Abstract
Inconsistency of the association of polymorphisms of XRCC7 with cancer is noted. Three commonly studied XRCC7 polymorphisms including rs7003908 (T>G), rs7830743 (A>G), and rs10109984 (T>C) were selected to explore their association with risk of development of cancer by meta-analysis of published case–control studies. The results showed that no significant associations with cancer risk were found in any model in terms of rs7003908, rs7830743 and rs10109984 when all studies were pooled into the meta-analysis. But when stratified by cancer type, statistically significantly elevated cancer risk was only found in prostate cancer for rs7003908 (GG vs. TT: OR = 1.845, 95 % CI = 1.178–2.888; dominant model: OR = 1.423, 95 % CI = 1.050–1.929; recessive model: OR = 1.677, 95 % CI = 1.133–2.482). In the subgroup analysis by ethnicity or study design, no significantly increased risks were found for all three polymorphisms. This meta-analysis suggests that XRCC7 rs7003908 polymorphism may contribute to cancer susceptibility for prostate cancer, which is recommended to be included in future large-sample studies and functional assays.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer is the leading cause of death in economically developed countries and the second leading cause of death in developing countries, which has become a major public health challenge [1]. New markers for identifying high-risk populations as well as novel strategies for early detection and preventive care are urgently needed. While the exact etiology of cancer is poorly understood, there are some recognized risk factors that may contribute to the development of cancer including age, ethnicity, lifestyle, as well as genetic factors. Low-penetrance susceptibility genes combining with environmental and heritable factors have been indicated to be important for carcinogenesis [2].
As the preservation of genomic integrity is essential in the prevention of tumor initiation and progression, mutations and variations in DNA repair genes may play a role in the genetic predisposition to cancer. One of the most detrimental forms of DNA damage is the double strand break (DSB), because the DNA loses physical integrity and information content on both strands. The DSBs may lead to genome instability, which in turn may enhance the development of cancer [3]. There are two DSB repair pathways in mammalian cells: the homologous recombination (HR) repair and non-homologous end-joining (NHEJ) [3]. The NHEJ is responsible for repairing most DSBs [4]. At present, several proteins involved in the NHEJ pathway have been identified; namely, the ligase IV and its associated protein XRCC4, the three components of the DNA dependent protein kinase (DNA-PK) complex, Ku70, Ku80, and the catalytic subunit PKcs [5]. DNA dependent protein kinase complex is encoded by the human X-ray repair cross-complementing group 7 (XRCC7) gene (Genbank accession no: NM_001469), also known as PRKDC/HYRC/HYRC1, which is located on chromosome 8q11. In recent years, some original publications [6–19] have reported the role of XRCC7 polymorphisms in cancer risk. Three polymorphic variants, rs7003908 (T>G), rs7830743 (A>G), and rs10109984 (T>C) have been the research focus in scientific community and have drawn increasing attention. In the studies mentioned above, some showed these polymorphisms were risk factors for developing cancer, while the others showed no such association. Since the results are inconsistent and inconclusive, probably due to the possible small effect of the polymorphism on cancer risk or the relatively underpowered sample size in each of published studies, it is reasonable for us to perform this meta-analysis to derive a more precise estimation.
Materials and methods
Search strategy
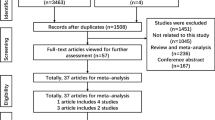
The databases, PubMed, Medline, Embase, Ovid, Springer, Cochrane and Web of Science, were searched (updated to Jun 6, 2012) using the terms: “XRCC7”, “PRKDC”, “DNA-PKcs” “polymorphism” and “cancer”). All the searched studies were retrieved, and their references were checked as well for other relevant publications. Review articles were also searched to find additional eligible studies. Only those published in English language with full text articles were included. For overlapping studies, only the first published one was selected; for republished studies, only the one with the largest sample numbers was included.
Eligible studies and data extraction
Selection criteria were: (a) case–control studies of XRCC7 polymorphisms with complete genotypes distribution data; (b) the diagnosis of cancer patients was confirmed pathologically and controls were confirmed as free from cancer; (c) written in English; (d) the number of case and control was more than 100; and (e) fulfilling Hardy–Weinberg equilibrium (HWE) in the control group (P > 0.05 was eligible).
The following variables were extracted from each study if available: first author’s name, publication year, cancer type, country of origin, study design, genotype distributions, and HWE of controls, respectively.
Different ethnicity descents were categorized as Asian or Caucasian. Study design was stratified into hospital-based studies and population-based studies. Data was extracted independently by two investigators and consensuses were reached on all items. If they could not come to an agreement, a third investigator (Yu Gan) adjudicated the disagreements.
Statistics
The studies whose allele frequencies in controls exhibited significant deviation from the HWE were excluded from this analysis according to inclusion criteria, given that the deviation may denote bias. For the assessment of the deviation from HWE, the appropriate goodness-of-fit Chi-square test was performed [20, 21]. For the interpretation of the goodness-of-fit Chi-square test, statistical significance was defined as P < 0.05.
Based on the genotype frequencies in cases and controls, crude odds ratios (ORs) as well as their 95 % confidence intervals (CIs) were calculated. For each polymorphism, three different ORs were calculated: (a) heterozygous carriers versus ‘wild type’ (b) homozygous carriers versus ‘wild type’ (c) dominant model, i.e. heterozygous and homozygous carriers grouped together versus wild type and (d) recessive model, i.e. homozygous carriers versus ‘wild type’ and heterozygous carriers grouped together. Separate ethnicity-specific, study-design-specific and cancer-type-specific analyses were considered if relevant data were available.
The fixed-effects model (Mantel–Haenszel method), or the random effects (DerSimonian Laird) model, were appropriately used to calculate the pooled OR. Between-study heterogeneity and between-study inconsistency were assessed by using Cochran Q statistic and by estimating I2, respectively [22]. In case significant heterogeneity was detected, the random effects model was chosen. Meta-analysis was performed using the ‘metan’ STATA command.
Evidence of publication bias was determined using Egger’s [23] formal statistical test and by visual inspection of the funnel plot. For the interpretation of Egger’s test, statistical significance was defined as P < 0.10. The Egger’s test was performed using the ‘metabias’ STATA command.
Analyses were conducted using STATA 10.0 (STATA Corp., College Station, TX, USA).
Results
Study characteristics
The related studies were characterized and listed in Table 1, including first author, publication year, cancer type, country of origin, ethnicity, study design, genotype distributions, and HWE of controls, respectively. The studies of Liu et al. [9], Hu et al. [10] and Long et al. [17] did not fulfill HWE in the control group for rs7003908 and were excluded from this meta-analysis. Thus, 3,323 cases and 4,744 controls for XRCC7 rs7003908 (T>G) polymorphism, 1,965 cases and 2,284 controls for XRCC7 rs7830743 (A>G) polymorphism, and 1,939 cases and 2,417 controls for XRCC7 rs10109984 (T>C) polymorphism were involved in the meta-analysis, respectively.
Main results and publication bias
The pooled ORs of corresponding studies along with their 95 % CIs were presented in the Table 2.
In terms of rs7830743 and rs10109984, no significant associations with cancer risk were found in any model (co-dominant, dominant, or recessive model). Similar results were observed in term of rs7003908, but when stratified by cancer type, statistically significantly elevated cancer risk was found in prostate cancer (GG vs. TT: OR = 1.845, 95 % CI = 1.178–2.888; dominant model: OR = 1.423, 95 % CI = 1.050–1.929; recessive model: OR = 1.677, 95 % CI = 1.133–2.482). In the subgroup analysis by ethnicity or study design, no significantly increased risks were only found for all three polymorphisms. When the studies [9, 10, 17] in which controls were not in agreement with HWE were included or a single study involved in the meta-analysis was deleted each time to reflect the influence of the individual data-set to the pooled ORs, the corresponding pooled ORs were not materially altered (data not shown), indicating that our results were statistically robust.
Begg’s funnel plot and Egger’s test were performed to evaluate the publication bias of the literatures. The shape of the funnel plot did not reveal any evidence of obvious asymmetry (figures not shown), and the Egger’s test suggested the absence of publication bias (data not shown).
Discussion
Genes involved in NHEJ such as Ku70, Ku80 and XRCC7 are considered to be essential for genome stability and consequently for cell survival. Severe defects in these genes would result in cell death triggered by cell cycle checkpoint surveillance. However, small genetic variations such as SNPs might escape cell checkpoint surveillance. These variations can lead to suboptimal DNA repair which would allow DNA damage to accumulate and this could trigger tumor initiation [24]. In the present meta-analysis, the associations between XRCC7 rs7003908, rs7830743 and rs10109984 polymorphisms and cancer risks were explored with no positive results in any model. Actually, it might be not uncommon that the epidemiology results were not coincidence with the results of functional study. Because cancer is a complicated multi-genetic disease, different genetic backgrounds may contribute to the discrepancy [25]. The influence of the XRCC7 polymorphisms might be masked by the presence of other as-yet unidentified causal genes involved in cancer development.
In the subgroup analysis by cancer type, statistically significantly elevated cancer risk was only found for prostate cancer in term of XRCC7 rs7003908. Possibly, the variant genotype could be linked with a phenotype with suboptimal DSB repair which allows accumulation of mutations and promotes chromosomal instability and ultimately prostate cancer development. However, it is also likely that the observed differences in different cancer types may be due to chance because small sample size may lead to insufficient statistical power to detect a slight effect or may have generated a fluctuated risk estimate [26]. Considering all these factors, our results of this meta-analysis should be interpreted with caution.
In this meta-analysis, only studies published in English were included because the English language is generally perceived to be the universal language of science. However, the exclusive reliance on English-language studies may not represent all of the evidence. Excluding languages other than English may introduce a language bias and lead to erroneous conclusions [27]. Actually, through searching all database available, only one study published in Chinese investigated the relationship between XRCC7 rs7003908 and cancer risk and this would not influence our results because the study did not fulfill HWE in the control group and should be excluded.
Several limitations of this meta-analysis should be summarized and addressed. Firstly, the sample size was still relatively small for some SNPs and for some stratified analyses. Secondly, in our analysis, the controls were not uniformly defined. Although most of the controls were selected mainly from healthy populations, some had benign disease. Therefore, non-differential misclassification bias was possible because these studies may have included the control groups who have different risks of developing cancer. Finally, our results were based on unadjusted estimates, while a more precise analysis should be conducted if all individual raw data were available, which would allow for the adjustment by other co-variants including age, gender, smoking status, drinking status, obesity, environmental factors, and other lifestyle.
In spite of these limitations, our meta-analysis had several strengths. First, more cases and controls were pooled from different studies, which significantly increased the statistical power of the analysis. Second, no publication biases were detected, indicating that the whole pooled results may be unbiased.
In conclusion, this meta-analysis suggests that though no associations between XRCC7 rs7003908, rs7830743 and rs10109984 polymorphisms and cancer risks were found when all studies were pooled, XRCC7 rs7003908 polymorphism may contribute to cancer susceptibility for prostate cancer, which is recommended to be included in future functional assays. More consortia and international collaborative studies with homogeneous cancer patients and well matched controls, which may be a way to maximize study efficacy and overcome the limitations of individual studies, are needed to help further illuminate the landscape of these polymorphisms and cancer risks.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61:69–90
Lichtenstein P, Holm NV, Verkasalo PK et al (2000) Environmental and heritable factors in the causation of cancer—analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343:78–85
Karran P (2000) DNA double strand break repair in mammalian cells. Curr Opin Genet Dev 10:144–150
Hopfner KP, Putnam CD, Tainer JA (2002) DNA double-strand break repair from head to tail. Curr Opin Struct Biol 12:115–122
Jackson SP (2002) Sensing and repairing DNA double-strand breaks. Carcinogenesis 23:687–696
Wang LE, Bondy ML, Shen H et al (2004) Polymorphisms of DNA repair genes and risk of glioma. Cancer Res 64:5560–5563
Hirata H, Hinoda Y, Matsuyama H et al (2006) Polymorphisms of DNA repair genes are associated with renal cell carcinoma. Biochem Biophys Res Commun 342:1058–1062
Hirata H, Hinoda Y, Tanaka Y et al (2007) Polymorphisms of DNA repair genes are risk factors for prostate cancer. Eur J Cancer 43:231–237
Liu Y, Zhang H, Zhou K et al (2007) Tagging SNPs in non-homologous end-joining pathway genes and risk of glioma. Carcinogenesis 28:1906–1913
Hu Z, Liu H, Wang H et al (2008) Tagging single nucleotide polymorphisms in phosphoinositide-3-kinase-related protein kinase genes involved in DNA damage “checkpoints” and lung cancer susceptibility. Clin Cancer Res 14:2887–2891
Siraj AK, Al-Rasheed M, Ibrahim M et al (2008) RAD52 polymorphisms contribute to the development of papillary thyroid cancer susceptibility in Middle Eastern population. J Endocrinol Invest 31:893–899
Wang SY, Peng L, Li CP et al (2008) Genetic variants of the XRCC7 gene involved in DNA repair and risk of human bladder cancer. Int J Urol 15:534–539
Bhatti P, Struewing JP, Alexander BH et al (2008) Polymorphisms in DNA repair genes, ionizing radiation exposure and risk of breast cancer in U.S. Radiologic technologists. Int J Cancer 122:177–182
McKean-Cowdin R, Barnholtz-Sloan J, Inskip PD et al (2009) Associations between polymorphisms in DNA repair genes and glioblastoma. Cancer Epidemiol Biomarkers Prev 18:1118–1126
Gangwar R, Ahirwar D, Mandhani A, Mittal RD (2009) Do DNA repair genes OGG1, XRCC3 and XRCC7 have an impact on susceptibility to bladder cancer in the North Indian population? Mutat Res 680:56–63
Mandal RK, Kapoor R, Mittal RD (2010) Polymorphic variants of DNA repair gene XRCC3 and XRCC7 and risk of prostate cancer: a study from North Indian population. DNA Cell Biol 29:669–674
Long XD, Yao JG, Huang YZ et al (2011) DNA repair gene XRCC7 polymorphisms (rs#7003908 and rs#10109984) and hepatocellular carcinoma related to AFB1 exposure among Guangxi population, China. Hepatol Res 41:1085–1093
Al-Hadyan KS, Al-Harbi NM, Al-Qahtani SS, Alsbeih GA (2012) Involvement of single-nucleotide polymorphisms in predisposition to head and neck cancer in Saudi Arabia. Genet Test Mol Biomarkers 16:95–101
Nasiri M, Saadat I, Omidvari S, Saadat M (2012) Genetic variation in DNA repair gene XRCC7 (G6721T) and susceptibility to breast cancer. Gene 505:195–197
Thakkinstian A, McElduff P, D’Este C, Duffy D, Attia J (2005) A method for meta-analysis of molecular association studies. Stat Med 24:1291–1306
Rohlfs RV, Weir BS (2008) Distributions of Hardy–Weinberg equilibrium test statistics. Genetics 180:1609–1616
Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327:557–560
Egger M, Davey SG, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634
Fu YP, Yu JC, Cheng TC et al (2003) Breast cancer risk associated with genotypic polymorphism of the nonhomologous end-joining genes: a multigenic study on cancer susceptibility. Cancer Res 63:2440–2446
Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K (2002) A comprehensive review of genetic association studies. Genet Med 4:45–61
Wacholder S, Chanock S, Garcia-Closas M, El GL, Rothman N (2004) Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst 96:434–442
Gregoire G, Derderian F, Le Lorier J (1995) Selecting the language of the publications included in a meta-analysis: is there a Tower of Babel bias? J Clin Epidemiol 48:159–163
Acknowledgments
This research was supported by grants from the National Natural Science Foundation of China (81101499), Shanghai Natural Science Foundation (11ZR1407600), and Fudan University Science Foundation for Young Scholars (09FQ76).
Conflict of interest
None declared.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, J., Wu, Xh. & Gan, Y. Current evidence on the relationship between three polymorphisms in the XRCC7 gene and cancer risk. Mol Biol Rep 40, 81–86 (2013). https://doi.org/10.1007/s11033-012-2018-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-2018-9