitis–grapevine interaction. Ph.D. Thesis, Cornell University, Ithaca, NY
Herlache TC, Hotchkiss AT, Burr TJ et al (1997) Characterization of the Agrobacterium vitis pehA gene and comparison of the encoded polygalacturonase with the homologous enzymes from Erwinia carotovora and Ralstonia solanacearum. Appl Environ Microbiol 63:338–346
Herlache TC, Zhang HS, Ried CL et al (2001) Mutations that affect Agrobacterium vitis-induced grape necrosis also alter its ability to cause a hypersensitive response on tobacco. Phytopathol 91:966–972
Hoekema A, de Pater BS, Fellinger AJ et al (1984) The limited host range of an Agrobacterium tumefaciens strain extended by a cytokinin gene from a wide host range T-region. EMBO 3:3043–3047
Hooykaas PJJ (2000) Agrobacterium, a natural metabolic engineer of plants. In: Verpoorte R, Alfermann AW (eds) Metabolic engineering of plant secondary metabolism. Springer, The Netherlands, Dordrecht, pp 51–67
Huang S, Long M, Fu G et al (2015) Characterization of a new pathovar of Agrobacterium vitis causing banana leaf blight in China. J Basic Microbiol 55:129–134
Huss B, Tinland B, Paulus F et al (1990) Functional analysis of a complex oncogene arrangement in biotype III Agrobacterium tumefaciens strains. Plant Mol Biol 14:173–186
Ignatov AN, Khodykina MV, Vinogradova SV et al (2016) First report of Agrobacterium vitis causing crown galls of wine grape in Russia. Plant Dis 100:853
Irelan NA, Meredith CP (1996) Genetic analysis of Agrobacterium tumefaciens and A. vitis using randomly amplified polymorphic DNA. Am J Enol Vitic 47:145–151
Johnson KL, Zheng D, Kaewnum S et al (2013) Development of a magnetic capture hybridization real-time PCR assay for detection of tumorigenic Agrobacterium vitis in grapevines. Phytopathol 103:633–640
Johnson KL, Cronin H, Reid CL et al (2016) Distribution of Agrobacterium vitis in grapevines and its relevance to pathogen elimination. Plant Dis 100:791–796
Jones DA, Ryder MH, Clare BG et al (1988) Construction of a Tra− deletion mutant of pAgK84 to safeguard the biological control of crown gall. Mol Gen Genet 212:207–214
Jumas-Bilak E, Michaux-Charachon S, Bourg G et al (1998) Unconventional genomic organization in the alpha subgroup of the Proteobacteria. J Bacteriol 180:2749–2755
Jung S-M, Hur Y-Y, Preece JE et al (2016) Profiling of disease-related metabolites in grapevine internode tissues infected with Agrobacterium vitis. Plant Pathol J 32:489–499
Kado CI (1998) Origin and evolution of plasmids. Antonie Van Leeuwenhoek 73:117–126
Kaewnum S, Zheng D, Reid CL et al (2013) A host-specific biological control of grape crown gall by Agrobacterium vitis strain F2/5: its regulation and population dynamics. Phytopathol 103:427–435
Karimi M, Van Montagu M, Gheysen G (2000) Nematodes as vectors to introduce Agrobacterium into plant roots. Mol Plant Pathol 1:383–387
Kawaguchi A (2013) Biological control of crown gall on grapevine and root colonization by nonpathogenic Rhizobium vitis strain ARK-1. Microbes Environ 28:306–311
Kawaguchi A (2014) Reduction in pathogen populations at grapevine wound sites is associated with the mechanism underlying the biological control of crown gall by Rhizobium vitis strain ARK-1. Microbes Environ 29:296–302
Kawaguchi A (2015) Biological control agent Agrobacterium vitis strain ARK-1 suppresses expression of the virD2 and virE2 genes in tumorigenic A. vitis. Eur J Plant Pathol 143:789–799
Kawaguchi A, Inoue K (2009) Grapevine crown gall caused by Rhizobium radiobacter (Ti) in Japan. J Gen Plant Pathol 75:205–212
Kawaguchi A, Inoue K (2012) New antagonistic strains of non-pathogenic Agrobacterium vitis to control grapevine crown gall. J Phytopathol 160:509–518
Kawaguchi A, Inoue K, Nasu H (2005) Inhibition of crown gall formation by Agrobacterium radiobacter biovar 3 strains isolated from grapevine. J Gen Plant Pathol 71:422–430
Kawaguchi A, Inoue K, Nasu H (2007) Biological control of grapevine crown gall by nonpathogenic Agrobacterium vitis strain VAR03-1. J Gen Plant Pathol 73:133–138
Kawaguchi A, Inoue K, Ichinose Y (2008a) Biological control of crown gall of grapevine, rose, and tomato by nonpathogenic Agrobacterium vitis strain VAR03-1. Phytopathol 98:1218–1225
Kawaguchi A, Sawada H, Ichinose Y (2008b) Phylogenetic and serological analyses reveal genetic diversity of Agrobacterium vitis strains in Japan. Plant Pathol 57:747–753
Kawaguchi A, Inoue K, Tanina K (2014) Evaluation of the nonpathogenic Agrobacterium vitis strain ARK-1 for crown gall control in diverse plant species. Plant Dis 99:409–414
Kawaguchi A, Inoue K, Tanina K et al (2017) Biological control for grapevine crown gall using nonpathogenic Rhizobium vitis strain ARK-1. Proc Jpn Acad Ser B Phys Biol Sci 93:547–560
Kerr A (1972) Biological control of crown gall: seed inoculation. J Appl Bacteriol 35:493–497
Kerr A, Panagopoulos CG (1977) Biotypes of Agrobacterium radiobacter var. tumefaciens and their biological control. J Phytopathol 90:172–179
Khmel IA, Sorokina TA, Lemanova NB et al (1998) Biological control of crown gall in grapevine and raspberry by two Pseudomonas spp. with a wide spectrum of antagonistic activity. Biocontrol Sci Technol 8:45–57
Kim JG, Park BK, Kim SU et al (2006) Bases of biocontrol: sequence predicts synthesis and mode of action of agrocin 84, the Trojan horse antibiotic that controls crown gall. Proc Natl Acad Sci USA 103:8846–8851
Knauf VC, Panagopoulos CG, Nester EW (1982) Genetic factors controlling the host range of Agrobacterium tumefaciens. Phytopathol 72:1545–1549
Knauf VC, Panagopoulos CG, Nester EW (1983) Comparison of Ti plasmids from three different biotypes of Agrobacterium tumefaciens isolated from grapevines. J Bacteriol 153:1535–1542
Krastanova SV, Balaji V, Holden MR et al (2010) Resistance to crown gall disease in transgenic grapevine rootstocks containing truncated virE2 of Agrobacterium. Transgenic Res 19:949–958
Kuczmog A, Galambos A, Horvath S et al (2012) Mapping of crown gall resistance locus Rcg1 in grapevine. Theor Appl Genet 125:1565–1574
Kuzmanović N, Ivanović M, Prokić A et al (2014) Characterization and phylogenetic diversity of Agrobacterium vitis from Serbia based on sequence analysis of 16S-23S rRNA internal transcribed spacer (ITS) region. Eur J Plant Pathol 140:757–768
Kuzmanović N, Biondi E, Bertaccini A et al (2015) Genetic relatedness and recombination analysis of Allorhizobium vitis strains associated with grapevine crown gall outbreaks in Europe. J Appl Microbiol 119:786–796
Kuzmanović N, Biondi E, Ivanović M et al (2016) Evaluation of different PCR primers for identification of tumorigenic bacteria associated with grapevine crown gall. J Plant Pathol 98:311–319
Kuzmanović N, Puławska J, Smalla K et al (2018) Agrobacterium rosae sp. nov., isolated from galls on different agricultural crops. Syst Appl Microbiol
Lehoczky J (1968) Spread of Agrobacterium tumefaciens in the vessels of the grapevine, after natural infection. J Phytopathol 63:239–246
Lehoczky J (1971) Further evidences concerning the systemic spreading of Agrobacterium tumefaciens in the vascular system of grapevines. Vitis 10:215–221
Lehoczky J (1978) Root-system of the grapevine as a reservoir of Agrobacterium tumefaciens cells. In: 4th international conference on plant pathogenic bacteria, Angers, France, pp 239–243
Lehoczky J (1989) Inoculation experiment on trellised grapevines with Agrobacterium tumefaciens to study the process of crown gall disease. Acta Phytopathol Entomol Hung 24:283–291
Li Y, Gronquist MR, Hao G et al (2005) Chromosome and plasmid-encoded N-acyl homoserine lactones produced by Agrobacterium vitis wildtype and mutants that differ in their interactions with grape and tobacco. Physiol Mol Plant Pathol 67:284–290
Lim SH, Kim JG, Kang HW (2009) Novel SCAR primers for specific and sensitive detection of Agrobacterium vitis strains. Microbiol Res 164:451–460
López MM, Gorris MT, Montojo AM (1988) Opine utilization by Spanish isolates of Agrobacterium tumefaciens. Plant Pathol 37:565–572
Loubser JT (1978) Identification of Agrobacterium tumefaciens biotype 3 on grapevine in South Africa. Plant Dis Rep 62:730–731
Lowe N, Gan HM, Chakravartty V et al (2009) Quorum-sensing signal production by Agrobacterium vitis strains and their tumor-inducing and tartrate-catabolic plasmids. FEMS Microbiol Lett 296:102–109
Ma DQ, Yanofsky MF, Gordon MP et al (1987) Characterization of Agrobacterium tumefaciens strains isolated from grapevine tumors in China. Appl Environ Microbiol 53:1338–1343
Mahmoodzadeh H, Nazemieh A, Majidi I et al (2004) Evaluation of crown gall resistance in Vitis vinifera and hybrids of Vitis spp. Vitis 43:75–79
Mansouri H, Petit A, Oger P et al (2002) Engineered rhizosphere: the trophic bias generated by opine-producing plants is independent of the opine type, the soil origin, and the plant species. Appl Environ Microbiol 68:2562–2566
McCullen CA, Binns AN (2006) Agrobacterium tumefaciens and plant cell interactions and activities required for interkingdom macromolecular transfer. Annu Rev Cell Dev Biol 22:101–127
McGuire RG, Rodriguez-Palenzuela P, Collmer A et al (1991) Polygalacturonase production by Agrobacterium tumefaciens biovar 3. Appl Environ Microbiol 57:660–664
Meier-Kolthoff JP, Auch AF, Klenk H-P et al (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:1–14
Merlo DJ, Nester EW (1977) Plasmids in avirulent strains of Agrobacterium. J Bacteriol 129:76–80
Mohammadi M, Fatehi-Paykani R (1999) Phenotypical characterization of Iranian isolates of Agrobacterium vitis, the causal agent of crown gall disease of grapevine. Vitis 38:115–121
Momol EA, Burr TJ, Reid CL et al (1998) Genetic diversity of Agrobacterium vitis as determined by DNA fingerprints of the 5′-end of the 23S rRNA gene and random amplified polymorphic DNA. J Appl Microbiol 85:685–692
Moore LW, Chilton WS, Canfield ML (1997) Diversity of opines and opine-catabolizing bacteria isolated from naturally occurring crown gall tumors. Appl Environ Microbiol 63:201–207
Moore LW, Bouzar H, Burr TJ (2001) Agrobacterium. In: Schaad NW, Jones JB, Chun W (eds) Laboratory guide for identification of plant pathogenic bacteria, 3rd edn. APS Press, St Paul, Minnesota, pp 17–35
Mousavi SA, Osterman J, Wahlberg N et al (2014) Phylogeny of the Rhizobium-Allorhizobium-Agrobacterium clade supports the delineation of Neorhizobium gen. nov. Syst Appl Microbiol 37:208–215
Mousavi SA, Willems A, Nesme X et al (2015) Revised phylogeny of Rhizobiaceae: proposal of the delineation of Pararhizobium gen. nov., and 13 new species combinations. Syst Appl Microbiol 38:84–90
Moyer M (2013) Grapevine crown gall, disease management white paper. http://winewsuedu/wp-content/uploads/sites/66/2013/05/2013-CrownGallWhiteSheetpdf
Nascimento T, Oliveira H, Schulz T (1999) Identification of Agrobacterium vitis by amplification of molecular markers. Petria 9:81–84
Nautiyal CS, Dion P (1990) Characterization of the opine-utilizing microflora associated with samples of soil and plants. Appl Environ Microbiol 56:2576–2579
Nautiyal CS, Dion P, Chilton WS (1991) Mannopine and mannopinic acid as substrates for Arthrobacter sp. strain MBA209 and Pseudomonas putida NA513. J Bacteriol 173:2833–2841
Oger P, Farrand SK (2002) Two opines control conjugal transfer of an Agrobacterium plasmid by regulating expression of separate copies of the quorum-sensing activator gene traR. J Bacteriol 184:1121–1131
Oger P, Petit A, Dessaux Y (1997) Genetically engineered plants producing opines alter their biological environment. Nat Biotechnol 15:369–372
Ophel K, Kerr A (1990) Agrobacterium vitis sp. nov. for strains of Agrobacterium biovar 3 from grapevines. Int J Syst Bacteriol 40:236–241
Ophel K, Burr TJ, Magarey PA et al (1988) Detection of Agrobacterium tumefaciens biovar 3 in South Australian grapevine propagation material. Austral Plant Pathol 17:61–66
Ormeño-Orrillo E, Servín-Garcidueñas LE, Rogel MA et al (2015) Taxonomy of rhizobia and agrobacteria from the Rhizobiaceae family in light of genomics. Syst Appl Microbiol 38:287–291
Otten L, De Ruffray P (1994) Agrobacterium vitis nopaline Ti plasmid pTiAB4: relationship to other Ti plasmids and T-DNA structure. Mol Gen Genet 245:493–505
Otten L, Canaday J, Gerard JC et al (1992) Evolution of agrobacteria and their Ti plasmids—a review. Mol Plant Microbe Interact 5:279–287
Otten L, Gerard JC, De Ruffray P (1993) The Ti plasmid from the wide host range Agrobacterium vitis strain Tm4: Map and homology with other Ti plasmids. Plasmid 29:154–159
Otten L, Crouzet P, Salomone JY et al (1995) Agrobacterium vitis strain AB3 harbors two independent tartrate utilization systems, one of which is encoded by the Ti plasmid. Mol Plant Microbe Interact 8:138–146
Otten L, De Ruffray P, de Lajudie P et al (1996a) Sequence and characterisation of a ribosomal RNA operon from Agrobacterium vitis. Mol Gen Genet 251:99–107
Otten L, de Ruffray P, Momol EA et al (1996b) Phylogenetic relationship between Agrobacterium vitis isolates and their Ti plasmids. Mol Plant Microbe Interact 9:782–786
Otten L, Burr T, Szegedi E (2008) Agrobacterium: a disease-causing bacterium. In: Tzfira T, Citovsky V (eds) Agrobacterium: from biology to biotechnology. Springer, New York, USA, pp 1–46
Palacio-Bielsa A, Cambra MA, López MM (2009a) PCR detection and identification of plant-pathogenic bacteria: updated review of protocols (1989–2007). J Plant Pathol 91:249–297
Palacio-Bielsa A, González-Abolafio R, Álvarez B et al (2009b) Chromosomal and Ti plasmid characterization of tumorigenic strains of three Agrobacterium species isolated from grapevine tumours. Plant Pathol 58:584–593
Panagopoulos CG, Psallidas PG (1973) Characteristics of Greek isolates of Agrobacterium tumefaciens (E.F. Smith & Townsend). Conn J Appl Bacteriol 36:233–240
Panagopoulos CG, Psallidas PG, Alivizatos AS (1978) Studies on biotype 3 of Agrobacterium radiobacter var. tumefaciens. In: 4th international conference on plant pathogenic bacteria, Angers, France, pp 221–228
Pappas KM, Cevallos MA (2011) Plasmids of the Rhizobiaceae and their role in interbacterial and transkingdom interactions. In: Witzany G (ed) Biocommunication in soil microorganisms. Springer, Berlin Heidelberg, Berlin, Heidelberg, pp 295–337
Paulus F, Huss B, Bonnard G et al (1989a) Molecular systematics of biotype III Ti plasmids of Agrobacterium tumefaciens. Mol Plant-Microbe Interact 2:64–74
Paulus F, Ride M, Otten L (1989b) Distribution of two Agrobacterium tumefaciens insertion elements in natural isolates: evidence for stable association between Ti plasmids and their bacterial hosts. Mol Gen Genet 219:145–152
Paulus F, Canaday J, Otten L (1991a) Limited host range Ti plasmids: recent origin from wide host range Ti plasmids and involvement of a novel IS element, IS868. Mol Plant Microbe Interact 4:190–197
Paulus F, Canaday J, Vincent F et al (1991b) Sequence of the iaa and ipt region of different Agrobacterium tumefaciens biotype III octopine strains: reconstruction of octopine Ti plasmid evolution. Plant Mol Biol 16:601–614
Perović T, Renzi M, Mazzaglia A et al (2015) First report of Agrobacterium tumefaciens as a causal agent of crown gall on grapevine in Montenegro. Plant Dis 100:515
Perry KL, Kado CI (1982) Characteristics of Ti plasmids from broad-host-range and ecologically specific biotype 2 and 3 strains of Agrobacterium tumefaciens. J Bacteriol 151:343–350
Pinto UM, Pappas KM, Winans SC (2012) The ABCs of plasmid replication and segregation. Nat Rev Microbiol 10:755–765
Pitzschke A, Hirt H (2010) New insights into an old story: Agrobacterium-induced tumour formation in plants by plant transformation. EMBO J 29:1021–1032
Poppenberger B, Leonhardt W, Redl H (2002) Latent persistence of Agrobacterium vitis in micropropagated Vitis vinifera. Vitis 41:113–114
Pu X-A, Goodman RN (1993) Effects of fumigation and biological control on infection of indexed crown gall free grape plants. Am J Enol Vitic 44:241–248
Puławska J, Willems A, De Meyer SE et al (2012) Rhizobium nepotum sp. nov. isolated from tumors on different plant species. Syst Appl Microbiol 35:215–220
Puławska J, Kuzmanović N, Willems A et al (2016) Pararhizobium polonicum sp. nov. isolated from tumors on stone fruit rootstocks. Syst Appl Microbiol 39:164–169
Ramírez-Bahena MH, Vial L, Lassalle F et al (2014) Single acquisition of protelomerase gave rise to speciation of a large and diverse clade within the Agrobacterium/Rhizobium supercluster characterized by the presence of a linear chromid. Mol Phylogenet Evol 73:202–207
Richter M, Rossello-Mora R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131
Ridé M, Ridé S, Petit A et al (2000) Characterization of plasmid-borne and chromosome-encoded traits of Agrobacterium biovar 1, 2, and 3 strains from France. Appl Environ Microbiol 66:1818–1825
Rodriguez-Palenzuela P, Burr TJ, Collmer A (1991) Polygalacturonase is a virulence factor in Agrobacterium tumefaciens biovar 3. J Bacteriol 173:6547–6552
Roh JH, Yun HK, Park KS et al (2003) In vivo evaluation of resistance of grape varieties to crown gall disease. Plant Pathol J 19:235–238
Rouhrazi K, Rahimian H (2012a) Characterization of Iranian grapevine isolates of Rhizobium (Agrobacterium) spp. J Plant Pathol 94:555–560
Rouhrazi K, Rahimian H (2012b) Genetic diversity of Iranian Agrobacterium strains from grapevine. Ann Microbiol 62:1661–1667
Saito K, Watanabe M, Matsui H et al (2018) Characterization of the suppressive effects of the biological control strain VAR03-1 of Rhizobium vitis on the virulence of tumorigenic R. vitis. J Gen Plant Pathol 84:58–64
Salomone JY, Otten L (1999) Structure and function of a conserved DNA region coding for tartrate utilization in Agrobacterium vitis. FEMS Microbiol Lett 174:333–337
Salomone JY, Crouzet P, De Ruffray P et al (1996) Characterization and distribution of tartrate utilization genes in the grapevine pathogen Agrobacterium vitis. Mol Plant Microbe Interact 9:401–408
Salomone JY, Szegedi E, Cobanov P et al (1998) Tartrate utilization genes promote growth of Agrobacterium spp. on grapevine. Mol Plant Microbe Interact 11:836–838
Savka MA, Farrand SK (1997) Modification of rhizobacterial populations by engineering bacterium utilization of a novel plant-produced resource. Nat Biotechnol 15:363–368
Savka MA, Black RC, Binns AN et al (1996) Translocation and exudation of tumor metabolites in crown galled plants. Mol Plant Microbe Interact 9:310–313
Sawada H, Ieki H (1992) Crown gall of kiwi caused by Agrobacterium tumefaciens in Japan. Plant Dis 76:212
Sawada H, Ieki H, Takikawa Y (1990) Identification of grapevine crown gall bacteria isolated in Japan. Jap J Phytopathol 56:199–206
Schroth MN, McCain AH, Foott JH et al (1988) Reduction in yield and vigor of grapevine caused by crown gall disease. Plant Dis 72:241–246
Schulz TF, Bauer C, Lorenz D et al (1993) Studies on the evolution of Agrobacterium vitis as based on genomic fingerprinting and IS element analysis. Syst Appl Microbiol 16:322–329
Sciaky D, Montoya AL, Chilton M-D (1978) Fingerprints of Agrobacterium Ti plasmids. Plasmid 1:238–253
Sim ST, Golino D (2010) Micro- vs. macroshoot tip culture therapy for disease elimination in grapevines. FPS Grape Program Newsl Foundation Plant Services, University of California, Davis
Slater SC, Goldman BS, Goodner B et al (2009) Genome sequences of three Agrobacterium biovars help elucidate the evolution of multichromosome genomes in bacteria. J Bacteriol 191:2501–2511
Slater S, Setubal JC, Goodner B et al (2013) Reconciliation of sequence data and updated annotation of the genome of Agrobacterium tumefaciens C58, and distribution of a linear chromosome in the genus Agrobacterium. Appl Environ Microbiol 79:1414–1417
Smillie C, Garcillan-Barcia MP, Francia MV et al (2010) Mobility of plasmids. Microbiol Mol Biol Rev 74:434–452
Spencer PA, Tanaka A, Towers GHN (1990) An Agrobacterium signal compound from grapevine cultivars. Phytochem 29:3785–3788
Staphorst JL, van Zyl FGH, Strijdom BW et al (1985) Agrocin-producing pathogenic and nonpathogenic biotype-3 strains of Agrobacterium tumefaciens active against biotype-3 pathogens. Curr Microbiol 12:45–52
Stewart E, Wenner N, Long L et al (2013) Crown gall of grape: understanding the disease, prevention and management. Online Publication. http://plantpath.psu.edu/research/labs/grapes/publications/disease-fact-sheets/Crown-gall-grape.pdf
Stover EW, Swartz HJ, Burr TJ (1997a) Crown gall formation in a diverse collection of Vitis genotypes inoculated with Agrobacterium vitis. Am J Enol Vitic 48:26–32
Stover EW, Swartz HJ, Burr TJ (1997b) Endophytic Agrobacterium in crown gall-resistant and -susceptible Vitis genotypes. Vitis 36:21–26
Süle S (1978) Biotypes of Agrobacterium tumefaciens in Hungary. J Appl Bacteriol 44:207–213
Süle S (1986) Survival of Agrobacterium tumefaciens in Berlandieri x Riparia grapevine rootstock. Acta Phytopathol Entomol Hung 21:203–206
Süle S, Kado CI (1980) Agrocin resistance in virulent derivatives of Agrobacterium tumefaciens harboring the pTi plasmid. Physiol Plant Pathol 17:347–356
Süle S, Lehoczky J, Jenser G et al (1995) Infection of grapevine roots by Agrobacterium vitis and Meloidogyne hapla. J Phytopathol 143:169–171
Süle S, Cursino L, Zheng D et al (2009) Surface motility and associated surfactant production in Agrobacterium vitis. Lett Appl Microbiol 49:596–601
Suzuki K, Tanaka K, Yamamoto S et al (2009) Ti and Ri Plasmids. In: Schwartz E (ed) Microbial megaplasmids, vol 11. Microbiology monographs. Springer, Berlin, Heidelberg, pp 133–147
Szegedi E (1985) Host range and specific L(+)tartrate utilization of biotype 3 of Agrobacterium tumefaciens. Acta Phytopathol Acad Sci Hung 20:17–22
Szegedi E (2003) Opines in naturally infected grapevine crown gall tumors. Vitis 42:39–41
Szegedi E, Kozma P (1984) Studies on the inheritance of resistance to crown gall disease of grapevine. Vitis 23:121–126
Szegedi E, Otten L (1998) Incompatibility properties of tartrate utilization plasmids derived from Agrobacterium vitis strains. Plasmid 39:35–40
Szegedi E, Czakó M, Otten L et al (1988) Opines in crown gall tumours induced by biotype 3 isolates of Agrobacterium tumefaciens. Physiol Mol Plant Pathol 32:237–247
Szegedi E, Korbuly J, Otten L (1989) Types of resistance of grapevine varieties to isolates of Agrobacterium tumefaciens biotype 3. Physiol Mol Plant Pathol 35:35–43
Szegedi E, Otten L, Czakó M (1992) Diverse types of tartrate plasmids in Agrobacterium tumefaciens biotype III strains. Mol Plant Microbe Interact 5:435–438
Szegedi E, Czakó M, Otten L (1996) Further evidence that the vitopine-type pTi’s of Agrobacterium vitis represent a novel group of Ti plasmids. Mol Plant Microbe Interact 9:139–143
Szegedi E, Süle S, Burr TJ (1999) Agrobacterium vitis strain F2/5 contains tartrate and octopine utilization plasmids which do not encode functions for tumor inhibition on grapevine. J Phytopathol 147:554–558
Szegedi E, Bottka S, Mikulas J et al (2005) Characterization of Agrobacterium tumefaciens strains isolated from grapevine. Vitis 44:49–54
Szegedi E, Deak T, Forgács I et al (2014) Agrobacterium vitis strains lack tumorigenic ability on in vitro grown grapevine stem segments. Vitis 53:147–154
Tanaka K, Urbanczyk H, Matsui H et al (2006) Construction of physical map and mapping of chromosomal virulence genes of the biovar 3 Agrobacterium (Rhizobium vitis) strain K-Ag-1. Genes Genet Syst 81:373–380
Tarbah FA, Goodman RN (1986) Rapid detection of Agrobacterium tumefaciens in grapevine propagating material and the basis for an efficient indexing system. Plant Dis 70(70):566–568
Tarbah FA, Goodman RN (1987) Systemic spread of Agrobacterium tumefaciens biovar 3 in the vascular system of grapes. Phytopathol 77:915–920
Thies KL, Griffin DE, Graves CH Jr et al (1991) Characterization of Agrobacterium isolates from muscadine grape. Plant Dis 75:634–637
Thomashow MF, Panagopoulos CG, Gordon MP et al (1980) Host range of Agrobacterium tumefaciens is determined by the Ti plasmid. Nature 283:794–796
Thomashow MF, Knauf VC, Nester EW (1981) Relationship between the limited and wide host range octopine-type Ti plasmids of Agrobacterium tumefaciens. J Bacteriol 146:484–493
Tolba IH, Zaki MF (2011) Characterization of Agrobacterium vitis isolates obtained from galled grapevine plants in Egypt. Annals of Agricultural Sciences 56:113–119
Tremblay G, Gagliardo R, Chilton WS et al (1987) Diversity among opine-utilizing bacteria: identification of coryneform isolates. Appl Environ Microbiol 53:1519–1524
van Nuenen M, de Ruffray P, Otten L (1993) Rapid divergence of Agrobacterium vitis octopine-cucumopine Ti plasmids from a recent common ancestor. Mol Gen Genet 240:49–57
Vidal JR, Kikkert JR, Malnoy MA et al (2006) Evaluation of transgenic ‘Chardonnay’ (Vitis vinifera) containing magainin genes for resistance to crown gall and powdery mildew. Transgenic Res 15:69–82
Wabiko H, Kagaya M, Sano H (1990) Various nopaline catabolism genes located outside the Ti-plasmids in Agrobacterium tumefaciens strains. Microbiol 136:97–103
Wample RL, Bary A, Burr TJ (1991) Heat tolerance of dormant Vitis vinifera cuttings. Am J Enol Vitic 42:67–72
Warren JG, Kasun GW, Leonard T et al (2016) A phage display-selected peptide inhibitor of Agrobacterium vitis polygalacturonase. Mol Plant Pathol 17:480–486
Watanabe S, Sueda R, Fukumori F et al (2015) Characterization of flavin-containing opine dehydrogenase from bacteria. PLoS ONE 10:e0138434
Webster J, Dos Santos M, Thomson JA (1986) Agrocin-producing Agrobacterium tumefaciens strain active against grapevine isolates. Appl Environ Microbiol 52:217–219
Wetzel ME, Kim KS, Miller M et al (2014) Quorum-dependent mannopine-inducible conjugative transfer of an Agrobacterium opine-catabolic plasmid. J Bacteriol 196:1031–1044
Wetzel ME, Olsen GJ, Chakravartty V et al (2015) The repABC plasmids with quorum-regulated transfer systems in members of the rhizobiales divide into two structurally and separately evolving groups. Genome Biol Evol 7:3337–3357
White CE, Winans SC (2007) Cell–cell communication in the plant pathogen Agrobacterium tumefaciens. Philos Trans. R Soc B: Biol Sci 362:1135–1148
Yang YL, Li JY, Wang JH et al (2009) Mutations affecting chemotaxis of Agrobacterium vitis strain E26 also alter attachment to grapevine roots and biocontrol of crown gall disease. Plant Pathol 58:594–605
Yanofsky M, Lowe B, Montoya A et al (1985a) Molecular and genetic analysis of factors controlling host range in Agrobacterium tumefaciens. Mol Gen Genet 201:237–246
Yanofsky M, Montoya A, Knauf V et al (1985b) Limited-host-range plasmid of Agrobacterium tumefaciens: molecular and genetic analyses of transferred DNA. J Bacteriol 163:341–348
Young JM, Kerr A, Sawada H (2005) Genus Agrobacterium Conn 1942, 359AL. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM (eds) Bergey’s manual of systematics of archaea and bacteria. The Proteobacteria, vol 2C, 2ed edn. Springer, New York, pp 340–345
Zäuner S, Crespan JE, Burr TJ et al (2006) Inhibition of crown gall induction by Agrobacterium vitis strain F2/5 in grapevine and Ricinus. Vitis 45:131–139
Zheng D, Burr TJ (2013) An Sfp-type PPTase and associated polyketide and nonribosomal peptide synthases in Agrobacterium vitis are essential for induction of tobacco hypersensitive response and grape necrosis. Mol Plant Microbe Interact 26:812–822
Zheng D, Burr TJ (2016) Inhibition of grape crown gall by Agrobacterium vitis F2/5 requires two nonribosomal peptide synthetases and one polyketide synthase. Mol Plant Microbe Interact 29:109–118
Zheng D, Zhang H, Carle S et al (2003) A luxR homolog, aviR, in Agrobacterium vitis is associated with induction of necrosis on grape and a hypersensitive response on tobacco. Mol Plant Microbe Interact 16:650–658
Zheng D, Hao G, Cursino L et al (2012) LhnR and upstream operon LhnABC in Agrobacterium vitis regulate the induction of tobacco hypersensitive responses, grape necrosis and swarming motility. Mol Plant Pathol 13:641–652
Zhu J, Oger PM, Schrammeijer B et al (2000) The bases of crown gall tumorigenesis. J Bacteriol 182:3885–3895
Zidarič I (2009) Monitoring of bacteria of the genus Agrobacterium on grapevine in 2006 and 2007. In: 9th Slovenian conference on plant protection, Nova Gorica, Slovenia. Book of lectures and papers, pp 225–229
Zupan J, Muth TR, Draper O et al (2000) The transfer of DNA from Agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J 23:11–28
Acknowledgements
The research of T. J. Burr presented in this paper was partially funded by USDA Federal Capacity Fund grants, USDA-APHIS National Clean Plant Network, and by the New York Wine and Grape Foundation. N. Kuzmanović was supported by the Georg Forster Fellowship for postdoctoral researchers from the Alexander von Humboldt-Foundation, Bonn, Germany. We kindly thank Dr. Ernő Szegedi for his critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this chapter
Cite this chapter
Kuzmanović, N., Puławska, J., Hao, L., Burr, T.J. (2018). The Ecology of Agrobacterium vitis and Management of Crown Gall Disease in Vineyards. In: Gelvin, S. (eds) Agrobacterium Biology. Current Topics in Microbiology and Immunology, vol 418. Springer, Cham. https://doi.org/10.1007/82_2018_85
Download citation
DOI: https://doi.org/10.1007/82_2018_85
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-03256-2
Online ISBN: 978-3-030-03257-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)