Abstract
Equivalence testing of aerodynamic particle size distribution (APSD) through multi-stage cascade impactors (CIs) is important for establishing bioequivalence of orally inhaled drug products. Recent work demonstrated that the median of the modified chi-square ratio statistic (MmCSRS) is a promising metric for APSD equivalence testing of test (T) and reference (R) products as it can be applied to a reduced number of CI sites that are more relevant for lung deposition. This metric is also less sensitive to the increased variability often observed for low-deposition sites. A method to establish critical values for the MmCSRS is described here. This method considers the variability of the R product by employing a reference variance scaling approach that allows definition of critical values as a function of the observed variability of the R product. A stepwise CI equivalence test is proposed that integrates the MmCSRS as a method for comparing the relative shapes of CI profiles and incorporates statistical tests for assessing equivalence of single actuation content and impactor sized mass. This stepwise CI equivalence test was applied to 55 published CI profile scenarios, which were classified as equivalent or inequivalent by members of the Product Quality Research Institute working group (PQRI WG). The results of the stepwise CI equivalence test using a 25% difference in MmCSRS as an acceptance criterion provided the best matching with those of the PQRI WG as decisions of both methods agreed in 75% of the 55 CI profile scenarios.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Demonstration of equivalence in aerodynamic particle size distribution (APSD; Table I lists all abbreviations used in this manuscript) plays an important role in establishing bioequivalence (BE) of orally inhaled drug products (OIDPs) (1, 2). Multi-stage cascade impactors (CIs) such as the Andersen Cascade Impactor or the Next Generation Impactor can be used to assess the APSD of OIDPs. In order to achieve a more objective evaluation of APSD equivalence or comparability between test (T) and reference (R) OIDPs, the modified chi-square ratio statistic (mCSRS; Eq. 1) was introduced and evaluated for its properties in the first two articles of this series (3, 4). The computational form of mCSRS is summarized below.
where p represents the number of deposition sites, T ij and R ik represent the normalized deposition (i.e., by the dividing the absolute deposition on the ith site by the total deposition on all sites under consideration) on the ith site of the jth profile (j = 1,…, n T ) of the T sample and on the ith site of the kth profile (k = 1, …, n R ) of the R sample, respectively. n T and n R represent the number of samples that were obtained from the T and R product, respectively, and \( {\overline{R}}_i \) represents the sample mean on the ith site of all R profiles.
In the first article, the median of the distribution of 900 mCSRSs (MmCSRS; using a sample size of 30 T and 30 R products) was demonstrated to be equal to one when T and R products were identical, regardless of the shape and the number of deposition sites of CI profiles (3). This key finding, which represents a major improvement over the original CSRS, indicated that the MmCSRS could potentially be applied for comparing the CI profiles of T and R products for a reduced number of deposition sites. Specifically, deposition sites that do not have well-defined upper cutoff diameters in a CI setting can be excluded (e.g., pre-separator or induction port).
In the second article, the behavior of the MmCSRS when T and R profiles differ from each other on a single or multiple deposition site(s) was characterized (4). This analysis resulted in two key observations. First, the MmCSRS is more sensitive to differences between T and R profiles that occur on high-deposition sites, thus, reducing the influence of low-deposition sites (often associated with high variability) on the overall APSD equivalence evaluation. This characteristic of the MmCSRS may represent a distinct advantage over statistical methods that are based on site-by-site comparisons as those methods often fail to demonstrate equivalence when testing different batches of the same R product due to the high variability on low-deposition sites. Second, it was demonstrated that the MmCSRS is inversely proportional to the variability of the R product when T and R products differ from each other. Specifically, a perfect linear relationship was obtained when regressing the MmCSRS against the inverse square of coefficient of variation (CV) of the R product. Consequently, critical values for equivalence testing based on the MmCSRS require scaling on the variability of the R product for consistently discriminating equivalent from inequivalent CI profiles. Henceforth, this scaling of the MmCSRS on the variability of the R product is also referred to as reference variance scaling. When the original CSRS was evaluated for its properties by Product Quality Research Institute (PQRI) “Aerodynamic Particle Size Distribution Profile Comparisons Working Group” (WG), reference variance scaling was not considered (5–7).
In this article, a stepwise CI equivalence test that assesses the equivalence of single actuation content (step 1), impactor sized massFootnote 1 (ISM; step 2), and the APSD profiles on all ISM deposition sites through the MmCSRS is proposed (step 3). Henceforth, the third step of the proposed CI equivalence test is referred to as MmCSRS-APSD analysis. Since the MmCSRS-APSD analysis only compares relative differences between T and R products on the ISM sites, steps 1 and 2 were included rationally to provide an assessment of absolute mass-related metrics. The stepwise CI equivalence test is then applied to 55 characteristic CI profile scenarios that were published by the PQRI WG (6, 8, 9). The PQRI WG classified each of those 55 scenarios as either equivalent or inequivalent. The results of the CI equivalence test are then compared with those of the PQRI WG evaluations.
Before the stepwise CI equivalence test could be applied to the 55 PQRI CI profile scenarios, critical values for the MmCSRS-APSD test had to be derived. The derivation of these critical values is also outlined in the current manuscript. The scope of this article focuses on describing the key procedures and aspects of the proposed stepwise CI equivalence test and evaluating the overall performance of this test in relation to the PQRI WG assessments of the 55 CI profile scenarios.
CASCADE IMPACTOR EQUIVALENCE TEST
The proposed CI equivalence test that consists of three key steps (Fig. 1) is described below. This description is intended as a manual for users who would like to apply the proposed CI equivalence test to their data.
-
Step 1.
Perform a population bioequivalence (PBE (10)) analysis on single actuation content. Proceed to step 2 if equivalence of single actuation content is demonstrated; otherwise, conclude that T and R are inequivalent and do not proceed to step 2.
-
Step 2.
Perform a PBE analysis on ISM. Proceed to step 3 if equivalence of ISM is shown; otherwise, conclude that T and R are inequivalent and do not proceed to step 3.
-
Step 3.
Perform the MmCSRS-APSD analysis for the ISM deposition sites after normalization of the ISM data. Conclude equivalence of APSD if the result of the MmCSRS-APSD analysis meets its criterion (see below); otherwise, conclude that T and R are inequivalent.
Within the third step, the MmCSRS, a non-parametric confidence interval for the MmCSRS, and a metric for reference variance scaling that characterizes the observed variability of the R product need to be calculated. Details on each of mathematical operations are given below.
MmCSRS
The MmCSRS is calculated as described above (Eq. 1).
Construction of Non-parametric Confidence Intervals for the MmCSRS
A 90% non-parametric confidence interval for the MmCSRS is calculated using a bias corrected and accelerated bootstrapping (BCA) method (see Appendix).
Calculation of a Metric for Reference Variance Scaling
For scaling the MmCSRS on the variability of the R product, the variability of the R product is estimated as a single summary measure from the variability on each of the ISM sites. This is accomplished by applying the following metric to the normalized ISM profiles
where CV i represents the coefficient of variation (in %) of the ith deposition site of the R sample (after normalization on ISM); \( {\overline{R}}_i \)and p were defined above (Eq. 1). This metric for estimating the variability of the R product is similar to that used for calculating the pooled CV from multiple clinical studies (11). Details on the selection and performance of this metric for reference variance scaling will be discussed elsewhere (manuscript in preparation).
Evaluating Equivalence of MmCSRS-APSD Test
If the upper bound of the 90% non-parametric confidence interval (see above) is smaller than the critical value determined for the variance of respective R product, which is estimated by the metric described in Eq. 2, then equivalence is concluded. Details on the construction of critical values for the MmCSRS-APSD analysis are given below (see Construction of Critical Values for the MmCSRS-APSD Analysis).
METHODS
For both the derivation of critical values for the MmCSRS-APSD analysis and the application of the proposed stepwise CI equivalence test to the 55 PQRI CI profile scenarios, CI profiles were generated by Monte Carlo simulation methods. The R extension package “mvtnorm” was used for random sample generation from the multivariate normal distribution (12, 13). Details and a discussion about this simulation method were provided in the previous publication (4). A sample size of 30 T and 30 R CI profiles is assumed throughout this paper. Implications of assumed sample size are discussed below (see Discussion).
Construction of Critical Values for the MmCSRS-APSD Analysis
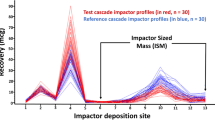
The CI profile in Fig. 2, which is identical to the profile M8 in the previous publication (4) and consists of eight deposition sites, represented the (population) mean vector for simulating R profiles. The profile M8 was selected since it resembles the general shape of real ISM profiles (after rank ordering the ISM sites by decreasing magnitude of their normalized deposition) across different inhalation products (i.e., dry powder inhalers, metered-dose inhalers, and inhalation suspension). The rank ordering of CI profiles is possible here as the MmCSRS, according to its computational form, is independent of the ordering of the deposition sites (3, 4). The impact of selecting profile M8 as R product for deriving critical values and on the overall assessment of the 55 PQRI CI profiles by the MmCSRS-APSD analysis is discussed below.
CI profile M8 (presented as rank-ordered by decreasing magnitude of normalized deposition). This profile was used for derivation of the acceptance limits for the MmCSRS-APSD analysis (Table II). Deposition (site) 35% (1, 2), 10% (3, 4), 4% (5, 6), and 1% (7, 8)
The behavior of the MmCSRS was then investigated for T profiles that differed from the specific R profile in each of the eight deposition sites by 10, 15, 20, 25, or 30%. To ensure mass balance between T and R products, the following procedure for generating T profiles, which is identical to that in the previous publication (4), was performed. While pairing the two sites of the mean R profile with identical depositions (Fig. 2), the T profiles were then constructed from the R profiles by having the deposition on four sites to be higher than those of the R profiles and the deposition on the other four sites to be lower than those of the R profiles. For instance, when a 10% difference between T and R was investigated, the (population) mean depositions on the eight sites of the T profile were 38.5, 31.5, 11, 9, 4.4, 3.6, 1.1, and 0.9%. The variability of both the T and R CI profiles was assigned by setting CVs of all deposition sites to 2.5, 5, 10, 15, 20, 25, 30, 35, 40, or 45%. Thus, the variability of T and R profiles, in terms of their CVs, was the same for deriving the critical values for the MmCSRS-APSD test.
In one aspect, the simulation method for generating T and R profiles that was used in the current paper for generating profiles suitable to construct critical values for the MmCSRS-APSD analysis represented an improvement over that one in the previous paper (4). Instead of simply assigning the CVs to the non-normalized profiles, a modified simulation procedure that consisted of additional three simulation cycles and incorporated inter-site correlations ensured that the CV of each deposition site remained the same after normalization (Fig. 3). As a result, these modifications improved the quantitative aspect that correlates MmCSRS with the variability of the R product, while they did not affect the qualitative findings of our previous investigation (4).
For each of the 50 scenarios (from combinations of five differences in mean deposition between T and R and ten variances investigated), 20,000 sets of 30 T and 30 R profiles were simulated, and subsequently, the MmCSRS was calculated. The averages of the 20,000 MmCSRSs were then regressed against the respective inverse-squares of the CV for each of the five investigated differences between T and R products (10, 15, 20, 25, or 30%), and the resulting estimated slope and intercept parameters were used for constructing critical values for the MmCSRS-APSD analysis. The linear regression was warranted since a perfect linear relationship between the inverse square of the CV of the R product and the MmCSRS was previously demonstrated (see above and (4)).
Application of Proposed CI Equivalence Test to the 55 PQRI CI Scenarios
The means and the standard deviations on all deposition sites of both the T and R products were publicly available for the 55 PQRI CI profile scenarios (8, 9). Those means and standard deviations were used as population means and population standard deviations for profile simulations, respectively. Since information on the inter-site correlation was not available, profile simulation was performed under the assumption of no inter-site correlation. For each of the 55 scenarios, Monte Carlo simulations were used to generate 20,000 independent sets of 30 T and 30 R profiles while assuming a multivariate normal distribution (see above).
For each of the 55 scenarios, the proposed CI equivalence test (Fig. 1) was applied to each of the 20,000 sets of 30 T and 30 R profiles. The total mass on all deposition sites was used as a surrogate for single actuation content at the first step of the proposed CI equivalence test. The deposition sites 6–13, 4–11, and 3–10 were defined as ISM sites for scenarios 1–44, 45–51, and 52–55, respectively. This selection appears to correspond to the definition of the ISM for an Andersen Cascade Impactor setup. For scenarios 52–55, the CI equivalence test was also applied when defining deposition sites 4–10 as ISM, which appear to correspond to the ISM definition for a Next Generation Impactor setup. These selections were in accordance with the evaluation of the PQRI WG (6, 8, 9).
For each of 55 scenarios, the percent of the 20,000 tests that resulted in a pass for steps 1 and 2 were recorded. At the third step as described above, a two-sided non-parametric 90% BCA bootstrapping confidence interval for the MmCSRS was calculated (14). Two thousand bootstrapping replicates were obtained for each of the 20,000 sets. The upper bound of the 90% confidence interval was then compared to the appropriate critical value for the APSD profile comparison within the ISM considering the variability of the R product determined with the specific metric for reference variance scaling (Eq. 2). For each of 55 scenarios, the percent of the 20,000 BCA confidence intervals for which the upper bound was below the respective critical value were recorded. The proposed CI equivalence test (Fig. 1) was applied in a stepwise fashion to each of the 20,000 sets of 30 T and 30 R profiles. The results were summarized as follows: (1) PQRI WG, a pass was assigned when >50% of the members classified a scenario as equivalent; (2) stepwise MmCSRS-APSD-test, a pass for each step of the CI equivalence test was assigned when >50% of 20,000 sets passed the PBE test at steps 1 and 2 and the MmCSRS-APSD profile test at step 3. The MmCSRS-APSD profile test was performed for acceptance limits of 10, 15, 20, 25, and 30%. For each acceptance limit, a pass for the CI equivalence test was assigned when all of the three steps resulted in a pass.
RESULTS
Construction of Critical Values for the MmCSRS-APSD Analysis
The estimated intercept and slope parameters for constructing critical values for the MmCSRS-APSD analysis as a linear function of the inverse-square of the CVs are given in Table II. Figure 4 provides a visualization how these parameters can be applied, together with variability of the R product, for constructing of critical values for the MmCSRS-APSD analysis.
Visualization of critical values for different acceptance limits and variability of the R product (critical value plot). The scaling of the x-axis is the inverse square of CV% while the labels of the x-axis refer to the actual CV%. The black bar represents a 90% BCA confidence interval for a sample of 30 T and 30 R products. Differently shaded areas and associated lines represent different acceptance limits. In this case, the estimated variability of the R product is 7.5% using the proposed metric for reference variance scaling (Eq. 2). Hence, the T product would pass acceptance limits ≥25% (upper bound of confidence interval below 25% acceptance line for given R variability) and fail acceptance limits ≤20% (upper bound of confidence interval above 20% acceptance line for given R variability)
Application of Proposed CI Equivalence Test to the 55 PQRI CI Scenarios
The results of applying the proposed CI equivalence test to the 55 PQRI CI profile scenarios are shown in Table III. A visualization of the results is provided in Fig. 5. Since the results for the scenarios 52–55 when defining the ISM over seven sites were very similar (and did not affect the overall assessment) to those when defining the ISM over eight sites, only the results of the eight ISM sites are presented. Application of the proposed CI equivalence test for the 55 scenarios yielded 58, 65, 71, 75, and 65% match to the assessment of the PQRI WG, when applying acceptance limits of 10, 15, 20, 25, and 30% for the MmCSRS-APSD analysis, respectively. Table IV lists those scenarios where the outcome (i.e., pass or fail) of the proposed CI equivalence test did not match that of the PQRI WG.
Categorization of 55 PQRI CI profile scenarios based on the proposed CI equivalence test. Point estimates (filled circles) and 90% BCA bootstrap confidence intervals (vertical bars) are displayed for those scenarios that received a pass in the first (total mass) and second step (ISM) in >50% of the 20,000 cases. Point estimates and upper and lower bounds of the confidence intervals are the averages over the 20,000 cases; numbers above the vertical bars represent the identifier of the 55 PQRI scenarios; green-colored scenarios were classified by ≥70% of the PQRI WG as equivalent; orange-colored scenarios were classified by ≥30 and <70% of the PQRI WG as equivalent; red-colored scenarios were classified by <30% of the PQRI WG as equivalent. The proposed metric (Eq. 2) was used for reference variance scaling. Differently shaded areas and associated lines represent different acceptance limits (e.g., 20% difference on all ISM sites). Results are presented in three graphs with different ranges of the CV% of the R product and MmCSRS to allow visualization of the entire collection with sufficient resolution. The scaling of the x-axis is the inverse square of CV% while the labels of the x-axis refer to the actual CV%. The CV% was estimated by the proposed metric for reference variance scaling (Eq. 2). The estimates are given in Table II
DISCUSSION
The proposed stepwise CI equivalence test utilizes the characteristics of the MmCSRS, which has its advantages over the original chi-square ratio method. Previous publications demonstrated that the MmCSRS is robust when T and R profiles are identical, as the MmCSRS was independent of the number of deposition sites and shape of the CI profiles (3). The second paper (4) provided evidence that the MmCSRS is sensitive to differences between T and R profiles. One of the key findings of that publication was that MmCSRS is more sensitive to differences between T and R profiles on high-deposition sites. This is considered as beneficial for equivalence testing of CI profiles as it decreases the likelihood of failing identical CI profiles by chance, in part, due to increasing analytical variability associated with lower-deposition sites. A second important finding of that paper was that critical values for APSD equivalence testing based on the MmCSRS need scaling on the variability of the R product. The latter is expected to be important for consistently being able to discriminate equivalent from inequivalent CI profiles.Footnote 2 In fact, the absence of such a metric for reference variance scaling and the use of a constant critical value in the original CSRS approach is one of the main reasons why it could not consistently classify the 55 CI profile scenarios in the previous investigation. The need for this reference variance scaling was incorporated in the proposed stepwise CI equivalence test by defining critical values for the MmCSRS-APSD analysis as a function of the variability of the R product. The critical value plot (Fig. 4) shows this perfect linear relationship between the MmCSRS and the inverse square of the CV of the R product. The critical value plot and/or the parameters given in Table II together with the metric for reference variance scaling in Eq. 2 are then the basis for performing the MmCSRS-APSD analysis. In order to show equivalence, the upper bound of a 90% confidence interval for the MmCSRS needs to be smaller than the specific critical value obtained through the critical value plot and/or the parameters in Table II. Since the distribution of the MmCSRS is unknown,Footnote 3 a non-parametric two-sided 90% confidence interval is calculated based on a BCA bootstrapping method (14). The upper bound of the confidence interval is then compared with the specific critical value. Since an acceptance limit for a profile comparison test is yet to be established, critical values were derived for various acceptance limits (i.e., 10, 15, 20, 25, and 30% difference between T and R products).
In the previous publication, the behavior of the MmCSRS when T and R differ from each other on eight deposition sites was evaluated in detail for various cases (4). It was demonstrated that a certain difference (e.g., 10%) between T and R profiles on all deposition sites resulted in a larger value for the MmCSRS when the profiles were maximally skewed compared with the case where the deposition on all sites is uniform. The logical explanation for this behavior is the fact that a 10% difference results in larger absolute differences between T and R when the profiles are maximally skewed and, thus, have sites with a large deposition. However, since visual analysis of real CI profiles (after rank ordering the ISM sites by decreasing magnitude of their normalized deposition) had displayed good agreement between different inhalation devices (i.e., dry powder inhalers, metered-dose inhalers, and inhalation suspensions), this finding became not relevant for defining critical values for the MmCSRS-APSD analysis (results are not shown, propriety information). Consequently, a single CI profile, which resembles the shape of real CI profiles after rank ordering the ISM sites by decreasing magnitude of their normalized deposition, could be applied for derivation of the critical values for the MmCSRS-APSD analysis. Since the CI profile M8, which is depicted in Fig. 2, did fulfill this criterion of resemblance to real CI profiles of inhalation products currently approved in the USA, it was selected for derivation of critical values for the MmCSRS-APSD analysis.
Using the critical value plots for real T and R profiles was complicated by the fact that the observed CVs for the R product differ across deposition sites.Footnote 4 As construction of the critical value plot is therefore contingent on a cumulative (“mean”) CV observed over all deposition sites, a metric that represents the overall variability of the included R sites while allowing a reduced impact of highly variable low deposition sites needed to be defined. Differing from the previous theoretical evaluations for which variability across stages was assumed to be similar, the here-described metric for reference variance scaling (Eq. 2) considers differences in the CV of individual sites while providing a normalized mean CV estimate that puts more weight on the higher deposition sites. More information on selection and behavior of this metric for reference variance scaling will be provided in a separated paper (manuscript in preparation).
The slopes of the critical value plots (Table II and Fig. 4) increase with increasing difference between T and R profiles. Thus, larger MmCSRS values are acceptable when larger difference between T and R products are allowed for APSD equivalence establishment.
The proposed stepwise CI equivalence test includes PBE analysis on single actuation content and ISM within the first two steps. This stepwise approach is important as MmCSRS-APSD analysis is based on relative site deposition (% of ISM) and as such only compares the relative shape of the CI profile without considering the absolute amount of drug deposited on a given site. Analysis of single actuation content and ISM ensure that T and R products emit an equivalent dose and deposit an equivalent amount on the sites to which the profile comparison test is applied, respectively. Moreover, the second step of the CI equivalence test is necessary since it allows the MmCSRS-APSD analysis (i.e., third step) to be applied only to deposition sites with a defined upper cutoff diameter (i.e., ISM sites). This stepwise approach incorporating the unique characteristics of the MmCSRS is expected to provide adequate sensitivity to product differences from an equivalence perspective, while preventing the failure of some products due to increased variability on low-deposition sites, like in the case of using univariate confidence intervals on each deposition site for equivalence evaluation.
The proposed CI equivalence test was applied to 55 characteristic CI profile scenarios (Fig. 5, Tables III and IV) that were published and categorized as equivalent or inequivalent by the PQRI WG (6, 8, 9). It should be remarked that the results of the PQRI WG evaluation were taken from the literature (6) and the specific methods and acceptance criteria that the PQRI WG applied for their evaluation were unknown. The results of the proposed CI equivalence test were compared with those of the PQRI WG. For acceptance limits greater than 10%, the proposed CI equivalence test produced results that were at least 65% (of the 55 CI profile scenarios) in agreement with those based on the PQRI WG evaluation. The smaller percentage of agreement for a 10% acceptance limit (58%, Table III) was attributed to scenarios 6, 20, 21, 24, 29, 33, 34, 35, 36, 39, 42, and 50, which all received a pass by >50% of the PQRI WG but failed to meet the 10% acceptance limit based on the proposed CI equivalence test. The agreement with the PQRI WG evaluation increased with increasing acceptance limit and eventually all of these scenarios received a pass for the proposed CI equivalence test (Tables III and IV). For scenarios 22, 26, 28, 30, 31, 32, 37, and 38 that all received a fail by ≥50% of the PQRI WG, the agreement between the results of the proposed CI equivalence and PQRI WG evaluation decreased with increasing acceptance limit and eventually all scenarios received a fail for the proposed CI equivalence test (Tables III and IV).
The best agreement (75%) was reached when the acceptance limit of 25% was used. For this acceptance limit, although a 100% agreement was not achieved between the results of the proposed CI equivalence test and the PQRI WG evaluation, the differences between these two assessments can generally be understood. Scenarios 1, 2, 14, 25, 45, 46, and 55 were classified as equivalent by >50% of the PQRI WG despite having an ISM difference of >10% (Table III and Supplemental Material). All of those scenarios failed the proposed CI equivalence test due to the inequivalence assessment at the second step (ISM PBE test; Table III). It should be noted that the acceptance limit of the PBE test for the mean difference (assuming equal variances) is generally about 10% (10).
Scenario 5 is considered a borderline MmCSRS-APSD case as simulated profiles passed individual equivalence tests (i.e., >50% of 20,000 sets passed the PBE tests for ISM and single actuation content and the APSD-MmCSRS analysis for 25 and 30% acceptance limits) but failed the stepwise approach (i.e., less than 50% of the data sets passed all three tests simultaneously). Scenarios 17 and 32 on other hand are borderline PQRI cases (i.e., very close to 50% limit), where PQRI WG evaluation resulted in a pass in 64 and 50%, respectively (Table III). For Scenarios 10 and 13, despite that the PQRI WG considered them as equivalent, the visual difference in the overall APSD profile within ISM stages was apparent, especially with respect to the highest deposition site. In contrast, for scenarios 26 and 38, the PQRI concluded inequivalence although the mean APSD profile within ISM appeared to be similar (i.e., the highest deposition site are highly similar and the lower deposition sites show minor differences between the R and T products). It should be emphasized that the results of MmCSRS-APSD analysis for scenarios 10, 13, 26, and 38 were consistent with the characteristics of MmCSRS, which showed greater sensitivity to changes in the high-deposition site.
The Supplemental Material contains figures of the mean ISM profiles (T and R) and the T/R ISM ratios for the 55 CI profile scenarios to allow the readers to make their independent equivalence evaluation.
Overall, the proposed CI equivalence test provides a classification for all 55 CI profile scenarios consistent with what was expected from the characteristics of MmCSRS. Interestingly, a different metric for reference variance scaling that was used in the preliminary evaluation of the appropriateness of the proposed CI equivalence test resulted in very similar qualitative classifications of the 55 PQRI CI profile scenarios as in the current assessment (15). Hence, the impact of the selected metric for reference variance scaling seems minor for the analysis of PQRI scenarios.
It should also be noted that our evaluation of the proposed CI equivalence test did not include an analysis of the impact of increasing or decreasing sample sizes on the MmCSRS. It is apparent from the properties of the constructed confidence intervals (without the need to perform a simulation based analysis) that an increase in sample size (e.g., 40 T and 40 R samples) would reduce the width of the confidence intervals and, thus, would lead to an increased power to conclude equivalence if T and R are indeed equivalent. Similarly, a decrease in sample size (e.g., 20 T and 20 R samples) would increase the width of the confidence intervals and, thus, would lead to a decreased power. In the same context, it is irrelevant from a numerical point of view if, e.g., three batches of ten samples or six batches of five samples are obtained as the overall variability in all samples determines the width of the confidence intervals for all three steps of the proposed CI equivalence test. On the other hand, a simulation-based analysis, which estimates powers to conclude equivalence when T and R products are identical or different within the acceptance limits as a function of the sample size and overall variability in all samples, would be appealing.
Finally, it is important to emphasize that the proposed stepwise CI equivalence test was designed to compare the APSD of a T with that of an R product. Conclusions about the in vivo equivalence (e.g., pharmacokinetic and pharmacodynamics endpoints) cannot be drawn from the proposed CI equivalence test as this is outside the scope of this investigation. The in vitro in vivo correlation of APSD and pharmacokinetic and/or pharmacodynamics endpoints remains poorly understood. More experimental work will be needed by all parties (academic and clinical researchers, regulators, and industry) to investigate whether it is possible to establish clinical relevance of APSD and quantitative correlations (or absence of correlations) between this in vitro measure and in vivo data.
CONCLUSIONS
The proposed stepwise CI equivalence test and scaling of critical values for the MmCSRS-APSD analysis (the third step of the proposed test) by the variability of the R product provides an adequate assessment for most of the 55 CI profile scenarios that is in accordance with the evaluation by the PQRI WG. The results indicated that the use of an acceptance criterion of 25% for the MmCSRS-APSD analysis best matches with the decisions made by PQRI WG. It is, however, important to emphasize that the PQRI WG assessment might not be based on a single method and could be a statistical or subjective analysis, and the exact equivalence criteria are not available in the literature. Moreover, R product inter-batch variability and change in R product performance over shelf life should also be considered during the selection of critical value, all of which could not be assessed with the PQRI profiles. Overall, the proposed CI equivalence test including the MmCSRS-APSD analysis has been demonstrated to be a promising approach for consistently discriminating equivalent from inequivalent CI profiles.
Notes
ISM is defined as the sum of the drug mass on all CI stages plus the filter, but excluding the initial stage because of its lack of a specified or well-defined upper cutoff diameter.
This was one of the main points, the PQRI working group criticized when evaluating the behavior of the original chi-square ratio method.
The distribution of the mCSRS when T and R products are identical is an approximate F distribution when certain conditions hold true (3).
This was not considered in previous work that was based on identifying general relationships between R variability and MmCSRS.
References
Lee SL, Adams WP, Li BV, Conner DP, Chowdhury BA, Yu LX. In vitro considerations to support bioequivalence of locally acting drugs in dry powder inhalers for lung diseases. AAPS J. 2009;11:414–23.
Evans C, Cipolla D, Chesworth T, Agurell E, Ahrens R, Conner D, et al. Equivalence considerations for orally inhaled products for local action-ISAM/IPAC-RS European workshop report. J Aerosol Med Pulm Drug Deliv. 2012;25:117–39.
Weber B, Hochhaus G, Adams W, Lionberger R, Li B, Tsong Y, et al. A stability analysis of a modified version of the chi-square ratio statistic: implications for equivalence testing of aerodynamic particle size distribution. AAPS J. 2013;15:1–9.
Weber B, Lee SL, Lionberger R, Li BV, Tsong Y, Hochhaus G. A sensitivity analysis of the modified chi-square ratio statistic for equivalence testing of aerodynamic particle size distribution. AAPS J. 2013;15:465–76.
Adams WP, Christopher D, Lee DS, Morgan B, Pan Z, Singh GJ, et al. Product Quality Research Institute evaluation of cascade impactor profiles of pharmaceutical aerosols, part 1: background for a statistical method. AAPS PharmSciTech. 2007;8:E32–7.
Christopher D, Adams W, Amann A, Bertha C, Byron PR, Doub W, et al. Product Quality Research Institute evaluation of cascade impactor profiles of pharmaceutical aerosols, part 3: final report on a statistical procedure for determining equivalence. AAPS PharmSciTech. 2007;8:65–74.
Christopher D, Adams WP, Lee DS, Morgan B, Pan Z, Singh GJ, et al. Product Quality Research Institute evaluation of cascade impactor profiles of pharmaceutical aerosols: part 2—evaluation of a method for determining equivalence. AAPS PharmSciTech. 2007;8:E39–48.
PQRI APSD Profile Comparisons Working Group. Realistic Scenarios 1–33 of 55. http://www.pqri.org/commworking/minutes/pdfs/dptc/psdpcwg/Addl/Realistic Scenarios 1–33 of 55.pdf. Accessed 08 June 2014
PQRI APSD Profile Comparisons Working Group. Realistic Scenarios 34–55 of 55. http://www.pqri.org/commworking/minutes/pdfs/dptc/psdpcwg/Addl/Realistic Scenarios 34–55 of 55.pdf. Accessed 08 June 2014
Hollander M, Wolfe DA. Nonparametric statistical methods. 2nd ed. New York: John Wiley & Sons; 1999. p. 178–86.
Labes D. Power and Sample size based on two one-sided t-tests (TOST) for (bio)equivalence studies. http://cran.r-project.org/web/packages/PowerTOST/PowerTOST.pdf. Accessed 08 June 2014
R Development Core Team. R: A language and environment for statistical computing. http://www.r-project.org/. Accessed 08 June 2014
Genz A, Bretz F, Miwa T, Mi X, Leisch F, Scheipl F, et al. Multivariate normal and t distributions. http://cran.r-project.org/web/packages/mvtnorm/mvtnorm.pdf. Accessed 08 June 2014
Efron B. Better bootstrap confidence intervals. JASA. 1987;82:171–85.
Weber B, Adams W, Lionberger R, Li B, Tsong Y, Hochhaus G, et al. Evaluation of statistical methods for determining equivalence of aerodynamic particle size distribution. Respir Drug Deliv. 2012;3:803–8.
Author information
Authors and Affiliations
Corresponding author
Additional information
Guest Editors: Lawrence Yu, Sau L. Lee, Guenther Hochhaus, Lana Lyapustina, Martin Oliver, and Craig Davies-Cutting
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Appendix
Appendix
Construction of 90% BCA Confidence Intervals for the MmCSRS
A sample size of 30 T and 30 R products will be assumed for simplicity. However, this method could be applied to any other sample size of T and R products. Moreover, this example assumes a bootstrapping sample size of 2000. For details and statistical background on the calculations, please refer to the original publication (14).
1. For both the T and R products obtain a bootstrapping sample with size equal to 30 by resampling with replacement.
2. Repeat (1) 2000 times.
3. Obtain the MmCSRS for each of the 2000 bootstrapping samples of 30 T and 30 R products.
4. For the distribution of 2000 MmCSRS that is obtained in (3), the Z L and Z U percentile of the 2000 MmCSRS are the lower and upper bounds of the BCA confidence interval, respectively. The computation of Z L and Z U is shown below.
where Φ is the cumulative distribution function of the standard normal distribution and \( {z}_{0.95} \) is the 95th percentile of the standard normal distribution
where
\( {\widehat{\theta}}_{-i} \) is the MmCSRS obtained for the original sample of 30 T and 30 R products by leaving a single T and R sample out (i.e., 900 MmCSRS based upon either 30 T and 29 R or 29 T and 30 R are obtained)
and
and
where I is an indicator function (i.e., equal to one if the condition is true and zero otherwise), \( {\widehat{\theta}}_b \) is the MmCSRS for bth bootstrapping sample, and \( \widehat{\theta} \) is the MmCSRS of the original sample.
Rights and permissions
About this article
Cite this article
Weber, B., Lee, S.L., Delvadia, R. et al. Application of the Modified Chi-Square Ratio Statistic in a Stepwise Procedure for Cascade Impactor Equivalence Testing. AAPS J 17, 370–379 (2015). https://doi.org/10.1208/s12248-014-9698-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1208/s12248-014-9698-0