Abstract
Azospirillum brasilense has the ability of swimming and swarming motility owing to the work of a constitutive polar flagellum and inducible lateral flagella, respectively. The interplay between these flagellar systems is poorly understood. One of the key elements of the flagellar export apparatus is the protein FlhB. Two predicted flhB genes are present in the genome of A. brasilense Sp245 (accession nos. HE577327–HE577333). Experimental evidence obtained here indicates that the chromosomal coding sequence (CDS) AZOBR_150177 (flhB1) of Sp245 is essential for the production of both types of flagella. In an flhB1:: Omegon-Km mutant, Sp245.1063, defects in polar and lateral flagellar assembly and motility were complemented by expressing the wild-type flhB1 gene from plasmid pRK415. It was found that Sp245.1063 lost the capacity for slight but statistically significant decrease in mean cell length in response to transfer from solid to liquid media, and vice versa; in the complemented mutant, this capacity was restored. It was also shown that after the acquisition of the pRK415-harbored downstream CDS AZOBR_150176, cells of Sp245 and Sp245.1063 ceased to elongate on solid media. These initial data suggest that the AZOBR_150176-encoded putative multisensory hybrid sensor histidine kinase–response regulator, in concert with FlhB1, plays a role in morphological response of azospirilla to changes in the hardness of a milieu.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Azospirilla are found worldwide in diverse habitats (Baldani et al. 2014 and references therein). All Azospirillum species swim by rotation of a constitutive polar flagellum (Fla). At a certain density of a medium (containing, e.g., ≥ 0.35% Bacto agar), some species, including Azospirillum brasilense, also assemble numerous lateral flagella (Laf) used for swarming motility (Moens et al. 1995). Assembly of both Fla and Laf is blocked in A. brasilense mutants in the rpoN gene of the σ54 factor of RNA polymerase (Milcamps et al. 1996) or in the flbD gene of the putative σ54-dependent transcriptional regulator (Chang et al. 2007). This indicates that some elements are shared in the regulation of Fla and Laf systems.
In several bacterial species with mixed flagellation, Fla is thought to control the assembly of Laf (McCarter et al. 1988; Moens et al. 1996). For example, in A. brasilense Sp7, expression of the laf1 flagellin gene is induced under conditions of difficulties in the Fla rotation (Moens et al. 1996). On the other hand, in A. brasilense Sp245 and in Rhodospirillum centenum, unswarming mutants have been isolated that still elaborate inducible Laf, although their Fla is paralyzed or absent (Jiang et al. 1998; Scheludko et al. 1998). Thus, the signaling mechanism used by A. brasilense to induce Laf assembly seems more complicated than just hindrance in the Fla rotation. Conversely, Fla− mutants of the gammaproteobacteria Vibrio parahaemolyticus and Aeromonas hydrophila are still able to swarm owing to Laf (McCarter et al. 1988; Canals et al. 2006).
Bacterial flagella consist of three main parts: basal body → hook → filament. Within the basal body is located a type III secretion system that exports proper flagellar proteins through a central channel of the flagellum. This export apparatus is encoded by the flhA, flhB, fliO, fliP, fliQ, fliR, fliH, fliI, and fliJ genes. Conserved protein FlhB consists of a transmembrane N-terminal domain, a cytoplasmic C-terminal domain, and a lithe linker between them. The C-terminal domain of FlhB is essential for substrate binding and for substrate-specificity switching in the export apparatus to ensure that the flagellar rod and hook proteins are exported before the proteins of the hook-filament linkage and the filament (Fraser et al. 2003; Ferris et al. 2005; Minamino 2014).
In the genome of A. brasilense Sp245, predicted flagellar genes are located on the chromosome and several plasmids (Wisniewski-Dyé et al. 2011). Previously, we obtained mutant Sp245.1063 with a single insertion of Omegon-Km (Fellay et al. 1989) in the chromosomal CDS AZOBR_150177 (flhB1 gene). The mutant was unexpectedly found to be defective in both Fla and Laf formation (despite the presence of another flhB gene in AZOBR_p4) and, thus, in swimming and swarming (Kovtunov et al. 2013). Downstream of flhB1, the CDS AZOBR_150176 is located, the product of which is a hypothetical hybrid sensor histidine kinase/response regulator (HSHK–RR). No other predicted genes of the two-component sensor-regulator systems are found in the vicinity of AZOBR_150176 (Wisniewski-Dyé et al. 2011).
In the chromosomes of some other A. brasilense strains (e.g., accession nos. CP012914 and CP007793), homologs of AZOBR_150177 and AZOBR_150176 are also contiguous and may be functionally connected. Because no experiments on the complementation of Sp245.1063 have been done, the importance of AZOBR_150177 or AZOBR_150176 for the assembly of Fla and Laf remained unproven. For this reason, the aim of our study was to assess the impact of the acquisition of the cloned Sp245 CDSs AZOBR_150177 and AZOBR_150176 on cell morphology, flagellation, and motility of the corresponding derivatives of Sp245.1063.
Materials and methods
Strains, plasmids, and primers used in this work are listed in Table 1. A scheme for the DNA region of Sp245 altered in Sp245.1063 is shown in Fig. 1a. DNA manipulations and E. coli DH5α transformation were performed according to standard procedures (Sambrook et al. 1989). The correctness of the constructs pRK415-150177 and pRK415-150176 was verified by DNA sequencing. The databases and the programs on the servers of the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) and InterPro (https://www.ebi.ac.uk/interpro/) were used to retrieve and analyze nucleotide and amino acid sequences.
a Scheme for the A. brasilense Sp245 DNA region, altered in Fla− Laf− mutant Sp245.1063. The thick arrows designate the CDS. The triangle shows the location of Omegon-Km (Ω) in Sp245.1063. The thin arrows indicate the segments cloned in pRK415. b Domain organization of the predicted products of AZOBR_150177 (FlhB1) and AZOBR_150176 (HSHK–RR). T3SS_SE_C type III secretion system substrate exporter, C-terminal (IPR029025), PAS PAS sensor domains (IPR000014), His_Kin histidine kinase domain (IPR005467), Resp_reg signal transduction response regulator, receiver domain (IPR001789). Black rectangles designate predicted transmembrane regions
Triparental matings of E. coli DH5α harboring pRK415, pRK415-150177, or pRK415-150176, E. coli K802 (pRK2013), and A. brasilense were used to transfer pRK415 or its derivative to azospirilla (helper narrow-host-range plasmid pRK2013 was not maintained in Azospirillum). Transconjugant clones were selected and purified on a solid malate-salt medium (MSM) (Döbereiner and Day 1976) supplemented with NH4Cl (1 g/L), Tc (25 μg/mL) and, in the case of Sp245.1063, kanamycin (Km; 30 μg/mL). Plasmid profiles of azospirilla were checked by the Eckhardt (1978) procedure.
Azospirilla were grown at 28 °C. For growth analyses, overnight broth cultures were inoculated in triplicate in flasks with a fresh medium containing appropriate supplements to an initial absorbance at 590 nm (A 590) of 0.055 and were incubated with shaking at 140 rpm. The A 590 value was recorded every 2 h.
Phase contrast and transmission electron microscopy of bacteria was done with a Leica DM6000 B (Leica Microsystems, Germany) and a Libra 120 (Carl Zeiss, Germany) microscopes at the Symbiosis Center for the Collective Use of Research Equipment (IBPPM RAS; Saratov, Russia). Flagellation was examined in at least 200 cells of each strain.
Bacterial movement in media was examined under a Jenaval phase-contrast microscope and was video recorded with a DCR-TRV900E digital camera (Sony, Japan). The mean swimming motility speed of 40–90 single cells was calculated as described earlier (Schelud’ko et al. 2009). Soft agar plates, supplemented with 0.4–0.6% Bacto agar, were point inoculated with 42-h bacterial cultures from solid media. The morphology of the bacterial spreading zones on the soft agar plates was inspected by eye and by phase contrast microscopy.
Data on the cell lengths of azospirilla were collected from at least three experiments. Transmission electron microscopy images of the cells were taken at random within an individual grid. Several grids were analyzed in each experiment. The lengths of all the cells within the field of view were measured only when both cell poles were clearly discernible. A minimum of 56 cells per sample were taken for measurements from different fields of view. Because of the technical obstacles, cell lengths of azospirilla grown in semisolid media were not estimated.
All quantitative studies were based on at least three experiments done in triplicate. Quantitative data (processed with Microsoft Office Excel 2010) are presented in Tables as means ± confidence intervals for a 95% significance level. Statistical analyses were done with unpaired Student’s t test. Differences were considered significant at the P < 0.05 level.
Results and discussion
In A. brasilense Sp245.1063, the Omegon-Km insertion is located after the 924th bp of flhB1 (translation product accession no. CCC98759) and the C-terminal domain of the mutant FlhB1 protein is expected to be truncated by 50 amino acids (Kovtunov et al. 2013). Although a minor percentage of Sp245.1063 cells somehow overcame the negative effect of the truncation of the C-end of FlhB1, managing to assemble a long or short single polar organelle (Table 2), any motile cells or cells with Laf were never found in this mutant or its derivatives harboring the empty vector pRK415.
The CDS AZOBR_150176 for the putative HSHK–RR (accession no. CCC98758) is located at a distance of 51 bp from the 3′-end of flhB1 and is transcribed in the same direction (Fig. 1a). The existence of three PAS domains within the sensory part of this HSHK–RR (Fig. 1b), which may sense a wide range of signals (IPR000014; Henry and Crosson 2011), suggests that this is a multisensory protein. The response regulator receiver domain (IPR001789) of HSHK–RR belongs to the CheY-like superfamily, which includes not only chemotaxis proteins but also a wide range of other regulators (IPR011006; Galperin 2006).
To prove that the Fla− Laf− phenotype of Sp245.1063 is caused by flhB1 truncation and not by a polar effect on AZOBR_150176, we complemented the mutant with pRK415 possessing the CDS AZOBR_150177. As a first step toward understanding whether AZOBR_150176 plays any role in flagellar assembly and/or bacterial adaptation to the stiffness of the milieu, we increased its dosage in azospirilla by transfer of plasmid pRK415-150176.
The flagellation and motility deficiencies in Sp245.1063 (pRK415-150176) resembled those in Sp245.1063 and Sp245.1063 (pRK415) and were clearly different from the flagellation and motility features of Sp245, Sp245 (pRK415), and Sp245 (pRK415-150176) (Fig. 2; Tables 2 and 3). Thus, the acquisition of an additional copy of AZOBR_150176 did not restore Fla or Laf assembly in Sp245.1063 and had no detectable effect on the swimming motility of Sp245 in liquids or in aquatic channels within semisolid media.
The acquisition of pRK415 or pRK415-150176 did not alter the swarming proficiency of Sp245 or the swarming deficiency of Sp245.1063. As compared to Sp245 and Sp245 (pRK415), strain Sp245 (pRK415-150176) tended to form somewhat wider swarm colonies on soft agar (Fig. 3).
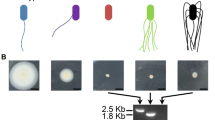
Behavior of A. brasilense strains on soft agar plates. Cultures of strains Sp245 (a), Sp245.1063 (b), Sp245.1063 (pRK415-150177) (c), Sp245.1063 (pRK415-150176) (d), Sp245 (pRK415-150176) (e), Sp245 (pRK415) (f), and Sp245.1063 (pRK415) (g) were spot inoculated on MSM with 0.6% Bacto agar and incubated for 72 h. Bar corresponds to 10 mm
In Sp245.1063 (pRK415-150177), normal constitutive Fla and inducible Laf were restored. On solid media, its cells also possessed numerous Laf in addition to Fla (Fig. 2, Table 2). After the introduction of pRK415-150177 into Sp245.1063, single-cell swimming speed (Table 3) and swarming of the bacteria on the media with 0.6% Bacto agar were completely repaired (Fig. 3). These data demonstrate that the chromosomal flhB1 gene is essential for the proper assembly of Fla and Laf in Sp245 and that the defects in flagellation and motility of Sp245.1063 are not due to a polar effect of Omegon-Km on AZOBR_150176.
The restoration of the wild-type Fla structure in Sp245.1063 (pRK415-150177) could repair the hypothetical mechanosensing apparatus necessary for the inducible assembly of Laf. On the other hand, the above-described effects of the acquisition of flhB1 on the flagellation of Sp245.1063 might also be explained by the involvement of FlhB1 in the construction of both types of flagella. Thus, the subject of future research will be to analyze the consequences of inactivation of another predicted flhB gene of Sp245 – AZOBR_p410073 (for a putative protein with 43% identity and 61% similarity to FlhB1), located on plasmid AZOBR_p4 within the predicted cluster of Laf genes (accession no. HE577331).
The acquisition of pRK415, pRK415-150176, or pRK415-150177 by Sp245 and Sp245.1063 did not affect their equal growth rates. As expected, the cell sizes of all strains under study grown on liquid or solid media ranged widely. However, on average, cells of Sp245 from broth cultures were slightly shorter than its cells from colonies formed on solids. This difference in average cell lengths of the bacteria from planktonic and immobilized communities was statistically significant. Regardless of the solidity of the milieu, flhB1 mutant Sp245.1063 produced cells whose mean length equaled that of Sp245 cells from solid media (Table 4).
For unclear reasons, cells of Sp245.1063 (pRK415) tended to become a bit shorter than the Sp245.1063 cells; however, their mean lengths still did not vary after the bacteria were transferred between media with different stiffness. Although cells of Sp245.1063 (pRK415-150177) from broth cultures remained a bit longer than the cells of Sp245 or Sp245 (pRK415), its ability to respond to transfer to solid media by a slight but statistically significant cell elongation was restored (Table 4).
The acquisition of the pRK415-borne CDS AZOBR_150176 affected cell size in both Sp245 and Sp245.1063 derivatives. Strain Sp245 (pRK415-150176) stopped to respond to the exterior stiffness by slight cell elongation, and so did Sp245.1063 (pRK415-150176), the mean cell length of which remained the same as in planktonic Sp245 cells independently of the solidity of the milieu (Table 4).
Thus, our initial work proposed that AZOBR_150176 and AZOBR_150177 play a role in the morphological response of A. brasilense Sp245 to shifts in the hardness of a milieu.
In the closely related strain A. brasilense Sp7, one of the four predicted chemotactic pathways, Che1, affects, via unknown mechanisms, not only transient increases in swimming speed during bacterial responses to chemoattractants, but also cell length (Bible et al. 2008). The histidine kinase CheA1 of the Che1 pathway was recently found to be produced in two isoforms (Gullett et al. 2017). A cytoplasmic isoform, which is similar to prototypical CheA, is localized at the cell poles of strain Sp7. A membrane-anchored isoform, which is produced as a fusion with an N-terminal conserved transmembrane domain of unknown function (TMX), is distributed throughout the cell surface. Results of the mutagenesis of the TMX domain showed its involvement in regulating changes in the average cell length. How the two CheA1 isoforms are formed in strain Sp7 is unknown (Gullett et al. 2017). The N-terminal TMX domain of the CheA1 isoform from A. brasilense Sp7 is a strain-specific feature not found in other Azospirillum strains with sequenced genomes, including A. brasilense Sp245. Moreover, mutations in the Sp245 cheA1 homolog had no effect on the average cell length of this strain (Gullett et al. 2017).
Conclusion
Here, we have proven that in A. brasilense Sp245, the chromosomal CDS AZOBR_150177 (flhB1) is essential for the production of both Fla and Laf. The acquisition of the pRK415-borne wild-type flhB1 gene by the flhB1 mutant Sp245.1063 led to the restoration of proper bacterial flagellation and motility, as well as slight cell shortening on liquid media. Initial experimental data on the contiguous CDS AZOBR_150176 suggested that it also affects bacterial responses to shifts in steadiness of the milieu. Further studies with mutants in AZOBR_150176 are expected to provide a deeper understanding of the Sp245’s adaptation to changes in the hardness and other features of a milieu.
References
Baldani JI, Videira SS, Teixeira KRDS et al. (2014) The family Rhodospirillaceae. In: Rosenberg E, DeLong EF, Lory S et al (eds) The prokaryotes: alphaproteobacteria and betaproteobacteria. Springer, Berlin, pp 533–618. doi:10.1007/978-3-642-30197-1_300
Baldani VLD, Baldani JI, Döbereiner J (1983) Effects of Azospirillum inoculation on root infection and nitrogen incorporation in wheat. Can J Microbiol 29:924–929. doi:10.1139/m83-148
Bible AN, Stephens BB, Ortega DR et al (2008) Function of a chemotaxis-like signal transduction pathway in modulating motility, cell clumping, and cell length in the alphaproteobacterium Azospirillum brasilense. J Bacteriol 190:6365–6375. doi:10.1128/JB.00734-08
Canals R, Ramirez S, Vilches S et al (2006) Polar flagellum biogenesis in Aeromonas hydrophila. J Bacteriol 188:542–555. doi:10.1128/JB.188.2.542–555.2006
Chang Y, Tang T, Li JL (2007) Isolation of a flagellar operon in Azospirillum brasilense and functional analysis of FlbD. Res Microbiol 158:521–528. doi:10.1016/j.resmic.2007.04.005
Döbereiner J, Day JM (1976) Associative symbiosis in tropical grass: characterization of microorganisms and dinitrogen fixing sites. In: Newton WE, Nijmans CJ (eds) Symposium on nitrogen fixation. Washington State University Press, Pullman, pp 518–538
Eckhardt T (1978) A rapid method for the identification of plasmid deoxyribonucleic acid in bacteria. Plasmid 1:584–588. doi:10.1016/0147-619X(78)90016-1
Fellay R, Krisch HM, Prentki P, Frey J (1989) Omegon-Km: a transposable element designed for in vivo insertional mutagenesis and cloning of genes in gram-negative bacteria. Gene 76:215–226. doi:10.1016/0378-1119(89)90162-5
Ferris HU, Furukawa Y, Minamino T et al (2005) FlhB regulates ordered export of flagellar components via autocleavage mechanism. J Biol Chem 280:41236–41242. doi:10.1074/jbc.M509438200
Figurski DH, Helinski DR (1979) Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A 76:1648–1652. doi:10.1073/pnas.76.4.1648
Fraser GM, Hirano T, Ferris HU et al (2003) Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol Microbiol 48:1043–1057. doi:10.1046/j.1365-2958.2003.03487.x
Galperin MY (2006) Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol 188:4169–4182. doi:10.1128/JB.01887-05
Gullett JM, Bible A, Alexandre G (2017) Distinct domains of CheA confer unique functions in chemotaxis and cell length in Azospirillum brasilense Sp7. J Bacteriol 199:e00189-17. doi:10.1128/JB.00189-17
Henry JT, Crosson S (2011) Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol 65:261–286. doi:10.1146/annurev-micro-121809-151631
Jiang Z-Y, Rushing BG, Bai Y et al (1998) Isolation of Rhodospirillum centenum mutants defective in phototactic colony motility by transposon mutagenesis. J Bacteriol 180:1248–1255
Keen NT, Tamaki S, Kobayashi D, Trollinger D (1988) Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191–197. doi:10.1016/0378-1119(88)90117-5
Kovtunov EA, Petrova LP, Shelud’ko AV, Katsy EI (2013) Transposon insertion into a chromosomal copy of flhB gene is concurrent with defects in the formation of polar and lateral flagella in bacterium Azospirillum brasilense Sp245. Russ J Genet 49:881–884. doi:10.1134/S1022795413080061
McCarter L, Hilmen M, Silverman M (1988) Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell 54:345–351. doi:10.1016/0092-8674(88)90197-3
Milcamps A, Van Dommelen A, Stigter J et al (1996) The Azospirillum brasilense rpoN gene is involved in nitrogen fixation, nitrate assimilation, ammonium uptake, and flagellar biosynthesis. Can J Microbiol 42:467–478. doi:10.1139/m96-064
Minamino T (2014) Protein export through the bacterial flagellar type III export pathway. Biochim Biophys Acta 1843:1642–1648. doi:10.1016/j.bbamcr.2013.09.005
Moens S, Michiels K, Keijers V et al (1995) Cloning, sequencing, and phenotypic analysis of laf1, encoding the flagellin of the lateral flagella of Azospirillum brasilense Sp7. J Bacteriol 177:5419–5426
Moens S, Schloter M, Vanderleyden J (1996) Expression of the structural gene, laf1, encoding the flagellin of the lateral flagella in Azospirillum brasilense Sp7. J Bacteriol 178:5017–5019
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, second edn. Cold Spring Harbor Laboratory, New York
Scheludko AV, Katsy EI, Ostudin NA et al (1998) Novel classes of Azospirillum brasilense mutants with defects in the assembly and functioning of polar and lateral flagella. Mol Gen Mikrobiol Virusol 4:33–37
Schelud’ko AV, Makrushin KV, Tugarova AV et al (2009) Changes in motility of the rhizobacterium Azospirillum brasilense in the presence of plant lectins. Microbiol Res 164:149–156. doi:10.1016/j.micres.2006.11.008
Wisniewski-Dyé F, Borziak K, Khalsa-Moyers G et al (2011) Azospirillum genomes reveal transition of bacteria from aquatic to terrestrial environments. PLoS Genet 7:e1002430. doi:10.1371/journal.pgen.1002430
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Filip’echeva, Y., Shelud’ko, A., Prilipov, A. et al. Chromosomal flhB1 gene of the alphaproteobacterium Azospirillum brasilense Sp245 is essential for correct assembly of both constitutive polar flagellum and inducible lateral flagella. Folia Microbiol 63, 147–153 (2018). https://doi.org/10.1007/s12223-017-0543-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-017-0543-6