Abstract
The aim of this work was to test the hypothesis that a putative multisensory hybrid histidine kinase—response regulator (HSHK–RR) and FlhB (a component of the type 3 secretion apparatus located in the basal body of Fla) encoded by Azospirillum baldaniorum Sp245 by adjacent chromosomal genes, are involved in mechanosensing and mechanotransduction. We used Sp245 strains and its Fla–Laf– mutant Sp245.1063 (flhB1::Omegon-Km). To construct A. baldaniorum derivatives with an increased dose of CDS AZOBR_150176 (HSHK–RR), this sequence of strain Sp245 was cloned in the pRK415 expression vector. The resulting structure was transferred to Sp245 and Sp245.1063 bacteria. Cell morphology was studied using phase-contrast and transmission electron microscopy. The relative amount of biofilm biomass was determined by staining the bacteria with a crystal violet. Derivatives of the Sp245 strain and its immotile mutant Sp245.1063 (flhB1::Omegon-Km) with an increased dose of HSHK–RR were obtained. It was found that a mutation in the gene encoding the FlhB protein or an increase in the copy number of the gene encoding HSHK–RR affect the motility and ultrastructure of cells, the dynamics of changes in cell size and the flagellation when changing the mechanical properties of the medium. Primary data have been obtained indicating that the cell membrane-associated FlhB proteins and HSHK–RR are involved in the perception of changes in the mechanical properties of the medium and in the transmission of the corresponding mechanical signals in A. baldaniorum cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Microorganisms live in different habitats, which are heterogeneous and characterized by dynamic physicochemical and mechanical properties [1]. Bacteria are subject to diverse mechanical stimuli (for example, in a fluid flow or in contact with biotic and abiotic viscous or solid surfaces). Bacterial responses to these stimuli include changes in motility, transition from free swimming to swarming, and adhesion to a surface, with formation of microcolonies and biofilms [1, 2].
The plant-growth-promoting bacteria of the genus Azospirillum used in agricultural biotechnology are adapted to inhabit diverse natural environments and display versatile genetic and phenotypic plasticity [3]. Thus, some Azospirillum strains, including Azospirillum baldaniorum Sp245 (originally identified as Azospirillum brasilense [4]), show a slight but statistically significant change in the cell length, the flagellation (synthesize only constitutive polar flagellum [Fla] or inducible lateral flagella [Laf]) and mode of living (swimming, swarming, or formation of colonies and biofilms) in response to a change in the medium density [5, 6]. However, the mechanisms involved in the perception of mechanical signals and the response to such stimuli have been studied insignificantly in these as well as other microorganisms. At the same time, some data indicate that the flagella, which first come into contact with the microenvironment of bacteria, perform mechanosensory functions [2]. A comparative analysis of biofilm formation by the A. baldaniorum Sp245 strain and its Fla– derivative strains revealed a negative effect of Fla loss on biomass accumulation in biofilms of mutants formed at the interphase between solid and liquid media, as well as on the stability of biofilms grown with hydrodynamic stress [7]. Restoration of Fla formation in the complemented Fla– mutant restored biomass accumulation and biofilm stability [6].
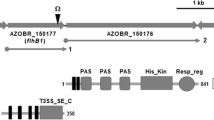
All bacterial flagella share a basic conserved structure and consist of a basal body, a hook, and a filament. The basal body is the anchoring site for a flagellum within the cell membrane, it acts as a rotary motor and export machinery. The basal body includes basal body rings, rod proteins, and a type 3 secretion system [1, 2]. The FlhB protein is a key component of the secretion system. The Sp245 strain genome contains two putative flhB genes located in the chromosome and the AZOBR p4 plasmid (accession nos. HE577327–HE577333). We established that the flhB1 chromosomal gene (the AZOBR_150177 locus) is necessary for the formation of both Fla and Laf [8]. It was found that the flhB1 insertion mutant Sp245.1063 lost the capacity to decrease the cell length in response to transfer from agar to liquid medium and vice versa, while this capacity to elicit a morphological response to changes in medium density was restored in the Sp245.1063 (pRK415–flhB1) complemented mutant [8].
We hypothesize that perception of changes in mechanical stimuli in medium and transmission of mechanical signals in A. baldaniorum cells involve integral membrane proteins: FlhB (a component of the type 3 secretion apparatus located in the basal body of Fla) and a putative multisensory hybrid histidine kinase—response regulator (HSHK–RR). These proteins are encoded by the adjacent coding sequences (CDSs) AZOBR_150177 (flhB1) and AZOBR_150176 (HSHK–RR) in a chromosome cluster of putative flagellar genes of the A. baldaniorum Sp245 type strain. Homologues of these CDSs have the same adjacent location in the genomes of a range of other Azospirillum strains.
MATERIALS AND METHODS
Bacterial Strains, Nutrient Media, and Cultivation Conditions
The bacterial strains and plasmids used in the work are shown in Table 1. Bacteria were grown in a malate-salt medium (MSM [12]) or Luria–Bertani medium (LB [13]) at 30°C. When necessary, kanamycin (Km, 30 μg/mL) and tetracycline (Tc, 25 μg/mL) antibiotics were added to the medium. The concentration of bacto agar in solid media was 20 g/L.
Planktonic cultures were grown in liquid media upon intensive stirring (140 rpm) and a temperature of 28°C using the Excella E24 incubator shaker (New Brunswick Scientific, United States). The OD590 of bacterial cultures was measured every 2 h. Phase contrast and transmission electron microscopy were performed.
Construction of A. baldaniorum Derivatives with an Increased Dose of CDS AZOBR_150176
To determine the significance of CDS AZOBR_ 150176 for the processes determining cell morphology, flagellation, and bacterial behavior, this sequence of the Sp245 strain was cloned into the pRK415 expression vector. Forward (F) 150176HF (CCAAGCTTCGAACGGCTGACCTGGAGT) and reverse (R) 150176ER (GGGAACCGAATTCTGGGCCTATCACG) primers were used for PCR, which contained HindIII (H) or EcoRI (E) restriction sites. CDS was cloned behind the lac promoter of the pRK415 plasmid. The resulting structure was transferred to A. baldaniorum using a three-parent crossing (Table 1).
Analysis of Formation and Microstructure of Azospirillum Biofilms
The 18-h Azospirillum cultures grown in a liquid MSM were diluted with a sterile medium to values of OD590 = 0.05–0.10, placed into glass tubes (2 mL in each) and incubated for 6 days at 30°C under stationary conditions. In Azospirillum, by the sixth day of incubation, the relative amount of biofilm biomass is stabilized and the formation is completed [7]. To estimate the relative amount of biomass in biofilms, bacteria were stained with crystal violet [14]. Phase-contrast and transmission electron microscopy of biofilms and individual cells was performed using the Leica DM6000 B (Leica-Microsystems, Germany) and Libra 120 microscopes (Carl Zeiss, Germany). Detailed protocols for the preparation of microscopy specimens and their analysis can be found in [7].
Statistical Analysis
At least three independent experiments were performed, at least in two repeats. Statistical data were analyzed using the Student’s t-test (95% confidence intervals are given) and one-factor analysis of variance (ANOVA) (with a significance level of p ≤ 0.05). The data were analyzed using the Microsoft Office Excel 2010 software.
RESULTS AND DISCUSSION
Dynamics of Changes in Cell Length and Flagellation after Transfer of Bacteria from Agar to Liquid Culture Medium
To find an answer to whether CDS AZOBR_ 150176 (HSHK–RR) plays any role in the assembly of flagella and/or adaptation of bacteria to medium density, we first increased the dose of this sequence in the cells of Sp245 and Sp245.1063 strains by introducing the pRK415-150176 plasmid. We compared the growth of A. baldaniorum Sp245 strains, its Fla–Laf– mutant Sp245.1063, and their derivatives (Table 1) in liquid media and showed that, upon intensive stirring, their planktonic cultures were in a stationary growth phase in 24 h of incubation. The density of bacterial culture doubled in 2.5–3 h. The Sp245 and Sp245(pRK415-150176) cells of daily cultures carry Fla at one of the cell poles, which about 86.2 ± 2.4% of bacteria use to swim along rectilinear trajectories at a swimming speed of 29.2 ± 1.9 μm/s and with random direction variations. The Sp245.1063 and Sp245.1063(pRK415-150176) cells were immotile. On agar media, all strains formed colonies in 48 h of cultivation. Bacteria Sp245 and Sp245(pRK415-150176) additionally synthesized numerous Laf (phenotype Fla+Laf+). The cells of the Sp245.1063 mutant and its derivative Sp245.1063 (pRK415-150176) lack both types of flagella regardless of the cultivation conditions.
Next, we investigated a change in flagellation upon transfer of Sp245 and Sp245(pRK415-150176) Fla+Laf+ cells from agar medium to liquid. In 3 h of cultivation, in Sp245 culture, 26% of bacteria retained Fla and Laf, 65% of bacteria synthesized only Fla, and 9% of cells had no flagella. In the case of Sp245(pRK415-150176), 67% of cells had Fla and Laf, 26% had only Fla, and 7% did not synthesize flagella. In 6 h of cultivation, the ratio of (Fla+Laf+)/(Fla+)/(Fla–) cells in Sp245(pRK415-150176) became similar to the values characteristic of Sp245 culture at 3 h of cultivation, i.e., 32% of the derivative cells had Fla and Laf, 61% carried only Fla, and 7% did not synthesize flagella. In the Sp245 strain, in 6 h, the ratio of (Fla+Laf+)/(Fla+)/(Fla–) cells was 0/92/8%, respectively. In 18-h (growing planktonic culture enters the stationary growth phase) and 24-h (stationary growth phase) cultures, 93% of Sp245 bacteria formed Fla and 7% had no flagella; in the case of Sp245(pRK415-150176), 94% of cells had Fla and 6% of cells lacked flagella. In 48 h of cultivation in planktonic cultures of Sp245 and Sp245(pRK415-150176), flagellation did not change significantly.
In addition, we evaluated a change in the length of intact cells upon transfer of bacteria from agar to liquid medium. In the Sp245.1063 strain, planktonic cells were 20% longer than Sp245 and Sp245(pRK415-150176) cells. On solid media, the size of Sp245 and Sp245.1063 cells did not differ, and the cell length of Sp245(pRK415-150176) was 30% shorter than that of the parent strain and the mutant. Three to six hours after inoculation from solid medium into a liquid culture, the cell size of all studied strains did not change significantly. In 18- and 24-h Sp245 planktonic cultures, the cell length was 18% shorter than the cell length of this strain grown in agar media. The Sp245(pRK415-150176)and Sp245.1063 cells did not change the cell size upon transfer from solid to liquid medium.
Dynamics of Biofilm Formation, Change in Cell Length, and Flagellation upon Transition from Planktonic Cell Growth to a Biofilm State (Biofilm Population)
The process of biofilm formation by the strains under study at the interface of liquid and solid hydrophilic surface includes cell adsorption and adhesion (2–3 days) and growth and stabilization of biofilm biomass (6–7 days, mature biofilms). At the stages of adsorption and adhesion, all the strains under study developed approximately equal amounts of bacterial biomass on the solid surface. However, in mature biofilms, in the case of Sp245.1063 and Sp245.1063(pRK415-150176), the biomass values were 40–50% lower than the values characteristic of Sp245 and Sp245(pRK415-150176).
We determined the length and width of biofilm-embedded bacteria cells at stages of their adsorption/adhesion (2-day biofilms) and in mature biofilms (7-day cultivation) and compared with the size of bacteria grown as planktonic cultures or in agar medium (Table 2). Integration of Sp245 planktonic cells into biofilms increased cell length as early as at the stage of bacterial adsorption/adhesion to the size of bacteria of this strain grown on a solid culture medium. In turn, the cell length and width of Sp245 cells did not change significantly in mature biofilms. The width of bacteria in biofilms did not change significantly over entire cultivation.
The Sp245 bacteria that exchanged a planktonic mode of growth for a biofilm lifestyle (interface at solid surface/liquid medium) increased the cell length to the sizes of cells grown in a solid agar medium (interface at solid medium/air) (Table 2). The length of Fla–Laf– flhB1::Omegon-Km cells of the Sp245.1063 mutant in biofilms, planktonic cultures, and those grown on a solid medium was equal to the size of Sp245 bacteria grown on a solid agar medium; in the case of Sp245(pRK415-150176) and Sp245.1063(pRK415-150176), the cell length was equal to the size of the parent-strain cells grown in a liquid medium (Table 2).
The change in flagellation upon transition of Fla+ cells of Sp245 and Sp245(pRK415-150176) strains from a planktonic culture to a biofilm lifestyle (with the cells being attached on a solid surface under a layer of liquid MSM) was investigated. It is noted that Laf was not detected on cells from biofilm of all the studied strains using transmission electron microscopy in both these experiments (daily planktonic cultures were used for inoculation into a liquid medium) and in our previous works [6, 7]. In the case in which Fla+Laf+ cells from a solid medium were used for inoculation into liquid MSM, bacteria with Laf were neither detected in mature biofilms. Fla was present in Sp245 and Sp245(pRK415-150176) cells in biofilms at all stages of biofilm population formation. Sp245.1063 and Sp245.1063(pRK415-150176) cells lacked Fla or Laf, similar to planktonic bacteria.
In summary, upon transition of Sp245 bacteria from planktonic cultures to a biofilm mode of life (at the interface of solid surface/liquid medium), the cell length increases to sizes characteristic of strains grown in agar culture medium (at the interface of solid medium/air). The lack of Laf is a significant difference of biofilm cells from bacteria grown on agar medium.
CONCLUSIONS
It is noted that the introduction of the pRK415 vector plasmid into Sp245 and Sp245.1063 did not influence cell morphology, motility, and capacity to form biofilms by these strains. The introduction of pRK415-150176 did not eliminate defects in flagellation and motility of the Sp245.1063 mutant strain. However, an increase in the HSHK–RR gene dose in Sp245 and Sp245.1063 (strains Sp245(pRK415-150176) and Sp245.1063(pRK415-150176)) influences the ability of cells to change size in response to a change of medium density. It was previously found that a Fla–Laf– flhB1::Omegon-Km Sp245.1063 mutant lost the ability to decrease cell size, and, in а Sp245.1063(pRK415–flhB1) mutant, this ability to elicit a morphological response to changes in medium density was restored [8]. In the case of Sp245, an additional copy of CDS AZOBR_150176 affects the dynamics of changes in flagellation upon transition of Fla+Laf+ cells to a planktonic lifestyle (Fla+ phenotype). Hence, FlhB1 and HSHK-RR proteins located in the cell membrane—are involved in the perception of changes in mechanical stimuli of the medium and transmit corresponding mechanical signals in A. baldaniorum cells.
REFERENCES
Dufrêne, Y.F. and Persat, A., Mechanomicrobiology: How bacteria sense and respond to forces, Nat. Rev. Microbiol., 2020, vol. 18, no. 4, pp. 227–240. https://doi.org/10.1038/s41579-019-0314-2
Fajardo-Cavazos, P. and Nicholson, W.L., Mechanotransduction in prokaryotes: A possible mechanism of spaceflight adaptation, Life, 2021, vol. 11, no. 1, p. 33. https://doi.org/10.3390/life11010033
Fibach-Paldi, S., Burdman, S., and Okon, Y., Key physiological properties contributing to rhizosphere adaptation and plant growth promoting abilities of Azospirillum brasilense, FEMS Microbiol. Lett., 2012, vol. 326, no. 2, pp. 99–108. https://doi.org/10.1111/j.1574-6968.2011.02407.x
Dos Santos Ferreira, N., Hayashi Sant’ Anna, F., Massena Reis, V., Ambrosini, A., Gazolla Volpiano, C., Rothballer, M., et al., Genome-based reclassification of Azospirillum brasilense Sp245 as the type strain of Azospirillum baldaniorum sp. nov., Int. J. Syst. Evol. Microbiol., 2020, vol. 70, no. 12, pp. 6203–6212. https://doi.org/10.1099/ijsem.0.004517
Moens, S., Michiels, K., Keijers, V., Van Leuven, F., and Vanderleyden, J., Cloning, sequencing and phenotypic analysis of laf1, encoding flagellin of the lateral flagella of Azospirillum brasilense Sp7, J. Bacteriol., 1995, vol. 177, no. 19, pp. 5419–5426. https://doi.org/10.1128/jb.177.19.5419-5426.1995
Shelud’ko, A.V., Filip’echeva, Y.A., Telesheva, E.M., Yevstigneeva, S.S., Petrova, L.P., and Katsy, E.I., Polar flagellum of the alphaproteobacterium Azospirillum brasilense Sp245 plays a role in biofilm biomass accumulation and in biofilm maintenance under stationary and dynamic conditions, World J. Microbiol. Biotechnol., 2019, vol. 35, no. 2, p. 19. https://doi.org/10.1007/s11274-019-2594-0
Shelud’ko, A.V., Filip’echeva, Yu.A., Shumilova, E.M., Khlebtsov, B.N., Burov, A.M., Petrova, L.P., and Katsy, E.I., Changes in biofilm formation in the nonflagellated flhB1 mutant of Azospirillum brasilense Sp245, Microbiology (Moscow), 2015, vol. 84, no. 2, pp. 144–151. https://doi.org/10.1134/S0026261715010129
Filip’echeva, Yu., Shelud’ko, A., Prilipov, A., Telesheva, E., Mokeev, D., Burov, A., et al., Chromosomal flhB1 gene of the alphaproteobacterium Azospirillum brasilense Sp245 is essential for correct assembly of both constitutive polar flagellum and inducible lateral flagella, Folia Microbiol., 2018, vol. 63, no. 2, pp. 147–153. https://doi.org/10.1007/s12223-017-0543-6
Baldani, V.L.D., Baldani, J.I., and Döbereiner, J., Effects of Azospirillum inoculation on root infection and nitrogen incorporation in wheat, Can. J. Microbiol., 1983, vol. 29, no. 8, pp. 924–929. https://doi.org/10.1139/m83-148
Figurski, D.H. and Helinski, D.R., Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans, Proc. Natl. Acad. Sci. U. S. A., 1979, vol. 76, no. 4, pp. 1648–1652. https://doi.org/10.1073/pnas.76.4.1648
Keen, N.T., Tamaki, S., Kobayashi, D., and Trollinger, D., Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria, Gene, 1988, vol. 70, no. 1, pp. 191–197. https://doi.org/10.1016/0378-1119(88)90117-5
Döbereiner, J. and Day, J.M., Associative symbiosis in tropical grass: Characterization of microorganisms and dinitrogen fixing sites, Proc. 1st International Symposium on Nitrogen Fixation, Pullman, WA, 1974, Newton, W.E. and Nyman, C.J., Eds., Washington, DC: Washington State Univ. Press, 1976, pp. 518–538.
Molecular Cloning: A Laboratory Manual, Sambrook, J., Fritsch, E.F., and Maniatis, T., Eds., New York: Cold Spring Harbor Laboratory Press, 1989.
O’Toole, G.A. and Kolter, R., Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: A genetic analysis, Mol. Microbiol., 1998, vol. 28, no. 3, pp. 449–461. https://doi.org/10.1046/j.1365-2958.1998.00797.x
ACKNOWLEDGMENTS
The authors thank the Simbioz Center for the Collective Use of Research Equipment in the Field of Physical-Chemical Biology and Nanobiotechnology, Institute of Biochemistry and Physiology of Plants and Microorganisms, Russian Academy of Sciences for kindly providing access to the Leica DM6000 B and Libra 120 equipment.
Funding
This study was partially supported by the Russian Foundation for Basic Research, project no. 20-04-00006_а.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest. The authors declare that they have no conflict of interest.
This article does not contain any studies involving animals or human beings performed by any of the authors.
Additional information
Translated by M. Novikova
About this article
Cite this article
Evstigneeva, S.S., Mokeev, D.I., Petrova, L.P. et al. Genetic Aspects of Mechanosensitivity in the Alphaproteobacteria Azospirillum baldaniorum with Mixed Flagellation. Mol. Genet. Microbiol. Virol. 37, 86–90 (2022). https://doi.org/10.3103/S0891416822020045
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0891416822020045