Abstract
Background
Micro-RNAs (miRNAs) have been reported as an emerging biomarker in many cancer types. They are used as diagnostic and prognostic biomarkers and could be considered therapeutic targets in treating the same.
Main body
Studies have proven that miRNAs play an essential role in molecular cancer pathophysiology, including oral squamous cell carcinoma. Distinct expression profiles of different miRNAs have been demonstrated in oral squamous cell carcinoma. Among the miRNAs, the miR-31 has strong potential as a unique biomarker in head and neck squamous cell carcinoma, and the increased expression was correlated to a poor clinical outcome with a likely contribution to oral carcinogenesis.
Short conclusion
The recent research on different aspects of miR-31 as a biomarker and also its potential application in the development of therapy for oral squamous cell carcinoma has been focused in this review.
Graphical abstract

Similar content being viewed by others
Background
Squamous cell carcinoma of the oral cavity is the sixth most common cancer globally. However, despite advanced diagnostic aids, the prognosis remains unfavorable. This might emphasize the requirement for immediate identification of biomarkers specific to Oral Squamous Cell Carcinoma (OSCC) for the early-stage diagnosis. Biomarkers are specialized molecular signatures specific to a disease [1]. Recent improvements in molecular diagnostic methods provide diverse biomarker identification specific to OSCC. The microRNAs (miRNAs) are considered attractive biomarkers since they are tissue-specific and continuously circulate in body fluids. They are small (18–22 nucleotides) endogenous, noncoding RNA molecules accountable for various cellular and metabolic pathways comprising cell proliferation, differentiation, and survive in normal and disease states. In addition, miRNAs usually participate in genomic instability, transcriptional control, epigenetic regulation changes and biogenesis machinery abnormalities [2]. miRNA genes are typically found in cancer-related chromosomal fragile areas, the minimal region of loss of heterozygosity, minimal regions of amplification or common breakpoint regions. Gene amplification, deletion or translocation are thought to produce changes in miRNA expression levels in malignant cells [3]. These miRNAs are frequently decreased in expression or deleted in cancer, suggesting that they are tumor suppressors. However, miRNAs have also been identified as potential oncogenes. Few miRNAs are repeatedly explored, and it has also been recognized as a helpful marker in clinical application for predicting, diagnosing and patient management. A recent systematic review revealed that a higher level of miR-21, miR-29b, miR-31, miR-155, miR-183 and miR-221 were associated with poor prognosis and patient survival and appeared to have significant predictive value in OSCC [4]. The miR-31 gene is found on the chromosome band 9p21.3, around 500 kb away from the CDKN2A and CDKN2B tumor suppressor genes, which code for the cell cycle inhibitor proteins p15 and p16, respectively.
Several types of cancers have expressed a genomic loss at this locus. Deletion of the miR-31 locus or epigenetic silencing to downregulate its gene expression are a few mechanisms observed in different cancers. In gastric, liver, breast, ovarian and prostate cancer, the miR-31 expressions were observed to be low, whereas the miR-31 expressions in lung, colorectal, esophagal and OSCC have shown elevated expression levels [5]. A latest study also indicated a positive association between the miR-31 expression and increased consumption of iron and vitamin C and an inverse association with increased intake of total sugar, cholesterol, vitamin-B9 and zinc in Head and Neck Squamous Cell Carcinoma (HNSCC) patients. The author also discusses the roles of miRNA in oral carcinogenesis with particular emphasis on miR-31 and its various possible functions in oral carcinogenesis and considers miR-31 as a diagnostic and prognostic biomarker [6]. Majorly miR-31 plays an essential role in regulating cell cycle and proliferation, hypoxia pathways and increasing the stemness in OSCC subjects (Fig. 1).
Main text
Role of miRNAs in oral carcinogenesis
The miRNAs are found in Caenorhabditis elegans and play a critical role in controlling target gene expression. miRNA clusters are groups of miRNA expressing genes driven by a single promoter, transcribed in the same direction, and not separated by a transcription unit [7]. Let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, let-7g, let7i, miRNA-98, and miRNA-202 are all members of the let-7 family of miRNAs. Let-7 decreases the expression of endoribonuclease dicer (DICER), high-mobility group AT-hook 2, and Ras in OSCC cells. Let-7d knockdown enhances epithelial-mesenchymal transition, migration, and invasion of OSCC cells, according [8]. In a study conducted by Maheswari et al., 167 patients with OSCC, 78 patients with OPMD, 147 healthy controls, and 20 patients with aphthous stomatitis were studied using saliva samples. In these investigations found that five miRNAs, miRNA-31, miRNA-24, miRNA-27b, miRNA-21, and miRNA-184, are elevated in OSCC, whereas 15 miRNAs, miRNA-200a, miRNA-125a, miRNA-11, miRNA-191, miRNA-136, miRNA-147, miRNA-1250, miRNA-632, miRNA-646, miRNA-668, miRNA-877, miRNA-503, miRNA-200a, mi-323-5, and miRNA-145, were downregulated in OSCC compared with healthy controls [9].
CpG hypermethylation induced downregulation of miR-34b, miR-137, miR-193a, and miR-203 expression in 18 OSCC cell lines, according to miRNA profiling. Two miRNAs, miR-218 and miR-585, were often silenced by DNA hypermethylation in OSCC. Oral cancer may be aided by epigenetic suppression of miR-218 and subsequent activation of the mTOR–Akt signaling pathway mediated by overexpression of RICTOR (a putative target of miR-218) [10]. A miRNA profiling of primary OSCC tissue specimens indicated that 46 differentially expressed miRNAs were able to activate PI3K/AKT signaling genes and alter the p53 signaling pathways. As a result, miRNAs play a crucial role in the AKT/mTOR pathway's regulation [11].
miRNA-375 was shown to be downregulated in OSCC and progressive oral leukoplakia compared to non-progressive lesions, indicating that it functions as a tumor suppressor by decreasing SLC7A11 expression during oral carcinogenesis [12]. Specific miRNAs can cause cancer cells to undergo ferroptosis by decreasing SLC7A11 expression [13]. Understanding these miRNA molecules and their control would be a significant step in designing a more tailored cancer therapy because oral cancer is complicated and linked to the host pathophysiology [14]. Of all the miRNAs studied in OSCC, miR-31 was shown to discriminate clearly between healthy subjects and oral cancer patients making it a potential biomarker [15].
miR31 as oncogenes and tumor suppressors
An in-vitro study revealed that cancer cell lines had significantly greater levels of miR-31-5p expression than regular epithelial cell lines (NOK-16 B). In addition, a miR-31-5p has also shown increased expression in cell lines that are more migratory and invasive than their matched cell lines, and the inhibitor significantly prevents the proliferation of the same. The miR-31-5p mimics considerably increased the ability of normal HACaT epithelial cells to proliferate [16]. CASC2 (cancer susceptibility 2) is a tumor suppressor gene that was associated with the chemoresistance of diverse tumors [17, 18]. The advancement of OSCC is said to be curbed by CASC2 [19,20,21]. Wang et al. [22] found that in (first-line chemotherapy drug cisplatin) DDP resistant OSCC cells CASC2 delivers its actions via miR-31-5p. A member of the KANK family (KN motif and ankyrin repeat domain-containing protein 1—KANK1) [23], mediates the actin polymerization cytoskeleton construction [24]. Additionally, in various tumors, it has been proved as a suppressor [25,26,27] and the advancement of nerve sheath tumors of a human malignant peripheral nerve cell is impeded by KANK1, posing their role in neurodegeneration [25]. A research study done by Wang and his colleagues investigated the role of KANK1, CASC2, and miR-31-5p in OSCC and found the following—there was a downregulation of KANK1 mRNA in OSCC tissues compared to the healthy ones. As a result the KANK1 protein expression was also downregulated substantially in the OSCC tissues. And in OSCC tissues the KANK1 expression has a negative correlation with miR-31-5p and potential binding sites for miR-31-5p were found in KANK1. Hence, miR-31-5p targets KANK1 in OSCC cells. The enhanced expression of KANK1 its mRNA and protein has been reversed by a miR-31-5p introduction which implements the fact that in DDP-resistant OSCC cells CASC2 implies its function by miR-31-5p [22], p. 2. And CASC2 modulates KANK1 by sponging miR-31-5p in OSCC cells that are DDP resistant. Data suggests that in vivo increased CASC2 improves DDP sensitivity and encourages cell apoptosis in OSCC cells that are DDP-resistant executing the overall fact that miR-31-5p plays its part through KANK1 in OSCC cells [22], p. 202. One of the new and potential treatment targets for OSCC can be LncRNAs [26] and these findings could lead to a possible new strategy for improving DDP-resistant in OSCC cells. In conclusion sensitivity towards DDP of OSCC cells is enhanced by CASC2 by the upregulation of KANK1 via miR-31-5p.
An adaptive protein that plays multifarious part in various cell functions including oncogenesis is NUMB [28, 29]. It is said to be a regulator of cell fate as it asymmetrically partitions at mitosis [30] and a tumor protein p53 regulator [31] and an endocytic protein as well [32]. NUMB can access various oncogenic signaling pathways with p53, NOTCH and Hedgehog pathway. The suppressor function of NUMB against OSCC have been uncovered by many studies and various roles mediated by six NUMB isoforms resulting from alternative splicing have been widely investigated. Isoform of NUMB like Numb1 to Numb4 have the potential to regulate the development of OSCC. NUMB can suppress NOTCH signaling by directly attaching to the NOTCH intracellular domain (NICD), preventing it from accessing the nucleus. NUMB can also directly inhibit NOTCH by recruiting Itch to polyubiquitinate and degrade Notch proteins [33]. Ectopic expression of miR-31 is remarkably upregulated in OSCC [33], which is consistently expressed in saliva and serum or plasma of OSCC patients [34]. miR-31 may bind to the 3′ untranslated region (UTR) of factor-inhibiting hypoxia-inducible factor (FIH), a hypoxia-inducible factor (HIF) regulatory factor that prevents HIF from functioning as a transcriptional regulator under normal environments [33]. Monocarboxylate Transporter (MCT) family members are responsible for metabolite transport in metabolic activities. MCT1 and/or MCT4 expression upregulation has also been demonstrated to be a prospective indicator of HNSCC. NUMB binds MCT1/MCT4 in OSCC cells, causing MCT1/MCT4 degradation, decreasing oncogenicity, and increasing glycolytic metabolism [33]. NUMB had an effect on lactate synthesis and glycolytic respiration in OSCC cells. MiR-31 has been demonstrated to target SIRT3 to impair mitochondrial function and ACOX1 to modify the lipidome profile in OSCC. The inhibition of NUMB by miR-31 has both proliferative and metabolic consequences [33]. Overall, Chou et al. [33] identified that the existence of the miR-31-NUMB-MCT1/MCT4 axis in modulating oncogenesis and metabolic switching, implying that disrupting this cascade may prevent tumor pathogenesis and aerobic glycolysis. NUMB activation through CRISPR/dCas-SAM mechanism and the regulatory axis miR-31-NUMB-MCT1/MCT4 appears to be a significant therapeutic target in OSCC [33]. As NUMB is a direct target of miR-31 and numerous other oncogenic miRNAs, the findings of a study confirmed that miR-31 is an oncogenic miRNA that also affects OSCC's complicated metabolic control. miR-31 inhibits NUMB, which has carcinogenic and metabolic consequences (Fig. 2). A twofold sgRNA-guided CRISPR/Cas9 cleavage technique is used to validate the efficiency of miR-31 deletion. This technique might be developed further to produce effective miR-31 inhibition in vivo tumor treatment [35], p. 2.
Mechanism of miR-31 in OSCC cell proliferation and cell cycle regulation
One of the most prevalent cellular hallmarks in malignancies is increased Wnt /catenin pathway activation. The transcription of several effector genes involved in carcinogenesis and cancer progression, such as c-MYC and cyclin D1, is triggered when canonical Wnt signaling is activated. Furthermore, in a subset of cancers, disruption of non-canonical Wnt signaling may contribute to tumorigenesis and cancer progression through metabolic and inflammatory changes. Recent research suggests that these Wnt signaling pathways may also influence malignant cancers in the oral cavity [35], p. 2. An experimental model study on Drosophila melanogaster explored the functional relationship between miR-31 and its putative target in-vivo. Overexpression of miR-31 resulted in a considerable reduction in tissue growth, implying that it functions as a tumor suppressor, and the level of Wnt less mRNA, a vital regulator of the Wnt signaling pathway, was significantly reduced. In parallel investigations, a comparable miR-31 dependent control of human Wnt less mRNA has been verified in OSCC cells. Furthermore, it has been proposed that miR-31 regulates the cell cycle and proliferation of OSCC cells by downregulating cyclin D1 and C-MYC, two of the vital transcriptional targets of Wnt signaling [36].
A recent study on OSCC tissue samples demonstrated that macrophage-derived exosomes (M2 exo) exhibited a high level of miR-31-5p and the M2 exo –mediated increase in OSCC cell proliferation and growth depended on this miRNA. M2 exo contributes to OSCC tumorigenesis via the miR-31-5p /LATS2 (large tumor suppressor 2) axis of the Hippo signaling pathway and facilitates their tumor inducing effect [37]. The miR-31 target gene CXCL12 is significantly changed in oral pre- and carcinoma tissues. This is critical for tumor growth and metastasis; moreover, hypermethylation of the promoter region has been associated with decreased CXCL12 expression in head and neck cancer [38]. Another study denoted that, while miR-31(*) expression is upregulated in OSCC tissues, it is less abundant than miR-31. The proliferation and migration of SAS and FaDu cells are inhibited by miR-31(*). Furthermore, miR-31(*) inhibits RhoA production by targeting the 3'UTR of the protein. OSCC cells have been shown to proliferate and migrate less when RhoA expression is knocked down. However, administration of pre-miR-31 increases the oncogenicity of OSCC by upregulating both miR-31 and miR-31(*) [39].
miR-31 as a hypoxia regulating pathway
The miR-31 functioned as an oncogene in HNSCC by downregulating ARID1A (AT-Rich interaction Domain 1A). In addition, in normoxic conditions, ectopic stable expression of miR-31 suppressed the tumorigenicity of HNSCC cells by inversely regulating FIH. The miR-31 causes hypoxia in OSCC by blocking FIH, which stimulates HIF1. MIR31HG interacts with miR-31 to form a complex with HIF1, which subsequently enhances HIF1/p300 binding to hypoxia response elements [33]. The increase in miR-31 in OSCC tissues facilitates oral oncogenesis through modulating hypoxia pathways in oral cancer cells by targeting hypoxia inducing factor inhibiting protein. miR-31 mutates the HIF pathway to increase tumor formation under normoxic circumstances by targeting the 3" UTR of FIH. FIH is a sensor that detects oxygen levels. Asparagine hydroxylates HIF-1α when the oxygen level is available. As a result, HIF-1α lacking the COOH terminal transactivation domain is incapable of causing the synthesis of vascular endothelial growth factor (VEGF). The inhibitory effect of FIH on growth is diminished in hypoxic conditions. There is a negative association between the amount of FIH and VEGF when oxygen levels are over the threshold. Under normoxic conditions, the miR-31 suppresses the deleterious effects of FIH while activating VEGF expression in HNSCC [40].
HIF-1α was a miR-31 downstream mRNA, and its dysregulation was linked to miR-31-mediated neuronal cell damage. VEGF produced by the endothelial traumatic neuron damage or HIF-1α overexpression increased α levels. In addition, HIF-1α boosted vascular endothelial growth factor-A expression, which increased neuronal cell damage [41]. In addition, the miR-31 FIH axis has been identified to influence glycogen metabolism keratinocyte development in corneal via Notch signaling [42]. According to [43], p. 2 miR-31-5p had a functional impact on oral cancer cell motility and direct modulation of the rate-limiting enzyme in peroxisomal β-oxidation, ACOX1. In addition, this mis regulated axis alters lipid metabolomes, including promigratory prostaglandin E2, by augmenting ERK-MMP9 signaling. Clinically increased levels of PGE2 in saliva were also observed, which is in association with local invasion [43], p. 2. Exosomes have been demonstrated to induce exosome release and cause considerable changes in cellular contents and activities in response to hypoxia, showing that exosomes play a vital role as regulators in distant intercellular communication [16]. OSCC cell metabolism and aggressiveness are both influenced by the miR-31-SIRT3 (Silent information regulator3) regulatory axis. SIRT3 expression attenuated the miR-31 enhanced tumor cell migration and invasion. The mitochondrial membrane potential and structural integrity were both harmed by miR-31-SIRT3. This axis was dysregulated, resulting in oxidative stress and a transition from aerobic to glycolytic metabolism in tumor cells [44].
mir-31 and increased stemness of cancer cells
Hsa-miR-31 is a known oncogene that has previously been linked to Nanog/OCT4/SOX 2 stemness factors and lower survival. The epithelial cell adhesion molecule (EPCAM) expression was also accountable for the stemness of cancerous tissue. When overexpressed, negative regulators of oncogenic pathways are inhibited, allowing OSCC cancer cells to proliferate and become tumorigenic. A possible mechanism is that the miR-31 collaborates with hTERT to immortalize NOKs and contributes to early-stage oral carcinogenesis by inhibiting FIH with VEGF and increasing cellular immortalization via p53 mutation and VEGF upregulation. In addition, the EGFR activation also contributed to the increase of miR-31 in oral cancer tissues [45]. Studies have revealed that the EGFR/AKT signaling cascade can promote miR-31 production by C/EBPβ, which is essential because EGFR overexpression is linked to OSCC development [46]. AT-rich interacting domain containing protein members play pluripotent roles in modulating chromatin accessibility of the transcription machinery for gene expression. A reverse expression between miR-31 ARID1A was noted in OSCC tumors, and there was also a reverse correlation between ARID1A and stem cell markers. It has been suggested that tumors carrying high miR-31 expression and low ARID1A expression had poor survival [47]. It has been proposed that high levels of miR-31 expression can immortalize or change oral keratinocytes into cancerous cells [48].
EMT and other prospective mechanisms rooting OSCC mediated by mir-31
One of the extensively accepted mechanisms that involves the development of cancer towards metastasis is EMT (Epithelial to mesenchymal transition), which takes part in tumorigenesis pathological process [49]. Reviews demonstrated EMT to be one of the important pathomechanisms rooting miR-31 mediated OSCC (Fig. 3) [42, 45, 50]. The acquirement of EMT by M310K1 cells was contributed by the up-regulation of miR-31 [45] and also stated that hippo signaling pathway exerts an important action in the tumorogenesis of OSCC mediated by miR-31. Along with above mentioned mechanisms, oxidative stress and glycolytic mechanism also exerts their role in multiple cancer progression [51]. miR-31 increases lactate creation in OSCC cells by disruption of mitochondrial structure [44]. Studies also found the unique mechanism by which miR-31 targets RhoA for progression of OSCC and the knockout of RhoA in SAS and Fadu cells results in miR-31 (passenger strand) upregulation [39]. Chemoresistance of OSCC cells was also observed towards the first line drug cisplatin, in which an upregulation of miR-31-5p was observed and it was said to be allied with miR-31 anti-apoptosis characteristic [22, 52]. By distributing miRNA, exosomes that are derivative of M2 macrophages were reported to encourage progression of cancer [53]. Likewise, Yuan et al. [37] stated that exosomal miR-31-5p that are derived from macrophage enables OSCC progression by inhibition of LATS2 (Large tumor suppressor kinase 2) tumor suppressor [37]. One of the factors that is concerned to play a vital role in the progression of OSCC is EGFR, by the initiation of various intrinsic signaling pathways such as intracellular networks and protein networks [54]. In an OSCC which is a malignant phenotype via AKT (protein kinase B) signaling cascade EGF upregulates the expression of miR-31 expression [55].
miR-31 as a diagnostic/prognostic biomarker
The miR-31 has been studied in cell cultures, tissues, scrapings, body fluids and extracellular vesicles like exosomes. Western blot analysis, in situ hybridization, real-time quantitative polymerase chain reaction (RT-qPCR), pre-amplification quantitative polymerase chain reaction (preamp qPCR), quantitative reverse transcription-polymerase chain reaction (qRT-PCR), digital droplet PCR, Taqman H low-density array qRT-PCR, miRNA-based microarray and next-generation sequencing are the few conventional diagnostic methods [56].
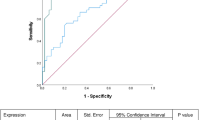
A meta-analysis study of deregulated miRNAs in oral cancer and matched non-cancerous tissue in the same patients revealed that eleven miRNAs were consistently upregulated in several studies, including miR-31-5p. These meta signatures might be considered a potential biomarker in distinguishing cancer tissues from regular [47]. According to Lu et al., miR-503 and miR-31 were downregulated in six oral cancer cell lines relative to normal keratinocytes [15]. A recent review article also found that 17 miRNAs were commonly dysregulated in OSCC and found to have clinical significance, including mir-31. In addition, upregulation of all examined miRNAs in OSCC samples was discovered in research compared to controls. A Receiver Operating Characteristics Curve study compared OSCC to controls showed that miR-31 has the most significant diagnostic accuracy of 0.9 [48]. With an area under the ROC curve of 0.776 (sensitivity = 76.8% and specificity = 73.6%), the logistic regression model, panel of five miRNAs (miR-99a-5p, miR-31-5p, miR-138-5p, miR-21-5p, miR-375-3p) discriminated oral cancer patients from healthy controls. There were substantial differences in serum miR-31-5p levels between oral cancer patients and healthy controls and between pre-and postoperative patients, and the levels were decreased after surgery [15].
A study revealed that the increased tissue/serum miR-31 expression was positively correlated with poor clinical variables and dismal prognosis. The serum miR-31 expression in HNSCC patients was markedly increased compared to normal controls. Tissue miR-31 levels in HNSCC tumor specimens exhibited higher than oral epithelial dysplasia samples and normal tissues. Tissue mir-31 was confirmed to be an independent prognostic factor in HNSCC. Salivary miR-31 was also proved as a potential biomarker for the detection and postoperative follow-up in oral cancer patients. Liu et al. found that saliva had a higher concentration of miR-31 than the plasma and remarkably higher in oral cancer patients' saliva. The study also revealed that salivary miR-31 was significantly reduced following oral carcinoma removal, indicating that most of the elevated salivary miR-31 derived from tumor tissues [57]. Al-Makey MK et al., in their study, also stated that the salivary miR-31 was significantly elevated in OC patients and can be used for early detection and postoperative follow-up [58].
A recent study on 40 Asian cohorts also proved that the salivary miR-31 levels were significantly increased in pre-operative patients than in postoperative OSCC patients [59]. Furthermore, according to Siow et al., miR-31 levels were significantly higher in early-stage tumors with no metastatic nodes and those from buccal mucosa with microarray profiling and real-time PCR. They also concluded that miR-31 could play an essential role in oral carcinogenesis [50]. A study on the k14-EGFP-miR-31 transgenic mouse model proved that miR-31 shows an oncogenic function in OSCC, and it is capable of promoting 4NQO –induced DNA damage via targeting Ku80, a repair gene. These might favor genomic instability and epithelial-mesenchymal transition [60]. Recent research stated that in HNSCC cells, both strands of pre-miR-31 behaved as carcinogenic miRNAs. In silico study revealed that 5 candidate tumor suppressor genes (CACNB2, IL34, CGNL1, CNTN3 and GAS 7) were independent predictive factors in HNSCC. This indicated that the upregulating miR-31 inhibited the expression of genes implicated in OSCC malignant transformation [61]. Furthermore, miR-31 is a reliable OSCC marker in biofluids such as plasma and saliva. Therefore, further identification of the effectors or co-players linked to miR-31 should aid in developing new therapeutics [62].
miR-31 as a therapeutic target
Several studies on animal models that investigated anti-miR knockdown or miRNA replacement therapy showed promising results for cancer therapies. Compared to protein-based drugs and plasmid DNA based gene therapy, miRNAs as natural antisense nucleotides have far less toxicity and immune response. The antisense oligonucleotides to block miRNAs, tumor and cancer stem cell focused nanoparticle therapy, and chemotherapeutic drug combination therapy have intriguing clinical uses. The intratumoral (local) and systemic delivery systems have been described. The exosome/microvesicle/liposome-mediated approaches have also been tried as novel therapeutic tools for miRNA delivery [63]. Exogenous miR-31 contributed to the immortalization of normal oral keratinocytes, implying a role in early-stage oral carcinogenesis. LNA-miR-31 therapy inhibited the proliferation of immortalized normal oral keratinocytes in vitro [64]. In antagomiR-31-5p- treated xenografts, p-AKT expression was considerably reduced, whereas PTEN expression was enhanced. They further proposed that miR-31-5p via PTEN/AKT pathway could be used as a therapeutic target miRNA in oral cancer [15].
Conclusion and future perspectives
MicroRNAs offer a vital and promising tool in gene control and a potential new class of therapeutic target and drug development. The relative stability of miRNA in body fluids makes them a convenient diagnostic sample. miR-31 has been studied extensively and proved to be one of the significant oncogenic miRNAs in participating in oral carcinogenesis. Due to its role in OSCC development, miR-31 could be used as a unique biomarker for oral cancer identification and/ prediction of disease progression. It has also been evidenced that miR-31 has a prospective role in OSCC by regulating the gene expression, signal transduction, cell proliferation, cell cycle regulation, cell survival and epithelial mesenchyme transition. Mainly involved in binding with target proteins, it is necessary to gain a better understanding of miR-31’s interactions with molecules. These studies are needed for the clinical utilization of miR-31 as a potential single or panel of biomarkers in oral cancer and could be considered as an alternative to other traditional circulating biomarkers. Apart from practical difficulties in designing and applying miR-31 as a therapeutic target, these would be a great source in developing a different class of drugs in cancer therapeutics and designing new diagnostic devices.
Availability of data and materials
Not applicable.
Abbreviations
- miRNA:
-
MicroRNA
- OSCC:
-
Oral Squamous Cell Carcinoma
- HNSCC:
-
Head and Neck Squamous Cell Carcinoma
- DICER:
-
Endoribonuclease dicer
- M2 exo:
-
Macrophage-derived exosomes
- LATS2:
-
Large tumor suppressor 2
- ARID1A:
-
AT-Rich interaction Domain 1A
- FIH:
-
Factor inhibiting HIF-1
- HIF:
-
Hypoxia-inducible factor
- VEGF:
-
Vascular endothelial growth factor
- SIRT3:
-
Silent information regulator3
- EPCAM:
-
Epithelial cell adhesion molecule
References
Khurshid Z, Zafar MS, Khan RS, Najeeb S, Slowey PD, Rehman IU (2018) Chapter Two—role of salivary biomarkers in oral cancer detection. In: Makowski GS (ed) Advances in clinical chemistry (vol. 86, pp 23–70). Elsevier. https://doi.org/10.1016/bs.acc.2018.05.002
John K, Wu J, Lee B-W, Farah CS (2013) MicroRNAs in head and neck cancer. Int J Dent 2013:e650218. https://doi.org/10.1155/2013/650218
Peng Y, Croce CM (2016) The role of MicroRNAs in human cancer. Signal Transduct Target Ther 1(1):1–9. https://doi.org/10.1038/sigtrans.2015.4
Yete S, Saranath D (2020) MicroRNAs in oral cancer: Biomarkers with clinical potential. Oral Oncol 110:105002. https://doi.org/10.1016/j.oraloncology.2020.105002
Yu T, Ma P, Wu D, Shu Y, Gao W (2018) Functions and mechanisms of microRNA-31 in human cancers. Biomed Pharmacother 108:1162–1169. https://doi.org/10.1016/j.biopha.2018.09.132
Ferreira TJ, de Araújo CC, Lima AC, Matida LM, Griebeler AF, Coelho AS, Gontijo AP, Cominetti C, Vêncio EF, Horst MA (2022) Dietary intake is associated with miR-31 and miR-375 expression in patients with head and neck squamous cell carcinoma. Nutr Cancer 74(6):2049–2058. https://doi.org/10.1080/01635581.2021.1990972
Kandettu A, Radhakrishnan R, Chakrabarty S, Sriharikrishnaa S, Kabekkodu SP (2020) The emerging role of miRNA clusters in breast cancer progression. Biochim et Biophys Acta BBA - Rev Cancer 1874(2):188413. https://doi.org/10.1016/j.bbcan.2020.188413
Domingues CSDC, Serambeque BP, Laranjo Cândido MS, Marto CMM, Veiga FJDB, Sarmento Antunes Cruz Ribeiro AB, Figueiras ARR, Botelho MFR, Dourado MDARF (2018) Epithelial-mesenchymal transition and microRNAs: challenges and future perspectives in oral cancer. Head Neck 40(10):2304–2313. https://doi.org/10.1002/hed.25381
Maheswari TNU, Venugopal A, Sureshbabu NM, Ramani P (2018) Salivary micro RNA as a potential biomarker in oral potentially malignant disorders: a systematic review. Tzu-Chi Med J 30(2):55–60. https://doi.org/10.4103/tcmj.tcmj_114_17
Manasa V, Kannan S (2017) Impact of microRNA dynamics on cancer hallmarks: an oral cancer scenario. Tumor Biology. https://doi.org/10.1177/1010428317695920
Harsha C, Banik K, Ang HL, Girisa S, Vikkurthi R, Parama D, Rana V, Shabnam B, Khatoon E, Kumar AP, Kunnumakkara AB (2020) Targeting AKT/mTOR in oral cancer: mechanisms and advances in clinical trials. Int J Mol Sci 21(9):3285. https://doi.org/10.3390/ijms21093285
Niklander S, Guerra D, Contreras F, González-Arriagada W, Marín C (2022) MicroRNAs and their role in the malignant transformation of oral leukoplakia: a scoping review. Med Oral Patol Oral Cir Bucal 27(1):e77–e84. https://doi.org/10.4317/medoral.24975
Zhi Y, Gao L, Wang B, Ren W, Liang KX, Zhi K (2021) Ferroptosis holds novel promise in treatment of cancer mediated by non-coding RNAs. Front Cell Dev Biol 9:5. https://doi.org/10.3389/fcell.2021.686906
Rishabh K, Khadilkar S, Kumar A, Kalra I, Kumar AP, Kunnumakkara AB (2021) MicroRNAs as modulators of oral tumorigenesis: a focused review. Int J Mol Sci 22(5):2561. https://doi.org/10.3390/ijms22052561
Lu Z, He Q, Liang J, Li W, Su Q, Chen Z, Wan Q, Zhou X, Cao L, Sun J, Wu Y, Liu L, Wu X, Hou J, Lian K, Wang A (2019) MiR-31-5p Is a potential circulating biomarker and therapeutic target for oral cancer. Mol Therapy - Nucleic Acids 16:471–480. https://doi.org/10.1016/j.omtn.2019.03.012
Yu F, Liang M, Huang Y, Wu W, Zheng B, Chen C (2021) Hypoxic tumor-derived exosomal miR-31-5p promotes lung adenocarcinoma metastasis by negatively regulating SATB2-reversed EMT and activating MEK/ERK signaling. J Exp Clin Cancer Res 40(1):179. https://doi.org/10.1186/s13046-021-01979-7
Feng Y, Zou W, Hu C, Li G, Zhou S, He Y, Ma F, Deng C, Sun L (2017) Modulation of CASC2/miR-21/PTEN pathway sensitizes cervical cancer to cisplatin. Arch Biochem Biophys 623–624:20–30. https://doi.org/10.1016/j.abb.2017.05.001
Liao Y, Shen L, Zhao H, Liu Q, Fu J, Guo Y, Peng R, Cheng L (2017) LncRNA CASC2 interacts with miR-181a to modulate glioma growth and resistance to TMZ through PTEN pathway. J Cell Biochem 118(7):1889–1899. https://doi.org/10.1002/jcb.25910
Pan L, Chen H, Bai Y, Wang Q, Chen L (2019) Long non-coding RNA CASC2 serves as a ceRNA of microRNA-21 to promote PDCD4 expression in oral squamous cell carcinoma. Onco Targets Ther 12:3377–3385. https://doi.org/10.2147/OTT.S198970
Yan P, Su Z, Zhang Z, Gao T (2019) LncRNA NEAT1 enhances the resistance of anaplastic thyroid carcinoma cells to cisplatin by sponging miR-9-5p and regulating SPAG9 expression. Int J Oncol 55(5):988–1002. https://doi.org/10.3892/ijo.2019.4868
Zhang B, Pan X, Cobb GP, Anderson TA (2007) MicroRNAs as oncogenes and tumor suppressors. Dev Biol 302(1):1–12. https://doi.org/10.1016/j.ydbio.2006.08.028
Wang J, Jia J, Zhou L (2020) Long non-coding RNA CASC2 enhances cisplatin sensitivity in oral squamous cell cancer cells by the miR-31-5p/KANK1 axis. Neoplasma 67(6):1279–1292. https://doi.org/10.4149/neo_2020_191029N1102
Chen NP, Sun Z, Fässler R (2018) The Kank family proteins in adhesion dynamics. Curr Opin Cell Biol 54:130–136. https://doi.org/10.1016/j.ceb.2018.05.015
Bouchet BP, Gough RE, Ammon Y-C, van de Willige D, Post H, Jacquemet G, Altelaar AM, Heck AJ, Goult BT, Akhmanova A (2016) Talin-KANK1 interaction controls the recruitment of cortical microtubule stabilizing complexes to focal adhesions. Elife 5:e18124. https://doi.org/10.7554/eLife.18124
Cui Z, Shen Y, Chen KH, Mittal SK, Yang J-Y, Zhang G (2017) KANK1 inhibits cell growth by inducing apoptosis through regulating CXXC5 in human malignant peripheral nerve sheath tumors. Sci Rep 7(1):40325. https://doi.org/10.1038/srep40325
Gu Y, Zhang M (2018) Upregulation of the Kank1 gene inhibits human lung cancer progression in vitro and in vivo. Oncol Rep 40(3):1243–1250. https://doi.org/10.3892/or.2018.6526
Barnes L (2005) World Health Organization classification of tumours. Pathol Genet Head Neck Tumours. https://cir.nii.ac.jp/crid/1573668924085792512
Choi HY, Seok J, Kang GH, Lim KM, Cho SG (2021) The role of NUMB/NUMB isoforms in cancer stem cells. BMB Rep. 54(7):335
Flores AN, McDermott N, Meunier A, Marignol L (2014) NUMB inhibition of NOTCH signalling as a therapeutic target in prostate cancer. Nat Rev Urol 11(9):499–507
Roegiers F, Jan YN (2004) Asymmetric cell division. Curr Opin Cell Biol 16(2):195–205. https://doi.org/10.1016/j.ceb.2004.02.010
Colaluca IN, Tosoni D, Nuciforo P, Senic-Matuglia F, Galimberti V, Viale G, Pece S, Di Fiore PP (2008) NUMB controls p53 tumour suppressor activity. Nature 451(7174):76–80. https://doi.org/10.1038/nature06412
Santolini E, Puri C, Salcini AE, Gagliani MC, Pelicci PG, Tacchetti C, Di Fiore PP (2000) Numb is an endocytic protein. J Cell Biol 151(6):1345–1352. https://doi.org/10.1083/jcb.151.6.1345
Chou C-H, Chiang C-YF, Yang C-C, Liu Y-C, Chang S-R, Chang K-W, Lin S-C (2021) MiR-31-NUMB cascade modulates monocarboxylate transporters to increase oncogenicity and lactate production of oral carcinoma cells. Int J Mol Sci 22(21):11731. https://doi.org/10.3390/ijms222111731
Mazumder S, Datta S, Ray JG, Chaudhuri K, Chatterjee R (2019) Liquid biopsy: MiRNA as a potential biomarker in oral cancer. Cancer Epidemiol 58:137–145. https://doi.org/10.1016/j.canep.2018.12.008
Kawakita A, Yanamoto S, Yamada S, Naruse T, Takahashi H, Kawasaki G, Umeda M (2014) MicroRNA-21 promotes oral cancer invasion via the Wnt/β-catenin pathway by targeting DKK2. Pathol Oncol Res 20(2):253–261. https://doi.org/10.1007/s12253-013-9689-y
Jung JE, Lee JY, Kim IR, Park SM, Kang JW, Kim YH, Park HR, Lee JH (2020) MicroRNA-31 regulates expression of Wntless in both Drosophila melanogaster and human oral cancer cells. Int J Mol Sci 21(19):7232. https://doi.org/10.3390/ijms21197232
Yuan Y, Wang Z, Chen M, Jing Y, Shu W, Xie Z, Li Z, Xu J, He F, Jiao P, Wang J, Xu J, Xia Y, Liu S, Du H, Li H, Dai L, Dai Y, Zhang Y (2021) Macrophage-derived exosomal miR-31-5p promotes oral squamous cell carcinoma tumourigenesis through the large tumor suppressor 2-mediated hippo signalling pathway. J Biomed Nanotechnol 17(5):822–837. https://doi.org/10.1166/jbn.2021.3066
Chattopadhyay E, Singh R, Ray A, Roy R, De Sarkar N, Paul RR, Pal M, Aich R, Roy B (2016) Expression deregulation of mir31 and CXCL12 in two types of oral precancers and cancer: Importance in progression of precancer and cancer. Sci Rep. 6(1):1–7
Chang K-W, Kao S-Y, Wu Y-H, Tsai M-M, Tu H-F, Liu C-J, Lui M-T, Lin S-C (2013) Passenger strand miRNA miR-31∗ regulates the phenotypes of oral cancer cells by targeting RhoA. Oral Oncol 49(1):27–33. https://doi.org/10.1016/j.oraloncology.2012.07.003
Liu C-J, Tsai M-M, Hung P-S, Kao S-Y, Liu T-Y, Wu K-J, Chiou S-H, Lin S-C, Chang K-W (2010) MiR-31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma. Can Res 70(4):1635–1644. https://doi.org/10.1158/0008-5472.CAN-09-2291
Qian Y, Li X, Fan R, Li Q, Zhang Y, He X, Yang W, Sun W, Lv S (2021) MicroRNA-31 inhibits traumatic brain injury-triggered neuronal cell apoptosis by regulating hypoxia-inducible factor-1A/vascular endothelial growth factor A axis. NeuroReport 33(1):1–12. https://doi.org/10.1097/WNR.0000000000001741
Peng H, Kaplan N, Hamanaka RB, Katsnelson J, Blatt H, Yang W, Hao L, Bryar PJ, Johnson RS, Getsios S, Chandel NS, Lavker RM (2012) MicroRNA-31/factor-inhibiting hypoxia-inducible factor 1 nexus regulates keratinocyte differentiation. Proc Natl Acad Sci 109(35):14030–14034. https://doi.org/10.1073/pnas.1111292109
Lai Y-H, Liu H, Chiang W-F, Chen T-W, Chu LJ, Yu J-S, Chen S-J, Chen H-C, Tan BC-M (2018) MiR-31-5p-ACOX1 axis enhances tumorigenic fitness in oral squamous cell carcinoma via the promigratory prostaglandin E2. Theranostics 8(2):486–504. https://doi.org/10.7150/thno.22059
Kao Y-Y, Chou C-H, Yeh L-Y, Chen Y-F, Chang K-W, Liu C-J, Fan Chiang C-Y, Lin S-C (2019) MicroRNA miR-31 targets SIRT3 to disrupt mitochondrial activity and increase oxidative stress in oral carcinoma. Cancer Lett 456:40–48. https://doi.org/10.1016/j.canlet.2019.04.028
Hung P-S, Tu H-F, Kao S-Y, Yang C-C, Liu C-J, Huang T-Y, Chang K-W, Lin S-C (2014) MiR-31 is upregulated in oral premalignant epithelium and contributes to the immortalization of normal oral keratinocytes. Carcinogenesis 35(5):1162–1171. https://doi.org/10.1093/carcin/bgu024
Lu W-C, Liu C-J, Tu H-F, Chung Y-T, Yang C-C, Kao S-Y, Chang K-W, Lin S-C (2016) MiR-31 targets ARID1A and enhances the oncogenicity and stemness of head and neck squamous cell carcinoma. Oncotarget 7(35):57254–57267. https://doi.org/10.18632/oncotarget.11138
Zeljic K, Jovanovic I, Jovanovic J, Magic Z, Stankovic A, Supic G (2018) MicroRNA meta-signature of oral cancer: evidence from a meta-analysis. Upsala J Med Sci 123(1):43–49. https://doi.org/10.1080/03009734.2018.1439551
Lin S-C, Liu C-J, Ji S-H, Hung W-W, Liu Y-C, Chang S-R, Tu H-F, Chang K-W (2022) The upregulation of oncogenic miRNAs in swabbed samples obtained from oral premalignant and malignant lesions. Clin Oral Invest 26(2):1343–1351. https://doi.org/10.1007/s00784-021-04108-y
Moloudizargari M, Rahmani J, Asghari MH, Goel A (2022) The prognostic role of miR-31 in colorectal cancer: the results of a meta-analysis of 4720 patients. Epigenomics 14(2):101–112. https://doi.org/10.2217/epi-2021-0277
Siow M, Karen Ng L, Vincent Chong V, Jamaludin M, Abraham M, Abdul Rahman Z, Kallarakkal T, Yang Y-H, Cheong S, Zain R (2014) Dysregulation of miR-31 and miR-375 expression is associated with clinical outcomes in oral carcinoma. Oral Dis 20(4):345–351. https://doi.org/10.1111/odi.12118
Mossenta M, Busato D, Dal Bo M, Toffoli G (2020) Glucose metabolism and oxidative stress in hepatocellular carcinoma: role and possible implications in novel therapeutic strategies. Cancers 12(6):1668. https://doi.org/10.3390/cancers12061668
Florea A-M, Büsselberg D (2011) Cisplatin as an anti-tumor drug: cellular mechanisms of activity, drug resistance and induced side effects. Cancers 3(1):1351–1371. https://doi.org/10.3390/cancers3011351
Wei K, Ma Z, Yang F, Zhao X, Jiang W, Pan C, Li Z, Pan X, He Z, Xu J, Wu W, Xia Y, Chen L (2022) M2 macrophage-derived exosomes promote lung adenocarcinoma progression by delivering miR-942. Cancer Lett 526:205–216. https://doi.org/10.1016/j.canlet.2021.10.045
Berndt A, Büttner R, Gühne S, Gleinig A, Richter P, Chen Y, Franz M, Liebmann C (2014) Effects of activated fibroblasts on phenotype modulation, EGFR signalling and cell cycle regulation in OSCC cells. Exp Cell Res 322(2):402–414. https://doi.org/10.1016/j.yexcr.2013.12.024
Lu W-C, Kao S-Y, Yang C-C, Tu H-F, Wu C-H, Chang K-W, Lin S-C (2014) EGF Up-Regulates miR-31 through the C/EBPβ Signal Cascade in Oral Carcinoma. PLoS ONE 9(9):e108049. https://doi.org/10.1371/journal.pone.0108049
Dave VP, Ngo TA, Pernestig A-K, Tilevik D, Kant K, Nguyen T, Wolff A, Bang DD (2019) MicroRNA amplification and detection technologies: Opportunities and challenges for point of care diagnostics. Lab Invest 99(4):452–469. https://doi.org/10.1038/s41374-018-0143-3
Liu C-J, Lin S-C, Yang C-C, Cheng H-W, Chang K-W (2012) Exploiting salivary miR-31 as a clinical biomarker of oral squamous cell carcinoma. Head Neck 34(2):219–224. https://doi.org/10.1002/hed.21713
Al-Malkey M (2015) Expression analysis of salivary microRNA-31 in oral cancer patients. Int J Curr Microbiol App Sci 4:375–382
Kumari P, Syed SA, Wahid M, Qureshi MA, Kumar R (2021) Expression of miR-31 in saliva-liquid biopsy in patients with oral squamous cell carcinoma. J Taibah Univ Med Sci 16(5):733–739. https://doi.org/10.1016/j.jtumed.2021.03.007
Tseng S-H, Yang C-C, Yu E-H, Chang C, Lee Y-S, Liu C-J, Chang K-W, Lin S-C (2015) K14-EGFP-miR-31 transgenic mice have high susceptibility to chemical-induced squamous cell tumorigenesis that is associating with Ku80 repression. Int J Cancer 136(6):1263–1275. https://doi.org/10.1002/ijc.29106
Oshima S, Asai S, Seki N, Minemura C, Kinoshita T, Goto Y, Kikkawa N, Moriya S, Kasamatsu A, Hanazawa T, Uzawa K (2021) Identification of tumor suppressive genes regulated by miR-31-5p and miR-31-3p in head and neck squamous cell carcinoma. Int J Mol Sci 22(12):6199. https://doi.org/10.3390/ijms22126199
Chang K-W, Hung W-W, Chou C-H, Tu H-F, Chang S-R, Liu Y-C, Liu C-J, Lin S-C (2021) LncRNA MIR31HG drives oncogenicity by inhibiting the limb-bud and heart development gene (LBH) during oral carcinoma. Int J Mol Sci 22(16):8383. https://doi.org/10.3390/ijms22168383
Rupaimoole R, Slack FJ (2017) MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discovery 16(3):203–222. https://doi.org/10.1038/nrd.2016.246
D’Souza W, Kumar A (2020) microRNAs in oral cancer: moving from bench to bed as next generation medicine. Oral Oncol 111:104916. https://doi.org/10.1016/j.oraloncology.2020.104916
Acknowledgements
MK would like to thank the Vinayaka Mission's Research Foundation, deemed to be University, India, for supplying her with all of the materials she needed to write this paper, and VB wishes to thank Bharathiar University for providing the facilities required to write this paper.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
MK conceptualized and wrote the manuscript. JD was involved in conceptualizing and supervision. AY, AE and SM was in charge of data acquisition and writing the manuscript. AV, HW and MYP wrote the manuscript and did the necessary editing. AN supervised and helped in the editing of the manuscript. VB was involved in the conceptualization of the paper, supervising and editing of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kavitha, M., Jayachandran, D., Aishwarya, S.Y. et al. A new insight into the diverse facets of microRNA-31 in oral squamous cell carcinoma. Egypt J Med Hum Genet 23, 149 (2022). https://doi.org/10.1186/s43042-022-00361-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43042-022-00361-2