Abstract
Although the specific causes of colorectal cancer (CRC) are not known, a robust DNA repair capacity may decrease the risk of this malignancy. DNA repair capacity may be reduced by alterations of genes involved in DNA repair process. This may affect susceptibility to carcinogenesis. It is hypothesized that single nucleotide polymorphisms (SNPs) of several DNA repair genes may be a risk factor for CRC susceptibility and prognosis. Using PCR–RFLP method, we conducted a case-control study to genotype 291 patients with CRC and 140 healthy individuals to determine variants in the PRKDC, XPD and XRCC1 genes. Results showed that the genotypes of XRCC1 c.580C>T polymorphism were associated with the risk of CRC. Compared with CC, CT (odds ratio (OR) = 5.35, P < 0.001) and CT/TT (OR = 4.74, P < 0.001) as well as T allele (OR = 4.95, P < 0.001) were overrepresented among the CRC patients. Variant genotype CC (OR = 2.37; P = 0.042) and C allele of XPD c.2251A>C (OR = 1.37; P = 0.028) polymorphism, enhanced the risk of CRC cases. Compared with GG, positive association was also obtained for all genotypes (GT, TT, GT/TT) of PRKDC rs7003908; 6721G>T polymorphism with CRC. Moreover, T allele of PRKDC demonstrated significant risk for CRC (OR = 5.61; P < 0.001). Besides, significant relevance of the PRKDC rs7003908; 6721G>T variations to smoking as well as XPD c.2251A>C variations to smoking and alcohol consumption in individuals with CRC was observed. Our findings indicated that genetic polymorphisms of PRKDC, XRCC1, XPD genes may influence susceptibility of CRC in the Iranian population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
DNA repair pathways have a crucial role in maintaining the genome integrity against mutations caused by general DNA replication errors as well as environmental insults. There are multiple DNA repair pathways. Bulky DNA lesions were removed by Nucleotide Excision Repair (NER); damaged bases were repaired by Base excision repair (BER) which links to single strand break (SSB) repair. DNA non-homologous end-joining (NHEJ) as well as Homologous Recombination (HR) pathways repair DNA double strand breaks (DSBs); mismatched base pairs were corrected by Mismatch repair (MMR) [1]. The x-ray repair cross-complementation group 1 (XRCC1) mapped to chromosome 19q13.2–13.3 has an essential role in the BER repair pathway. Although no known enzymatic activity has been reported for XRCC1, there are several crucial repair proteins that it can interact well with through its different domains, such as, DNA polymerase β, DNA ligase III, and PARP (ADP-ribose polymerase) in a complex, to coordinate the processes of BER repair pathway. The BER pathway effectively involves in DNA damage repair caused by a wide range of exogenous and endogenous factors, such as ionizing radiation, oxidation, and alkylating agents [2, 3]. Excision repair cross-complementing group 2/xeroderma pigmentosum group D (ERCC2/XPD), mapped to chromosome 19q13.3, has a vital role in the NER repair pathway via eliminating bulky DNA adducts caused by xenobiotics and environmental toxins [4]. The DNA-dependent protein kinase catalytic subunit (DNA-PKcs; encoded by PRKDC) has a pivotal role in DSBs repair via the NHEJ pathway [5].
The DNA repair capacity may be influenced by single nucleotide polymorphisms (SNPs) of DNA repair genes which, in turn, can be associated with an increase in cancer risk. Amongst the known repair genes’ polymorphisms, SNPs in XRCC1, XPD and PRKDC genes were studied most frequently in most cancer types. Three SNPs including p.Arg399Gln (exon 10, G to A substitution), p.Arg194Trp (exon 6, C to T substitution) and p.Arg280His (exon 9, G to A substitution), more often found in XRCC1' conserved sites. It has been identified that these polymorphisms are related to cancer susceptibility [6]. As an example, the XRCC1 p.Arg399Gln variant has been associated mainly with colorectal, gastric, breast, esophageal, head and neck, and lung cancers [2, 7, 8]. Also, the XRCC1 c.580C>T (p.Arg194Trp) polymorphism was related to various cancers such as head and neck, skin, colorectal, and gastric tumors [9–11]. Although the functional effects of XRCC1 polymorphisms have not been revealed yet, it is proposed that XRCC1 function may be altered by amino acid changes [1]. Furthermore, polymorphism at codon 751 of XPD is the most extensively studied SNPs in XPD which results in lysine to glutamine substitution. Although the functional effects of XPD p.Lys751Gln polymorphism is still unclear, the A to C base substitution leads to reduced DNA repair capacity of the protein [12]. In the past decade, there are a number of molecular epidemiological reports about the relation between XPD c.2251A>C (p.Lys751Gln) polymorphism and various types of cancer risk such as lung, breast, head and neck, and colorectal in different populations [12–15]. Moreover, among many polymorphisms in PRKDC reported as risk factors for cancers, the intronic polymorphism (rs7003908; 6721G>T) is the most widely studied one. Several studies had investigated the relation between the risk of cancers and PRKDC 6721G>T polymorphism. However, the results were not consistent [16, 17].
Colorectal cancer (CRC) has been a grand challenge in global health within the past few decades. Molecular epidemiology has confirmed that genetic susceptibility may have a main role it CRC carcinogenesis. The relation between polymorphisms of several DNA repair genes including XRCC1, XPD and PRKDC and CRC risk has been evaluated in some populations [18–22].
At present, three common polymorphisms including XRCC1 c.580C>T (p.Arg194Trp), XPD c.2251A>C (p.Lys751Gln) and PRKDC (rs7003908; 6721G>T) have been found in the Iranian population. But, studies focused on the association between CRC susceptibility and these polymorphisms in this population were few [18, 23, 24]. More importantly, due to different patient population as well as relatively small sample size, the results published from foreign population about these polymorphisms were inconsistent or even contradictory. Therefore, this study was stablished in the Iranian population in order to reveal the relation among the genetic polymorphisms of XRCC1 c.580C>T (p.Arg194Trp), XPD c.2251A>C (p.Lys751Gln) and PRKDC (rs7003908; 6721G>T) genes, age, smoking or drinking habit and CRC susceptibility.
MATERIALS AND METHODS
Study Subjects and Sample Collection
This study of colorectal cancer has been conducted from 2014 to 2017. The case group was comprised of 291 patients (153 males and 138 females) with a mean age of 60.93 years and a confirmed invasive adenocarcinoma of the colon through colonoscopy and histology of biopsies. They had no known types of inherited cancer syndrome. The control group was consisted of 140 healthy individuals (67 males and 73 females) with no identified cancer profiles, with a mean age of 59.09 years. All demographic data were recorded by the specialists. Five milliliters of peripheral blood was placed into ethylene diamine tetraacetic acid (EDTA)-coated tubes and stored at –70°C until analyzed.
XRCC1, XPD and PRKDC Genotyping
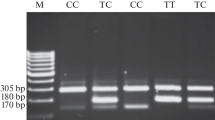
Extraction of the genomic DNA from whole blood was based on salting out method [25]. Genotyping for XRCC1 c.580C>T (p.Arg194Trp), XPD c.2251A>C (p.Lys751Gln) and PRKDC (rs7003908; 6721G>T) polymorphisms was based upon polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay using primers shown in Table1. PCR was conducted in a total volume of 25 μL reaction mixture consisting of 50 ng genomic DNA, 0.5 μM of each primer (GeneScript, Germany), and 12.5 μL 2× Taq DNA Polymerase Mix-Mgcl2 1.5 mM (Amplicon, Denmark). The thermal cycling programs conducted were 2 min at 95°C followed by 30 cycles of 95°C for 45 s, annealing temperature for 30 s at 62°C for XRCC1 c.580C>T and PRKDC rs7003908;6721G>T as well as 63°C for XPD c.2251A>C, and extension at 72°C for 20 s. A final extension was set at 72°C for 5 min. For confirmation of PCR amplicons of 485, 436 and 368 bp for XRCC1 c.580C>T, XPD c.2251A>C and PRKDC 6721G>T were resolved through a 1% agarose gel respectively. In about 10% of the samples, the sequences of PCR products were confirmed utilizing DNA sequencing. Amplicons were then digested with PvuII for XRCC1 c.580C>T and PRKDC rs7003908;6721G>T PCR products as well as PstI for XPD c.2251A>C PCR products at 37°C and were separated using 3% agarose gel electrophoresis. The genotypes were determined as listed in Table 1.
Statistical Analysis
Using Chi square (X2) test, the Hardy-Weinberg equilibrium (HWE) for any deviation from expected allele frequencies was evaluated before association study was performed. χ2 test was used to compare nonquantitative (allelic and genotypic) variables across groups. To conclude the crude odds ratio (OR) and 95% confidence interval (CI) between each of XRCC1 c.580C>T (p.Arg194Trp), XPD c.2251A>C (p.Lys751Gln) and PRKDC (rs7003908; 6721G>T) polymorphisms and CRC, the logistic regression was considered separately. All analyses were carried out by using SPSS version 22.0 (SPSS Inc, Chicago, Illinois). Major characteristics of study groups were as mean and percent. Statistical significance was set at P < 0.05.
RESULTS
In this study, a total of 431 subjects in two groups, control and case, were enrolled. The mean age was 60.93 ± 11.3 years for the colorectal cancer patients. Among those, 153 were male and 138 were female. The mean age of the control group at baseline was 59.09 ± 12.3 years comprising 67 male and 73 female. We found no statistically significant difference with regard to gender and age between cases and controls (P > 0.05), indicating a well-matched study population. All single-nucleotide polymorphisms were consistent with Hardy–Weinberg equilibrium for controls.
The genotype distribution of polymorphisms in XRCC1 c.580C>T (p.Arg194Trp), XPD c.2251A>C (p.Lys751Gln) and PRKDC (rs7003908; 6721G>T) genes for both groups are shown in Table 2. The frequencies of genotypes including CC, CT, and TT for XRCC1 polymorphism were 82.5, 15.5, 2.1% and 95.7, 4.3, 0% among cases and controls, respectively. The frequency of XPD polymorphism was found to be 10% for AA, 67.7% for AC, and 22.3% for CC in CRC population while among control group the frequency was 15.7% for AA, 72.1% for AC, and 12.1% for CC. The frequencies of genotypes including GG, GT, and TT for PRKDC polymorphism were 18.6, 53.3, 28.2% and 73.6, 17.1, 9.3% among cases and controls, respectively.
In this study, the relation of XRCC1 c.580C>T, XPD c.2251A>C and PRKDC rs7003908; 6721G>T polymorphisms to colorectal cancer susceptibility was investigated. According to the results listed in Table 2, we set individuals with XRCC1 CC, PRKDC GG and XPD AA genotypes as the baseline for statistical analysis of association between CRC and genetic polymorphisms. Based on obtained results, when compared with the CC wild-type, the XRCC1 CT heterozygous genotype (OR = 5.35; 95% CI, 2.13–13.38) as well as XRCC1 CT/TT (OR = 4.74; 95% CI, 1.98–11.3) appeared to be significantly relevant to CRC while the XRCC1 TT homozygote was not significantly related to an increased risk of colorectal cancer. Also, the frequencies of heterozygous and homozygote variant genotypes of PRKDC rs7003908; 6721G>T polymorphism were not significantly different between the CRC patients and control group. Carriers of the GT PRKDC or TT PRKDC polymorphisms, had 11.57-fold (95% CI [6.33–21.15], p < 0.001) and 14.06-fold (95% CI [6.75–29.27], p < 0.001) increased risk to develop CRC, respectively. Moreover, the risk of CRC for individuals carrying GT/TT genotypes was increased, and there was 12.21-fold compared with the individuals carrying GG genotype (Table 2), suggesting that PRKDC GT/TT genotype may increase the risk of CRC. In XPD c.2251A>C polymorphism, the risk (OR) to develop CRC was significantly increased to 2.37 (p = 0.042) in association with the homozygous CC-XPD genotype. The risk for CRC was not significantly different for individuals featuring the XPD AC heterozygous genotype (OR = 1.21; 95% CI, 0.631–2.33), or the XPD AC/CC genotypes (OR = 1.68; 95% CI, 0.92–3.05) (Table 2). Among cases and controls, the allele frequency distributions are shown in Table 2. The allele frequencies of XRCC1 c.580C>T (p.Arg194Trp), XPD c.2251A>C (p.Lys751Gln) and PRKDC (rs7003908; 6721G>T) genes polymorphisms were statistically different between CRC patients and normals. The prevalence of the G allele of the PRKDC rs7003908;6721G>T polymorphism among the case and control groups was 45 and 82.1% respectively. The T allele frequency of this polymorphism were 55 and 17.9% in patients and healthy individuals respectively. So colorectal cancer risk showed a significant relationship with the T allele (OR = 5.61; 95% CI, 3.97–7.94). The frequencies for the T (XRCC12 p.Arg194Trp) and C (XPD p.Lys751Gln) alleles amongst the CRC were, respectively, 9.8 and 56.2%. In addition, the distribution of these alleles were 2.1 and 48.2% in controls respectively. Therefore, the T allele of the XRCC1 (p.Arg194Trp) (OR = 4.95; 95% CI, 2.11–11.6) and C allele of the XPD (p.Lys751Gln) (OR = 1.37; 95% CI, 1.03–1.83) polymorphisms may increase the risk of CRC (Table 2).
Analysis of PRKDC, XPD and XRCC1 genotypes with demographic and risk parameters showed a significant association between the PRKDC (rs7003908;6721G>T) variations and smoking in colorectal cancer patients (Table 3). XPD (c.2251A>C) variations were also relevant to smoking and alcohol consumption in individuals with CRC (Table 3). No significant association was noted between XRCC1 genotypes and these two risk parameters (Table 3). Also, significant relationships between the XRCC1 and PRKDC variant genotypes and CRC risk stratified by age factor were shown here.
DISCUSSION
CRC is one of the most commonly diagnosed malignancies worldwide whose development and progression can be associated with a range of factors including environmental and lifestyle. Many environmental factors such as smoking, diet and radiation result in DNA damage. Unrepaired DNA leads to gene mutations, and genomic instability. Therefore, DNA repair genes have a pivotal role in protecting against mutations. Several studies have revealed that the polymorphisms of repair genes can affect the repair capability of them. There is increasing evidence to support the important role of DNA-repair genes' polymorphisms in CRC susceptibility. For example, the correlation between RAD51, XRCC2, and XRCC3 polymorphisms and CRC was conducted by Krupa R et al. in 100 Polish patients [29]. They reported an increase in risk of CRC when XRCC2 (p.Thr 241 Met) was in combination with XRCC3 (p.Arg188 His). They also showed that the risk of CRC can be decreased when XRCC2 (p.Thr 241 Met) was combined with RAD51 (c.135C>C). Another study investigated the role of the XRCC2 (p.Arg188His) in Turkish population reported that in CRC patients, the frequency of XRCC2 polymorphism was two-fold higher than in control group [30]. Also, Larijani et al. (2018) has reported a novel polymorphism (p.Ser150Arg) in the XRCC2 gene in Iranian population and showed that the 150Arg in XRCC2 is associated with the development of CRC [31].
Here, we established a case-control study to determine the relationship between XRCC1 c.580C>T (p.Arg194Trp), XPD c.2251A>C (p.Lys751Gln) and PRKDC (rs7003908; 6721G>T) polymorphisms and the CRC risk in an Iranian population. This investigation included 291 patients with CRC and 140 age and gender matched controls. Many studies with the aim of identifying the role of PRKDC (rs7003908;6721G>T), XPD (p.Lys751Gln) and XRCC1 (p.Arg194Trp) polymorphisms on the risk of various cancers have been established. For example, previously, a significant association was found between XRCC1 (p.Arg194Trp) and PRKDC (rs7003908;6721G>T) polymorphisms and prostate cancer [16]. Moreover, results obtained from a meta-analysis assessed the effect of XPD (p.Lys751Gln) polymorphism on various cancer types, reported a significantly association between this polymorphism and increased risk for cancers such as esophageal, breast and lung [12].
We revealed that the variant allele (Trp) in heterozygous (Arg/Trp) genotype of XRCC1 gene was related to the increased risk for CRC (OR = 5.35; 95% CI, 2.13–13.38) in an Iranian population, what was consistent with data published by Ye Li. They showed the relationship between XRCC1 (p.Arg194Trp) polymorphism and an elevated colorectal cancer risk (OR = 1.45, 95% CI 1.12–1.88) [19]. In studies established by Nissar et al. and Abdel Rehman et al., although significant associations between XRCC1 (p.Arg194Trp) polymorphism and CRC were reported but they showed that individuals inheriting the 194Trp variant allele (in both homozygous (Trp/Trp) and heterozygous (Arg/Trp) conditions) were more likely to develop CRC in Kashmiri and Egyptian populations respectively [10, 32]. Despite our data, few studies suggested that SNP variants in XRCC1 gene possibly did not have a pivotal role in the CRC development. For example, Muñiz-Mendoza et al. revealed that XRCC1 (p.Arg194Trp) SNP did not has association with risk of CRC in Mexican population [33]. In another study established in a Malaysian population, no positive relationship was reported between XRCC1 (p.Arg194Trp) polymorphism and CRC risk [34]. Besides, Mehrzad et al. failed to show any significant relationship between p.Arg194Trp SNP and risk of CRC in Iranian population (north east of Iran) [18].
Here, it was also revealed that variant allele (Gln) in homozygous (Gln/Gln) genotype of XPD c.2251A>C (p.Lys751Gln) polymorphism could increase a possible risk of CRC. This observation was consistant with the results reported by Jelonek et al. [21], but contrary results were reported by Sliwinski et al. [35], Skjelbred et al. [36], and Bigler et al. [37]. Only one previous study has experimentally examined the role of the XPD c.2251A>C (p.Lys751Gln) polymorphism in CRC in an Iranian population reported no evidence of a significant relationship between this polymorphism and an elevated risk of CRC. In Rezaei et al. study, although compared to control group, the frequency of heterozygous genotype (XPD p.Lys 751 Gln) was more in cancer patients, the results were not statistically significant [24].
The PRKDC gene had long been investigated about the association with the risk of cancers; however, the results remained controversial [16, 17]. Here, an intronic variant was analyzed in PRKDC gene. This variant (rs7003908;6721G>T) not leading to any amino-acid substitution or an out-of-frame mutation. But, it is known that this single-nucleotide polymorphism might affect slightly the PRKDC mRNA splicing and, thus, decrease the protein expression level in splicing stages, which can affect DNA repair pathway [38]. Our results demonstrated that individuals with a variant allele (T) in both conditions including heterozygous (GT) genotype (OR = 11.57; 95% CI, 6.33–21.15) and homozygous (TT) genotype (OR = 14.06, 95% CI, 6.75–29.27) had an elevated susceptibility to CRC, when compared to the individuals with GG genotype of XRCC1 gene, which was inconsistent with the findings published by Saadat et al. [23]. Based on their obtained results, no statistically significant relationship was reported between the variant allele (T) in PRKDC gene and risk of CRC in an Iranian population. They suggest that PRKDC TT genotype can be an important risk factor for the CRC development among persons with positive FH (family history) [23]. Due to results obtained here, T allele in PRKDC may be suggested as a marker for the CRC susceptibility.
Here, the role of environmental risk factors as modulating elements for CRC risk in the presence of various genetic variations in XRCC1, XPD and PRKDC genes in an Iranian population was also investigated. While the role of DNA repair genes (XRCC1, XPD and PRKDC) polymorphisms as risk factors for CRC was considered by Mehrzad et al., Saadat et al., and Rezaei et al. in Iranian population, no study has evaluated the role of interaction between these polymorphisms and environmental risk factors such as alcohol intake and smoking behavior in colorectal carcinogenesis in this population. To the best of authors’ knowledge, it is the first Iranian study that investigated the role of DNA repair genes variants as modulators of the effect of the environmental risk factors on CRC risk. The role of interaction between 194Trp allele and environment in colorectal carcinogenesis was evaluated in other populations. For example, in the United States, the effect of alcohol consumption in the risk of CRC was shown to be modulated by 194Trp allele [20]. Moreover, one meta-analysis published in 2013 suggested that the XRCC1 p.Arg194Trp polymorphism might be modifier of alcohol consumption and smoking effects on CRC risk in Chinese population in Singapore [39]. However, our results suggested that there was no relationship between effects of XRCC1 p.Arg194Trp polymorphism in the risk of CRC and alcohol consumption or smoking. In line with our study, no significant role for interaction between XRCC1 p.194Trp allele and environmental exposures in elevating the risk of CRC was reported by Nissar et al. [32]. Besides, our results showed that p.Lys751Gln XPD genetic variations were related to an increase in the risk of colorectal cancer with smoking and alcohol consumption. Consistently, according to results published by Procopciuc et. al., the association between p.Lys751Gln XPD genetic variations and smoking was a risk factor for late-onset colorectal cancer [40]. Also, we found that the association between alcohol intake and PRKDC rs7003908;6721G>T genetic variations did not increase the risk of CRC. But, individuals positive for PRKDC rs7003908; 6721G>T genetic variations, with smoking habit had a higher risk to develop CRC. Furthermore, here, results showed significant associations between the XRCC1 and PRKDC variant genotypes and CRC risk stratified by age factor. Our results were in agreement with those obtained by Datkhile et al. when reported that PRKDC gene polymorphisms represented association with an elevated risk of oral cancer when examined by stratifying age factor [41].
This discrepancy between studies established in Iranian population and ours is likely due to lower sample sizes of cases and controls in those studies when compared to our investigation. Also Mehrzad et al. study was restricted to a north east population of Iran [18]. The divergence in single nucleotide polymorphisms results obtained from different population may be due to different levels of carcinogen exposure. Other than polymorphism, environmental exposures or other genetic factors affect cancer formation which are inconsistent among different populations. Moreover, confounding factors including diet, age and gender may attribute to such discrepancies. In addition, the genotyping method may also play important role in contradictory findings among various studies [42]. So further studies incorporating another ethnic population or/and a larger sample size are necessary to confirm the role of these polymorphisms as regards CRC susceptibility in an Iranian population.
CONCLUSIONS
In Conclusions, our evidences showed that T alleles of XRCC1 c.580C>T and PRKDC2 (6721G>T) as well as C allele of XPD c.2251A>C can be relevant to a higher risk of developing CRC among Iranian population. However, further studies with detailed data on environmental exposure and larger sample size from different ethnicities are needed to confirm these initial encouraging results.
REFERENCES
Salimi, S., Mohammadoo-Khorasani, M., Tabatabai, E., Sandoughi, M., Zakeri, Z., and Naghavi, A., XRCC1 Arg399Gln and Arg194Trp polymorphisms and risk of systemic lupus erythematosus in an Iranian population: a pilot study, Biomed. Res. Int., 2014, vol. 2014.
Mirsane, S.A. and Shafagh, S., The relationship between XRCC1 Arg399Gln polymorphism, alcohol consumption and colorectal cancer: one of the alcohol forbidding reasons in Islam, Gene.Cell. Tissue, 2016, vol. 3, e40 607.
Huang, Y., Li, X., He, J., Chen, L., et al., Genetic polymorphisms in XRCC1 genes and colorectal cancer susceptibility, World. J. Surg. Oncol., 2015, vol. 13, p. 244. https://doi.org/10.1186/s12957-015-0650-2
King, C., Yu, J., Freimuth, R., et al., Interethnic variability of ERCC2 polymorphisms, Pharmacogenomics J., 2005, vol. 5, p. 54.
Xiao, M., Shen, Y., Chen, L., et al., The rs7003908 (T> G) polymorphism in the XRCC7 gene and the risk of cancers, Mol. Biol. Rep., 2014, vol. 41, pp. 3577–3582.
Trabulus, S., Guven, G.S., Altiparmak, M.R., et al., DNA repair XRCC1 Arg399Gln polymorphism is associated with the risk of development of end-stage renal disease, Mol. Biol. Rep., 2012, vol. 39, pp. 6995–7001.
Guo, S., Mao, X., and Ming, L., XRCC1 Arg399Gln polymorphism is not associated with breast cancer in Chinese, Int. J. Clin. Exp. Med., 2015, vol. 8, p. 10429.
Yang, H-Y., Yang, S-Y., Shao, F-Y., et al., Updated assessment of the association of the XRCC1 Arg399Gln polymorphism with lung cancer risk in the Chinese population, Asian Pac. J. Cancer Prev., 2015, vol. 16, pp. 495–500.
Duarte, M.C., Colombo, J., Rossit, A.R.B., et al., Polymorphisms of DNA repair genes XRCC1 and XRCC3, interaction with environmental exposure and risk of chronic gastritis and gastric cancer, World. J. Gastroenterol., 2005, vol. 11, p. 6593.
Abdel-Rahman, S.Z., Soliman, A.S., Bondy, M.L., et al., Inheritance of the 194Trp and the 399Gln variant alleles of the DNA repair gene XRCC1 are associated with increased risk of early-onset colorectal carcinoma in Egypt, Cancer Lett., 2000, vol. 159, pp. 79–86.
Olshan, A.F., Watson, M.A., Weissler, M.C., and Bell, D.A., XRCC1 polymorphisms and head and neck cancer, Cancer. Lett., 2002, vol. 178, pp. 181–186.
Wu, K-G., He, X-F., Li, Y-H., et al., Association between the XPD/ERCC2 Lys751Gln polymorphism and risk of cancer: evidence from 224 case–control studies, Tumour Biol., 2014, vol. 35, pp. 11 243–11 259.
Benhamou, S. and Sarasin, A., ERCC2/XPD gene polymorphisms and cancer risk, Mutagenesis, 2002, vol. 17, pp. 463–469.
Qiu, L-X., Yao, L., Zhang, J., et al., XPD Lys751Gln polymorphism and breast cancer susceptibility: a meta-analysis involving 28 709 subjects, Breast Cancer Res. Treat., 2010, vol. 124, pp. 229–235.
Smith, G.D. and Egger, M., Meta-analysis of randomised controlled trials, Lancet, 1997, vol. 1350, p. 1182.
Mandal, R.K., Kapoor, R., and Mittal, R.D., Polymorphic variants of DNA repair gene XRCC3 and XRCC7 and risk of prostate cancer: a study from North Indian population, DNA Cell Biol., 2010, vol. 29, pp. 669–674.
Zhang, J., Wu, X-h., and Gan, Y., Current evidence on the relationship between three polymorphisms in the XRCC7 gene and cancer risk, Mol. Biol. Rep., 2013, vol. 40, pp. 81–86.
Mehrzad, J., Mohammaditabr, M., Khafi, A. S., and Erfanian Khorasanian, M., Association of XRCC1 gene polymorphisms with colorectal cancer risk, Int. J. Biosci., 2014, vol. 5, pp. 199–205.
Li, Y., Li, S., Wu, Z., Hu, F., et al., Polymorphisms in genes of APE1, PARP1, and XRCC1: risk and prognosis of colorectal cancer in a northeast Chinese population, Med. Oncol., 2013, vol. 30, p. 505.
Curtin, K., Samowitz, W.S., Wolff, R.K., et al., Assessing tumor mutations to gain insight into base excision repair sequence polymorphisms and smoking in colon cancer, Cancer Epidemiol. Biomarkers. Prev., 2009, vol. 18, pp. 3384–3388.
Jelonek, K., Gdowicz-Kłosok, A., Pietrowska, M., et al., Association between single-nucleotide polymorphisms of selected genes involved in the response to DNA damage and risk of colon, head and neck, and breast cancers in a Polish population, J. Appl. Genet., 2010, vol. 51, pp. 343–352.
Stern, M.C., Butler, L.M., Corral, R., et al., Polyunsaturated fatty acids, DNA repair single nucleotide polymorphisms and colorectal cancer in the Singapore Chinese Health Study, J. Nutrigen. Nutrigenom., 2009, vol. 2, pp. 273–279.
Saadat, M., and Rabizadeh-Hafshenjani, A., DNA repair gene XRCC7 G6721T variant and susceptibility to colorectal cancer, EJMHG, 2016, vol. 17, pp. 373–376.
Rezaei, H., Motovali-Bashi, M., Khodadad, K., et al., Relationship between XPD Lys 751 Gln polymorphism and colorectal cancer risk: a case-control study in a population-based study, Gastroenterol. Hepatol. Bed. Bench., 2013, vol. 6, p. 18.
Miller, S., Dykes, D., and Polesky, H., A simple salting out procedure for extracting DNA from human nucleated cells, Nucleic Acids Res., 1988, vol. 16, p. 1215.
Li, W-Q., Zhang, L., Ma, J-L., et al., Association between genetic polymorphisms of DNA base excision repair genes and evolution of precancerous gastric lesions in a Chinese population, Carcinogenesis, 2009, vol. 30, pp. 500–505.
Kang, S., Sun, H-Y., Zhou, R-M., et al., DNA repair gene associated with clinical outcome of epithelial ovarian cancer treated with platinum-based chemotherapy, APJCP, 2013, vol. 14, pp. 941–946.
Zhi, Y., Yu, J., Liu, Y., et al., Interaction between polymorphisms of DNA repair genes significantly modulated bladder cancer risk, Int. J. Med. Med. Sci., 2012, vol. 9, p. 498.
Krupa, R., Sliwinski, T., Wisniewska-Jarosinska, M., et al., Polymorphisms in RAD51, XRCC2 and XRCC3 genes of the homologous recombination repair in colorectal cancer–a case control study, Mol. Biol. Rep., 2011, vol. 38, pp. 2849–2854.
Cetinkunar, S., Gok, I., Celep, R.B., et al., The effect of polymorphism in DNA repair genes RAD51 and XRCC2 in colorectal cancer in Turkish population, Int. J. Clin. Exp. Med., 2015, vol. 8, p. 2649.
Sadat-Larijani, M., Derakhshani, S., Keshavarz-Pakseresht, B., et al., Impact of a missense variation (p.S150R: AGC > AGG) in the XRCC2 gene on susceptibility to colorectal cancer, Clin. Lab., 2018, vol. 64, pp. 233–237.
Nissar, S., Sameer, A.S., Rasool, R., et al., Polymorphism of the DNA repair gene XRCC1 (Arg194Trp) and its role in colorectal cancer in Kashmiri population: a case control study, APJCP, 2015, vol. 16, pp. 6385–6390.
Muniz-Mendoza, R., Ayala-Madrigal, M., Partida-Perez, M., et al., MLH1 and XRCC1 polymorphisms in Mexican patients with colorectal cancer, Genet. Mol. Res., 2012, vol. 11, pp. 2315–2320.
Cheah, P.L., Looi, L.M., Roslani April, C., et al., Lack of correlation between X-ray repair cross-complementing group 1 gene polymorphisms and the susceptibility to coloretal cancer in a Malaysian cohort, Eur. J. Cancer, 2017, vol. 26, pp. 506–510.
Sliwinski, T., Krupa, R., Wisniewska-Jarosinska, M., et al., No association between the Arg194Trp and Arg399Gln polymorphisms of the XRCC1 gene and colorectal cancer risk and progression in a Polish population, Exp. Oncol., 2008, vol. 30, pp. 253–254.
Skjelbred, C.F., Sæbø, M., Wallin, H., et al., Polymorphisms of the XRCC1, XRCC3 and XPD genes and risk of colorectal adenoma and carcinoma, in a Norwegian cohort: a case control study, BMC Cancer, 2006, vol. 6, p. 67.
Bigler, J., Ulrich, C.M., Kawashima, T., et al., DNA repair polymorphisms and risk of colorectal adenomatous or hyperplastic polyps, Cancer Epidemiol. Biomark. Prev., 2005, vol. 14, pp. 2501–2508.
Jahantigh, D. and Hosseinzadeh Colagar, A.J.I.j.o.e., XRCC5 VNTR, XRCC6-61C> G, and XRCC7 6721G> T gene polymorphisms associated with male infertility risk: evidences from case-control and in silico studies, Int. J. Endocrinol., 2017, vol. 2017.
Mao, D., Zhang, Y., Lu, H., and Fu, X., Association between X-ray repair cross-complementing group 1 Arg194Trp polymorphism and colorectal cancer risk, Tumor. Biol., 2013, vol. 34, pp. 2529–2538.
Procopciuc, L.M. and Osian, G., Interaction between lifestyle factors and the XRCC1, XPD, and XRCC3 genetic variations modulates the risk for sporadic colorectal cancer, Rev. Romana. Med. Lab., 2014, vol. 22, pp. 129–141.
Datkhile, K., Vhaval, R., Patil, M., et al., Role of genetic polymorphisms in DNA repair genes ((XRCC1, XRCC2, XRCC3, XRCC4, XRCC5, XRCC6, and XRCC7) in head and neck cancer susceptibility in rural Indian population: a hospital based case-control study from south-western Maharashtra, Int. J. Curr. Res., 2016, vol. 8, pp. 25482–25492.
Saadat, M. and Ansari-Lari, M., Polymorphism of XRCC1 (at codon 399) and susceptibility to breast cancer, a meta-analysis of the literatures, Breast. Cancer. Res. Treat., 2009, vol. 115, pp. 137–144.
ACKNOWLEDGMENTS
This study was financially supported by the 92/7815 grant from Islamic Azad University (Varamin-Pishva Branch) of Iran.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest. The authors declare that they have no conflict of interest.
Statement of compliance with standards of research involving humans as subjects. All procedures performed in studies involving human participants were approved by the Institutional Ethics Committee and were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants involved in the study.
About this article
Cite this article
Atieh Hashemi, Baghbani-arani, F. & Larijani, M.S. Genetic Polymorphisms of Three DNA-Repair Genes (PRKDC, XPD, XRCC1) are Related to Colorectal Cancer Susceptibility. Cytol. Genet. 54, 363–371 (2020). https://doi.org/10.3103/S0095452720040040
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.3103/S0095452720040040